Abstract
Leptospirosis is a widespread zoonosis caused by invasive spirochaetes belonging to the genus Leptospira. Pathogenic leptospires disseminate via the bloodstream to colonize the renal tubules of reservoir hosts. Little is known about leptospiral outer-membrane proteins expressed during the dissemination stage of infection. In this study, a novel surface-exposed lipoprotein is described; it has been designated LipL46 to distinguish it from a previously described 31 kDa peripheral membrane protein, P31LipL45, which is exported as a 45 kDa probable lipoprotein. The lipL46 gene encodes a 412 aa polypeptide with a 21 aa signal peptide. Lipid modification of cysteine at the lipoprotein signal peptidase cleavage site FSISC is supported by the finding that Leptospira interrogans intrinsically labels LipL46 during incubation in medium containing [14C]palmitate. LipL46 appears to be exported to the leptospiral outer membrane as a 46 kDa lipoprotein, based on Triton X-114 solubilization and phase partitioning studies, which included the outer and inner membrane controls LipL32 and LipL31, respectively. Surface immunoprecipitation and whole-cell ELISA experiments indicate that LipL46 is exposed on the leptospiral surface. Immunohistochemistry studies demonstrated expression of LipL46 by leptospires found in the bloodstream of acutely infected hamsters. Leptospires expressing LipL46 were also found in the intercellular spaces of the liver, within splenic phagocytes, and invading the glomerular hilum of the kidney. Infection-associated expression is supported by the finding that LipL46 is a major antigen recognized by sera from infected hamsters. These findings indicate that LipL46 may be important in leptospiral dissemination, and that it may serve as a useful serodiagnostic antigen.
INTRODUCTION
The genus Leptospira contains a spectrum of spirochaetes ranging from free-living saprophytes to invasive pathogens (Levett, 2001). Leptospiral pathogens have the capacity to adapt to both the ambient environment and mammalian host tissues. Cattle and feral rodents are the most important reservoir hosts, although pathogenic Leptospira species have been isolated from hundreds of mammalian species (Babudieri, 1958). Leptospiral colonization of reservoir host kidneys is considered a commensal infection, as there appears to be little or no adverse effects on renal histology or function (Faine et al., 1999). Transmission occurs either through direct contact with an infected animal, or through indirect contact via soil or water contaminated with urine from an infected animal. Entry into the body is gained through cuts and abrasions, or through mucous membranes, such as the conjunctival, oral or genital surfaces. Leptospires disseminate haematogenously to many organs through migration from the vascular lumen across endothelial barriers into host tissues (Inada et al., 1916). In the reservoir host, organisms are cleared from the bloodstream, and persist in the lumen of the renal tubules, resulting in shedding of organisms into the urine. However, in accidental hosts, including humans, the numbers of organisms in the circulation may reach critical levels, and the dissemination stage may be followed by potentially fatal systemic complications and multiorgan dysfunction, including hepatic and renal failure, with or without severe pulmonary haemorrhage syndrome (McBride et al., 2005; Segura et al., 2005).
The pathogenesis of leptospirosis would be elucidated by a molecular understanding of the components of the leptospiral outer membrane (OM) and their interactions with host factors. The leptospiral OM has been defined by detergent-extraction methods, and by isolation of OM vesicles on a sucrose density gradient (Cullen et al., 2002; Haake et al., 1991; Haake & Matsunaga, 2002; Nally et al., 2005b; Zuerner et al., 1991). Some of these approaches have been validated using well-characterized markers for the periplasm, inner membrane and cytoplasm. The leptospiral OM is known to contain phospholipids, LPS, and three different types of outer-membrane proteins (OMPs): transmembrane OMPs, lipoproteins, and the peripheral membrane protein P31LipL45. Unlike transmembrane OMPs found in other spirochaetes, leptospiral transmembrane OMPs appear to have the β-barrel structure typically found in Gram-negative organisms. The best-studied example of the leptospiral transmembrane OMPs is OmpL1, which has been shown to form channels in lipid bilayers, and is predicted to contain 10 transmembrane domains and 5 surface-exposed loops (Haake et al., 1993; Shang et al., 1995). Twenty per cent of OmpL1 proteins are chimeras, in which the largest surface-exposed loop is derived from other leptospiral species (Haake et al., 2004). Transmembrane OMPs are relatively minor components of the leptospiral OM compared with the lipoproteins, most notably LipL32, which is the most abundant protein in the leptospiral protein profile (Haake et al., 2000). LipL32 has also been referred to as the leptospiral major OM (Haake et al., 2000) and haemolysis-associated protein 1 (Branger et al., 2001). Additional OM lipoproteins for which there are localization data and experimental evidence of fatty acid modification include LipL21, LipL36, LipL41 and LigB (Cullen et al., 2003; Haake et al., 1998; Matsunaga et al., 2003; Shang et al., 1995). A number of other less well-characterized OM components have also been identified (Cullen et al., 2002, 2005; Koizumi & Watanabe, 2003; Nally et al., 2005b).
Key issues in assessing the relevance of OMPs to leptospiral pathogenesis are expression during mammalian infection, and exposure on the leptospiral surface. Serological evidence, combined with immunohistochemistry studies, has indicated that LPS, OmpL1, LipL32 and LipL41 are expressed during leptospiral infection of the kidney (Barnett et al., 1999; Haake et al., 2000). In contrast, LipL36 is an example of a prominent OM lipoprotein in cultivated organisms that is neither surface exposed, nor expressed during experimental infection of hamsters (Barnett et al., 1999; Haake et al., 1998; Shang et al., 1996). Leptospiral LPS expression appears to be either dramatically altered or downregulated in the liver of acutely infected animals, compared with organisms in culture, or those in the kidneys of chronically infected animals, indicating that surface-antigen expression undergoes changes during the different stages of leptospiral infection (Nally et al., 2005a). Surface immunoprecipitation studies have demonstrated that OmpL1 and LipL41 are accessible to antibody on the surface of intact leptospires, but that LipL36 can only be immunoprecipitated in disrupted organisms (Haake et al., 1991; Shang et al., 1996). The application of the surface-biotinylation reagent Sulfo-NHS-LC-Biotin to the characterization of the leptospiral surfaceome has provided evidence of surface exposure of several additional OMPs, including LipL21 and LipL32 (Cullen et al., 2003, 2005). The surface exposure of OmpL1, LipL41 and LipL32 has been confirmed by whole-cell immunoelectron microscopy (IEM) (Cullen et al., 2005; Haake et al., 1993), and, in the case of the Lig proteins, by thin-section IEM (Matsunaga et al., 2003). Additional techniques developed for assessing surface exposure include whole-cell ELISA and surface-immunofluorescence (Cullen et al., 2005). Use of multiple approaches and subsurface controls is essential before reaching conclusions regarding surface exposure of novel OMPs. Prior Triton X-114 and OM vesicle isolation studies identified a 46 kDa OMP (Haake et al., 1991; Haake & Matsunaga, 2002), which has been partially sequenced during proteomic analysis of leptospiral OMPs (Cullen et al., 2002; Nally et al., 2005b). Availability of leptospiral genome sequences (Nascimento et al., 2004; Ren et al., 2003) has provided the nucleotide sequence of the gene encoding the 46 kDa protein. In this study, we examine the lipidation, localization, surface exposure and in vivo expression of LipL46. Because little is known about the expression of leptospiral antigens during the early dissemination stage of leptospirosis, we examined the expression of LipL46 in the bloodstream, liver, spleen and kidney during this initial acute stage of leptospirosis.
METHODS
Bacterial strains and cultivation
Leptospira kirschneri serovar Grippotyphosa strain RM52 was isolated from an outbreak of porcine abortion in the USA (Thiermann et al., 1984). Leptospira interrogans serovar Copenhageni strain Fiocruz L1-130 is a human blood isolate obtained during an outbreak of leptospirosis in Salvador, Brazil (Matsunaga et al., 2003). L. interrogans serovar Pomona type kennewicki strain 11000-74A is a cattle isolate. All experiments were performed with virulent low-passage forms of these strains, which were obtained by infection and reisolation from Golden Syrian hamsters (Harlan Sprague Dawley). The spirochaetes were maintained in Ellinghausen–McCullough–Johnson–Harris (EMJH) medium, pH 7·2, supplemented with 1 % rabbit serum and 100 mg 5-fluoro-uracil ml−1 (Sigma), and incubated at 30 °C (Johnson & Harris, 1967). Albumin was purchased from Intergen (catalogue no. 31-003-3) and Sigma (catalogue no. A7906) for cultivation of L. kirschneri RM52 and L. interrogans Fiocruz L1-130, respectively.
Plasmid DNA
The lipL46 gene was identified by searching the L. interrogans serovar Copenhageni genome (Nascimento et al., 2004) for a gene encoding the peptide sequences [F/M]TG[L/I]NADEATK and YNTAATGTYK, which were obtained by Cullen et al. (2002) in a global analysis of leptospiral OMPs. The portion of the lipL46 gene beginning from the codon following the segment encoding the signal peptide was amplified by PCR with Phusion DNA polymerase (Finnzyme), using the forward primer 5′-ATCAGATCTGGTTCTTCCGGTTCCACTCGTGGTAAA-3′, and the reverse primer 5′-GCGCCATGGGTGCGAAGTTAGAATTTATTTCAAAGGT-3′. The primers included a BglII site and an NcoI site, respectively (underlined), and L. interrogans Fiocruz L1-130 genomic DNA was used as the template for PCR. The amplified lipL46 gene fragment was digested with BglII and NcoI, and ligated to the His6 vector pRSET B (Invitrogen) with T4 DNA ligase to generate the plasmid pRSETb-LipL46. Restriction and modifying enzymes were purchased from New England Biolabs.
Antisera and immunoblot analysis
LipL31, LipL32, LipL36, LipL41, LipL46 and GroEL antisera were obtained from New Zealand White rabbits immunized with recombinant His6 proteins, as described (Haake et al., 1998, 2000; Haake & Matsunaga, 2002; Matsunaga et al., 2002; Shang et al., 1996). Sera from infected hamsters were obtained by intraperitoneal challenge of animals (in groups of three) with serial 10-fold dilutions of L. interrogans serovar Copenhageni, ranging from 102–107 organisms per hamster. Hamsters surviving 28 days after challenge were euthanized, and serum was harvested for immunoblot studies. For immunoblot analysis, 1×109 leptospires were collected by centrifugation for 4 min at 9000 g in a Beckman Coulter Microfuge 18 centrifuge. The cell pellet was washed once in 100 mM PBS, pH 7·4, containing 5 mM MgCl2, and resuspended in 100 μl final sample buffer (FSB) consisting of 50 mM Tris/HCl (pH 6·8), 100 mM DTT, 2 % SDS, 0·1 % bromophenol blue and 20 % (v/v) glycerol, and boiled for 3 min. Unless otherwise indicated, 1×108 leptospires or whole-cell equivalents were loaded per lane. Electrophoresis and immunoblotting were performed as previously described (Matsunaga et al., 2005).
[14C]palmitate radiolabelling and immunoprecipitation of native LipL46
A 12 ml culture sample containing 1×108 ml−1 L. interrogans in the exponential phase of growth was intrinsically labelled by addition of 50 μCi (1·85 MBq) [U-14C]palmitate (GE Amersham), followed by further incubation in a shaker incubator at 30 °C for 48 h until the bacterial concentration reached 1×109 ml−1. A sample for immunoprecipitation containing 1·2×1010 L. interrogans was resuspended in 1·2 ml 50 mM Tris/HCl, pH 8·0, 100 mM NaCl, 2 mM EDTA and 0·2 % SDS, and boiled for 5 min. The insoluble material was removed by centrifugation at 16 000 g for 10 min. A 200 μl volume of the supernatant was added to 5 μl LipL46 rabbit antiserum and 795 μl 50 mM Tris/HCl, pH 8·0, 100 mM NaCl, 2 mM EDTA and 0·2 % Triton X-100, and incubated on ice overnight. The next day, 25 μl of a slurry of EZview Red Protein A Affinity Gel (Sigma) was added, and the suspension was gently agitated for 2 h. The affinity-gel–antibody–antigen complexes were washed twice in 0·01 % Triton X-100 in 10 mM Tris/HCl (pH 8·0) and 400 mM NaCl, once in 0·01 % Triton X-100 in 10 mM Tris/HCl, pH 8·0, and resuspended in FSB. After SDS-PAGE, gels were prepared for autoradiography by being soaked in Amplify (GE Amersham), dried, and exposed to Hyperfilm (GE Amersham).
Triton X-114 extraction of Leptospira
L. interrogans was extracted with 0·5 % Triton X-114, as described previously (Haake et al., 2000; Zuerner et al., 1991). In brief, L. interrogans cells were washed in PBS containing 5 mM MgCl2, and extracted in the presence of 0·5 % protein-grade Triton X-114 (Calbiochem), 150 mM NaCl, 10 mM Tris, pH 8, and 2 mM EDTA, at 4 °C. The insoluble material was removed by centrifugation at 16 000 g for 10 min. After centrifugation, 1 M CaCl2 was added to the supernatant, to a final concentration of 20 mM. Phase separation was performed by warming the supernatant to 37 °C, and subjecting it to centrifugation for 10 min at 2000 g. The detergent and aqueous phases were separated, and precipitated with acetone.
Surface immunoprecipitation
The surface immunoprecipitation technique was a modification of a previously described method (Shang et al., 1996). A 980 μl volume of leptospiral culture containing 1·2×109 bacteria (>99 % actively motile) was mixed with 20 μl rabbit antiserum to L. kirschneri strain RM52. The suspension was then gently shaken for 1 h at 30 °C. Agglutinated leptospires were pelleted at 1000 g for 10 min, resuspended in PBS containing 5 mM MgCl2, pelleted again at 1000 g for 10 min, and then resuspended in 450 μl 10 mM Tris/HCl, pH 8·0, 100 mM NaCl, 2 mM EDTA and 1 mM PMSF. To this suspension was added 50 μl 10 % protein-grade Triton X-100 (Calbiochem), followed by gentle agitation for 30 min at 4 °C. The insoluble material was removed by centrifugation at 16 000 g for 10 min. To the supernatant was added 50 μl 2 % deoxycholate, 5 μl 10 % SDS, and 50 μl of a slurry of EZview Red Protein A Affinity Gel. This mixture was gently agitated for 2 h at 4 °C. The affinity-gel–antibody–antigen complexes were washed once in 0·01 % Triton X-100 in 10 mM Tris/HCl, pH 8·0, and 100 mM NaCl, once in 0·01 % Triton X-100 in 10 mM Tris/HCl, pH 8·0, and resuspended in FSB. Samples were then processed by SDS-PAGE, and immunoblotting with antisera specific for LipL36, LipL41 and LipL46, at dilutions of 1 : 2000, 1 : 5000, and 1 : 5000, respectively. Immunoblots were developed using protein A conjugated to horseradish peroxidase (GE Amersham) at a dilution of 1 : 1500. An immunoprecipitation control experiment was performed in parallel using the same conditions described above, except that immunoprecipitation was performed after solubilization in Triton X-100. In some experiments, antiserum to LipL36 at a dilution of 1 : 200 was added during the initial immunoprecipitation step.
Whole-cell ELISA
Whole-cell ELISA experiments were performed using a modification of a previously described method (Cullen et al., 2005). Flat-bottom polystyrene high-binding microtitre plates (Corning) were coated overnight at 4 °C with 100 μl per well of 109 ml−1 whole or sonicated L. interrogans serovar Copenhageni strain Fiocruz L1-130, which had been grown overnight in serum-free EMJH medium (to reduce the background signal from rabbit immunoglobulin), immobilized with 75 mM sodium azide, centrifuged at 1000 g, and resuspended in 0·05 M sodium carbonate buffer (pH 9·6). Plates were blocked for 3 h at room temperature, and washed three times with 200 μl Leptospira Enrichment EMJH (Difco). Wells were incubated for 1 h at room temperature with 100 μl per well of primary antiserum diluted in Leptospira Enrichment EMJH, and washed three times with 200 μl PBS containing 0·05 % Tween 20 (PBS-T). Wells were incubated with 100 μl per well of a 1 : 5000 dilution of horseradish-peroxidase-linked donkey whole-antibody anti-rabbit IgG (GE Amersham) for 1 h at room temperature, followed by two washes with 200 μl PBS-T, and three washes with PBS. ELISA plates were developed by adding 50 μl per well of 0·01 % (w/v) 3,3′,5,5′-tetramethylbenzidine in substrate buffer [0·03 % (v/v) hydrogen peroxide, 25 mM citric acid, 50 mM Na2HPO4, pH 5·0] for 30 min in the dark, at room temperature. The reaction was stopped by adding a 25 μl volume of 1 M H2SO4, and the absorbance at 450 nm was measured.
Immunohistochemistry
The methods used to obtain tissues from infected hamsters, and to perform immunohistochemistry, were modified from those previously described (Barnett et al., 1999). Golden Syrian hamsters were inoculated intraperitoneally with 105 virulent L. interrogans serovar Pomona strain 11000-74A. Moribund hamsters were euthanized, and liver, lung, spleen and kidney tissues were removed, fixed in 10 % buffered formalin, and paraffin embedded. Tissue sections were stained with haematoxylin and eosin (H/E) or a modified periodic acid Schiff (PAS)/Steiner silver stain (Steiner & Steiner, 1944).
Serial 4 μm sections of kidney, liver, lung and spleen from hamsters infected with L. interrogans were cut. Tissue sections were placed on Probe-On Plus Slides (Fisher). Paraffin was removed from sections with xylene and ethanol, using standard procedures. Non-specific staining of tissue sections was blocked using 10 % normal horse serum at room temperature for 60 min, prior to incubation overnight at 4 °C with primary antibody. Anti-LipL32 and Anti-LipL46 antisera were used at a 1 : 200 dilution. Controls included no primary antibody, normal horse serum and preimmune serum on all sections from both infected and uninfected hamsters. Sections were washed with PBS to remove unbound antibody, and then incubated for 60 min at room temperature in the dark with goat anti-rabbit Alexa Fluor 488 secondary antibody (Molecular Probes). After washing, sections were incubated for 5 min with 4′,6-diamidino-2-phenylindole (DAPI; 1·5 mg ml−1) for nuclear staining. Slides were mounted with SlowFade Light antifade kit (Molecular Probes). All images were captured on a Spot RT colour CCD camera mounted on a Nikon Eclipse E800 microscope using a ×40 Plan Fluor objective (not under oil) and B-2A filter (excitation, 450–490 nm; emission, 520 nm). Higher-magnification images were crops of the original image. All fluorescent images, regardless of antibody, were obtained using the same exposure times. DAPI images were captured separately, and then merged with the Alexa Fluor 488 image using Spot Advanced Software. H/E and PAS/Steiner silver stain images were obtained using a filter that provides ×1·5 magnification.
RESULTS
Sequence analysis of the lipL46 gene
Amino acid sequence data obtained through proteomic analyses (Cullen et al., 2002; Nally et al., 2005b) allowed identification of the gene loci encoding LipL46 as LIC11885 and LA2024 in L. interrogans serovar Copenhageni strain Fiocruz L1-130 and serovar Lai strain 56601, respectively. In L. interrogans serovar Copenhageni strain Fiocruz L1-130, the lipL46 structural gene consists of 1236 bases encoding a protein of 412 aa. A consensus ribosome-binding site (AGGAGA) is present 11 nt upstream of the initiation codon. An inverted repeat located 24 bp downstream from the termination codon may function as a rho-independent transcription terminator. The deduced amino acid sequences of the LipL46 protein are identical in L. interrogans serovars Copenhageni and Lai (Nascimento et al., 2004; Ren et al., 2003). LipL46 is predicted to be a lipoprotein by the LipoP and SpLip algorithms (Juncker et al., 2003; Setubal et al., 2006). The 21-residue signal peptide consists of a basic amino-terminal region (aa 1–8), a hydrophobic core (aa 9–17), and a carboxy-terminal signal peptidase II cleavage site. The LipL46 FSISC lipobox sequence is very similar to the LSISC lipobox sequence of Borrelia burgdorferi OspD (Norris et al., 1992), and the FFISC lipobox of LruA (Verma et al., 2005). A serine in the highly constrained −1 position relative to cysteine also occurs in leptospiral LigB (Matsunaga et al., 2003). Although serine at this position is uncommon in enteric Gram-negative bacteria, it is predicted by the spirochaetal lipoprotein algorithm SpLip to be the most common amino acid at that position in leptospiral lipoproteins (Setubal et al., 2006).
After cleavage of the 21 aa signal peptide by leptospiral signal peptidase II, the mature polypeptide would have a predicted molecular mass of 42·4 kDa, and an isoelectric point of 5·7. Secondary structure analysis predicts a mature protein consisting of 60 % α-helix and 30 % β-sheet. The predicted amino acid sequences following isoleucine residue 252 and lysine residue 326 are identical to those obtained by electrospray ionization MS of leptospiral OMP pL45 (Cullen et al., 2002). The LipL46 amino acid sequence appears to be unique to leptospires: no significant paralogues, homologues or domains were found in any of the online databases.
L. interrogans acylates LipL46
Intrinsic labelling of L. interrogans with [14C]palmitate resulted in acylation of LPS and a number of proteins in a profile consistent with known leptospiral lipoproteins, including LipL32 (Haake et al., 2000), LipL36 (Haake et al., 1998) and LipL41 (Shang et al., 1996) (Fig. 1). Immunoprecipitation of a whole-cell lysate of [14C]palmitate-labelled L. interrogans using LipL46 antiserum confirmed that LipL46 is one of the proteins acylated by L. interrogans. Immunoprecipitation with antiserum to LipL32 was included as a positive control.
Fig. 1.
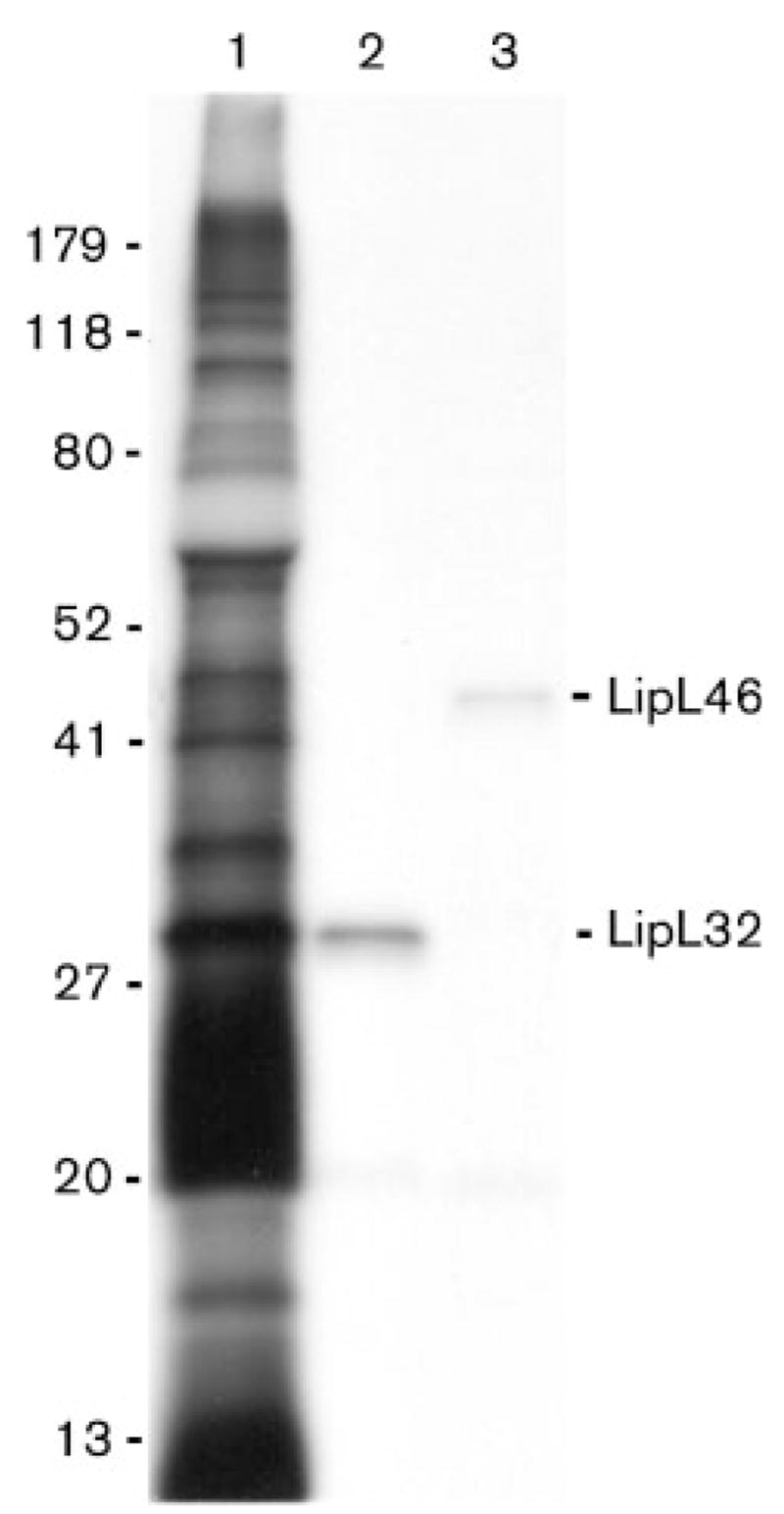
LipL46 is acylated by L. interrogans. An autoradiogram of L. interrogans proteins intrinsically labelled by [14C]palmitate, and separated by SDS-PAGE is shown. Lane 1, total L. interrogans proteins; lanes 2 and 3, material immunoprecipitated by addition of LipL32 and LipL46 antiserum to a whole-cell lysate of L. interrogans. Locations of molecular size standards (kDa) are shown on the left.
Behaviour of LipL46 during Triton X-114 extraction and phase partitioning
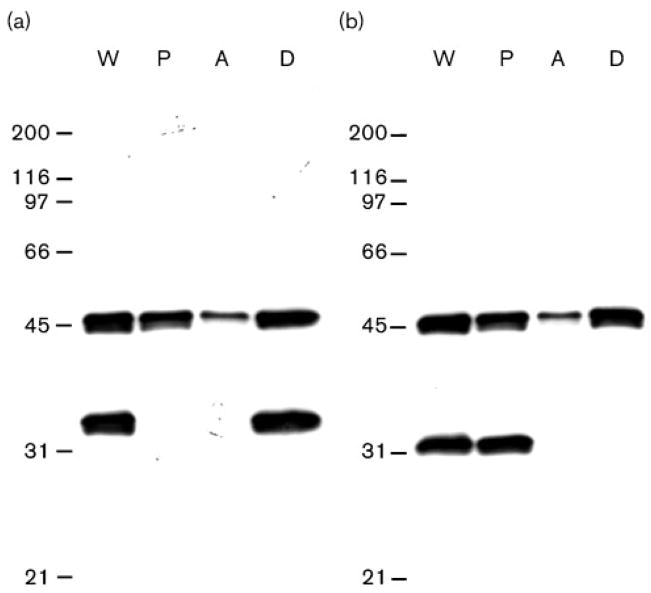
We analysed the behaviour of LipL46 in the nonionic detergent Triton X-114. Triton X-114 extraction selectively solubilizes the leptospiral OM, including LipL32, but not cytoplasmic membrane components, such as LipL31 (Cullen et al., 2003; Haake & Matsunaga, 2002). LipL46 was found in both the Triton X-114 soluble and insoluble fractions, suggesting that this lipoprotein may be present in both the outer and inner membranes. However, it should be noted that incomplete solubilization by Triton X-114 has also been observed for the leptospiral OMPs OmpL1 and LipL41 (Haake et al., 1993; Shang et al., 1996). Although the His6–LipL46 fusion protein was highly soluble in aqueous buffers lacking detergent (data not shown), fractionation of native LipL46 by Triton X-114 revealed partitioning of LipL46 primarily into the detergent-phase fraction (Fig. 2). The amphiphilic behaviour of LipL46 is consistent with modification by fatty acids (Haake, 2000).
Fig. 2.
Behaviour of LipL46 in Triton X-114. Triton X-114 fractions of L. interrogans organisms were separated by SDS-PAGE, and probed with LipL46 and LipL32 antisera (a), and LipL46 and LipL31 antisera (b). LipL32 and LipL31 are markers for the leptospiral outer and cytoplasmic membranes, respectively. (a, b) The fractions analysed are shown in the lanes labelled as follows: W, whole organism; P, Triton-X-114-insoluble pellet; A, aqueous-phase material; D, detergent-phase material. The number of cell equivalents was the same in all four lanes. Locations of molecular size standards (kDa) are shown on the left. Most of the LipL46 protein solubilized by Triton X-114 partitioned into the detergent phase.
Assessment of LipL46 surface exposure by whole-cell ELISA
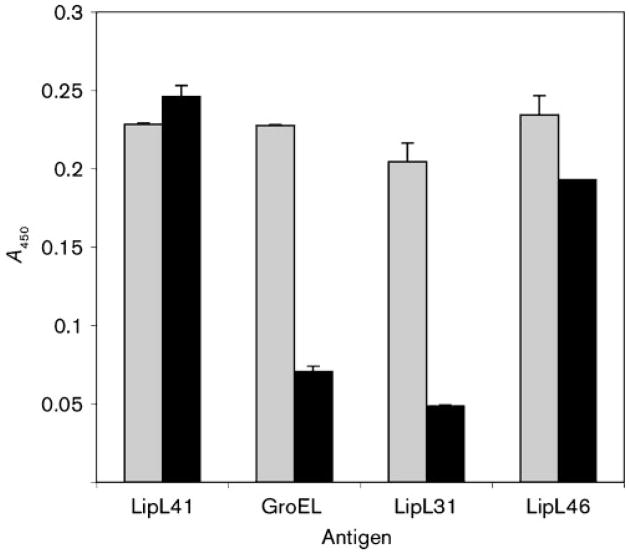
We have developed an ELISA assay for assessing surface exposure of leptospiral proteins (Cullen et al., 2005). In this assay, we compare binding of antibody to leptospiral sonicates and whole cells. Antisera to the OM lipoprotein LipL41, the cytoplasmic membrane protein LipL31 (Haake & Matsunaga, 2002) and the cytoplasmic protein GroEL were included as positive and negative controls. As expected, binding of antibodies to the subsurface control proteins GroEL and LipL31 was considerably higher to sonicated material than to whole cells (Fig. 3). This result indicates that the leptospiral OM remained intact during the analysis. In contrast, LipL41 antibodies bound in relatively equal amounts to leptospiral sonicates and whole cells, consistent with prior studies demonstrating exposure of LipL41 on the leptospiral surface (Cullen et al., 2005; Shang et al., 1996). Comparable levels of reactivity to leptospiral sonicates and whole cells were also observed when antiserum to LipL46 was applied to the wells, indicating that LipL46 is a surface-exposed lipoprotein.
Fig. 3.
Detection of L. interrogans surface labelling by LipL41 and LipL46 antisera and whole-cell ELISA. Wells of microtitre plates were coated with either L. interrogans sonicates (grey bars) or whole-cells (black bars). Antisera were diluted to give ELISA reactivity to L. interrogans sonicates in the 0·2–0·25 absorbance range at 450 nm. Antisera dilutions were identical in the sonicate and whole-cell ELISA experiments. Sonicate and whole-cell ELISA reactivities of antisera to LipL46 and the surface-exposed LipL41 were comparable, while sonicate ELISA reactivity exceeded whole-cell ELISA reactivity using antisera to the cytoplasmic heat-shock protein GroEL and the cytoplasmic membrane protein LipL31. The A450 values were obtained after subtracting the value for the control without antibody.
LipL46 is exposed on the leptospiral surface
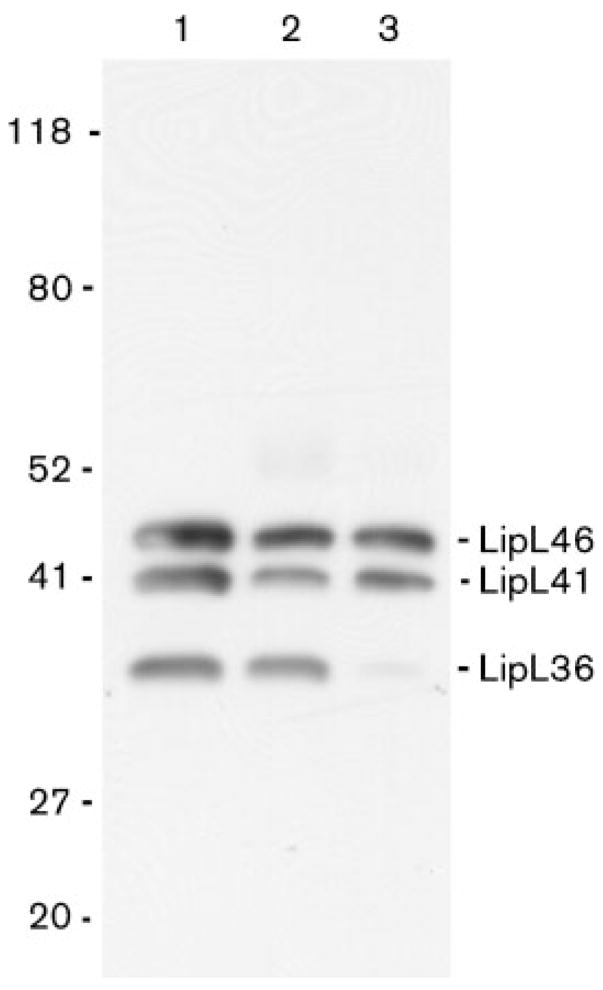
Previous surface-immunoprecipitation studies identified OmpL1, LipL41 and a protein antigen with a molecular mass of approximately 46 kDa on the leptospiral surface (Haake et al., 1991; Shang et al., 1996). We re-examined the surface-immunoprecipitation technique using antisera specific for LipL46, LipL41 and LipL36 to determine whether LipL46 was exposed on the leptospiral surface. In this procedure, antiserum raised to whole L. kirschneri is allowed to bind to motile intact organisms. Unbound antibody is removed prior to solubilization and immunoprecipitation. To evaluate the specificity of the surface-immunoprecipitation technique for surface antigens, immunoprecipitation was performed both before and after Triton X-100 extraction. The surface and Triton-soluble immunoprecipitates were analysed by SDS-PAGE and immunoblot with antisera specific for LipL46, LipL41 and LipL36. As shown in Fig. 4, LipL46, LipL41 and LipL36 were immunoprecipitated from Triton-X-100-solubilized extracts. In contrast, only LipL46 and LipL41 were accessible to antibody on the surface of intact organisms. The surface-immunoprecipitation experiments were also performed with LipL36 antiserum added during the incubation with intact bacteria, and they showed that the inability to identify LipL36 on the leptospiral surface was not due to insufficient LipL36 antibodies in antiserum to whole bacteria (data not shown). These data indicate that LipL46 and LipL41 are surface exposed, and confirm that LipL36 is a subsurface protein.
Fig. 4.
Immunoprecipitation of LipL41 and LipL46 on the surface of L. kirschneri. Total L. kirschneri proteins (lane 1), and proteins immunoprecipitated by antiserum to L. kirschneri after (lane 2) and before (lane 3) solubilization with Triton X-100, were separated by SDS-PAGE, and probed with LipL36, LipL41 and LipL46 antisera. Both LipL41 and LipL46 were found to be present in the surface-immunoprecipitated material. Although LipL36 was immunoprecipitated by the antiserum after solubilization by Triton X-100, it was not accessible to antibody on the surface of L. kirschneri. The locations of the molecular size (kDa) standards are shown on the left.
Humoral immune response to LipL46 in hamsters
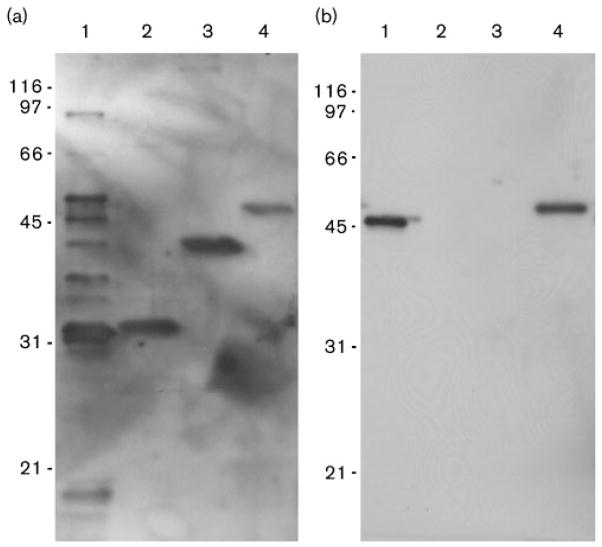
We have previously examined the humoral immune response to leptospiral proteins in the hamster model of leptospirosis (Barnett et al., 1999). In the previous studies, leptospiral antigens with molecular masses of 37, 41 and approximately 46 kDa were recognized by sera from hamsters surviving to day 28 after infection, with the 41 kDa protein identified as LipL41. In the current study, only 2 of 18 hamsters challenged with L. interrogans serovar Copenhageni survived to day 28. Serum from the first hamster demonstrated a similar pattern of reactivity to native leptospiral antigens to that found in the prior study, with reactivity to the same 37, 41 and 46 kDa antigens, and additional bands of 32 and 49 kDa also observed in Fig. 5(a). Antibodies to the 32, 41 and 46 kDa antigens in the serum of the first hamster were consistent with reactivity with recombinant His6-tagged LipL32, LipL41 and LipL46. Serum from the second hamster reacted uniquely with LipL46 (Fig. 5b). These studies suggest that LipL46 is a prominent immunogen during leptospiral infection of hamsters, and that it may be useful as a recombinant antigen in the serodiagnosis of leptospirosis.
Fig. 5.
Reactivity of infected hamster sera with native and recombinant leptospiral proteins. (a, b) Immunoblots of total L. interrogans proteins (lane 1, 5×107 cells per lane) and recombinant LipL32, LipL41 and LipL46 (lanes 2–4, respectively, 200 μg per lane) separated by SDS-PAGE and probed with sera from hamsters infected with L. interrogans. His6-tagged recombinant proteins migrate slightly slower than native proteins. Serum from the first hamster (a) contained antibodies to all three proteins, while serum from the second hamster (b) contained antibodies to LipL46 only.
Immunohistochemistry with LipL32 and LipL46 antisera
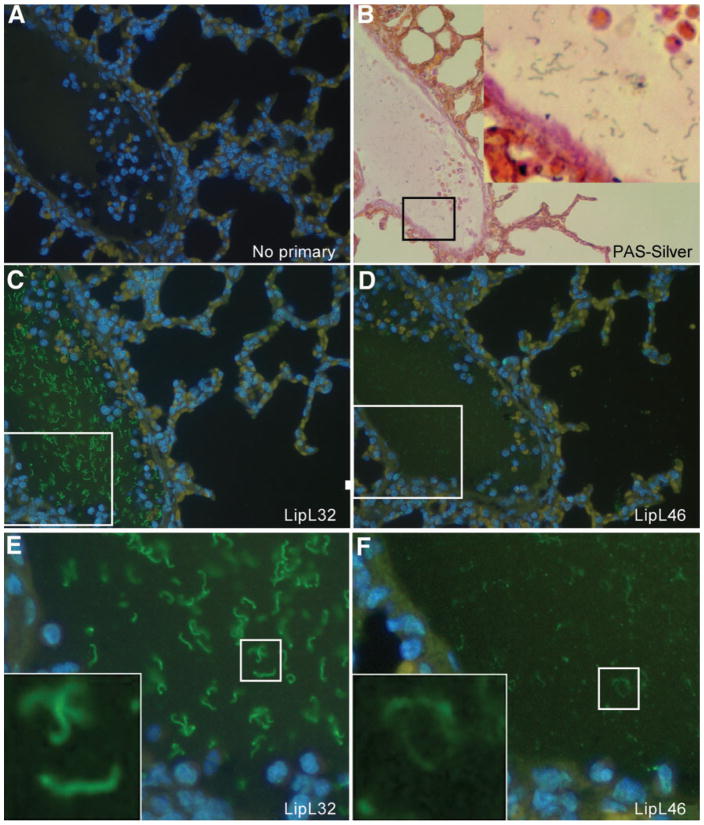
Previous immunohistochemistry studies of the hamster model of leptospirosis have focused on detection of leptospiral antigens expressed by organisms within renal tubules (Barnett et al., 1999; Haake et al., 2000). In this study, we examined antigen expression by leptospires in blood vessels and a variety of organs, including the lung, liver, spleen and kidney. PAS/Steiner silver staining and immunohistochemistry with antisera to leptospiral lipoproteins provided new insights into protein expression and distribution of organisms during the initial acute stage of infection. All tissues examined revealed organisms staining positively with antisera to LipL32 and LipL46, indicating that these lipoproteins are expressed during the early dissemination stage of leptospirosis. Examination of lung tissue revealed a high density of organisms in the lumen of a vein (Fig. 6), consistent with a haematogenous route of dissemination. Organisms were also found within the pulmonary parenchyma (data not shown). Infection in the liver was primarily extracellular, with large numbers of organisms found in the sinusoids between hepatocytes (supplementary Fig. S1). In the red pulp of the spleen, disrupted leptospiral antigen was primarily observed within phagocytes (supplementary Fig. S2). In contrast, in the white pulp of the spleen, organisms were primarily extracellular. Kidney sections revealed organisms within glomeruli, often with the highest density of leptospires located in the glomerular hilum at the confluence of the afferent and efferent arterioles (supplementary Fig. S3). Organisms were also found in the lumena of proximal and distal tubules, as well as within collecting ducts (data not shown). PAS/Steiner silver staining and immunohistochemistry were shown to be specific for leptospiral structures by use of negative-control liver (supplementary Fig. S4) and kidney (supplementary Fig. S5) tissues from uninfected hamsters.
Fig. 6.
Serial sections of hamster lung tissue showing a high density of L. interrogans serovar Pomona strain 11000-74A within the lumen of a blood vessel. Tissue sections were stained with PAS/Steiner silver stain (B), or by immunohistochemistry using no primary antibody (A), LipL32 antiserum (C, E), or LipL46 antiserum (D, F). Magnification ×30 (B) and ×40 (A, C, D). (E, F) Enlargements of the boxed areas shown in (C) and (D), respectively. (B, E, F) Insets are enlargements of boxed areas in the same panels.
DISCUSSION
In this study, we examined the lipid modification, export and expression of a novel 46 kDa lipoprotein during leptospiral bloodstream and organ dissemination. The carboxy-terminal region of the signal peptide contains an FSISC lipoprotein signal peptidase cleavage site, consistent with intrinsic labelling of LipL46 with [14C]palmitate by L. interrogans. Experimental evidence is presented indicating that LipL46 is exported to the leptospiral OM, and is accessible to antibody on the leptospiral surface. Relatively few of the 157–164 lipoproteins predicted to be encoded by the L. interrogans genome have been characterized (Setubal et al., 2006). Consequently, little is known about the sequence determinants of the leptospiral lipobox, lipoprotein export signals, and types of lipoproteins expressed during infection of the mammalian host. Previous immunohistochemistry studies using antisera to leptospiral proteins have examined leptospiral expression in the kidney during the later tubular stages of infection (Barnett et al., 1999; Haake et al., 2000). This is the first study we are aware of to examine lipoprotein expression in multiple organs during the acute dissemination phase of leptospirosis.
Haematogenous dissemination from sites of infection acquisition to the kidney is an essential step in the leptospiral life cycle in reservoir hosts. New insights into the pathogenesis of acute leptospirosis were gained by utilizing a strain of L. interrogans serovar Pomona, obtained by serial passage from the liver of Golden Syrian hamsters 3–4 days after intraperitoneal challenge. Silver staining of tissues from infected animals showed that this organism achieved high densities in the bloodstream and liver during the acute stage of leptospirosis. Lower densities of organisms were also found in the spleen, lung and kidney. Immunohistochemistry using antisera specific for LipL46 and LipL32 demonstrated that both lipoproteins were expressed by L. interrogans at all investigated sites of infection. Although pathogenic leptospires have been shown to have the ability to penetrate within host cells in tissue culture (Merien et al., 1997), organisms observed in the liver appeared to be primarily extracellular. Although LipL46 antigen was observed within splenic phagocytes, the intracellular antigen distribution suggests that organisms within phagocytes had undergone phagocytosis, rather than intracellular penetration. However, active penetration of tissue barriers did appear to be occurring because organisms were found within both the glomerular hilum and the capsule, suggesting glomerular vessels as an important site for leptospires to gain access to the proximal tubule. An alternative interpretation is that leptospiral invasion may be affected by the glomerular haemorrhage observed in supplementary Fig. S3.
LipL46 is one of three surface-exposed leptospiral OMPs originally identified by Triton X-114 fractionation and surface immunoprecipitation (Haake et al., 1991). Two of these surface proteins have been recognized to be the porin OmpL1 (Haake et al., 1993) and the lipoprotein LipL41 (Shang et al., 1996). A study of leptospiral proteins recognized by the humoral immune response identified a highly conserved 46 kDa protein recognized by a majority of sera from human leptospirosis patients. Immunoblots of 2D gels determined that the 46 kDa protein antigen had a slightly more acidic isoelectric point than LipL41 (Guerreiro et al., 2001). This same 46 kDa protein antigen has been recognized as a component of leptospiral OM vesicles isolated by alkaline plasmolysis and sucrose density gradient ultracentrifugation (Haake & Matsunaga, 2002). Subsequent characterization of pL45 by MALDI-TOF analysis and partial sequencing allowed identification of the gene encoding the 46 kDa protein (Cullen et al., 2002; Nally et al., 2005b).
LipL46 joins a growing list of leptospiral lipoproteins for which there is experimental evidence of lipidation (Table 1). Lipidation studies are extremely useful for prediction algorithms based on weight matrix approaches because they enable more accurate identification of genes encoding lipoproteins in the genomes of Leptospira species and other spirochaetes (Setubal et al., 2006). Cellular localization studies may also provide insight into the amino acid requirements for export of lipoproteins to the OM. In double-membrane bacteria, the amino acids near the amino-terminal cysteine of the mature lipoprotein are thought to determine sorting to the outer or inner membrane by strengthening or weakening, respectively, interactions with the Lol export pathway. The leptospiral genome contains homologues of the genes encoding the LolCDE sortase complex and the periplasmic LolA shuttle protein (Nascimento et al., 2004; Ren et al., 2003). Table 1 compares the amino-terminal sequences of known or probable lipoproteins for which cellular localization data are available. The LipL46 sequence is consistent with an emerging pattern in which all the known surface-exposed OM lipoproteins (LipL21, LipL32, LipL41 and LigB) have neutral amino acids in the +2 to +4 positions. In contrast, the inner membrane lipoprotein LipL31 has the negatively charged amino acid aspartate in the +3 position, and the lipoproteins LruA and LruB have the negatively charged amino acid glutamate in the +4 position. The subsurface OM lipoprotein LipL36 is an exception to this pattern, both in terms of its location and surface exposure, and in terms of its sequence: a positively charged amino acid, lysine, in the +2 position, and negatively charged aspartate residues in the +4 to +9 positions. Recently, site-directed mutagenesis studies in B. burgdorferi have shown that negatively charged amino acids placed near the amino terminus of OspA can function as inner membrane retention signals (Schulze & Zuckert, 2006). As genetic systems are developed for Leptospira species, site-directed mutagenesis studies will be required to validate the role of specific amino acids and positions as determinants of lipoprotein localization.
Table 1.
Membrane location and surface exposure of leptospiral lipoproteins
| Designation | Mass (kDa) | Membrane location★ | Surface exposure† | Lipobox and amino-terminal sequence | LIC locus tag‡ |
|---|---|---|---|---|---|
| LipL21§ | 21 | OM | + | IFAACSSTDTGQKDATTV | LIC10011 |
| LipL31 | 31 | IM | − | FFASCGDNSEVIETLDGN | LIC11456 |
| LipL32§ | 32 | OM | + | SITACGAFGGLPSLKSSF | LIC11352 |
| LipL36§ | 36 | OM | − | ALTACKSDDDDDDVVMLA | LIC13060 |
| LipL41§ | 41 | OM | + | FLGNCAATVDVEYPVFPK | LIC12966 |
| LipL46§ | 46 | OM | + | FSISCGSSGSTRGKKKEF | LIC11885 |
| LipL48 | 48 | OM | ? | SFINCKEDKDDNSMLLLL | LIC10898 |
| LruB | 50 | IM | − | LFSNCKSEANNDSERLAL | LIC10713 |
| LruA§ | 71 | IM | − | FFISCGAELPIEELSDAK | LIC11003 |
| LigB | 220 | OM | + | FFQSCMSWPLLTSLAGLA | LIC10464 |
Membrane localization as determined by membrane fractionation; IM, inner membrane.
Surface exposure determined by surface biotinylation, surface immunofluorescence, whole-cell ELISA, surface immunoprecipitation and/or immunoelectronmicroscopy.
The LIC locus tag corresponds to the L. interrogans serovar Copenhageni strain L1-130 genome.
Strains for which there is published experimental evidence of lipidation (this study and Cullen et al., 2003; Haake et al., 1998, 2000; Shang et al., 1996; Verma et al., 2005).
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grant AI-34431 (to D. A. H.) from the National Institute of Allergy and Infectious Diseases, and by VA Medical Research Funds (to J. M. and D. A. H.). The authors thank Paul Hauer, Mary Rasmusson and Sue Whitaker for supplying tissue from infected hamsters, Judy Stasko for histology support and providing new staining techniques, and William Stoffregen for expertise in veterinary pathology.
Abbreviations
- DAPI
4′,6-diamidino-2-phenylindole
- H/E
haematoxylin and eosin
- IEM
immunoelectron microscopy
- OM
outer membrane
- OMP
outer-membrane protein
- PAS
periodic acid Schiff
Footnotes
Supplementary figures are available with the online version of this paper.
References
- Babudieri B. Animal reservoirs of leptospires. Ann N Y Acad Sci. 1958;70:393–413. doi: 10.1111/j.1749-6632.1958.tb35398.x. [DOI] [PubMed] [Google Scholar]
- Barnett JK, Barnett D, Bolin CA, Summers TA, Wagar EA, Cheville NF, Hartskeerl RA, Haake DA. Expression and distribution of leptospiral outer membrane components during renal infection of hamsters. Infect Immun. 1999;67:853–861. doi: 10.1128/iai.67.2.853-861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branger C, Sonrier C, Chatrenet B, Klonjkowski B, Ruvoen-Clouet N, Aubert A, Andre-Fontaine G, Eloit M. Identification of the hemolysis-associated protein 1 as a cross-protective immunogen of Leptospira interrogans by adenovirus-mediated vaccination. Infect Immun. 2001;69:6831–6838. doi: 10.1128/IAI.69.11.6831-6838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PA, Cordwell SJ, Bulach DM, Haake DA, Adler B. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect Immun. 2002;70:2311–2318. doi: 10.1128/IAI.70.5.2311-2318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PA, Haake DA, Bulach DM, Zuerner RL, Adler B. LipL21 is a novel surface-exposed lipoprotein of pathogenic Leptospira species. Infect Immun. 2003;71:2414–2421. doi: 10.1128/IAI.71.5.2414-2421.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PA, Xu X, Matsunaga J, Sanchez Y, Ko AI, Haake DA, Adler B. Surfaceome of Leptospira spp. Infect Immun. 2005;73:4853–4863. doi: 10.1128/IAI.73.8.4853-4863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faine S, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. 2. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- Guerreiro H, Croda J, Flannery B, Mazel M, Matsunaga J, Galvao Reis M, Levett PN, Ko AI, Haake DA. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect Immun. 2001;69:4958–4968. doi: 10.1128/IAI.69.8.4958-4968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA. Spirochaetal lipoproteins and pathogenesis. Microbiology. 2000;146:1491–1504. doi: 10.1099/00221287-146-7-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Matsunaga J. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect Immun. 2002;70:4936–4945. doi: 10.1128/IAI.70.9.4936-4945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Walker EM, Blanco DR, Bolin CA, Miller MN, Lovett MA. Changes in the surface of Leptospira interrogans serovar Grippotyphosa during in vitro cultivation. Infect Immun. 1991;59:1131–1140. doi: 10.1128/iai.59.3.1131-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Champion CI, Martinich C, Shang ES, Blanco DR, Miller JN, Lovett MA. Molecular cloning and sequence analysis of the gene encoding OmpL1, a transmembrane outer membrane protein of pathogenic Leptospira spp. J Bacteriol. 1993;175:4225–4234. doi: 10.1128/jb.175.13.4225-4234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Martinich C, Summers TA, Shang ES, Pruetz JD, McCoy AM, Mazel MK, Bolin CA. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late-log-phase growth and mammalian infection. Infect Immun. 1998;66:1579–1587. doi: 10.1128/iai.66.4.1579-1587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Chao G, Zuerner RL, Barnett JK, Barnett D, Mazel M, Matsunaga J, Levett PN, Bolin CA. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infect Immun. 2000;68:2276–2285. doi: 10.1128/iai.68.4.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake DA, Suchard MA, Kelley MM, Dundoo M, Alt DP, Zuerner RL. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J Bacteriol. 2004;186:2818–2828. doi: 10.1128/JB.186.9.2818-2828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada R, Ido Y, Hoki R, Kaneko R, Ito H. The etiology, mode of infection, and specific therapy of Weil’s disease (spirochaetosis icterohaemorrhagica) J Exp Med. 1916;23:377–402. doi: 10.1084/jem.23.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RC, Harris VG. Differentiation of pathogenic and saprophytic letospires. I. Growth at low temperatures. J Bacteriol. 1967;94:27–31. doi: 10.1128/jb.94.1.27-31.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Watanabe H. Molecular cloning and characterization of a novel leptospiral lipoprotein with OmpA domain. FEMS Microbiol Lett. 2003;226:215–219. doi: 10.1016/S0378-1097(03)00619-0. [DOI] [PubMed] [Google Scholar]
- Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J, Young TA, Barnett JK, Barnett D, Bolin CA, Haake DA. Novel 45-kilodalton leptospiral protein that is processed to a 31-kilodalton growth-phase-regulated peripheral membrane protein. Infect Immun. 2002;70:323–334. doi: 10.1128/IAI.70.1.323-334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J, Barocchi MA, Croda J, et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol Microbiol. 2003;49:929–945. doi: 10.1046/j.1365-2958.2003.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga J, Sanchez Y, Xu X, Haake DA. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect Immun. 2005;73:70–78. doi: 10.1128/IAI.73.1.70-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AJ, Athanazio DA, Reis MG, Ko AI. Leptospirosis. Curr Opin Infect Dis. 2005;18:376–386. doi: 10.1097/01.qco.0000178824.05715.2c. [DOI] [PubMed] [Google Scholar]
- Merien F, Baranton G, Perolat P. Invasion of Vero cells and induction of apoptosis in macrophages by pathogenic Leptospira interrogans are correlated with virulence. Infect Immun. 1997;65:729–738. doi: 10.1128/iai.65.2.729-738.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally JE, Chow E, Fishbein MC, Blanco DR, Lovett MA. Changes in lipopolysaccharide O antigen distinguish acute versus chronic Leptospira interrogans infections. Infect Immun. 2005a;73:3251–3260. doi: 10.1128/IAI.73.6.3251-3260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nally JE, Whitelegge JP, Aguilera R, Pereira MM, Blanco DR, Lovett MA. Purification and proteomic analysis of outer membrane vesicles from a clinical isolate of Leptospira interrogans serovar Copenhageni. Proteomics. 2005b;5:144–152. doi: 10.1002/pmic.200400880. [DOI] [PubMed] [Google Scholar]
- Nascimento AL, Ko AI, Martins EA, et al. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J Bacteriol. 2004;186:2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris SJ, Carter CJ, Howell JK, Barbour AG. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren SX, Fu G, Jiang XG, et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- Schulze RJ, Zuckert WR. Borrelia burgdorferi lipoproteins are secreted to the outer surface by default. Mol Microbiol. 2006;59:1473–1484. doi: 10.1111/j.1365-2958.2006.05039.x. [DOI] [PubMed] [Google Scholar]
- Segura ER, Ganoza CA, Campos K, et al. Clinical spectrum of pulmonary involvement in leptospirosis in a region of endemicity, with quantification of leptospiral burden. Clin Infect Dis. 2005;40:343–351. doi: 10.1086/427110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setubal JC, Reis MG, Matsunaga J, Haake D. Lipoprotein computational prediction in spirochaetal genomes. Microbiology. 2006;152:113–121. doi: 10.1099/mic.0.28317-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang ES, Exner MM, Summers TA, Martinich C, Champion CI, Hancock RE, Haake DA. The rare outer membrane protein, OmpL1, of pathogenic Leptospira species is a heat-modifiable porin. Infect Immun. 1995;63:3174–3181. doi: 10.1128/iai.63.8.3174-3181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang ES, Summers TA, Haake DA. Molecular cloning and sequence analysis of the gene encoding LipL41, a surface-exposed lipoprotein of pathogenic Leptospira species. Infect Immun. 1996;64:2322–2330. doi: 10.1128/iai.64.6.2322-2330.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner G, Steiner G. New simple silver stain for demonstration of bacteria, spirochetes and fungi in sections of paraffin embedded tissue blocks. J Lab Clin Med. 1944;29:868–871. [Google Scholar]
- Thiermann AB, McClellan RD, Hill HT. Improved techniques for the isolation of leptospires from swine abortion cases. Ann Proc Am Assoc Vet Lab Diagn. 1984;27:233–244. [Google Scholar]
- Verma A, Artiushin S, Matsunaga J, Haake DA, Timoney JF. LruA and LruB, novel lipoproteins of pathogenic Leptospira interrogans associated with equine recurrent uveitis. Infect Immun. 2005;73:7259–7266. doi: 10.1128/IAI.73.11.7259-7266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuerner RL, Knudtson W, Bolin CA, Trueba G. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar Pomona. Microb Pathog. 1991;10:311–322. doi: 10.1016/0882-4010(91)90014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.