Abstract
Transcription-coupled repair (TCR) is a pathway dedicated to the removal of damage from the template strands of actively transcribed genes. Although the detailed mechanism of TCR is not yet understood, it is believed to be triggered when a translocating RNA polymerase is arrested at a lesion or unusual structure in the DNA. Conventional assays for TCR require high doses of DNA damage for the statistical analysis of repair in the individual strands of DNA sequences ranging in size from a few hundred bases to 30 kb. The single cell gel electrophoresis (Comet) assay allows detection of single-or double-strand breaks at a 10 to 100-fold higher level of resolution. Fluorescence in situ hybridization (FISH) combined with the Comet assay (Comet-FISH) affords a heightened level of sensitivity for the assessment of repair in defined DNA sequences of cells treated with physiologically relevant doses of genotoxins. This approach also reveals localized susceptibility to chromosomal breakage in cells from individuals with hypersensitivity to radiation or chemotherapy. Several groups have reported preferential repair in transcriptionally active genes or chromosomal domains using Comet-FISH. The prevailing interpretation of the behavior of DNA in the Comet assay assumes that the DNA is arranged in loops and matrix-attachment sites; that supercoiled, undamaged loops are contained within the nuclear matrix and appear in Comet “heads”, and that Comet “tails” consist of relaxed DNA loops containing one or more breaks. According to this model, localization of FISH probes in Comet heads signifies that loops containing the targeted sequences are free of damage. This implies that preferential repair as detected by Comet-FISH might encompass large chromosomal domains containing both transcribed and non-transcribed sequences. We review the existing evidence and discuss the implications in relation to current models for the molecular mechanism of TCR.
Keywords: transcription-coupled repair, Comet-FISH, DNA repair deficient disease
1. Introduction
The genomes of living organisms are continuously at risk for endogenous and environmentally induced structural alterations. DNA lesions at specific genomic sites can lead to changes in nucleotide sequence, through processes such as translesion synthesis or recombination, causing mutagenesis and cellular responses that result in apoptosis.
For a few types of lesions, the altered DNA can be directly reversed to the original sequence, but in most cases the damaged DNA is processed by one or more of the several cellular excision repair mechanisms: nucleotide excision repair (NER), base excision repair (BER), and mismatch excision repair (MMR). DNA strand breaks are repaired rapidly when affecting only one strand, but more complex repair mechanisms have evolved that process double strand breaks [1]. In addition to these general pathways that operate throughout the genome, certain lesions on the transcribed strands of active genes are subject to a dedicated repair subpathway. Preferential repair of UV-induced cyclobutane pyrimidine dimers (CPD) in active genes was discovered in our laboratory for mammalian cells [2]. We then showed that enhanced repair in active genes was largely due to the selective repair of the transcribed DNA strands [3]. This phenomenon, termed transcription-coupled repair (TCR), was subsequently shown to occur in other organisms, including bacteria [4] and yeast [5–7].
The studies cited above were carried out using the so-called Southern blot method [3], an assay suitable for investigating damage and repair in the individual DNA strands of specific restriction fragments up to 30 kb in length. This method requires induction of an average of one lesion in each strand of the fragment of interest for Poisson statistical analysis. Methods such as the ligation-mediated polymerase chain reaction have been used for analysis of damage and repair at the nucleotide level in much shorter sequences [8]. Both approaches for measuring TCR necessitate relatively high exposures to damaging agents that might be too toxic to the cells or the organism under study; in addition, they are time- and labor-intensive. In this review we will evaluate the Comet assay combined with fluorescence in situ hybridization (FISH), or Comet-FISH, for analysis of TCR in cells exposed to low doses of genotoxic agents.
2. Repair deficient diseases
The localization of damage and the efficiency of repair in certain active DNA sequences may have different critical effects upon biological endpoints such as mutation, transformation or cell death, than damage distributed throughout the genome or its repair [9]. Cells that are genetically deficient in any of the several repair pathways provide valuable systems for investigating the roles of the proteins involved, their interactions with each other, and their functional associations with other cellular components. For example, deficiencies in NER cause xeroderma pigmentosum (XP), Cockayne syndrome (CS), and UV-sensitive syndrome (UVSS). These syndromes comprise several complementation groups and exhibit a broad variety of symptoms, but all cause hypersensitivity to sunlight.
XP patients are extraordinarily affected by skin cancer; appearance of the first skin tumor occurs by age 8 on average. The incidence of internal tumors is also elevated, and 20% of XP patients suffer from progressive neurological abnormalities. CS and UVSS patients are prone to sunburn and exhibit skin dryness, freckles, pigment anomalies and telangiectasia [10–12]. UVSS patients have no indications of neurological or developmental abnormalities, whereas CS patients present with dwarfism, hypogonadism, mental retardation, and premature aging in addition to photosensitivity; their average life expectancy is only 12 years. At the molecular level, CS and UVSS cells are deficient in TCR of UV-induced lesions [13,14], while cells belonging to XP complementation groups C or E (XP-C, XP-E) exhibit the opposite paradigm: photoproducts are removed only from transcribed strands but not from the global genome [15]. The other XP complementation groups exhibit defective repair through both the GGR and TCR pathways (XP-A, -B, -D, -F and -G), or defective replicative lesion bypass by DNA polymerase η (XP -V). Mutations in XPB, XPD or XPG, may result in combined XP/CS phenotypes.
The cellular and biochemical responses of UVSS cells to UV are indistinguishable from those of CS (reviewed in [16]). What could account for the striking differences between symptoms of CS and UVSS? Our hypothesis is that UVSS cells are proficient in TCR or in transcriptional bypass of lesions caused by reactive oxygen species in actively transcribed DNA strands while CS cells are deficient in such process(es). In support of this hypothesis, we have found that while both CS-B and UVSS cells are defective in host-cell reactivation (HCR) of UV-irradiated shuttle vectors, only CS cells exhibit defective HCR of vectors containing the oxidized bases thymine glycol (Tg) or 8-oxoG; we also found that CS-A and CS-B cells exhibit enhanced sensitivity to treatment with hydrogen peroxide (H2O2), while UVSS cells behave like wild type cells in that regard [17]. Previous reports claiming to document TCR of oxidative DNA lesions in wild type human cells, and the absence of that pathway in CS cells, have been retracted [18–21]. To date there is no direct biochemical evidence for the existence of a dedicated mechanism for removal of oxidative lesions from DNA strands that are templates for transcription. The Comet-FISH assay provides a novel approach to investigate this proposed preferential repair of oxidative lesions.
3. Comet and Comet-FISH assays
The Comet assay has been employed since the mid-1980s to study the effects of environmental pollutants and occupational hazards, the safety of therapeutic compounds, toxicology, and to assess DNA repair capacity in human, animal and plant populations; for reviews see [22,23]. In the Comet assay, cells are mixed with agarose and layered on microscope slides, where they are lysed and subjected to electrophoresis; staining with fluorescent dyes such as DAPI or ethidium bromide permits microscopic visualization of the “Comets”. DNA containing breaks unwinds and migrates away from the “head” (the nucleus), forming a “tail”; quantification of the amount of DNA in tails and in heads of Comets provides an estimate of the frequency of strand breaks.
Electrophoresis can be performed in alkaline or in neutral solutions, which detect single- and double-strand DNA breaks, respectively, although this has been disputed by some researchers. The early experiments of Östling and Johanson using low-dose gamma rays suggested that DNA breaks, either single- or double-stranded, relaxed DNA supercoils, which formed Comet tails during electrophoresis in neutral buffer [24]; at higher doses of irradiation, the increased frequency of double strand breaks caused fragmentation, thus Comet tails obtained after neutral electrophoresis consisted of detached DNA fragments [25]. However, Olive and coworkers have clearly shown that the neutral Comet assay detects only double-strand breaks [26]. Adding alkali prior to and/or during electrophoresis changes the appearance of Comets, a likely consequence of partial or complete denaturation of DNA fragments between breaks; this may increase the sensitivity of the assay. In addition, alkali-sensitive sites such as those caused by alkylation can be detected [27].
The basic method can be combined with lesion-specific glycosylases to convert lesions into strand breaks, greatly enhancing the range of genotoxic agents that can be tested [28]. Comets prepared at different times after infliction of DNA damage contain less DNA in the tails as the lesions are repaired; however, prolonged induction of damage such as that occurring with certain chemical treatments, or delayed incision might result in different patterns of repair, while DNA fragmentation in apoptotic cells could obscure observation of DNA lesions and their repair.
The molecular events that occur during processing of the cells and DNA to generate Comets have been discussed in several papers. The general rationale is that the DNA in chromatin is arranged in “matrix attachment sites” and “loops”; in undamaged, non-dividing cells, the loops are tightly supercoiled. Upon mild detergent treatment to release histones and other DNA binding proteins, the DNA remains supercoiled within the skeleton of the nuclear membrane. In principle, one single-strand break is sufficient to release the superhelix tension in a loop, which is then free and can extend out from the nucleus; when the amount of damage is such that several loops have been affected, they form a “halo” that can be seen around the more intensely stained nucleus [29,30]. It should be mentioned here that the constitution, and even the existence of the nuclear matrix has been the subject of considerable controversy; it has been proposed that the nuclear matrix or scaffold is an operationally defined entity, dependent upon the experimental conditions [31,32]. Treatment of cells with salt and detergent has been used to prepare “nucleoids” [29]; the effects of incubation and electrophoresis at high pH on these structures are not known but it has been proposed that a nuclear membrane skeleton is retained, to which scaffold- or matrix-associated regions of DNA remain attached [33].
FISH was first used in combination with cells electrophoresed within an agarose gel to study the spatial localization of various chromosomal structures along stretched DNA fibers [34]. Soon thereafter, several groups applied FISH to Comets for research on DNA damage in specific DNA sequences; the term “Comet-FISH” was proposed by C. Bock and colleagues [35]. Examples of Comet-FISH applied to DNA damage and repair include the use of a probe for human chromosome 1 in doxorubicin-treated human lymphocytes [36], various heterochromatic and euchromatic chromosomal domain probes in the plant Vicia faba treated with endonucleases [37], whole chromosome probes in UVA-irradiated human lymphocytes [38], whole chromosome paints for measurement of DNA breaks in human oropharyngeal mucosa cells exposed to benzo[a]pyrene-diol epoxide [39], peptide nucleic acid (PNA) probes to detect telomeric repeats in peripheral blood cells treated with bleomycin or mitomycin C [40], the HER-2 and p53 loci in breast cancer cells [41], telomeres in human leukocytes treated with bleomycin and cisplatin [42], the Ret, c-Abl and Trp53 loci in peripheral blood cells to study gene fragmentation resulting from X-ray irradiation of mice [43], the 5q31 and 11q23 loci, susceptible to breakage in acute myeloid leukemia [44], and the APC, KRAS and p53 regions in human colon cells treated with agents thought to be relevant to the etiology of colon cancer [45]. Some probes used for FISH in the examples cited above were purchased from commercial sources as DNA fragments [36,39,41,44] or PNA [40,42] pre-labeled with fluorescent dyes to allow direct detection, or with digoxygenin, which requires a second step with enzyme-coupled anti-digoxygenin [38]. In some cases the probes were made in-house by PCR, using digoxygenin-11-dUTP and amplification with 3 rounds of antibodies [37], or labeling PCR fragments with biotin by nick-translation and detection with FITC-avidin [43].
An imaginative modification of the Comet-FISH assay can be used to detect induction and repair of DNA interstrand crosslinks: by using the crosslinking treatment in combination with a treatment that induces single-strand breaks, the extent of crossslinking, which inhibits the separation of the complementary DNA strands, can be assessed by the reduction in migration induced by the strand-breaking agent. Using this methodogy, it was shown that DNA interstrand crosslinks induced by mytomicin C were repaired faster in the p53 domain than in the genome overall [46].
A technique similar to Comet-FISH was used to examine repair of UV-induced CPD in human skin fibroblasts, by incubation of the Comets with a CPD-specific antibody and then with a fluorescent, Cy3-labeled secondary antibody. This method allowed simultaneous visualization of the CPD-specific signal and the ethidium bromide-stained DNA in the heads and tails of the Comets [47].
The work of Horvâthovâ and colleagues [33] involved small, oligonucleotide-sized probes that were used to detect 5′ and 3′ sequences (near or at a distance from the promoter) within genes; this approach adds a higher degree of sensitivity to the assay. Their results are consistent with the idea that for genes not containing matrix attachment sites, both the 3′ and 5′ regions of a gene will be found either in the head or in the tail, unless a break occurs between those regions.
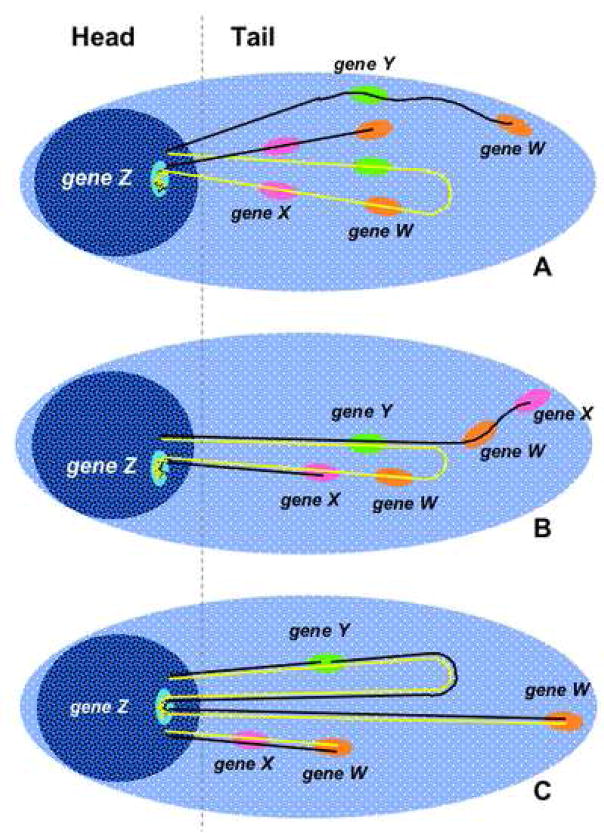
An interesting question is whether the complementary strands in a loop migrate into the tail independently or together upon alkaline denaturation and electrophoresis. Comet-FISH with a probe for the p53 gene was applied to cells that had been damaged by ionizing radiation; using parameters such as percent DNA in the tails, number of breaks, approximate loop size and numbers of probe-specific “spots” in heads and tails, the investigators found that their results favored a model in which both strands in a loop migrate into the tail, but separately, even in cases in which one strand is broken and the complementary strand is intact [46]. In support of this interpretation, Collins and coworkers stained Comets with acridine orange and observed that neutralization after alkaline electrophoresis allowed intact supercoils in Comet heads to reanneal, while DNA in tails remained single-stranded [48]. The diagrams in figure 1 (adapted from [33]) depict the DNA attached to the matrix or extended in loops, and the possible patterns of migration of sequence-specific spots, which may depend on the location of the breaks within or outside of the probed sequences and with respect to the attachment sites.
Figure 1. Diagram showing the structure of a Comet with FISH.
Duplex DNA is arranged in loops between matrix-attachment sites (MAS). Intact loops are supercoiled and remain within the head; loops containing one or more strand breaks are relaxed and extend forming the tail. In undamaged cells containing a diploid set of chromosomes, the two loops containing each of the pair of alleles for each gene will usually result in two separate spots; each loop can sustain damage, potentially increasing the number of spots. For simplicity we represent only one allele for each gene, thus the total number of spots could be higher than that given here. For alkaline Comet-FISH, two possible models are proposed for the distribution of domain-specific signals: in panel A, the complementary DNA strands in each loop migrate separately, thus the double-stranded probes annealing to each DNA strand appear in two or more spots regardless of the location of the break; however, a nick within the probed sequence would result in an additional spot (gene W). In panel B, the denatured strands migrate close to each other up to the nick, then the nicked strand unwinds away from its partner strand, thus signals could appear in one or more spots, depending on the location and number of nicks. Gene Z, near a MAS, is always found in heads. Panel C depicts a neutral Comet-FISH. Although the presence of a single nick is sufficient to relax the loop, the complementary DNA strands remain annealed. Intact sequences and those containing single-strand breaks appear in one spot (genes X and Y); genes containing a double strand break may appear in two spots (gene W).
4. Discussion
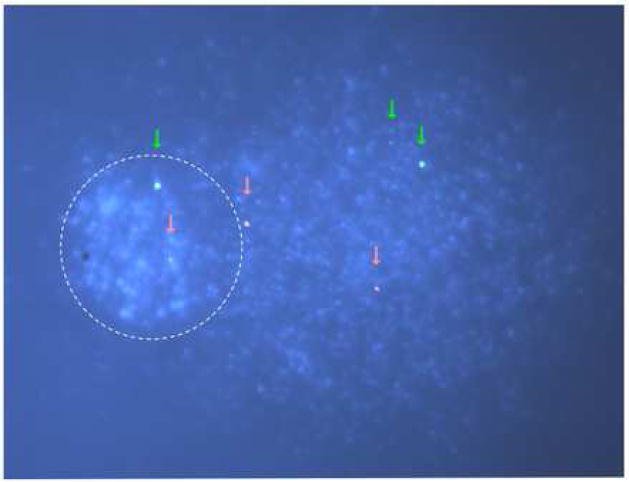
Conventional methods for determination of TCR such as the Southern-blot and ligation-mediated PCR require high doses of DNA damage. The alkaline Comet assay provides an extremely sensitive assay for detection of DNA lesions that can be converted to single strand breaks, allowing examination of damage and repair by genotoxic agents at subtoxic, physiologically relevant exposures. Therefore, combining the Comet assay with FISH should be a workable alternative for measuring TCR at doses of DNA–damaging agents that are 10 to 100-fold lower than those used in the traditional methods for analysis of TCR. As discussed above, the Comet-FISH assay allows detection of specific genes or domains in Comet tails heads; FISH signals in heads presume repair of all the lesions lying anywhere within the “loop” containing the fragment of interest and resumption of supercoiling. Quantification of repair in specific domains can be expressed as the percent of spots in heads and tails. In the example depicted in figure 2, a cell damaged with UVC has most of its DNA in the Comet tail; FISH probes for the transcriptionally active p53 and the inactive chromosome 7 centromere regions appear mostly in the tail as well.
Figure 2. Representative image from a Comet-FISH assay.
Primary human skin fibroblasts were treated with 0.1 J/m2 UVC, harvested, processed for alkaline Comet essentially as described in [33] and for FISH as described in [53]. Fluorescent probes were obtained from commercial sources, labeled with SpectrumOrange for a 145 kb domain containing the p53 gene, and with Direct-Green for the centromeric region of chromosome 7. One XP-C cell is shown from a slide prepared with cells collected immediately after UV-irradiation; the cells were treated with T4 endonuclease V to cut the DNA at the sites of CPD. The Comet head shows more intense DAPI staining than the tail. The signals from the FISH probes are shown as captured by the software, without enhancements. Red and green arrows indicate the positions of the signals for the p53 and chromosome 7 centromeric regions respectively.
Several groups have reported preferential repair of active genes or domains using Comet-FISH; their results are listed in Table I. TCR of UV-induced CPD in mammalian cells requires several proteins, including CSA, CSB and XAB2 in addition to all the factors in the GGR pathway, except those involved in lesion recognition, XPC and XPE/DDB2. It also requires active transcription by RNA polymerase II. Our Comet-FISH experiments using human primary skin fibroblasts (to be reported elsewhere) reveal a requirement for CSB, but not for XPC, for efficient repair of UVC-induced CPD in the active p53 gene. Our observation of lack of preferential repair in the centromeric region of chromosome 7 can result from the absence of transcription in the region, or from its heterochromatic structure.
Table I.
Reports of preferential repair of actively transcribed genes or domains detected by Comet-FISH.
| Cells | Agent | Probe | Reference |
|---|---|---|---|
| Human bladder carcinoma | X-rays | p53 | [54] |
| Human bladder carcinoma | MMC | p53 | [46] |
| Human bladder carcinoma | gamma-rays | p53 | [53] |
| Chinese hamster ovary | UV, H2O2, RO+VL | DHFR, MGMT | [33] |
| Normal human lymphocytes | H2O2, RO+VL | p53 | [33] |
| Breast cancer cells | gamma-rays | p53, HER-2 | [41] |
MMC, mitomycin C; DHFR, dihydrofolate reductase; MGMT, O6-methyguanine DNA methyltransferase; RO+VL, RO 19-8022 and visible light induce mostly oxidation of guanine.
The meaning of the reported preferential (faster) repair of active genes observed using Comet-FISH (see Table I) is unclear: the DNA regions detected by the probes usually contain both transcribed and silent genes, and generally only one strand of an active gene serves as template for transcription. The model depicted in figure 1, supported by the results of McKenna et al. and Horvâthovâ et al. [33,46], suggests that, aside from sequences attached to the nuclear matrix and always found in Comet heads, localizing any sequence of any size in the heads of Comets requires that the entire loop containing such sequences be free of damage. This presents us with a conundrum: if the two strands of a loop will continue to migrate to the tail until all the lesions and breaks are repaired, it might not be possible to detect TCR at the gene level by counting signal spots in Comet heads and tails.
Our results are consistent with TCR; however, it is difficult to reconcile the hypothesis that TCR is initiated when an elongating RNAPII is blocked at an obstacle, with the low frequency of lesions intrinsic to the Comet assay. If TCR is confined to the transcribed strand of active genes, it cannot be solely responsible for the faster rate of repair of entire loops. Is there a second-order category with respect to lesion recognition and access by DNA repair complexes in chromatin, such that if a loop contains at least one gene subject to TCR, the entire loop will be repaired by this pathway? The idea that TCR may affect the rate of repair of lesions in sequences that are not templates for transcription is supported by the observations of G. Kantor and colleagues, who found that after UV-irradiation, preferentially repaired DNA contained both active and inactive genes [49], and that fragments within a ~50kb genomic domain containing the p53 gene were preferentially repaired, although only the ~20kb p53 template strand was actively transcribed [50]. Additionally, repair of UV-induced CPD in the non-template strands of the DHFR, p53, AFP and ada genes in wild type human cells is faster than in either DNA strand in TCR-deficient CS-B or UVSS cells, suggesting a role for TCR in repair of non-transcribed DNA strands [14,51]. However, in terminally differentiated cells in which overall repair of CPD is depressed, repair of transcriptionally silent DNA sequences depends on transcription of the template strand complementary to or in the vicinity of the strand under analysis, but does not require TCR factors. This mode of repair, termed domain-associated repair, is carried out through the GGR pathway [52].
Further experiments will be necessary to assess the roles of transcription and/or chromatin structure in TCR of active genes as detected by Comet-FISH. Refinements of the technique, such as development of fluorescent single-stranded probes, could aid the clarification of ambiguities in the interpretation of data provided by Comet-FISH assays.
Acknowledgments
We are grateful to Marcela Gona and Gayle Campbell for technical and administrative assistance, and to Patricia Escobar, Declan McKenna, Andrew Collins, and members of the Comet Assay Interest Group (http://www.cometassay.com) for advice and encouragement. This work was supported by a grant CA91456 from NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedberg E, Walker G, Siede W, Wood R, Schultz R, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington DC: 2006. [Google Scholar]
- 2.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient that in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 3.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 4.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 5.Leadon SA, Lawrence DA. Strand-selective repair of DNA damage in the yeast gal7 gene requires RNA polymerase-II. J Biol Chem. 1992;267:23175–23182. [PubMed] [Google Scholar]
- 6.Smerdon MJ, Thoma F. Site-specific DNA-repair at the nucleosome level in a yeast minichromosome. Cell. 1990;61:675–684. doi: 10.1016/0092-8674(90)90479-x. [DOI] [PubMed] [Google Scholar]
- 7.Sweder KS, Hanawalt PC. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription, Proc. Natl Acad Sci USA. 1992;89:10696–10700. doi: 10.1073/pnas.89.22.10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeifer GP, Drouin R, Riggs AD, Holmquist GP. In vivo mapping of a DNA adduct at nucleotide resolution: detection of pyrimidine (6-4) pyrimidone photoproducts by ligation-mediated polymerase chain reaction, Proc. Natl Acad Sci USA. 1991;88:1374–1378. doi: 10.1073/pnas.88.4.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanawalt PC. Controlling the efficiency of excision repair, Mutat. Res. 2001;485:3–13. doi: 10.1016/s0921-8777(00)00071-9. [DOI] [PubMed] [Google Scholar]
- 10.Itoh T, Ono T, Yamaizumi M. A new UV-sensitive syndrome not belonging to any complementation groups of xeroderma pigmentosum or Cockayne syndrome: siblings showing biochemical characteristics of Cockayne syndrome without typical clinical manifestations, Mutat. Res DNA Repair. 1994;314:233–248. doi: 10.1016/0921-8777(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 11.Itoh T, Fujiwara Y, Ono T, Yamaizumi M. UVs syndrome, a new general category of photosensitive disorder with defective DNA repair, is distinct from xeroderma pigmentosum variant and rodent complementation group I, Am. J Hum Genet. 1995;56:1267–1276. [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh T, Yamaizumi M, Ichihashi M, Hiro-Oka M, Matsui T, Matsuno M, Ono T. Clinical characteristics of three patients with UVs syndrome, a photosensitive disorder with defective DNA repair, Brit. J Dermat. 1996;134:1147–1150. [PubMed] [Google Scholar]
- 13.Venema J, Mullenders LHF, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne Syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA, Proc. Natl Acad Sci USA. 1991;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spivak G, Itoh T, Matsunaga T, Nikaido O, Hanawalt P, Yamaizumi M. Ultraviolet-sensitive syndrome cells are defective in transcription-coupled repair of cyclobutane pyrimidine dimers. DNA Repair. 2002;1:629–643. doi: 10.1016/s1568-7864(02)00056-3. [DOI] [PubMed] [Google Scholar]
- 15.Venema J, van Hoffen A, Karcagi V, Natarajan AT, van Zeeland AT, Mullenders LHF. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes, Mol. Cell Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spivak G. UV-sensitive syndrome, Mutat. Res. 2005;577:162–169. doi: 10.1016/j.mrfmmm.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome cells. DNA Repair. 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Le Page F, Kwoh EE, Avrutskaya A, Gentil A, Leadon SA, Sarasin A, Cooper PK. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell. 2005;123:711. doi: 10.1016/j.cell.2005.11.005. [retraction of Le Page F, Kwoh EE, Avrutskaya A, Gentil A, Leadon SA, Sarasin A, Cooper PK Cell 2000 Apr 14;101(2):159–71; PMID: 10786832] [DOI] [PubMed] [Google Scholar]
- 19.Cooper PK, Nouspikel T, Clarkson SG. Retraction. Science. 2005;308:1740. doi: 10.1126/science.308.5729.1740b. [retraction of Cooper PK, Nouspikel T, Clarkson SG, Leadon SA. Science. 1997 Feb 14;275(5302):990–3; PMID: 9020084] [DOI] [PubMed] [Google Scholar]
- 20.Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA. Retraction. Science. 2003;300:1657. doi: 10.1126/science.300.5626.1657b. [retraction of Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA. Science. 1998 Aug 14;281(5379):1009–12; PMID: 9703501] [DOI] [PubMed] [Google Scholar]
- 21.Leadon SA. Retraction. DNA Repair. 2003;2:361. doi: 10.1016/s1568-7864(02)00236-7. [erratum appears in DNA Repair (Amst) 2003 Oct 7;2(10):1157][retraction of Leadon SA, Avrutskaya AV Mutat Res 1998 Mar;407(2):177–87; PMID: 9637246] [DOI] [PubMed] [Google Scholar]
- 22.Moller P. Genotoxicity of environmental agents assessed by the alkaline comet assay. Basic & clinical pharmacology & toxicology. 2005;96:1–42. [PubMed] [Google Scholar]
- 23.Moller P. The alkaline comet assay: towards validation in biomonitoring of DNA damaging exposures. Basic & clinical pharmacology & toxicology. 2006;98:336–345. doi: 10.1111/j.1742-7843.2006.pto_167.x. [DOI] [PubMed] [Google Scholar]
- 24.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochemical & Biophysical Research Communications. 1984;123:291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 25.Ostling O, Johanson KJ. Bleomycin, in contrast to gamma irradiation, induces extreme variation of DNA strand breakage from cell to cell. International Journal of Radiation Biology & Related Studies in Physics, Chemistry & Medicine. 1987;52:683–691. doi: 10.1080/09553008714552201. [DOI] [PubMed] [Google Scholar]
- 26.Olive PL, Wlodek D, Banath JP. DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Research. 1991;51:4671–4676. [PubMed] [Google Scholar]
- 27.Klaude M, Eriksson S, Nygren J, Ahnstrom G. The comet assay: mechanisms and technical considerations. Mutation Research. 1996;363:89–96. doi: 10.1016/0921-8777(95)00063-1. [DOI] [PubMed] [Google Scholar]
- 28.Collins AR, Duthie SJ, Dobson VL. Direct enzymic detection of endogenous oxidative base damage in human lymphocyte DNA. Carcinogenesis. 1993;14:1733–1735. doi: 10.1093/carcin/14.9.1733. [DOI] [PubMed] [Google Scholar]
- 29.Cook PR, Brazell IA, Jost E. Characterization of nuclear structures containing superhelical DNA. Journal of cell science. 1976;22:303–324. doi: 10.1242/jcs.22.2.303. [DOI] [PubMed] [Google Scholar]
- 30.Mullenders L, van Zeeland A, Natarajan A. The localization of ultraviolet-induced excision repair in the nucleus and the distribution of repair events in higher order chromatin loops in mammalian cells. J Cell Sci Suppl. 1987;6:243–262. doi: 10.1242/jcs.1984.supplement_6.17. [DOI] [PubMed] [Google Scholar]
- 31.Jack RS, Eggert H. The elusive nuclear matrix. European Journal of Biochemistry. 1992;209:503–509. doi: 10.1111/j.1432-1033.1992.tb17314.x. [DOI] [PubMed] [Google Scholar]
- 32.Nickerson J. Experimental observations of a nuclear matrix. Journal of Cell Science. 2001;114:463–474. doi: 10.1242/jcs.114.3.463. [DOI] [PubMed] [Google Scholar]
- 33.Horvâthovâ E, Dusinskâ M, Shaposhnikov S, Collins AR. DNA damage and repair measured in different genomic regions using the comet assay with fluorescent in situ hybridization. Mutagenesis. 2004;19:269–276. doi: 10.1093/mutage/geh030. [DOI] [PubMed] [Google Scholar]
- 34.Santos SJ, Singh NP, Natarajan AT. Fluorescence in situ hybridization with comets, Exp. Cell Res. 1997;232:407–411. doi: 10.1006/excr.1997.3555. [DOI] [PubMed] [Google Scholar]
- 35.Bock C, Rapp A, Dittmar H, Monajembashi S, Greulic K-O. Localization of specific sequences and DNA single-strand breaks in individual UV-A-irradiated human lymphocytes by COMET FISH. In: Bigio I, Schneckenburger H, Slavik J, Svanberg K, Viallet P, editors. Optical Biopsies and Microscopic Techniques III, Proc SPIE. 1999. pp. 207–217. [Google Scholar]
- 36.Anderson D, Yu TW, Browne MA. The use of the same image analysis system to detect genetic damage in human lymphocytes treated with doxorubicin in the Comet and fluorescence in situ hybridisation (FISH) assays. Mutation research. 1997;390:69–77. doi: 10.1016/s0165-1218(96)00167-x. [DOI] [PubMed] [Google Scholar]
- 37.Menke M, Angelis KJ, Schubert I. Detection of specific DNA lesions by a combination of comet assay and FISH in plants, Envir. Molec Mut. 2000;35:132–138. doi: 10.1002/(sici)1098-2280(2000)35:2<132::aid-em8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 38.Rapp A, Bock C, Dittmar H, Greulich KO. UV-A breakage sensitivity of human chromosomes as measured by COMET-FISH depends on gene density and not on the chromosome size. J Photochem Photobiol B, Biology. 2000;56:109–117. doi: 10.1016/s1011-1344(00)00052-x. [DOI] [PubMed] [Google Scholar]
- 39.Harreus UA, Kleinsasser NH, Zieger S, Wallner B, Reiter M, Schuller P, Berghaus A. Sensitivity to DNA-damage induction and chromosomal alterations in mucosa cells from patients with and without cancer of the oropharynx detected by a combination of Comet assay and fluorescence in situ hybridization, Mutat. Res. 2004;563:131–138. doi: 10.1016/j.mrgentox.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 40.Arutyunyan R, Gebhart E, Hovhannisyan G, Greulich KO, Rapp A. Comet-FISH using peptide nucleic acid probes detects telomeric repeats in DNA damaged by bleomycin and mitomycin C proportional to general DNA damage. Mutagenesis. 2004;19:403–408. doi: 10.1093/mutage/geh049. [DOI] [PubMed] [Google Scholar]
- 41.Kumaravel TS, Bristow RG. Detection of genetic instability at HER-2/neu and p53 loci in breast cancer cells sing Comet-FISH. Breast cancer research and treatment. 2005;91:89–93. doi: 10.1007/s10549-004-5780-0. [DOI] [PubMed] [Google Scholar]
- 42.Arutyunyan R, Rapp A, Greulich KO, Hovhannisyan G, Haroutiunian S, Gebhart E. Fragility of telomeres after bleomycin and cisplatin combined treatment measured in human leukocytes with the Comet-FISH technique, Exp. Oncol. 2005;27:38–42. [PubMed] [Google Scholar]
- 43.Amendola R, Basso E, Pacifici PG, Piras E, Giovanetti A, Volpato C, Romeo G. Ret. Abl1 (cAbl) and Trp53 gene fragmentations in comet-FISH assay act as in vivo biomarkers of radiation exposure in C57BL/6 and CBA/J mice. Radiation Res. 2006;165:553–561. doi: 10.1667/RR3544.1. [DOI] [PubMed] [Google Scholar]
- 44.Escobar PA, Smith MT, Vashista A, Hubbard AE, Zhang L. Leukemia-specific chromosome damage detected by comet with fluorescence in situ hybridization (comet-FISH) Mutagenesis advance access. 2007 doi: 10.1093/mutage/gem020. [DOI] [PubMed] [Google Scholar]
- 45.Glei M, Schaeferhenrich A, Claussen U, Kuechler A, Liehr T, Weise A, Marian B, Sendt W, Pool-Zobel BL. Comet fluorescence in situ hybridization analysis for oxidative stress-induced DNA damage in colon cancer relevant genes. Toxicological sciences: an official journal of the Society of Toxicology. 2007;96:279–284. doi: 10.1093/toxsci/kfl197. [DOI] [PubMed] [Google Scholar]
- 46.McKenna DJ, Gallus M, McKeown SR, Downes CS, McKelvey-Martin VJ. Modification of the alkaline Comet assay to allow simultaneous evaluation of mitomycin C-induced DNA cross-link damage and repair of specific DNA sequences in RT4 cells. DNA repair. 2003;2:879–890. doi: 10.1016/s1568-7864(03)00086-7. [DOI] [PubMed] [Google Scholar]
- 47.Sauvaigo S, Serres C, Signorini N, Emonet N, Richard MJ, Cadet J. Use of the single-cell gel electrophoresis assay for the immunofluorescent detection of specific DNA damage, Anal. Biochem. 1998;259:1–7. doi: 10.1006/abio.1998.2628. [DOI] [PubMed] [Google Scholar]
- 48.Collins AR, Dobson VL, Dusinska M, Kennedy G, Sttina R. The comet assay: what can it really tell us? Mutation Research. 1997;375:183–193. doi: 10.1016/s0027-5107(97)00013-4. [DOI] [PubMed] [Google Scholar]
- 49.Shanower GA, Kantor GJ. A difference in the pattern of repair in a large genomic region in UV-irradiated normal human and Cockayne syndrome cells, Mutat. Res. 1997;385:127–137. doi: 10.1016/s0921-8777(97)00038-4. [DOI] [PubMed] [Google Scholar]
- 50.Tolbert DM, Kantor GJ. Definition of a DNA repair domain in the genomic region containing the human p53 gene. Cancer Res. 1996;56:3324–3330. [PubMed] [Google Scholar]
- 51.van Hoffen A, Natarajan AT, Mayne LV, van Zeeland AA, Mullenders LHF, Venema J. Deficient repair of the transcribed strand of actives genes in Cockayne’s syndrome cells, Nucl. Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nouspikel TP, Hyka-Nouspikel N, Hanawalt PC. Transcription domain-associated repair in human cells. Molecular and cellular biology. 2006;26:8722–8730. doi: 10.1128/MCB.01263-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKenna DJ, Rajab NF, McKeown SR, McKerr G, McKelvey-Martin VJ. Use of the comet-FISH assay to demonstrate repair of the TP53 gene region in two human bladder carcinoma cell lines. Radiation Res. 2003;159:49–56. doi: 10.1667/0033-7587(2003)159[0049:uotcfa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 54.McKelvey-Martin VJ, Ho ET, McKeown SR, Johnston SR, McCarthy PJ, Rajab NF, Downes CS. Emerging applications of the single cell gel electrophoresis (Comet) assay. I Management of invasive transitional cell human bladder carcinoma II Fluorescent in situ hybridization Comets for the identification of damaged and repaired DNA sequences in individual cells. Mutagenesis. 1998;13:1–8. doi: 10.1093/mutage/13.1.1. [DOI] [PubMed] [Google Scholar]