Abstract
Bone sialoprotein (BSP) and bone osteopontin (OPN) are members of the SIBLING (small integrin-binding ligand, N-linked glycoproteins) family of proteins commonly found in mineralized tissues. Previously, OPN was shown to exhibit a preferential orientation for MC3T3-E1 cell adhesion when it was specifically bound to collagen. In this work, the orientation of BSP under similar circumstances is examined and compared with OPN. Radiolabeled adsorption isotherms were obtained for BSP bound to both tissue culture polystyrene (TCPS) and collagen-coated TCPS. The results show that collagen has the capacity to bind almost twice as much OPN under identical conditions. An in vitro MC3T3-E1 cell adhesion assay was then performed to compare the cell binding ability of BSP on either TCPS or collagen-coated TCPS with identical amounts of adsorbed protein. It was found that there is no significant difference in the cell binding ability of BSP on either of the substrates. For cell binding studies on collagen-coated TCPS, it was shown that there are a greater number of cells bound to substrates with adsorbed OPN as compared with BSP. The preferable orientation of OPN for cell binding coupled with the higher binding capability of collagen for OPN indicates that OPN is more important than BSP for osteoblast adhesion to the collagen matrix. In addition, a cell inhibition assay was performed to show that all of the cell binding that occurred throughout these studies was dependent upon integrin interactions with the RGD cell binding moiety.
Keywords: bone sialoprotein, bone osteopontin, collagen I, orientation, conformation
INTRODUCTION
The extracellular matrix (ECM) is a complex mixture of structural proteins, proteoglycans, glycoproteins, and growth factors which, among its many functions, regulates cell behaviors within various tissues.1 One of the primary structural proteins present in the ECM is collagen. Collagen provides strength, structural stability, and organization to the matrix. In bone tissue, for example, 10–30% of the tissue mass is protein with the remaining 70–90% being hydroxyapatite (HA). Of the protein components, 90% has been shown to be collagenous.2,3 During bone formation, cells first lay down a collagenous matrix composed of primarily type 1 collagen. After this collagen network has been formed, noncollagenous proteins bind to the matrix and then the matrix is mineralized to form bone.2,4 While a number of proteins, including bone sialoprotein (BSP), osteopontin (OPN), osteonectin, osteocalcin (OC), and bone morphogenic protein,2,4–6 have been found within bone tissue, only a limited number of these have been localized within the collagenous matrix ahead of the mineralization front in developing bone, particularly OPN and BSP. This indicates that these two proteins may play a role in the osteoblast adhesion to the matrix and the subsequent mineralization of the matrix during bone formation.5,7 Additionally, both proteins have been found to be enriched at bone–implant interfaces and are hypothesized to play an important role in cellular adhesion and osseointegration at this interface as well.8
OPN and BSP are both members of the SIBLING (small integrin-binding ligand, N-linked glycoproteins) family of proteins found in many mineralized tissues. They are known to share a number of properties, including that they are secreted, phosphorylated, and sulfated sialoproteins that are acidic in nature.9 OPN is composed of 260–317 amino acids, with a molecular weight of 45–75 kDa10,11 and BSP is composed of 281–327 amino acids, with a molecular weight of 60–80 kDa.12 Both proteins were originally isolated from bone matrix. Since then OPN has been found within other tissues throughout the body, while BSP has been found to have a limited distribution outside of mineralized tissue and the immediate surroundings. The functions of these two proteins within the body have been reviewed extensively in the literature.10–15 OPN and BSP are both known to contain an arginine–glycine–aspartic acid (RGD) motif that mediates cell binding through direct interactions with a number of transmembrane integrin pairs.16 Additionally, both proteins have been shown to have an affinity for both calcium and collagen,17,18 which is not unexpected based on their localization in developing and developed bone, and bone–implant interfacial zones.
It is well known that within seconds of implantation, biomaterials are coated by an adsorbed layer of protein and that it is this layer that modulates the subsequent cellular response to the implant, especially at early interaction times.19,20 Recent efforts in biomaterials design have focused on understanding and controlling the composition, conformation, and orientation of this adsorbed protein layer to improve the wound healing process and subsequent integration of biomaterials into the body.8,21 However, controlling the conformation and orientation of adsorbed proteins has proven to be challenging. Some success has been shown for controlling protein orientation with charged self-assembled monolayers (SAMs).22–24 In one such study, Liu et al.23 showed that OPN had a preferential orientation for endothelial cell adhesion when adsorbed to a positively charged NH2-terminated SAM as compared with a negatively charged COOH-terminated SAM. Other efforts have focused on using specific protein–protein binding interactions to induce a natural orientation of one of the proteins.25,26 Our previous work26 examined MC3T3-E1 osteoblast-like cell binding onto OPN covered surfaces when OPN was either oriented through its specific binding interaction with collagen I or randomly adsorbed onto tissue culture polystyrene (TCPS) substrates. It was found that when equal amounts of protein were adsorbed to the different surfaces, there were statistically significant differences in the MC3T3-E1 adhesion levels on the two surfaces, indicating a preferential orientation or conformation of OPN for MC3T3-E1 adhesion when it was specifically bound to collagen.
The foci of this work are to (a) probe the orientation of BSP when it is specifically bound to collagen relative to randomly adsorbed BSP on TCPS and (b) compare the cell-binding abilities of OPN and BSP to each other when they are specifically bound to collagen. These studies provide valuable information regarding the relative importance of each of these proteins for cellular adhesion during bone formation. The role of these two proteins in cell adhesion and mineralization has not been directly compared in this manner. It was hypothesized that BSP would have a favorable orientation for cell binding when specifically bound to collagen, similar to OPN.26 For a fair comparison of protein orientation or cellular adhesion ability, it is essential to ensure that there are identical amounts of adsorbed proteins. This is achieved in this work via radiolabeling assays. Additionally, a cell inhibition assay was performed with a GRGDSP peptide to test the importance of the RGD moiety for the cell-binding interactions. It was found in these studies that BSP does not have a more favorable orientation for cell binding when specifically bound to collagen and that OPN is more important than BSP for mediating MC3T3-E1 osteoblast adhesion to collagen-coated substrates.
EXPERIMENTAL PROCEDURES
Materials
TCPS flasks were purchased from Corning (Corning, NY), scored into 25 mm2 squares using a drill press, and broken apart prior to use in experiments. Tris buffer was prepared by dissolving 25 mM Tris–HCl (Sigma) and 125 mM NaCl (Sigma) in 18.2 MΩ H2O and adjusting the pH to 7.4 with NaOH (Sigma). Phosphate buffered saline (PBS) buffer was purchased from Sigma and prepared in 18.2 MΩ H2O. Both buffers were filter sterilized with 0.22 μm vacuum filters before use. Type 1 collagen from rat tail with a purity of >90% was purchased from BD Biosciences (Bedford, MA). Bovine serum albumin (BSA) was purchased from Sigma (St. Louis, MO). Heat denatured BSA was prepared by dissolving BSA in Tris buffer and heating it at 60°C for 30 min. Paraformaldehyde was purchased from Sigma. A 4% paraformaledehyde solution was made by dissolving paraformaldehyde in PBS at 60°C until the solution became clear, followed by 0.22 μm vacuum filtering before use. A soluble GRGDSP peptide was purchased from Calbiochem (La Jolla, CA). All other cell culture supplies including medium, serum, and trypsin-EDTA (0.05%/0.53 mM) were purchased from Gibco (Gaithersburg, MD). The osteoblast cell line MC3T3-E1 was a gift from Dr. P. Stayton (University of Washington).
Isolation of OPN and BSP from rat long bone
The noncollagenous ECM proteins were extracted from the tibiae of ~10-week-old rats by standard procedures as described previously.27,28 Briefly, the tibiae were extracted with 0.5 M EDTA in 4 M guanidium-HCl containing protease inhibitors (GdmCl; Acros Organics, Fairlawn, NJ). Next, the extracts were subjected to gel chromatography on Sephacryl S-200 in 4 M GdmCl. Using this procedure, the high-molecular weight protein fraction (designated as ES1) was separated from smaller sized proteins like OC. ES1 was next chromatographed on DEAE-Sephacel, eluted with a linear gradient formed from 750 mL each of starting buffer (50 mM Tris–HCl, 6 M urea, pH 7.2) and 50 mM Tris–HCl, 6 M urea containing 0.7 M NaCl (pH 7.2). The DEAE-Sephacel chromatography separated ES1 of bone extracts into seven major fractions.28 The DEAE-Sephacel fractions rich in OPN and BSP were passed separately over a Bio-Gel A50m column (BIO-RAD, Hercules, CA) to further purify the OPN and BSP. For Bio-Gel A50m chromatography, a buffer consisting of 6 M urea and 50 mM Tris–HCl at pH 7.2 was used. Isolated OPN and BSP were analyzed by polyacrylamide gel electrophoresis (SDS-PAGE) with Stains-All staining to ensure their purity. The identities of OPN and BSP were further confirmed by Western immunoblots using antibodies that were specifically reactive to OPN or BSP.
Protein adsorption isotherms
BSP was radiolabeled with 125I using the iodine mono-chloride method.29 125I radiolabeled BSP was added to 0.5 mg/mL unlabeled BSP to obtain a solution with a specific activity of 281.77 counts per minute (cpm) per nanogram protein. BSP adsorption isotherms were determined for two different substrates: TCPS and collagen-coated TCPS. TCPS substrates were prepared by soaking the substrates in 1 mL of Tris buffer overnight, followed by rinsing with 18.2 MΩ H2O before incubation with BSP overnight at 4°C in a humidified atmosphere. Collagen-coated TCPS substrates were prepared by soaking the substrates in 1 mL of a 50 μg/mL collagen solution in Tris buffer overnight. These substrates were then rinsed with 18.2 MΩ H2O before being soaked in a 1 mg/mL solution of heat denatured BSA for 5 h. Finally, the substrates were again rinsed with 18.2 MΩ H2O before they were incubated with BSP overnight at 4°C in a humidified atmosphere. Following the protein adsorption step, all of the surfaces were rinsed three times with Tris buffer to remove loosely adhered proteins. The cpm radioactivity of all of the samples was measured with a TM Analytic 1185R Gamma Trac Gamma Counting System (Elk Grove, IL). The amount of BSP bound to each of the surfaces was calculated by relating the cpm of each sample to its specific activity exposure amount and its surface area. Previous work has been completed to obtain the 125I adsorption isotherms for OPN in an identical fashion and those adsorption isotherms were used in this work.26
Cell culture
MC3T3-E1 cells were maintained in continuous growth on TCPS flasks in alpha Minimum Essential Medium (α-MEM), supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin solution in a humidified atmosphere at 37°C and 5% CO2. To passage, cells were rinsed twice with 10 mL of Tris buffer followed by incubation in 2 mL of trypsin/EDTA. After the cells detached from the TCPS flask, they were resuspended in supplemented medium and replated onto new TCPS flasks. Cells were passaged once a week and passages 5–10 were used for experiments.
Cell adhesion assay
The cell adhesion assay is similar to that described previously23,26,30 with minor modifications. The protein adsorption procedure was similar to that described above for the radiolabeled protein adsorption isotherm. However, only one specific protein concentration was used for each substrate and protein combination that was examined and only native (nonradiolabeled) protein was used. TCPS substrates were prepared and then exposed to 10 μg/mL of either OPN or BSP overnight at 4°C in a humidified atmosphere. Collagen-coated TCPS substrates were prepared and then exposed to either 35 μg/mL of OPN or 50 μg/mL of BSP overnight at 4°C in a humidified atmosphere. Control samples were prepared for both the TCPS and collagen-coated TCPS substrates by incubating the substrates with 1 mg/mL of heat denatured BSA. Following the overnight protein adsorption, the samples were transferred to a 48-well culture plate where they were rinsed three times with 1 mL of Tris buffer and then blocked with 1 mL of heat denatured BSA for 30 min. In the meantime, freshly confluent MC3T3-E1 cells were detached with 2 mL of trypsin/EDTA and resuspended in 5 mL of 5 mg/mL soybean trypsin inhibitor in PBS. The cells were centrifuged at 1000 rpm for 5 min, after which the supernatant was removed and the cells were washed twice with 10 mL of 5 mg/mL BSA in nonsupplemented (serum free) α-MEM. Following this step, the cells were resuspended in serum free α-MEM and diluted to a final concentration of 1 × 105 cells/mL, as determined with a hemocytometer. The cells were incubated for 15 min before use in the adhesion assay. Following the BSA blocking step, the solution was removed and the samples were rinsed three times with 1 mL of Tris buffer. Following this rinsing step, 1 mL of the cell solution was added to each well and the samples were incubated for 2 h in a humidified atmosphere at 37°C and 5% CO2. At least three samples were prepared for each protein and substrate combination during each cell adhesion assay and the assay was repeated five times.
Cell inhibition assay
The cell inhibition assay was completed in a similar fashion to the cell adhesion assay with one exception. Before the addition of the diluted cells to the samples, the cells were first incubated with 1 mM of a soluble GRGDSP peptide31 in α-MEM for 15 min. This incubation step replaced the final cell incubation step in the adhesion assay procedures. Five total samples were prepared for each protein and substrate combination over two separate occasions.
Cell fixing and staining
Following the cellular adhesion and inhibition assays, the cell solution was removed from the well plate and the samples were rinsed three times with warm Tris buffer (37°C) to remove nonadherent cells. Following this rinse step the samples were fixed with 1 mL of 4% paraformaldehyde for 5 min. The samples were then rinsed three times with 1 mL of warm Tris buffer and then stained with 0.75 mL of hematoxylin for 5 min. Following staining, the samples were rinsed extensively with 18.2 MΩ H2O and then exposed to 1 mL of Tris buffer for 3 min. Finally, the samples were rinsed three times with 1 mL of 18.2 MΩ H2O and dried in air. Four 10× brightfield images from each sample were randomly selected and captured using an Olympus IX70 inverted microscope (Center Valley, PA) equipped with an Olympus OLY-105 video camera (Center Valley, PA) and Hauppauge WinTV2000 software (Hauppauge, NY).
Data analysis
The number of adhered cells was used to compare the response of the MC3T3-E1 cells to each of the substrate and protein combinations. The total number of cells that adhered to each sample were physically counted using ImageJ from each of the images that were captured.32 A total of 64 images (n = 16) were analyzed for both BSP and OPN adsorbed to TCPS and collagen-coated TCPS, a total of 60 images (n = 15) were analyzed for both of the BSA control cases, and a total of 20 images (n = 5) were analyzed for each of the four inhibition assay protein/substrate combinations. The sample data are presented as the average of all of the images obtained and the error bars represent the standard error of the mean (SE). Sample results were analyzed using one-way analysis of variance and they were considered statistically significant when they had a probability value less than 0.05 (p < 0.05). Statistical analysis was performed using OriginPro 7.0 (OriginLab Corporation, MA).
RESULTS AND DISCUSSION
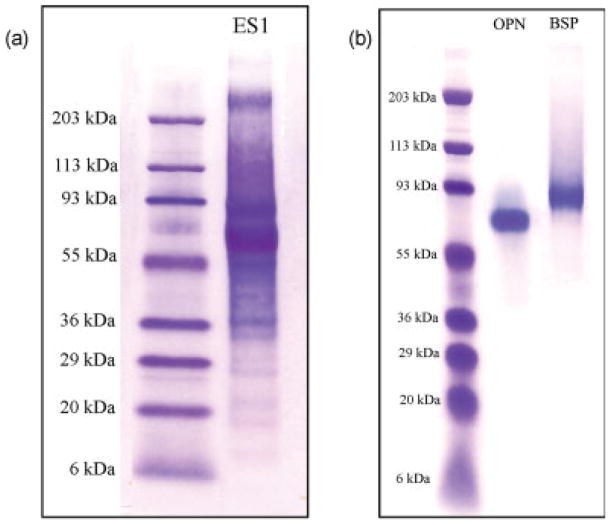
The purity of the OPN and BSP used in this investigation is suggested by Figure 1. Figure 1(a) shows the impure, high-molecular weight protein fraction ES1. This impure fraction can be compared with the protein bands obtained for purified OPN and BSP, which are shown in Figure 1(b). Neither protein band was seen following staining with Coomassie brilliant blue, as these highly acidic, phosphorylated proteins react poorly or do not react at all with Coomassie brilliant blue.33 The identities of OPN and BSP were further confirmed by Western immunoblots using antibodies specific to OPN and BSP, respectively (data not shown).
Figure 1.
SDS-PAGE and Stains-All staining of protein fractions: (a) Stains-All staining of the impure, high-molecular weight protein fraction (ES1), obtained after the removal of smaller molecular weight bone proteins; (b) Stains-All staining of OPN (lane 2) and BSP (lane 3). Lane 1 in both (a) and (b) was loaded with molecular weight standards. The lanes were loaded with 15 μg of ES1 or 6 μg of OPN and BSP, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
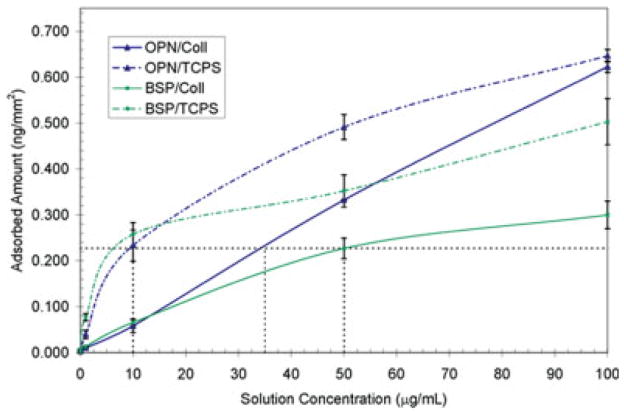
To compare the cellular adhesion properties of proteins that are in various orientations and conformations, substrates with equal amounts of adsorbed proteins must be used. Radiolabeled adsorption isotherms were obtained for BSP by determining the adsorbed amount of BSP from four different solution concentrations on both TCPS and collagen-coated TCPS substrates. The adsorbed amount of protein was calculated based on the specific radioactivity of BSP before adsorption and the cpm of each of the samples following the adsorption and rinsing steps. The resulting adsorption isotherms are shown in Figure 2. The isotherms for BSP were compared with those obtained previously for OPN26 to determine the solution concentrations of OPN and BSP on TCPS and collagen-coated TCPS that result in equal adsorption amounts. The equal surface levels of adsorbed proteins allow for a direct comparison of the cell binding behaviors of these two proteins on both of the substrates examined using cell adhesion assays. Specifically, 10 μg/mL of BSP or OPN was adsorbed to the TCPS substrates, 35 μg/mL of OPN was adsorbed to collagen-coated TCPS, and 50 μg/mL of BSP was adsorbed to collagen-coated TCPS for the cellular assays. These conditions resulted in ~0.22 ng/mm2 of adsorbed protein on each of the substrates examined. It should be pointed out that this is a lower concentration of adsorbed protein than that used by Liu et al. (0.38 ng/mm2) in the examination of the cell binding behavior imparted to OPN when it was specifically bound to collagen26 because BSP appeared to be near the peak of its adsorption isotherm at an exposure level of 50 μg/mL.
Figure 2.
125I radiolabel adsorption isotherms for BSP (squares) and OPN (triangles) on two substrates: TCPS substrates (dot–dash lines) and collagen-coated TCPS substrates (solid lines). The dotted lines represent the protein exposure concentrations used for the cell adhesion assays. The data for the OPN cases were taken from Liu et al.26 [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
BSP had a maximal adsorption level of ~0.3 ng/mm2 on the collagen-coated TCPS at an exposure level of 100 μg/mL. OPN had a maximal adsorption level that was twice this amount, ~0.6 ng/mm2, under the same exposure conditions as shown in the work of Liu et al.26 This difference in collagen binding behavior between OPN and BSP is similar to that obtained previously under different experimental conditions.17 This is consistent with the possibility that there are a different number of binding sites for the two proteins on collagen or that the OPN is forming a multilayered or aggregated structure. This later explanation is supported by the largely linear binding curve that was seen in the adsorption isotherm over the range of concentrations examined. It is possible that the OPN structure undergoes conformation changes upon binding to collagen that allows for the molecule to bind with itself. It has been shown that OPN has a fivefold increase in its binding level to collagen when it is in an aggregated and cross-linked form as compared with its monomeric form.34 Thus, the OPN conformation would be more favorable for aggregating after it is specifically bound to collagen.
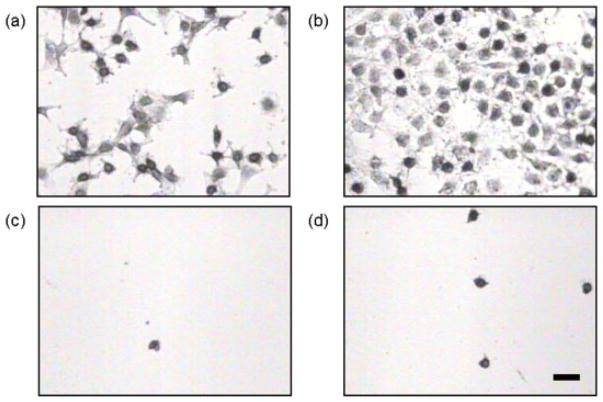
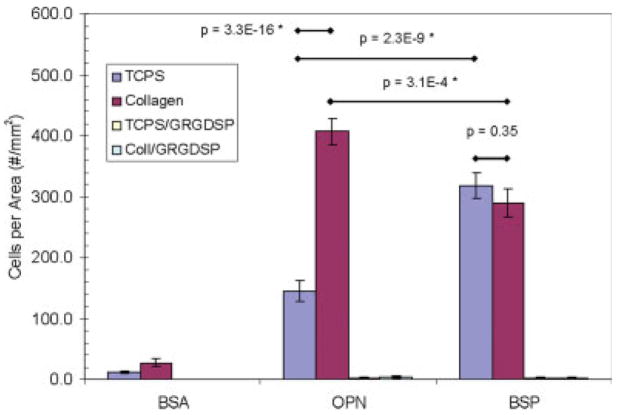
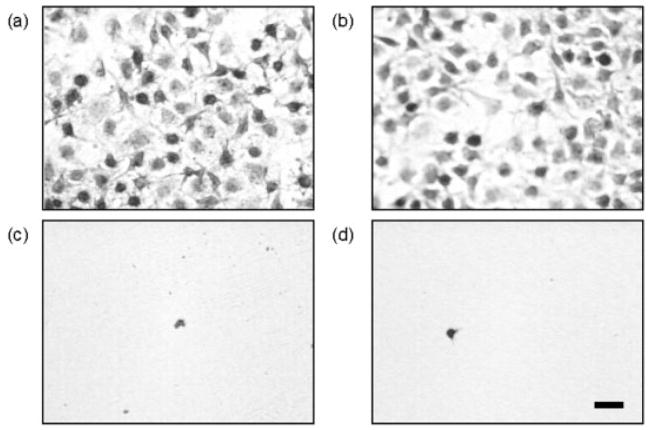
After determining protein solution concentrations that lead to identical amounts of adsorbed protein, the impact of orientation and conformation of both OPN and BSP was examined. Since there are no specific binding interactions between proteins and TCPS, the adsorbed proteins are considered to have a random orientation on this substrate. At the same time, both BSP and OPN are known to have specific binding interactions with collagen. It is believed that these interactions will impart a specific orientation or conformation to BSP and OPN when bound to the collagen-coated TCPS substrates. Insight can be gained into the orientation and conformation of OPN and BSP by comparing the adhesion of MC3T3-E1 cells to both of these substrates after protein adsorption. Previously, Liu et al. found that OPN has a preferential orientation for MC3T3-E1 adhesion when it is specifically bound to collagen-coated TCPS as compared with OPN randomly adsorbed to TCPS alone.26 The results of this work confirm those results, as can be seen in the optical microscopy images of the MC3T3-E1 cells in Figure 3 and the statistical analysis in Figure 6. It should be mentioned that there are fewer cells per unit area in this study due to lower amounts of adsorbed OPN. However, there are still almost three times more cells bound to the collagen-coated TCPS versus the TCPS alone and the trend in the MC3T3-E1 adhesion levels from this work is consistent with that found by Liu et al. Controls of heat denatured BSA adsorbed to TCPS and to collagen-coated TCPS were performed to show that the differences in cell binding were a direct result of the orientation or conformation of the adsorbed OPN, not the underlying substrates. Optical microscopy images of these control samples are shown in Figure 4. Finally, the results from the inhibition assay shown in Figure 3 and the statistical analysis in Figure 6 indicate that all of the cellular binding on both substrates appears to be RGD mediated. Essentially all of the cell binding can be prevented when the RGD-specific cell integrins are blocked with a soluble GRGDSP peptide before cell seeding. Together, these results indicate that the RGD cell binding sequence in OPN is more accessible for cell binding when OPN is specifically bound to collagen than when OPN is randomly adsorbed on TCPS. This accessibility is most likely related to the orientation and conformation of OPN on these two surfaces that, for MC3T3-E1 adhesion, is more favorable when the OPN is specifically bound to collagen I.
Figure 3.
Optical microscopy images of MC3T3-E1 cells on different surfaces: (a) 10 μg/mL OPN adsorbed on TCPS, (b) 35 μg/mL OPN adsorbed to collagen-coated TCPS, (c) 10 μg/mL OPN adsorbed on TCPS in the presence of 1.0 mM GRGDSP, and (d) 35 μg/mL OPN adsorbed to collagen-coated TCPS in the presence of 1.0 mM GRGDSP. The scale bar represents 50 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 6.
Average number of MC3T3-E1 cells (cells/mm2) that adhered to the TCPS and collagen-coated TCPS substrates with adsorbed BSA, OPN, or BSP in the presence or absence of 1.0 mM GRGDSP. The adhesion data are presented as the mean ± SE from at least 15 samples completed on five separate occasions. Inhibition data are presented as the mean ± SE from a total of five samples completed on two separate occasions. *Represents a statistically significant difference between the surfaces being compared (p < 0.05). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 4.
Optical microscopy images of MC3T3-E1 cells on different control surfaces: (a) 1 mg/mL heat denatured BSA adsorbed on TCPS and (b) 1 mg/mL heat denatured BSA adsorbed to collagen-coated TCPS. The scale bar represents 50 μm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
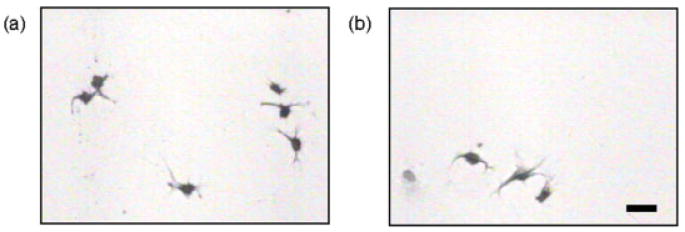
BSP demonstrates a trend different from OPN. The results indicate that there are an approximately equal number of cells that bind to both the BSP–TCPS and BSP–collagen-coated TCPS substrates as shown in the microscopy images in Figure 5 and the statistical analysis shown in Figure 6. In fact, the statistical analysis indicated that there are no differences between these two cases (p = 0.35). The results of the inhibition assay show that the MC3T3-E1 cell binding to both substrates is again RGD mediated, as the cellular binding was prevented when the soluble GRGDSP peptide was included in the cell medium prior to cell seeding. The results obtained for BSP indicate that the RGD moiety is equally accessible when BSP is randomly adsorbed to TCPS or when it is specifically bound to collagen-coated TCPS. Therefore, unlike OPN, there is no favorable BSP orientation/conformation for MC3T3-E1 adhesion when it is specifically bound to collagen I.
Figure 5.
Optical microscopy images of MC3T3-E1 cells on different surfaces: (a) 10 μg/mL BSP adsorbed on TCPS, (b) 50 μg/mL BSP adsorbed to collagen-coated TCPS, (c) 10 μg/mL BSP adsorbed on TCPS in the presence of 1.0 mM GRGDSP, and (d) 50 μg/mL BSP adsorbed to collagen-coated TCPS in the presence of 1.0 mM GRGDSP. The scale bar represents 50 μm.
An explanation for the fact that OPN is sensitive to the environment in which it is bound while BSP is not, lies within the conformational flexibility of the two proteins around their RGD cell binding sequence. In the predicted secondary structure for OPN, the RGD moiety is located between two sequences that are expected to line up with a sheet structure and this structure could be oriented either into or away from a surface.10 On the other hand, the RGD cell binding moiety in BSP has been predicted to be within a portion of the protein that has a random coil structure.12 These secondary structures indicate the relative flexibility of each of the peptide chains around the RGD moiety upon binding to a substrate. While it has been shown that both of these proteins are somewhat unstructured in solution, Fisher et al.9 did propose that these proteins would adopt specific structures based upon their binding partners and it is likely that the sheet structure surrounding the RGD moiety in OPN is less likely to change conformations upon a specific binding interaction then the random coil surrounding the RGD sequence in BSP. Additionally, nuclear magnetic resonance studies on a 59-amino acid peptide from BSP that contained the RGD sequence and circular dichroism and small angle X-ray scattering on both native and recombinant BSP show that the protein has an unfolded and open structure.35,36 The theory that these cellular adhesion differences are due to conformational flexibility differences is also supported by the cell binding ability of each of these proteins when randomly adsorbed to TCPS. Neither protein has a specific interaction with the polymer substrate, yet TCPS with adsorbed BSP has twice as many cells bound to it as compared to TCPS with adsorbed OPN after 2 h (p = 2.3 × 10−9). This difference is almost as significant as that between the two OPN cases, where there is believed to be a specific orientation or conformation imparted to the protein. Higher conformational flexibility of BSP is also supported by observations that unlike OPN,23 BSP does not appear to have a charge driven orientation effect when adsorbed onto charged SAMs (Bernards and Jiang, unpublished observation). Another possible reason for the lack of charge driven orientation could be related to the distribution of the charged groups throughout these two proteins. The differences in cell adhesion could be also be influenced by the presence of a second cell binding domain within the BSP structure.35,37 However, this is not likely since cell binding to this secondary adhesion domain was observed to be unaffected by the presence of RGD blocking peptides in those studies that identified its presence. In this study, the RGD peptide effectively eliminated all of the cellular binding involving adsorbed BSP.
Both OPN and BSP have been implicated as key proteins for mediating cellular adhesion at bone–implant interfaces.8 It is also feasible, based on immunolocalization studies, that either of these two proteins could play important roles in cell binding to the collagen matrix of developing bone.4,7 For these reasons, it is relevant to compare the MC3T3-E1 osteoblast binding abilities of OPN and BSP when they have been specifically bound to collagen. As can be seen in Figure 6, there are a statistically greater number of cells that bind to collagen-coated TCPS with adsorbed OPN than to collagen-coated TCPS with adsorbed BSP (p = 3.1 × 10−4). Although BSP may have more conformational flexibility around its RGD moiety, it does not appear to be as important for osteoblast binding to a collagen matrix. This is an important observation to consider in elucidating the roles that these two proteins play in bone development and at bone–implant interfaces. It is also supported by the localization of these proteins throughout the body. While BSP is almost exclusively expressed in mineralized tissues and its immediate surroundings, OPN is found within the ECM of many different tissues throughout the body. It is reasonable to believe that OPN may play a role in mediating cell binding to collagen throughout the ECM of the body, based on the capacity of collagen to bind OPN as seen in the adsorption isotherm26 and the availability of the RGD moiety for cell binding after OPN has been specifically bound to collagen, as shown in this work. Many cell types other than MC3T3-E1 cells have integrin receptor pairs that bind to the RGD moiety, thus these observations should not be limited to osteoblast cell adhesion alone. The lower magnitude role for BSP in cell adhesion to collagen may be related to the commonly perceived role for BSP, mediating the mineralization process of developing bone.2,5,12 Recent data (Huang and Qin, unpublished observations) have shown that OPN dissociates more easily from rat bone ECM than BSP, and this indicates that BSP may bind more tightly to the collagen matrix than OPN. These observations suggest that BSP must bind tightly to the collagen matrix to fulfill its role in mediating the mineralization of the collagen matrix. The overall conformational flexibility has also been implicated as important for BSP to play multiple biological roles,9,36) allowing for maximal exposure of the BSP molecule for binding interactions with collagen, HA, and cells simultaneously. While OPN has been implicated as important for preventing the over mineralization of developing bone,14,38 this study indicates that it may also play an important role in mediating cellular adhesion to the collagen matrix of developing bone and in the bone–implant interfacial zone.
CONCLUSIONS
In this work, BSP adsorption isotherms on both TCPS and collagen-coated TCPS substrates were obtained by radiolabeling and these isotherms were compared with those obtained previously for OPN. Under conditions with identical amounts of adsorbed protein specifically bound to collagen I, OPN was shown to have a significantly greater number of bound cells as compared to BSP. The differences in binding affinity to collagen and in the cell adhesiveness of BSP and OPN under the conditions explored here imply unique and specialized roles for these molecules in bone and other tissues.
Acknowledgments
Contract grant sponsor: National Science Foundation; contract grant numbers: CTS-0092699, EEC-9529161
Contract grant sponsor: National Institute of Health; contract grant number: DE005092
The authors acknowledge Drs. L. Cao and T. Horbett at the University of Washington for their assistance with the radiolabeling studies, Colleen Irvin at the University of Washington Engineered Biomaterials (UWEB) for her assistance with cell culture, and Dr. J. Berg at the University of Washington for the use of a microscope in his group.
References
- 1.Bornstein P, Sage EH. Matricellular proteins: Extracellular modulators of cell function. Curr Opin Cell Biol. 2002;14:608–616. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 2.Gokhale JA, Boskey AL, Robey PG. The biochemistry of bone. In: Marcus R, Feldman D, Kelsey J, editors. Osteoporosis. Vol. 1. San Diego: Academic Press; 2001. pp. 107–188. Chapter 4. [Google Scholar]
- 3.LeGeros RZ. Properties of osteoconductive biomaterials: Calcium phosphates. Clin Orthop Rel Res. 2002;395:81–98. doi: 10.1097/00003086-200202000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Veis A. Mineral–matrix interactions in bone and dentin. J Bone Miner Res. 1993;8:S493–S497. doi: 10.1002/jbmr.5650081312. [DOI] [PubMed] [Google Scholar]
- 5.Roach HI. Why does bone matrix contain non-collagenous proteins? The possible roles of osteocalcin, osteonectin, osteopontin, and bone sialoprotein in bone mineralization and resorption. Cell Biol Int. 1994;18:617–628. doi: 10.1006/cbir.1994.1088. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg HA, Warner KJ, Li MC, Hunter GK. Binding of bone sialoprotein, osteopontin and synthetic polypeptides to hydroxyapatite. Connective Tissue Res. 2001;42:25–37. doi: 10.3109/03008200109014246. [DOI] [PubMed] [Google Scholar]
- 7.Nanci A. Content and distribution of noncollagenous matrix proteins in bone and cementum: Relationship to speed of formation and collagen packing density. J Struct Biol. 1999;126:256–269. doi: 10.1006/jsbi.1999.4137. [DOI] [PubMed] [Google Scholar]
- 8.Puleo DA, Nanci A. Understanding and controlling the bone–implant interface. Biomaterials. 1999;20:2311–2321. doi: 10.1016/s0142-9612(99)00160-x. [DOI] [PubMed] [Google Scholar]
- 9.Fisher LW, Torchia DA, Fohr B, Young MF, Fedarko NS. Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem Biophys Res Commun. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- 10.Sodek J, Ganss B, McKee MD. Osteopontin. Crit Rev Oral Biol Med. 2000;11:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 11.Butler WT, Ridall AL, McKee MD. Osteopontin. In: Bilezikian JP, Raisz LG, Rodan GA, editors. Principles of Bone Biology. San Diego: Academic Press; 1996. pp. 167–182. Chapter 13. [Google Scholar]
- 12.Ganss B, Kim RH, Sodek J. Bone sialoprotein. Crit Rev Oral Biol Med. 1999;10:79–98. doi: 10.1177/10454411990100010401. [DOI] [PubMed] [Google Scholar]
- 13.Gorski JP. Is all bone the same? Distinctive distributions and properties of non-collagenous matrix proteins in lamellar vs. woven bone imply the existence of different underlying osteogenic mechanisms. Crit Rev Oral Biol Med. 1998;9:201–223. doi: 10.1177/10454411980090020401. [DOI] [PubMed] [Google Scholar]
- 14.Giachelli CM, Steitz S. Osteopontin: A versatile regulator of inflammation and biomineralization. Matrix Biol. 2000;19:615–622. doi: 10.1016/s0945-053x(00)00108-6. [DOI] [PubMed] [Google Scholar]
- 15.Mazzali M, Kipari T, Ophascharoensuk V, Wesson JA, John-son R, Hughes J. Osteopontin—A molecule for all seasons. QJM: An Int J Med. 2002;95:3–13. doi: 10.1093/qjmed/95.1.3. [DOI] [PubMed] [Google Scholar]
- 16.Siebers MC, ter Brugge PJ, Walboomers XF, Jansen JA. Integrins as linker proteins between osteoblasts and bone replacing materials. A critical review Biomaterials. 2005;26:137–146. doi: 10.1016/j.biomaterials.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Bal BS, Gorski JP. Calcium and collagen binding properties of osteopontin, bone sialoprotein, and bone acidic glycoprotein-75 from bone. J Biol Chem. 1992;267:24871–24878. [PubMed] [Google Scholar]
- 18.Fujisawa R, Kuboki Y. Affinity of bone sialoprotein and several other bone and dentin acidic proteins to collagen fibrils. Calcif Tissue Int. 1992;51:438–442. doi: 10.1007/BF00296677. [DOI] [PubMed] [Google Scholar]
- 19.Castner DG, Ratner BD. Biomedical surface science: Foundations to frontiers. Surf Sci. 2002;500:28–60. [Google Scholar]
- 20.Tirrell M, Kokkoli E, Biesalski M. The role of surface science in bioengineered materials. Surf Sci. 2002;500:61–83. [Google Scholar]
- 21.Wilson CJ, Clegg RE, Leavesley DI, Pearcy MJ. Mediation of biomaterial-cell interactions by adsorbed proteins: A review. Tissue Eng. 2005;11:1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Liu L, Zhou J, Jiang S. Controlling antibody orientation on charged self-assembled monolayers. Langmuir. 2003;19:2859–2864. [Google Scholar]
- 23.Liu L, Chen S, Giachelli CM, Ratner BD, Jiang S. Controlling osteopontin orientation on surfaces to modulate endothelial cell adhesion. J Biomed Mater Res. 2005;74A:23–31. doi: 10.1002/jbm.a.30221. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Castner DG, Ratner BD, Jiang S. Probing the orientation of surface immobilized immunoglobulin G by time-of-flight secondary ion mass spectrometry. Langmuir. 2004;20:1877–1887. doi: 10.1021/la035376f. [DOI] [PubMed] [Google Scholar]
- 25.Calonder D, Matthew HWT, Tassel PRV. Adsorbed layers of oriented fibronectin: A strategy to control cell-surface interactions. J Biomed Mater Res A. 2005;75A:316–323. doi: 10.1002/jbm.a.30417. [DOI] [PubMed] [Google Scholar]
- 26.Liu L, Qin C, Butler WT, Ratner BD, Jiang S. Controlling the orientation of bone osteopontin via its specific binding with collagen I to modulate osteoblast adhesion. J Biomed Mater Res A. 2007;80A:102–110. doi: 10.1002/jbm.a.30858. [DOI] [PubMed] [Google Scholar]
- 27.Prince CW, Oosawa T, Butler WT, Tomana M, Bhown AS, Bhown M, Schrohenloher RE. Isolation, characterization, and biosynthesis of a phosphorylated glycoprotein from rat bone. J Biol Chem. 1987;262:2900–2907. [PubMed] [Google Scholar]
- 28.Qin C, Brunn JC, Jones J, George A, Ramachandran A, Gorski JP, Butler WT. A comparative study of sialic acid-rich proteins in rat bone and dentin. Eur J Oral Sci. 2001;109:133–141. doi: 10.1034/j.1600-0722.2001.00001.x. [DOI] [PubMed] [Google Scholar]
- 29.Horbett TA. Techniques of Biocompatibility Testing. Boca Raton, FL: CRC; 1986. [Google Scholar]
- 30.Freitas F, Jeschke M, Majstorovic I, Mueller DR, Schindler P, Voshol H, Oostrum JV, Susa M. Fluoroaluminate stimulates phosphorylation of p130 Cas and Fak and increases attachment and spreading of preosteoblastic MC3T3-E1 cells. Bone. 2002;30:99–108. doi: 10.1016/s8756-3282(01)00625-1. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert M, Giachelli CM, Stayton PS. Biomimetic peptides that engage specific integrin dependant signaling pathways and bind to calcium phosphate surfaces. J Biomed Mater Res A. 2003;67A:69–77. doi: 10.1002/jbm.a.10053. [DOI] [PubMed] [Google Scholar]
- 32.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophoton Int. 2004;11:36–42. [Google Scholar]
- 33.Myers JM, Veis A, Sabsay B, Wheeler AP. A method for enhancing the sensitivity and stability of Stains-All for phosphoproteins separated in sodium dodecyl sulfate-poly-acrylamide gels. Anal Biochem. 1996;240:300–302. doi: 10.1006/abio.1996.0361. [DOI] [PubMed] [Google Scholar]
- 34.Kaartinen MT, Pirhonen A, Linnala-Kankkunen A, Maenpaa PH. Cross-linking of osteopontin by tissue transglutaminase increases its collage binding properties. J Biol Chem. 1999;274:1729–1735. doi: 10.1074/jbc.274.3.1729. [DOI] [PubMed] [Google Scholar]
- 35.Stubbs JT, Mintz KP, Eanes ED, Torchia DA, Fisher LW. Characterization of native and recombinant bone sialoprotein: Delineation of the mineral-binding and cell adhesion domains and structural analysis of the RGD domain. J Bone Miner Res. 1997;12:1210–1222. doi: 10.1359/jbmr.1997.12.8.1210. [DOI] [PubMed] [Google Scholar]
- 36.Tye CE, Rattray KR, Warner KJ, Gordon JAR, Sodek J, Hunter GK, Goldberg HA. Delineation of the hydroxyapatite-nucleating domains of bone sialoprotein. J Biol Chem. 2003;278:7949–7955. doi: 10.1074/jbc.M211915200. [DOI] [PubMed] [Google Scholar]
- 37.Mintz KP, Grzesik WJ, Midura RJ, Robey PG, Termine JD, Fisher LW. Purification and fragmentation of nondenatured bone sialoprotein: Evidence for a cryptic, RGD-resistant cell attachment domain. J Bone Miner Res. 1993;8:985–995. doi: 10.1002/jbmr.5650080812. [DOI] [PubMed] [Google Scholar]
- 38.Boskey AL, Maresca M, Ullrich W, Doty SB, Butler WT, Prince CW. Osteopontin–hydroxyapatite interactions in vitro: Inhibition of hydroxyapatite formation and growth in a gelatin-gel. Bone Miner. 1993;22:147–159. doi: 10.1016/s0169-6009(08)80225-5. [DOI] [PubMed] [Google Scholar]