Abstract
N-Linked glycosylation begins with the formation of a dolichol-linked oligosaccharide in the endoplasmic reticulum (ER). The first two steps of this pathway lead to the formation of GlcNAc2-PP-dolichol, whose synthesis is sequentially catalyzed by the Alg7p GlcNAc phosphotransferase and by the dimeric Alg13p/Alg14p UDP-GlcNAc transferase on the cytosolic face of the endoplasmic reticulum. Here, we show that the Alg7p, Alg13p, and Alg14p glycosyltransferases form a functional multienzyme complex. Coimmunoprecipitation and gel filtration assays demonstrate that the Alg7p/Alg13p/Alg14p complex is a hexamer with a native molecular weight of ∼200 kDa and an Alg7p:Alg13:Alg14p stoichiometry of 1:1:1. These results highlight and extend the striking parallels that exist between these eukaryotic UDP-GlcNAc transferases and their bacterial MraY and MurG homologs that catalyze the first two steps of the lipid-linked peptidoglycan precursor. In addition to their preferred substrate and lipid acceptors, these enzymes are similar in their structure, chemistry, temporal, and spatial organization. These similarities point to an evolutionary link between the early steps of N-linked glycosylation and those of peptidoglycan synthesis.
Keywords: endoplasmic reticulum, glycosylation, lipid-linked oligosaccharide, peptidoglycan, UDP-GlcNAc glycosyltransferase
Introduction
Most secreted and membrane-bound eukaryotic proteins are modified by the addition of an asparagine (N)-linked oligosaccharide. This modification is important for intracellular protein folding and quality control, and for the maintenance of the extracellular scaffold that regulates a cell's surface properties and shape. Synthesis of the N-linked glycan begins in the endoplasmic reticulum (ER) with the assembly of a lipid-linked oligosaccharide (LLO) containing 14 sugars (Glc3Man9GlcNAc2-PP-dol). The first seven sugars are added on the cytosolic face of the ER by membrane-bound glycosyltransferases that produce Man5GlcNAc2-PP-dol. After flipping into the lumen of the ER, this Man5GlcNAc2-PP-dol intermediate is extended by another seven sugars in the ER lumen (Helenius and Aebi 2004). The resulting Glc3Man9GlcNAc2 oligosaccharide is transferred from dolichol to select asparagine residues of nascent proteins. All of the 12 different membrane-bound ER glycosyltransferases that catalyze these sequential sugar transfer reactions have been identified, and all are highly conserved, from yeast to man (Weerapana and Imperiali 2006). The biological importance of the LLO is underscored by the finding that even mild defects in its assembly manifest as severe phenotypes in humans that are afflicted with congenital disorders of glycosylation (Aebi and Hennet 2001).
The first two steps of LLO assembly form the chitobiose disaccharide “core” that consists of two GlcNAc residues attached to pyrophosphoryldolichol. It has been suggested that these first two steps are important regulatory sites that influence N-glycan substrate availability and thus flux through the glycosylation pathway (Lehrman 1991), but our understanding of the mechanisms that regulate these reactions are limited. The first GlcNAc is added by the GlcNAc-1-phosphate (P) transferase Alg7p (also known as GPT) that adds GlcNAc-1-P from UDP-GlcNAc to produce GlcNAc-PP-dol (Lehrman 1991). Alg7p has multiple transmembrane spanning domains and a catalytic site that faces the cytosol (Dan and Lehrman 1997). In the second step, the Alg13p/Alg14p UDP-GlcNAc transferase adds a second GlcNAc from UDP-GlcNAc to produce GlcNAc2-PP-dol (Bickel et al. 2005; Chantret et al. 2005; Gao et al. 2005). The Alg13p/Alg14p UDP-GlcNAc transferase is an unusual glycosyltransferase because its catalytic and membrane-anchoring domains reside on two separate polypeptides. Alg13p is the soluble, cytosolic subunit that contains the catalytic domain; Alg14p is the membrane-spanning subunit that is required to recruit Alg13p to the cytosolic face of the ER (Bickel et al. 2005; Chantret et al. 2005; Gao et al. 2005; Averbeck et al. 2007).
Unlike the other ER glycosyltransferases involved in LLO assembly, both Alg7p and Alg13p/Alg14p display significant sequence similarity to bacterial proteins involved in the early stages of peptidoglycan synthesis. Alg7p has long been recognized as a member of the conserved family of UDP-HexNAc-1-P transferases (Lehrman 1994; Price and Momany 2005) that includes the bacterial MraY glycosyltransferase that catalyzes the formation of the first lipid-linked intermediate of peptidoglycan. Like Alg7, MraY is a membrane-associated phospho-sugar transferase that acts on a polyprenyl lipid-linked acceptor, transferring phospho-N-acetyl muramyl pentapeptide (MurNAc) from UDP-MurNAc to undecaprenyl-phosphate. MraY exists as part of a membrane-associated protein complex that includes MurG (Mohammadi et al. 2007), a peripherally associated glycosyltransferase that extends MurNAc with a single GlcNAc from UDP-GlcNAc (Mengin-Lecreulx et al. 1991; Mohammadi et al. 2007) to form the peptidoglycan precursor, GlcNAc-MurNAc-lipid. Remarkably, Alg13 and Alg14 are similar in sequence and predicted structure to MurG, which has a distinct two-domain structure (Ha et al. 2000). Alg13p is homologous to the C-terminal catalytic domain of MurG, while Alg14p is homologous to the N-terminal domain of MurG that contains the putative lipid-acceptor motif (Bickel et al. 2005; Chantret et al. 2005; Gao et al. 2005; Averbeck et al. 2007).
Here, we provide evidence that like their bacterial counterparts, the eukaryotic Alg7p, Alg13p, and Alg14p UDP-GlcNAc transferases also physically interact to form a functional hetero-oligomeric glycosyltransferase complex. These results demonstrate that the parallels between the early steps of N-glycosylation and those of peptidoglycan synthesis extend beyond the sequence similarities of the prokaryotic and eukaryotic catalytic enzymes. The evolutionary conservation of these protein–protein interactions also provides new insights that may impact on the design of specific antibiotics that target the essential early steps of peptidoglycan synthesis.
Results
Alg14p fractionates in a large protein complex
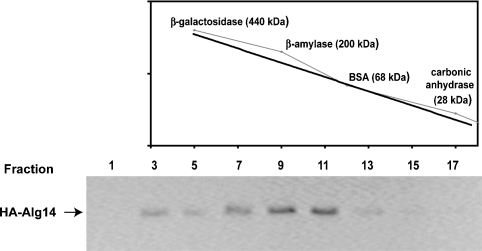
As a first step toward the biochemical characterization of the Alg13p/Alg14p UDP-GlcNAc hetero-oligomeric transferase, we investigated the native molecular weight of the Alg13p/Alg14p complex using gel filtration chromatography. Previous studies demonstrated that the Alg13p/Alg14p complex remains stable in the presence of 0.1% Triton X-100 (Averbeck et al. 2007) so buffers containing 0.1% Triton were used to solubilize the Alg13p/Alg14p complex and during its fractionation over a Superose 12 column (see Materials and methods). These detergent extracts were prepared from a yeast strain that produces HA-tagged Alg14p. In this strain (XGY151), the normal ALG14 gene was replaced with a GAL1-promoter-driven HA-tagged ALG14 (Gao et al. 2005). This strain grows normally in galactose-containing media, has no glycosylation defects, and grows in a manner that is indistinguishable from a wild-type strain, demonstrating that HA-tagged Alg14p functions normally (Averbeck et al. 2007). After size fractionation, aliquots from each fraction were analyzed by SDS–PAGE and subjected to immunoblotting with anti-HA antibodies to identify fractions that contain HA-Alg14p. The predicted molecular weight of HA-Alg14p is −30 kDa and that of Alg13p is −23 kDa. Therefore, if this complex was a simple hetero-dimer, we expected that Alg14p would cofractionate with molecular weight markers of −53 kDa. However, the majority of HA-Alg14p (and Alg13p, see below) cofractionated with molecular weight protein markers in the 160–200 kDa range (Figure 1). The leading edge of the Alg14p-containing fractions leaned toward the larger molecular weight markers (−200 kDa), and Alg14p was not detectable in fractions containing the lower molecular weight markers. These results suggested that the native molecular weight of the Alg14p-containing oligomer is significantly larger than the predicted mass of the Alg13p/Alg14p dimer and that Alg14p exists in a large molecular weight complex.
Fig. 1.
Gel filtration chromatography of the Alg14p membrane subunit of the ER UDP-GlcNAc transferase. Detergent extracts from cells expressing HA-ALG14 (XGY151) (Gao et al. 2005) were prepared in the presence of 0.1% Triton X-100 and fractionated over a Superose 12 column (see Materials and methods). An aliquot of each fraction was subjected to SDS–PAGE and analyzed by immunoblotting with the anti-HA-antibody. Molecular weight protein calibration markers, whose fractionation profile is graphed in the upper panel, were fractionated in parallel under identical conditions.
Alg7 and Alg14 coimmunoprecipitate
The large size of the Alg14p-containing protein complex implied that it contains multiple copies of Alg14p and/or Alg13p, or that this complex contains additional components. Several observations prompted us to examine whether or not Alg7p associates with Alg14p. Hamster Alg7p/GPT has been reported to form oligomers (Dan and Lehrman 1997) and also has a larger than expected native molecular weight (Shailubhai et al. 1988). Furthermore, a precedent exists for the idea that related ER glycosyltransferases function as hetero-oligomeric complexes: the Alg1, Alg2, and Alg11 mannosyltransferases that elongate GlcNAc2-PP-dol to Man5GlcNAc2-PP-dol on the cytosplasmic face of the ER do so as part of hetero-oligomeric enzyme complexes (Gao et al. 2004).
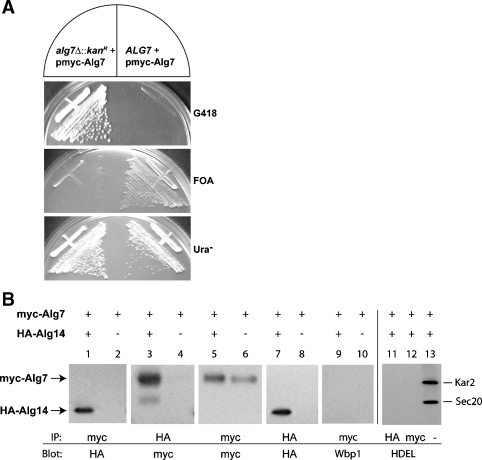
To explore the possibility that the related ER UDP-GlcNAc transferases (Alg7p, Alg14p, and Alg13p) also form hetero-oligomeric complexes as is shown by the Alg1, Alg2, and Alg11 mannosyltransferases, we performed experiments to determine if Alg14p interacts with Alg7p. For this purpose, yeast strains that solely express these epitope-tagged versions of Alg14 and Alg7 were assayed to ensure that these tagged alleles do not affect normal growth or glycosylation. We found that placing an epitope tag at the carboxyl terminus of Alg7p resulted in a nonfunctional protein (data not shown), a result that is consistent with earlier studies that showed that the C-terminal domain of the hamster Alg7p is required for function (Zara and Lehrman 1994). On the other hand, our results demonstrated that an N-terminal myc-tagged Alg7 is fully functional. The functionality of this N-terminal myc-Alg7p was assayed by complementation of the glycosylation defect and lethality that ensues from the loss of ALG7 function (Figure 2A). An ALG7/alg7Δ heterozygote (ALG7/alg7Δ::kanR) containing this plasmid-borne copy of ALG7 was sporulated and dissected. The kanamycin (G418)-resistant alg7Δ haploids harboring plasmid-borne ALG7 were viable, grew with the same rate at their wild-type sisters, and failed to grow on a medium containing 5′-fluoro-orotic acid (FOA) (Figure 2A). This plasmid-borne myc-ALG7 allele also complemented the glycosylation defect of a conditional alg7 mutant strain (not shown). Together, these results demonstrated that this plasmid-borne myc-ALG7 allele is functional. This ALG7 plasmid was thus used for the studies described below.
Fig. 2.
Coimmunoprecipitation of Alg7p and Alg14p. (Panel A) Functionality of plasmid-borne myc-ALG7. A viable strain (CNY2) deleted for ALG7 was produced by dissection of an ALG7/alg7Δ::kanR heterozygote that harbored myc-ALG7 on a 2 μ, URA3-containing plasmid (yEp352-GAP-myc-ALG7). The resulting haploid alg7Δ + pmyc-ALG7 strain was streaked on YPAD (+G418), 5′-FOA, and SD (-Ura) plates to assay the functionality of this plasmid-borne ALG7 in a strain in which this is the sole source of Alg7p. (Panel B) Coimmunoprecipitation assays in extracts from strains that express myc-ALG7 (W303 1A + yEpGAP-mycALG7) (lanes 2, 4, 6, 8, and 10) or that coexpress HA-ALG14 and myc-ALG7 (XGY151 + yEpGAP-mycAlg7) (lanes 1, 3, 5, 7, and 9). Detergent extracts were analyzed for coimmunoprecipitation of myc-Alg7p and HA-Alg14 by precipitating myc-Alg7p with the anti-myc antibody, fractionating samples (10 μg) by 12% SDS–PAGE, and detecting HA-Alg14p after immunoblotting with anti-HA antibodies (lane 1), or by precipitating HA-Alg14p with the anti-HA antibody and detecting myc-Alg7p after immunoblotting with the anti-myc antibody (lane 3). The relative amount of myc-Alg7p or HA-Alg14p in the detergent lysates was determined by immunoblotting the precipitate with the same antibody used for the immunoprecipitation (lanes 5–8). Control samples shown in lanes 9, 10, 11, and 12 were loaded on gels directly (lane 13) or subjected to immunoprecipitation with the anti-myc or anti-HA antibody, and blotted with anti-Wbp1 (lanes 9 and 10) or anti-HDEL antiserum (lanes 11, 12, and 13).
To determine if Alg7p interacts with Alg14p, we performed coimmunoprecipitation assays. Detergent extracts containing 0.1% Triton X-100 were prepared from strains that coexpress myc-ALG7 and HA-ALG14 (XGY151) to solubilize these membrane proteins. Alg7p was precipitated with an anti-myc antibody, precipitated proteins were resolved by SDS–PAGE, and subjected to immunoblotting with an anti-HA antibody to detect HA-Alg14p. This experiment demonstrated that Alg14 did indeed coprecipitate with Alg7 (Figure 2B, lane 1). Several control experiments demonstrated the specificity of this interaction. First, this interaction was antibody independent since the same result was observed with the opposite combination of antibodies, using the anti-HA antibody for precipitation of HA-Alg14p and anti-myc for Western blotting of coprecipitated myc-Alg7p (Figure 2B, lane 3). Second, these antibodies are specific; there was no detectable cross-reactivity of either antibody with any protein other than the cognate epitope (Figure 2B, lanes 5–8). Third, precipitation of myc-Alg7p is dependent on the expression of HA-Alg14p since myc-Alg7p did not coprecipitate in control strains that do not coexpress HA-Alg14p (Figure 2B, lane 2). Finally, this interaction is not the result of nonspecific aggregation since other ER membrane or soluble proteins (including Wbp1p (Figure 2B, lanes 9 and 10), Sec20p (Figure 2B, lanes 11 and 12), Kar2p (Figure 2B, lanes 11 and 12), Alg1p, and Alg2p (not shown)) did not coprecipitate with Alg7p or Alg14p. Taken together, these results demonstrate a high-affinity, specific interaction between Alg14p and Alg7p.
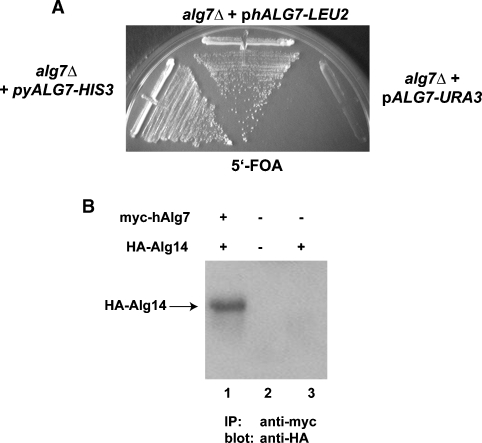
Although the hamster and yeast Alg7 proteins are similar to one another (42% identity), sequence alignments of fungal and mammalian Alg7/GPTs indicate the presence of large gaps in which their sequences have diverged significantly. To provide additional evidence for the relevance of the coimmunoprecipitation interactions, we analyzed whether or not the mammalian Alg7p counterpart (GPT) interacts with yeast Alg14p. To test this idea, the hamster (h) Alg7/GPT cDNA (Zhu and Lehrman 1990) was cloned into a yeast vector that places myc-hALG7 gene under the transcriptional control of the constitutive TDH3 (GAP) promoter. Consistent with earlier findings that demonstrate the functional conservation between human GPT and yeast Alg7p (Eckert et al. 1998), we found that hamster GPT completely complemented the lethality associated with the loss of ALG7 (Figure 3A). If the interaction between Alg7p and pAlg14p is of functional relevance, then we expect to observe a physical interaction between hamster GPT and yeast Alg14. To test this, coimmunoprecipitation assays were performed. Extracts from alg7Δ yeast strains that coexpress HA-Alg14p and myc-hAlg7p were constructed (see Materials and methods) and subjected to coimmunoprecipitation assays, as described above. The result of this experiment, shown in Figure 3B, demonstrated that hamster myc-GPT quantitatively coprecipitated with yeast Alg14p, providing further evidence for the functional relevance of these interactions.
Fig. 3.
Functional conservation of the hamster and yeast Alg7p. (Panel A) The hamster ALG7/GPT cDNA in an LEU2 yeast vector (yEpGAP-myc-hALG7) was tested for complementation of an alg7Δ strain (CNY2) that carried ALG7 on a URA3 plasmid (yEpGAP-mycALG7) strain by growth on a medium containing 5′-FOA, which forces elimination of the URA3 plasmid (CNY4). As a positive control, CNY2 harboring the yeast ALG7 on an HIS3 plasmid was analyzed in parallel. (Panel B) Coimmunoprecipitation of hamster Alg7p and yeast Alg14p. Detergent extracts containing 0.1% Triton X-100 were prepared from yeast strains coexpressing myc-tagged hamster Alg7p (yEpGAp-myc-hALG7) and HA-tagged Alg14. After immunoprecipitation with anti-myc antibodies, samples were resolved by 12% SDS–PAGE and subjected to immunoblotting with anti-HA antibodies.
Alg7p, Alg13p, and Alg14p cofractionate during gel filtration chromatography
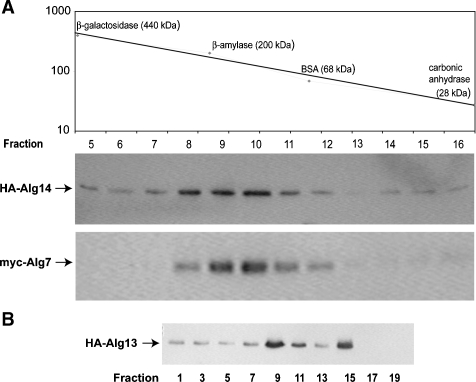
If Alg7p, Alg13p, and Alg14p exist together in an oligomeric complex, then all three proteins should cofractionate with the same predicted native molecular weight. To test this idea and provide an independent means of confirming the existence of this protein complex, the physical characteristics of Alg14p, Alg13p, and Alg7p were analyzed by gel filtration chromatography. This experiment was performed with two different strains. One strain coexpressed HA-ALG13 and myc-ALG14 (NAY13 + yEpGAP-myc-Alg14), while the other strain coexpressed HA-ALG14 and myc-ALG7 (XGY151 +yEpGAP-myc-Alg7). Several nonionic detergents, including digitonin, Triton X-100, Tween-20, and NP-40, were tested for their ability to solubilize Alg7p and Alg14p. Since the efficiency of coprecipitation of Alg7p and Alg14p was the same in all of these detergents (data not shown), further analyses used extracts prepared with Triton X-100. Detergent extracts prepared from each of these strains were fractionated in parallel with protein molecular weight markers on a Superose 12 column, using FPLC. Aliquots of fractions were assayed for the presence of HA-Alg14p, myc-Alg7p, and HA-Alg13p by immunoblotting. The results from these experiments demonstrated that all three proteins coeluted, with a peak position (fractions 9 and 10) that corresponds to a molecular weight of about 165–180 kDa (Figure 4A and B). Based on fractionation behavior of these proteins on Superose, we can estimate that the vast majority of cellular Alg7 and Alg14 interact together and that at least half of Alg13 cofractionates in this complex. The fractionation behavior of HA-Alg13p was notably distinct from HA-Alg14p and myc-Alg7p in that Alg13p displayed an additional elution peak (fraction 15) that cofractionated with lower molecular weight markers of <40 kDa (Figure 4B). Alg13p is a small soluble protein that exists in two pools. One pool is bound to the ER membrane through an association with Alg14p while the other pool is free in the cytosol (Bickel et al. 2005; Gao et al. 2005; Averbeck et al. 2008). The fractionation behavior of Alg13p is consistent with the interpretation that the two elution peaks represent these two pools of Alg13p. The second peak of Alg13p fractionated with a molecular weight that is larger than the predicted monomeric molecular weight (−23), but smaller than the predicted dimeric molecular weight (46). We have not further investigated the oligomeric properties of the cytosolic (inactive) pool of Alg13p. Nevertheless, taken together with the coimmunoprecipitation data described above, these results demonstrate that Alg13p, Alg14p, and Alg7p cofractionate with one another and are consistent with the model that these proteins interact as part of an oligomeric complex.
Fig. 4.
Alg13p, Alg14p, and Alg7p cofractionate during gel filtration chromatography. Detergent extracts prepared from strains coexpressing myc-Alg7p with HA-Alg14p (XGY151 + yEpGAP-mycAlg7) (Panel A) or with HA-Alg13 (NAY13 + yEpGAP-mycAlg7) (Panel B) were clarified by centrifugation, filtered, and fractionated by FPLC on a Superose 12 column. Two hundred microliters of fractions were collected; aliquots subjected to SDS–PAGE and immunoblotted with the anti-HA or anti-myc antibody. The elution profile of protein molecular weight standards run in parallel fractions is indicated graphically.
Evidence for a hexameric UDP-GlcNAc transferase complex
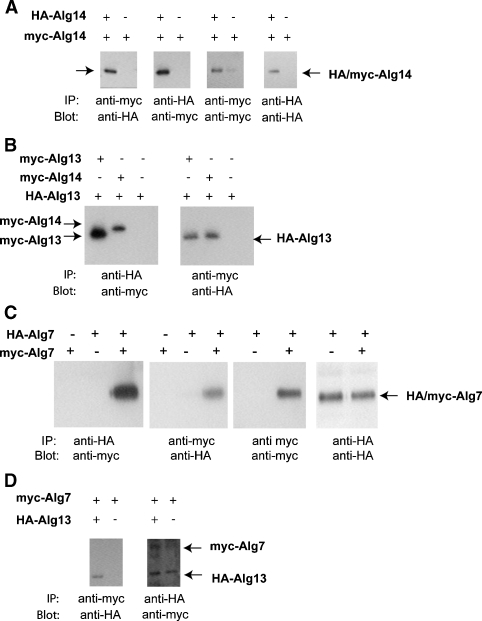
The estimated molecular weight of the Alg14p, Alg13p, and Alg7p complex by size chromatography is about 165–200 kDa. This size is significantly larger than the combined monomeric molecular weight of the three proteins, about 100 kDa. (The monomeric molecular weights of these proteins are as follows: Alg13p = 22.6 kDa, Alg14p = 27 kDa, and Alg7p = 50.3 kDa.) This discrepancy suggests that one or more of these proteins exist in more than one copy within the complex, or that the complex contains additional members. To determine if Alg13, Alg14, or Alg7 is present in more than one copy within the complex, we performed a series of coimmunoprecipitation assays using extracts from yeast that coexpress myc- and HA-tagged alleles of each gene. The rationale behind this approach is based on the idea that coimmunoprecipitation of the myc- and HA-tagged protein, for instance HA-Alg14p and myc-Alg14p, is only possible by homo-oligomerization or by being bridged to itself through another member of the complex. To perform this experiment, yeast strains were constructed that coexpress HA-Alg13p and myc-Alg13p, HA-Alg14p and myc-Alg14p, or HA-Alg7p and myc-Alg7p. Detergent extracts were prepared as described above and subjected to coimmunoprecipitation assays. As controls, each experiment was carried out with antibodies in both directions, precipitating with either anti-HA or anti-myc and blotting for the corresponding partner. In addition, control coprecipitations were performed in parallel using strains that express only one of the tagged alleles. From these experiments, the following conclusions were reached. First, we found evidence for high-affinity interactions between Alg14p and itself (Figure 5A), between Alg13p and itself (Figure 5B), and between Alg7p and itself (Figure 5C). myc-Alg14p quantitatively coprecipitated with HA-Alg14p, as was the case for myc- and HA-Alg13p, and myc- and HA-Alg7p. These results imply that Alg7p, Alg13p, and Alg14p are each present in two or more copies in the complex. In contrast, by this assay we found only a very weak interaction between Alg13p and Alg7p (Figure 5D). Moreover, this weak interaction between Alg13p and Alg7p was detected only if Alg7p was immunoprecipitated but not vice versa. When the order of this reaction was reversed and Alg13p was immunoprecipitated, bound Alg7p was barely detectable (Figure 5D). One explanation for this result is that Alg7p and Alg13p are not directly in contact but rather associate weakly through their mutual interaction with Alg14p. Together with our gel filtration data that estimate a native molecular weight of the complex between 165 and 200 kDa, these coimmunoprecipitation results suggest a 2:2:2 stoichiometry of Alg7p:Alg14p:Alg13p in the complex, resulting in a combined predicted molecular weight of −200 kDa.
Fig. 5.
Evidence for homo-oligomeric interactions between Alg14p, Alg13p, and Alg7p. Coimmunoprecipitation assays were performed from extracts from cells expressing or coexpressing HA- and myc-Alg14p (Panel A), HA- and myc-Alg13p (Panel B), HA-Alg7p and myc- Alg7p (Panel C), and HA-Alg13p and myc-Alg7p (Panel D). Expression (+) or the absence of expression (−) is indicated above each lane. Immunoprecipitations were performed as described in Figure 2 (and see Materials and methods) with anti-myc or anti-HA, and then immunoblotted with anti-HA or anti-myc antibodies as indicated.
Discussion
The Alg7p and Alg13p/Alg14p UDP-GlcNAc transferases together mediate the earliest steps of the N-linked glycosylation pathway. Here, we present data that demonstrate these temporally related UDP-GlcNAc glycosyltransferases that together catalyze the synthesis of GlcNAc2-PP-dol are organized as a multienzyme complex. As discussed below, the physical interactions between these proteins have implications for proposed mechanisms by which N-linked glycosylation is regulated, as well as for models of how this pathway evolved.
An emergent theme in biological systems is that proteins are organized into machines in which partners collaborate to carry out a process. The ER mannosyltransferases (i.e., Alg1, Alg2, and Alg11) that act sequentially to produce the Man5GlcNAc2-PP-dol intermediate on the cytosolic face of the ER form such machines (Gao et al. 2004). The fact that the related Alg7p, Alg13, and Alg14 UDP-GlcNAc transferases also do so demonstrates that all of the ER glycosyltransferases involved in the cytosolic steps of LLO function are organized into multienzyme complexes. This raises the possibility that hetero-oligomerization may be a general feature of ER glycosyltransferases and that the physical association of these related enzymes contributes to the efficiency of LLO synthesis. A major advantage of spatially linking related glycosyltransferases is that this arrangement would allow the channeling of LLO intermediates between successive active sites, without diffusion. Such a channeling mechanism is consistent with and indeed predicts the reported feedback relationship that exists between the products of the reactions catalyzed by Alg7p and Alg13/Alg14; the second intermediate, GlcNAc2-PP-dol, inhibits the formation of the first, GlcNAc-PP-dol (Kean et al. 1999). This feedback inhibition has been proposed to be an important factor that potentially regulates flux through the dolichol pathway (Kean et al. 1999). An investigation of these proteins will be required to determine whether or not their oligomeric interactions contribute to their regulated activity.
The dolichol-linked glycosylation pathway is highly conserved among eukaryotes, so a strong prediction is that the Alg7/Alg13/Alg14 UDP-GlcNAc transferase complex is organized in mammals as it is in yeast. In support of this idea, we found that mammalian GPT and yeast Alg7p are functionally interchangeable and can partner equally well with yeast Alg14p (Eckert et al. 1998) despite their low sequence similarity. Two other pieces of data are consistent with the idea that the organization of the UDP-GlcNAc transferase complex has been conserved in mammals. First, mammalian GPT has a high native molecular weight (Shailubhai et al. 1988; Dan and Lehrman 1997), suggesting that it homo- or hetero-oligomerizes. Second, elevated levels of GPT (Alg7p) cause a dominant negative phenotype in mammalian cells but this phenotype is independent of GPT catalytic activity (Gao, Shang, et al. 2008). Instead, this phenotype appears to be caused by the competitive binding of GPT to a putative scaffold required for LLO biosynthesis (Gao et al. 2008). Although speculative, our data are consistent with the idea that this scaffold may work through or be Alg14. Testing this idea will require a better characterization of the mammalian UDP-GlcNAc transferase enzymes.
A very important aspect of this study is that it provides a compelling argument for the idea that the lipid-linked cycles of peptidoglycan and N-linked glycoprotein synthesis have a common evolutionary origin. This idea, first described over 10 years ago, was based on parallels between Alg7p and MraY (Bugg and Brandish 1994). Alg7p and MraY are both membrane proteins that are related in sequence and that catalyze a chemically similar reaction. Instead of UDP-GlcNAc, MraY transfers the structurally related phospho-MurNAc-pentapeptide to undecaprenyl-P, a polyisoprenoid lipid that is similar to dolichol-P. The recent identification of Alg13p and Alg14p as the eukaryotic homologs of MurG revealed parallels between the dolichol and peptidoglycan lipid cycles far beyond what was first imagined since these enzymes catalyze a temporally related reaction that is chemically identical. Although the sequence identity between Alg13/Alg14 and MurG is low, secondary structure predictions (Chantret et al. 2005), molecular models of Alg13/Alg14 (Gao, Moriyama, et al. 2008) based on the MurG crystal structure (Ha et al. 2000), and the NMR structure of Alg13 (Wang et al. 2008) demonstrate that Alg13/Alg14 and MurG are remarkably similar in structure.
As shown in this study, the eukaryotic Alg7/Alg13/Alg14 proteins physically interact in a membrane-bound complex in the same way as their MraY/MurG bacterial counterparts (Mohammadi et al. 2007). Thus, the bacterial and eukaryotic enzymes are similar in structure, function, and organization. They sequentially catalyze similar chemical bonds, recognize structurally related polyprenyl lipid acceptors, and associate with one another in a similar hetero-oligomeric membrane-associated protein complex. Together, these data provide the most compelling evidence for an evolutionary link between these two lipid-linked cycles. From a practical point of view, these data also provide an explanation for the longstanding puzzle of why many inhibitory drugs that target the MraY and MurG bacterial enzymes lack host cell specificity. This new knowledge will allow us to define the features that distinguish the eukaryotic and prokaryotic enzymes, and enable development of drugs that inhibit the latter.
Material and methods
Yeast strains and genetic methods
Standard yeast media and genetic techniques were used (Guthrie and Fink 1991). Yeast strains were grown at 30°C in YPA (1% yeast extract, 2% peptone, 50 mg/L adenine sulfate) or in synthetic media that contained 0.67% yeast nitrogen base supplemented with the appropriate auxotrophic requirements, with 2% glucose (D) or galactose (G). Diploid strains were induced to sporulate by growth on agar plates containing 1% potassium acetate.
All yeast strains were derived from W303-1A (MATa his3-11 leu2-3, 112 ura3-1 trp1-1 ade2-1 can1-100 ssd1-d), W303-1B (MATα his3-11 leu2-3, 112 ura3-1 trp1-1 ade2-1 can1-100 ssd1-d), SEY6210 (MATα his3-11 trp1-1 lys2 leu2-3, 112 ura3-1) or the diploid BY4743 (MATa/α ura3Δ/ ura3Δ leu2Δ/ leu2Δ his3Δ/ his3Δ MET15/met15Δ LYS2/ lys2Δ). XGY151 contains a replacement of the chromosomal ALG14 locus with an HA3 epitope-tagged allele under the control of the glucose repressible GAL1/10 promoter (PGAL1) (Gao et al. 2005). NAY13 contains a replacement of the chromosomal ALG13 locus with a PGAL1-driven HA3 epitope-tagged ALG13 allele (Averbeck et al. 2008). CNY2 contains a deletion of the ALG7 gene (alg7Δ::kanR) and harbors a URA3 plasmid-borne copy of myc-ALG7 that maintains the viability of this otherwise inviable strain. This haploid strain was produced by sporulation and dissection of the heterozygote ALG7/alg7Δ::kanR BY4743 strain harboring yEpGAP-myc-ALG7. CNY4 and CNY8 contain a deletion of the chromosomal ALG7 gene and harbor a plasmid-borne yeast (CNY8) or hamster ALG7/GPT gene (CNY4). These strains were made by transforming CNY2 with plasmids containing an HIS3-marked yeast ALG7 (pRS315myc-ALG7) or LEU2-marked hamster-ALG7 cDNA (yEp 351-hALG7) and plating the transformants on FOA, a compound that is toxic to uracil prototrophs and thereby forces the elimination of the URA3-marked plasmid-borne yeast ALG7 gene in CNY2. CNY4 is the parent of CNY6, which coexpresses myc-tagged hamster ALG7, and HA-tagged ALG14, integrated at the normal chromosomal ALG14 locus. It should be noted that C-terminal epitope tagging of Alg7p, Alg13p, or Alg14p results in partial or complete loss of protein function (Gao et al. 2005; Averbeck et al. 2007; and results of this study). In addition, none of these proteins can be reliably detected at single copy (C. Noffz, N. Averbeck, unpublished observation). For these technical reasons, all epitope-tagged alleles encoded N-terminally tagged proteins whose expression was driven by the GAL1 or GAP promoter. Careful analyses of all strains used in this study were conducted to ensure that these alleles are completely functional, localize correctly, and have no detrimental effect on glycosylation, cell wall biosynthesis, or growth rates (C. Noffz, N. Averbeck, unpublished observation and see Results).
Plasmids
Standard methods were used for all cloning. Fragments obtained by PCR and cloned into vectors were verified by DNA sequence analysis. The hamster ALG7/GPT cDNA was amplified by PCR, using pJB20-TRG22 as the template (Zhu and Lehrman 1990). pJB20-TRG22 contains the entire GPT cDNA and was kindly provided by Mark Lehrman. The yeast ALG7 gene was amplified by PCR from genomic DNA (from W303-1A). These amplified yeast and hamster (h) PCR products were cloned into yEp352GAP (Yoko-o et al. 1998) for expression as N-myc-tagged products in a URA3, 2μ yeast vector (yEp352GAP-myc-ALG7 and yEp352GAP-myc hALG7), or into yEp351 for production of an N-terminally tagged product in an LEU2, 2μ plasmid (yEp351GAP-HA-ALG7 and yEp351GAP-myc hALG7). Plasmid maps and primer sequences are available on request.
pRS306-HA-ALG14p contains PALG14-HA-ALG14 in the URA3 integration vector, pRS306 (Sikorski and Hieter 1989). This plasmid was linearized by a partial NcoI digest to target its integration in the chromosomal ALG14 promoter.
Preparation of cell-free protein lysates
Exponentially growing cells (OD600 of 1–3) were harvested and converted to spheroplasts with lyticase as described (Gao and Dean 2000). 10 OD units of spheroplasted cells were washed and resuspended in 500 μL PBS containing 0.1% Triton-X 100 and 1 mM phenylmethylsulfonyl fluoride (PMSF) to solubilize membrane proteins. Cell debris and unlysed cells were removed by centrifugation at 14,000 × g for 15 min at 4°C. These detergent lysates were used directly for immunoprecipitation or clarified by centrifugation at 10,000 × g for 15 min for chromatography (see below).
Coimmunoprecipitations and Western immunoblotting
Detergent lysates were prepared as described above and 200 μL (representing −2–4 OD units of cells) were used for each immunoprecipitation, performed as described (Gao and Dean 2000). Epitope-tagged proteins were immunoprecipitated with anti-HA 12CA5 monoclonal antibodies (culture supernatants diluted 1:3), anti-myc 9E10 monoclonal antibodies (culture supernatants diluted 1:10), anti-HDEL rabbit polyclonal antibodies (diluted 1:500), or anti-myc A-14 rabbit polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) (diluted 1:130), and protein A-Sepharose. After precipitation, proteins were separated by 12% SDS–PAGE, transferred to the Immobilon−P PVDF membrane (Millipore, Santa Cruz, CA), and subjected to immunoblotting with anti-HA, anti-myc, anti-HDEL, or anti-Wbp1 antibodies as described previously (Gao and Dean 2000). To avoid cross-reaction with the dissociated IgG heavy and light chains, whose molecular weights are very close to HA-Alg13p (−26 kDa) and myc-Alg7p (−54 kDa), the combination of antibodies used in the immunoprecipitation and in Western analyses was derived from different animals. Primary antibodies were detected with secondary anti-rabbit or anti-mouse IgG conjugated to horseradish peroxidase, followed by chemiluminescence (ECL, GE Health Care, Inc., Santa Cruz, CA).
Gel filtration chromatography
Detergent lysates were prepared as described above. Lysates (300–500 μL) were filtered through a 20 μm membrane just prior to fast protein liquid chromatography (FPLC) on a Superose 12 HR 10/30 column (Amersham/GE Health Care, Inc., Santa Cruz, CA). The column was equilibrated with PBS containing 0.1% Triton-X 100. FPLC was performed with a flow rate of 0.4 mL/min, and 400 μL fractions were collected. Aliquots of each fraction were analyzed by SDS–PAGE and immunoblotted for the presence of epitope-tagged Alg7p, Alg14p, or Alg13p with anti-HA (12CA5) or anti-myc (9E10) antibodies.
Funding
The National Institutes of Health (RO1-GM048467 to N.D.)
Acknowledgments
We thank Mark Lehrman for plasmids.
Conflict of interest statement
None declared.
Abbreviations
- ER
endoplasmic reticulum
- FPLC
fast protein liquid chromatography
- FOA
5′-fluoro-orotic acid
- LLO
lipid-linked oligosaccharide
References
- Aebi M, Hennet T. Congenital disorders of glycosylation: Genetic model systems lead the way. Trends Cell Biol. 2001;11:136–141. doi: 10.1016/s0962-8924(01)01925-0. [DOI] [PubMed] [Google Scholar]
- Averbeck N, Gao XD, Nishimura SI, Dean N. Alg13p, the catalytic subunit of the ER UDP-GlcNAc glycosyltransferase, is a target for proteasomal degradation. Mol Biol Cell. 2008;19:2169–2178. doi: 10.1091/mbc.E07-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averbeck N, Keppler-Ross S, Dean N. Membrane topology of the Alg14 endoplasmic reticulum UDP-GlcNAc transferase subunit. J Biol Chem. 2007;282:29081–29088. doi: 10.1074/jbc.M704410200. [DOI] [PubMed] [Google Scholar]
- Bickel T, Lehle L, Schwarz M, Aebi M, Jakob CA. Biosynthesis of lipid-linked oligosaccharides in Saccharomyces cerevisiae: Alg13p and Alg14p form a complex required for the formation of GlcNAc(2)-PP-dolichol. J Biol Chem. 2005;280:34500–34506. doi: 10.1074/jbc.M506358200. [DOI] [PubMed] [Google Scholar]
- Bugg TD, Brandish PE. From peptidoglycan to glycoproteins: Common features of lipid-linked oligosaccharide biosynthesis. FEMS Microbiol Lett. 1994;119:255–262. doi: 10.1111/j.1574-6968.1994.tb06898.x. [DOI] [PubMed] [Google Scholar]
- Chantret I, Dancourt J, Barbat A, Moore SE. Two proteins homologous to the N- and C-terminal domains of the bacterial glycosyltransferase MurG are required for the second step of dolichyl-linked oligosaccharide synthesis in S. cerevisiae. J Biol Chem. 2005;280:9236–9242. doi: 10.1074/jbc.M413941200. [DOI] [PubMed] [Google Scholar]
- Dan N, Lehrman MA. Oligomerization of hamster UDP-GlcNAc:dolichol-P GlcNAc-1-P transferase, an enzyme with multiple transmembrane spans. J Biol Chem. 1997;272:14214–14219. doi: 10.1074/jbc.272.22.14214. [DOI] [PubMed] [Google Scholar]
- Eckert V, Blank M, Mazhari-Tabrizi R, Mumberg D, Funk M, Schwarz RT. Cloning and functional expression of the human GlcNAc-1-P transferase, the enzyme for the committed step of the dolichol cycle, by heterologous complementation in Saccharomyces cerevisiae. Glycobiology. 1998;8:77–85. doi: 10.1093/glycob/8.1.77. [DOI] [PubMed] [Google Scholar]
- Gao N, Shang J, Lehrman MA. Unexpected basis for impaired Glc3Man9GlcNAc2-P-P-dolichol biosynthesis by elevated expression of GlcNAc-1-P transferase. Glycobiology. 2008;18:125–134. doi: 10.1093/glycob/cwm109. [DOI] [PubMed] [Google Scholar]
- Gao XD, Dean N. Distinct protein domains of the yeast Golgi GDP-mannose transporter mediate oligomer assembly and export from the endoplasmic reticulum. J Biol Chem. 2000;275:17718–17727. doi: 10.1074/jbc.M909946199. [DOI] [PubMed] [Google Scholar]
- Gao XD, Moriyama S, Miura N, Dean N, Nishimura S. Interaction between the C termini of Alg13 and Alg14 mediates formation of the active UDP-N-acetylglucosamine transferase complex. J Biol Chem. 2008;283:32534–32541. doi: 10.1074/jbc.M804060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XD, Nishikawa A, Dean N. Physical interactions between the Alg1, Alg2, and Alg11 mannosyltransferases of the endoplasmic reticulum. Glycobiology. 2004;14:559–570. doi: 10.1093/glycob/cwh072. [DOI] [PubMed] [Google Scholar]
- Gao XD, Tachikawa H, Sato T, Jigami Y, Dean N. Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosamine transferase required for the second step of N-linked glycosylation. J Biol Chem. 2005;280:36254–36262. doi: 10.1074/jbc.M507569200. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:3–20. [PubMed] [Google Scholar]
- Ha S, Walker D, Shi Y, Walker S. The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci. 2000;9:1045–1052. doi: 10.1110/ps.9.6.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Kean EL, Wei Z, Anderson VE, Zhang N, Sayre LM. Regulation of the biosynthesis of N-acetylglucosaminylpyrophosphoryldolichol, feedback and product inhibition. J Biol Chem. 1999;274:34072–34082. doi: 10.1074/jbc.274.48.34072. [DOI] [PubMed] [Google Scholar]
- Lehrman MA. Biosynthesis of N-acetylglucosamine-P-P-dolichol, the committed step of asparagine-linked oligosaccharide assembly. Glycobiology. 1991;1:553–562. doi: 10.1093/glycob/1.6.553. [DOI] [PubMed] [Google Scholar]
- Lehrman MA. A family of UDP-GlcNAc/MurNAc:polyisoprenol-P GlcNAc/MurNAc-1-P transferases. Glycobiology. 1994;4:768–771. doi: 10.1093/glycob/4.6.768. [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D, Texier L, Rousseau M, van Heijenoort J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine:N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglu- cosamine transferase involved in the membrane steps of peptidoglycan synthesis. J Bacteriol. 1991;173:4625–4636. doi: 10.1128/jb.173.15.4625-4636.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, den Blaauwen T. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol. 2007;65:1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NP, Momany FA. Modeling bacterial UDP-HexNAc:polyprenol-P HexNAc-1-P transferases. Glycobiology. 2005;15:29R–42R. doi: 10.1093/glycob/cwi065. [DOI] [PubMed] [Google Scholar]
- Shailubhai K, Dong-Yu B, Saxena ES, Vijay IK. Purification and characterization of UDP-N-acetyl-D-glucosamine:dolichol phosphate N-acetyl-d-glucosamine-1-phosphate transferase involved in the biosynthesis of asparagine-linked glycoproteins in the mammary gland. J Biol Chem. 1988;263:15964–15972. [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Weldeghiorghis T, Zhang G, Imperiali B, Prestegard JH. Solution structure of Alg13: The sugar donor subunit of a yeast N-acetylglucosamine transferase. Structure. 2008;16:965–975. doi: 10.1016/j.str.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E, Imperiali B. Asparagine-linked protein glycosylation: From eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- Yoko-o T, Roy SK, Jigami Y. Differences in in vivo acceptor specificity of two galactosyltransferases, the gmh3+ and gma12+ gene products from Schizosaccharomyces pombe. Eur J Biochem. 1998;257:630–637. doi: 10.1046/j.1432-1327.1998.2570630.x. [DOI] [PubMed] [Google Scholar]
- Zara J, Lehrman MA. Role of the carboxyl terminus in stable expression of hamster UDP-GlcNAc:dolichol-P GlcNAc-1-P transferase. J Biol Chem. 1994;269:19108–19115. [PubMed] [Google Scholar]
- Zhu XY, Lehrman MA. Cloning, sequence, and expression of a cDNA encoding hamster UDP-GlcNAc:dolichol phosphate N-acetylglucosamine-1-phosphate transferase. J Biol Chem. 1990;265:14250–14255. [PubMed] [Google Scholar]