Summary
MicroRNAs are important players in stem cell biology. Among them, microRNA-9 (miR-9) is expressed specifically in neurogenic areas of the brain. Whether miR-9 plays a role in neural stem cell self-renewal and differentiation is unknown. We showed previously that nuclear receptor TLX is an essential regulator of neural stem cell self-renewal. Here we show that miR-9 suppresses TLX expression to negatively regulate neural stem cell proliferation and accelerate neural differentiation. Introducing a TLX expression vector lacking the miR-9 recognition site rescued miR-9-induced proliferation deficiency and inhibited precocious differentiation. In utero electroporation of miR-9 in embryonic brains led to premature differentiation and outward migration of the transfected neural stem cells. Moreover, TLX represses miR-9 pri-miRNA expression. MiR-9, by forming a negative regulatory loop with TLX, establishes a model for controlling the balance between neural stem cell proliferation and differentiation.
Keywords: nuclear receptor, TLX (NR2E1), microRNA, neural stem cells, proliferation, differentiation
Introduction
One of the most important issues in stem cell biology is to understand the molecular mechanisms underlying stem cell self-renewal and differentiation. Neural stem cells are a subset of undifferentiated precursors that retain the ability to proliferate and self-renew, and have the capacity to give rise to both neuronal and glial lineages1-4. Although the functional properties of neural stem cells have been studied extensively, how self-renewal and differentiation of neural stem cells is regulated is not completely understood.
Accumulating evidence indicates that both transcriptional and post-transcriptional regulation are important mechanisms for regulating genes that are essential for neural stem cell self-renewal and neurogenesis. MicroRNAs (miRNAs) are a recently identified large family of 20-22 nucleotide non-coding RNAs that are involved in numerous cellular processes, including development, proliferation, and differentiation5,6. MiRNAs are thus potentially key post-transcriptional regulators in stem cell self-renewal and differentiation. Distinct sets of miRNAs have been shown to be specifically expressed in embryonic stem cells7,8. Loss of Dicer1 causes embryonic lethality and loss of stem cell populations9,10. Argonaute family members, key components of the RNA-induced silencing complex (RISC), are required for maintaining germline stem cells in various species11. These observations together support a role for miRNAs in stem cell biology. Several brain-specific miRNAs have been identified recently. Among these miRNAs, miR-9 is expressed specifically in neurogenic regions of the brain during neural development and in adulthood12-15. Whether miR-9 plays a role in neural stem cell self-renewal and differentiation remains to be determined.
We have shown that TLX is an essential regulator of neural stem cell self-renewal16. TLX maintains adult neural stem cells in an undifferentiated and self-renewable state, in part through transcriptional repression of its downstream target genes, p21 and pten, by complexing with histone deacetylases17. Recently, TLX-positive neural stem cells have been shown to play a role in spatial learning and memory18. In addition to its function in adult brains, TLX also plays an important role in neural development by regulating cell cycle progression in neural stem cells of the developing brain19-21. TLX is therefore a key regulator that acts to establish the undifferentiated and self-renewable state of neural stem cells, though aspects of its regulation are enigmatic.
Here we demonstrate that miR-9 suppresses TLX expression by binding to the 3′ untranslated region (UTR) of TLX mRNA, which, in turn, regulates neural stem cell proliferation and differentiation. Increased expression of miR-9 led to reduced mouse neural stem cell proliferation and accelerated neural differentiation, whereas antisense-knockdown of miR-9 led to increased neural stem cell proliferation. Introducing a TLX expression vector lacking the endogenous TLX 3′ UTR rescued miR-9 overexpression-induced proliferation deficiency and reversed miR-9 promoted precocious differentiation. This result suggests that miR-9 regulates neural stem cell proliferation and differentiation, at least in part, through targeting TLX mRNA via its 3′ UTR. In utero electroporation of miR-9 into ventricular zone neural stem cells in embryonic mouse brains triggered premature differentiation and outward migration of the transfected cells, similar to that induced by electroporation of TLX siRNA21. Furthermore, TLX binds to the 3′ genomic sequences of miR-9-1 to inhibit its expression. MiR-9 and TLX thus form a feedback loop to regulate the switch of neural stem cell proliferation and differentiation.
Results
MiR-9 represses TLX expression by targeting its 3′ UTR
We hypothesized that TLX could be targeted by miRNAs to regulate its expression. Using the miRanda (http://www.microrna.org)22 and TargetScan (http://genes.mit.edu/targetscan)23 algorithms, miR-9 was predicted to have a target site in the TLX 3′ UTR. This target site is conserved in human, mouse, dog and chicken TLX (Supplementary Fig. 1a). Since TLX is specifically expressed in vertebrate forebrains and is an essential regulator of neural stem cell self-renewal, we first asked whether the candidate TLX-targeting miR-9 is expressed in the brain, and specifically, whether this miRNA is expressed in neural stem cells or in their differentiated progeny. As shown in Supplementary Fig. 1b, miR-9 is expressed specifically in the brain as revealed by Northern blotting, consistent with previous reports12,13,24. The size of the miRNA is as expected for the mature miR-9 (22 bp). MiR-9 is also expressed in neural stem cells (d0, Fig. 1a). Interestingly, the expression of miR-9 is upregulated during neural differentiation (Fig. 1a), in contrast to reduced expression of TLX (Fig. 1b).
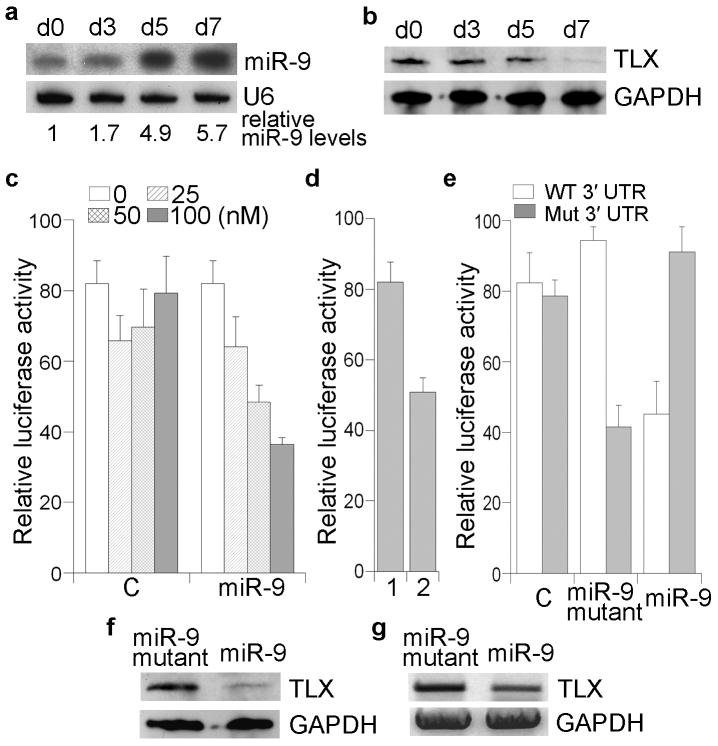
Fig. 1. miR-9-directed repression of TLX expression.
a. miR-9 expression in adult mouse neural stem cells during a differentiation time course. Day 0 (d0) represents the undifferentiated neural stem cell state. U6 was included as a loading control. Relative miR-9 levels, normalized to U6 levels, are indicated under the blots, with miR-9 level in d0 designated as 1. b. Western blot analysis of TLX expression in the same differentiation time course. GAPDH was included as a loading control. c. miR-9-mediated repression of luciferase reporter gene upstream of TLX 3′ UTR. The TLX 3′ UTR reporter gene was co-transfected with increasing amounts of miR-9 RNA duplexes or control siRNA duplexes into HEK 293 cells. d. Similar reporter assays were performed in cells transfected with MDH1-PGK-GFP control vector (1) or miR-9-1 expression vector (2). e. Mutation of the miR-9 target site in TLX 3′ UTR abolished miR-9-mediated repression. Luciferase reporter gene under the control of wild type (WT) or mutant (Mut) TLX 3′ UTR was transfected along with control siRNA (C), a miR-9 mutant with mutations in the seed region complementary to the mutant TLX 3′ UTR, or wild type miR-9 into HEK 293 cells. s.d. is indicated by error bars. f, g. miR-9-mediated repression of TLX expression in neural stem cells revealed by Western blot (f) and RT-PCR (g) analyses. A miR-9 mutant with mutations in the seed region was included as a control.
To validate whether miR-9 targets TLX, we made a luciferase reporter construct with mouse 1.4 kb TLX 3′ UTR containing the predicted miR-9 target site and flanking sequences inserted into the 3′ UTR of a Renilla luciferase reporter gene in a siCHECK vector. Increasing amounts of RNA duplexes of mature miR-9 were transfected into human embryonic kidney HEK 293 cells along with the corresponding reporter gene. Dose-dependent repression of the reporter gene was observed in miR-9-treated cells (Fig. 1c). In addition to the synthetic RNA duplexes, miR-9 was also expressed using a microRNA expressing vector, MDH1-PGK-GFP225, in which 489 neocleotide (nt) miR-9-1 genomic sequence, including the 89 nt miR-9 hairpin precusor and 200 nt genomic sequence flanking each side of the precursor, was cloned. The miR-9 expression vector repressed luciferase reporter gene activity (Fig. 1d), similar to the miR-9 RNA duplexes (Fig. 1c), suggesting that miR-9 suppresses TLX expression through its 3′ UTR. Base pairing between the miRNA seed sequence and its target gene is needed for miRNA to repress the target mRNA26,27. To test whether the predicted miR-9 target site in TLX 3′ UTR is critical for repression of TLX expression by miR-9, point mutations disrupting complementarity in the predicted miR-9 target site were introduced into TLX 3′ UTR in the luciferase reporter construct. Mutation of the miR-9 target site abolished the repression by miR-9 (Fig. 1e). A miR-9 mutant with compensatory mutations repressed a luciferase reporter gene with mutated TLX 3′ UTR (Fig. 1e). These results strongly suggest that miR-9 represses TLX expression through the predicted target site in TLX 3′ UTR.
Next we tested whether miR-9 targets TLX in neural stem cells. Mature miR-9 RNA duplexes were transfected into neural stem cells. TLX expression levels in transfected cells were examined by Western blot and Northern blot analyses. Marked reduction of TLX protein levels and mRNA levels were detected in miR-9 transfected cells (Fig. 1f, g), indicating that miR-9 down-regulates TLX expression.
MiR-9 regulates neural stem cell fate determination
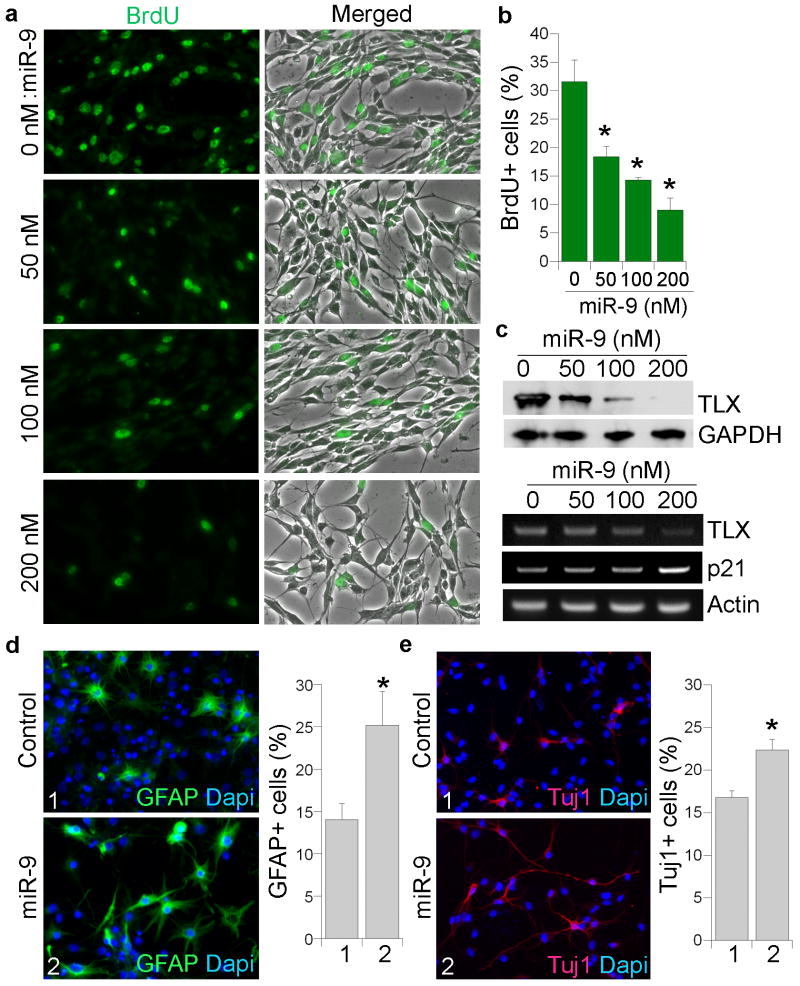
To examine whether miR-9 regulates neural stem cell proliferation, neural stem cells were transfected with increasing concentrations of miR-9 RNA duplexes. Cell proliferation was determined by 5-bromodeoxyuridine (BrdU) labeling of dividing cells. Transfection of miR-9 led to dose-dependent inhibition of cell proliferation (Fig. 2a, b) with a minimal effect on cell death (Supplementary Fig. 1c, d). Accordingly, reduced expression of TLX and increased expression of p21, a target gene that is repressed by TLX17, was observed in miR-9-transfected cells in a dose-dependent manner (Fig. 2c). This gene expression profile is consistent with miR-9-induced inhibition of neural stem cell proliferation (Fig. 2a, b), suggesting that miR-9 negatively regulates neural stem cell proliferation, presumably through downregulation of TLX signaling.
Fig. 2. Overexpression of miR-9 regulates neural stem cell proliferation and differentiation.
a. Cell proliferation in miR-9-transfected neural stem cells as revealed by BrdU labeling (green). Merged panels show BrdU staining along with phase contrast images. A miR-9 mutant with mutations in the seed region was included as a control, with a total of 200 nM miRNAs in each transfection. b. Quantification of BrdU-positive (BrdU+) cells in miR-9-treated neural stem cells with s.d. indicated by error bars. * p=0.0008 by one-way Anova. c. Top panel is Western blot analysis of TLX expression in miR-9 transfected neural stem cells. GAPDH was included as a loading control. Lower panel is RT-PCR analysis of TLX and p21 in miR-9-transfected neural stem cells. Actin was included as a loading control. d. Overexpression of miR-9 promotes glial differentiation. Control RNA or miR-9-transfected cells were induced into differentiation for 3 days and immunostained with a GFAP-specific antibody (green). Nuclear dapi staining is shown in blue. e. Overexpression of miR-9 promotes neuronal differentiation. Control RNA or miR-9 transfected neural stem cells were induced to differentiate for 3 days and immunostained with a Tuj1-specific antibody (red). Nuclear dapi staining is shown in blue. For both sections d and e, error bars are s.d. of the mean. * p=0.03 (d), 0.018 (e) by Student's t-test. About 4,000 cells were quantified for panels d and e, respectively.
To determine whether overexpression of miR-9 regulates neural stem cell differentiation, neural stem cells were transfected with miR-9 RNA duplexes and cultured under different conditions. Since complete withdrawal of EGF and FGF led to considerable cell death (data not shown), neural stem cells were cultured in N2-supplemented media with low EGF and FGF concentrations (1 ng ml− 1), which allowed cell viability with minimal cell proliferation. Over a 7-day time course, no difference in neuronal and glial differentiation could be detected between control RNA and miR-9 transfected cells (data not shown), suggesting that overexpression of miR-9 alone is not sufficient to trigger neuronal or glial differentiation. However, when neural stem cells were induced for differentiation using forskolin or retinoic acid, transfection of miR-9 promoted both astroglial and neuronal differentiation, leading to an increase in the percentage of GFAP-positive astrocytes and Tuj1-positive neurons at day 3 of differentiation (Fig. 2d, e), mimicking day 5 of differentiation in control cells (data not shown). These results indicate that miR-9 accelerates differentiation of neural stem cells that have been primed for differentiation.
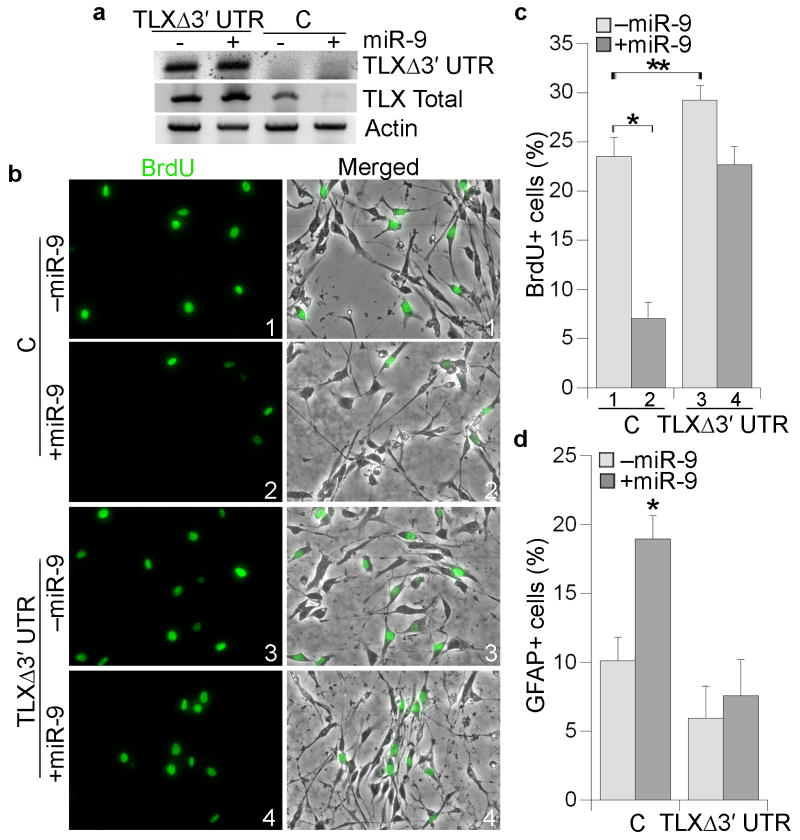
To determine whether the effect of miR-9 transfection on neural stem cell proliferation and differentiation is mediated through TLX, neural stem cells were stably transduced with a TLX-expressing vector (TLXΔ3′ UTR), which lacks the TLX 3′ UTR. Transfection of miR-9 had no effect on the expression of TLXΔ3′ UTR, although miR-9 downregulated endogenous TLX expression levels (Fig. 3a). Expression of TLXΔ3′ UTR led to 1.24-fold increase in neural stem cell proliferation. Co-transfection of TLXΔ3′ UTR and miR-9 reversed the proliferative deficiency induced by miR-9 substantially (Fig. 3b, c). Furthermore, while transfection of miR-9 increased astroglial differentiation in control neural stem cells, no appreciable increase in astrocyte differentiation was detected in TLXΔ3′ UTR-transduced cells upon miR-9 treatment (Fig. 3d). These results strongly suggest that miR-9 regulates neural stem cell proliferation and differentiation, at least in part, by inhibiting TLX expression through its 3′ UTR.
Fig. 3. TLXΔ3′ UTR rescues miR-9-induced neural stem cell proliferation deficiency.
a. RT-PCR analysis of TLXΔ3′ UTR and total TLX expression in control neural stem cells (C) and TLXΔ3′ UTR-expressing cells treated with control RNA (- miR-9) or miR-9 (+ miR-9). Actin was included as a loading control. b. Control (C) or TLXΔ3′ UTR-expressing cells were transfected with control RNA (-miR-9) or miR-9 followed by BrdU labeling (green). Merged panels show BrdU staining along with phase contrast images. c. Quantification of BrdU-positive (BrdU+) cells in control (C) and TLXΔ3′ UTR-expressing cells treated with control RNA (-miR-9) or miR-9. Error bars are s.d. of the mean. * p=0.003 by Student's t-test. ** p=0.04 by Student's t-test. d. Quantitation of GFAP-positive (GFAP+) cells in control (C) and TLXΔ3′ UTR-expressing cells treated with control RNA (-miR-9) or miR-9. Error bars are s.d. of the mean. * p=0.02 by Student's t-test. GFP siRNA was included as the control RNA.
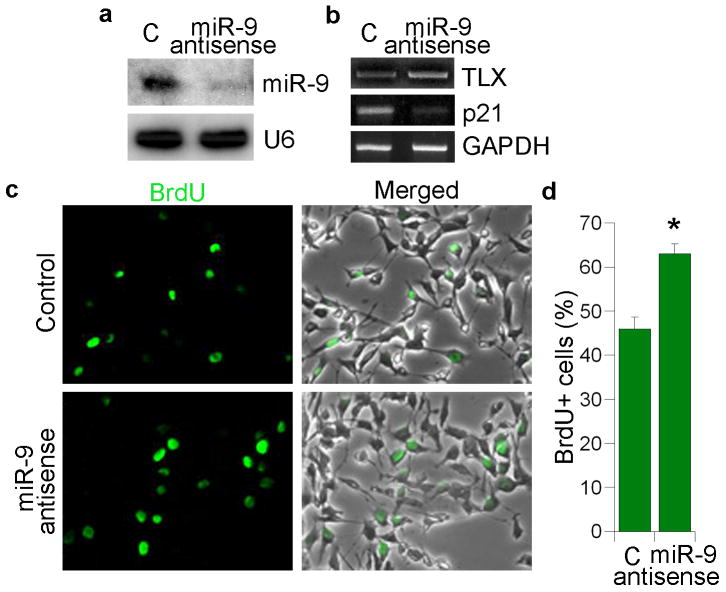
Using 2′-O-methyl antisense RNA oligonucleotides as small RNA inhibitors28,29, the role of miR-9 in neural stem cell proliferation was further investigated. 2′-O-methyl antisense oligonucleotide against miR-9 was synthesized and transfected into neural stem cells with 2′-O-methyl antisense oligonucleotide against green fluorescent protein (GFP) included as a negative control. Treatment of antisense oligonucleotides against miR-9 led to substantial knockdown of miR-9 mature forms (Fig. 4a). The expression of TLX was upregulated in miR-9 antisense RNA-treated neural stem cells, along with decreased expression of p21 (Fig. 4b). BrdU labeling analysis revealed that knockdown of miR-9 led to an increase in cell proliferation (1.37-fold, Fig. 4c, d), consistent with enhanced cell proliferation in TLXΔ3′ UTR-transduced neural stem cells (Fig. 3b, c).
Fig. 4. miR-9 antisense RNA promotes neural stem cell proliferation.
a. 2′-O-methyl miR-9 antisense RNA knocks down miR-9 mature form analyzed by Northern blot analysis. 2′-O-methyl antisense GFP RNA was included as a negative control (C) in all sections. U6 was included as a loading control. b. Expression of TLX and p21 in 2′-O-methyl miR-9 antisense RNA-treated neural stem cells analyzed by RT-PCR. GAPDH was included as a loading control. c. Neural stem cells were transfected with control RNA and 2′-O-methyl miR-9 antisense RNA. The transfected cells were labeled by BrdU staining (green). Merged panels show BrdU staining along with phase contrast images. d. Quantification of BrdU+ cells in control (C) and 2′-O-methyl miR-9 antisense RNA-treated neural stem cells. s.d. is represented by error bars. * p=0.03 by Student's t-test.
MiR-9 stimulates neural differentiation in the brain
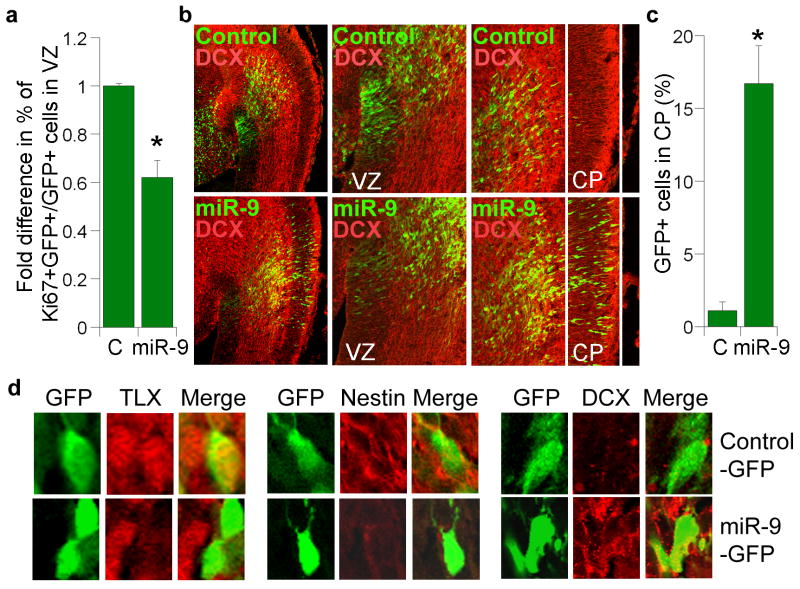
During development, neural stem cells reside in the ventricular zone and migrate out into cortical plate upon differentiation. To determine whether miR-9 influences neural stem cell proliferation and differentiation in vivo, miR-9 RNA duplexes were introduced into neural stem cells in the ventricular zone of E13.5 mouse brains by in utero electroporation. Electroporated brains were analyzed at E15.5. Cells that had taken up miR-9 were labeled green by co-expression of GFP. Transfection of miR-9 led to a marked decrease of cells that were positively labeled for Ki67, a proliferative marker (Fig. 5a), and a substantial increase in the number of cells that migrated from the ventricular zone to the cortical plate (Fig. 5b, c), suggesting that miR-9 negatively regulates neural stem cell proliferation and accelerates neural differentiation.
Fig. 5. In utero electroporation of miR-9 in embryonic neural stem cells.
a. In utero electroporation of miR-9 decreased cell proliferation in the ventricular zone (VZ) of embryonic brains. Proliferating cells were labeled by Ki67. Percent of Ki67-positive cells out of GFP-positive cells (Ki67+GFP+/GFP+) in miR-9-electroporated brains was calculated and normalized with the percent of Ki67-positive cells in control RNA (C)-electroporated brains. s.d. is indicated by error bars. * p=0.002 by Student's t-test. b. Electroporation of miR-9 led to precocious outward cell migration. The transfected cells were shown green due to the expression of GFP marker. Control: control RNA; DCX: double cortin; VZ: ventricular zone; CP: cortical plate. The left panels are 10× images, the middle and right panels are 20× images. c. Quantification of control RNA (C) and miR-9-electroporated cells (GFP+ cells) that migrated to the cortical plate (CP). Error bars are s.d. of the mean. * p=0.02 by Student's t-test. d. Immunostaining of cells from control-GFP or miR-9-GFP-electroporated brains.
Immunostaining revealed that transfection of miR-9 led to decreased TLX expression (Fig. 5d, Supplementary Fig. 2). The miR-9-transfected cells that migrated to the cortical plate lost the neural progenitor marker nestin (Fig. 5d). Instead, these cells expressed the neuronal marker, double cortin (DCX, Fig. 5d), indicating neuronal differentiation. In contrast, the control small RNA-transfected cells that remained in the ventricular zone were nestin-positive and lacked DCX expression (Fig. 5d). These results are similar to that obtained from in utero electroporation of a TLX siRNA21. Furthermore, in utero electroporation of TLXΔ3′ UTR along with miR-9 rescued the precocious migration induced by miR-9 transfection (Supplementary Fig. 3). These results strongly suggest that miR-9 regulates neural stem cell differentiation through targeting TLX expression in vivo.
In addition to neuronal differentiation, we also examined whether miR-9 plays a role in glial differentiation in vivo. Gliogenesis occurs from late embryonic stage through to the early postnatal stage30. MiR-9 RNA duplexes were electroporated into ventricular zone of E14.5 brains. Glial differentiation was analyzed at E17.5 by GFAP staining. Increased GFAP-positive cells were detected in miR-9-transfected cells, compared to that in miR-9 mutant-transfected cells (Supplementary Fig. 4), suggesting that miR-9 overexpression also promotes astroglial differentiation in vivo.
Regulation of miR-9 gene expression by TLX
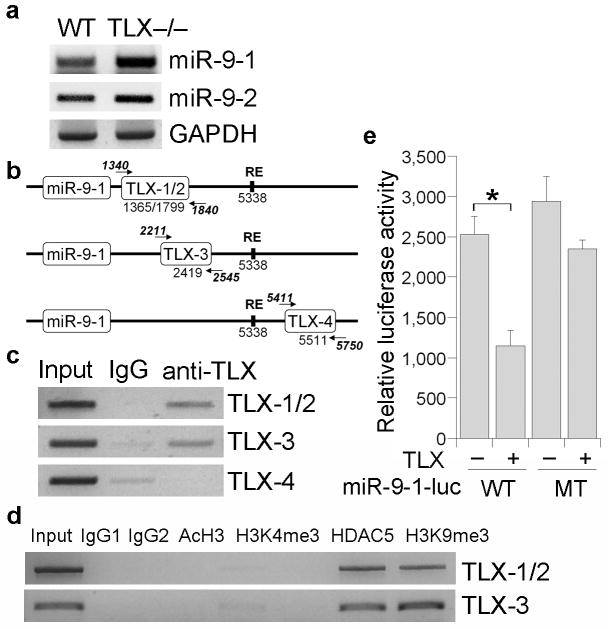
Three genes, miR-9-1, miR-9-2 and miR-9-3, encode miR-9 in the mouse genome. Both miR-9-1 and miR-9-2 have been shown to be expressed in mammalian brains14, whereas miR-9-3 has not been detected in vertebrate brains31. To determine whether miR-9 gene expression is affected by TLX expression, RT-PCR analysis was performed to assess the expression levels of miR-9-1 and miR-9-2 pri-miRNAs in brains of wild type and TLX-null mice. Interestingly, the expression of both miR-9-1 and miR-9-2 pri-miRNAs was upregulated in brains of TLX null mice (Fig. 6a), suggesting that expression of miR-9 precursors can be repressed by TLX.
Fig. 6. Regulation of miR-9 pri-miRNA expression by TLX.
a. RT-PCR analysis of miR-9 pri-miRNAs, miR-9-1 and miR-9-2, in wild type and TLX-null brains. GAPDH was included as a loading control. b. Schematics of miR-9-1 gene with consensus TLX binding sites (TLX-1/2, 3, 4) and REST responsive element (RE). The numbers are relative to miR-9 hairpin precursor ending site. The locations of the PCR primers for ChIP assays are indicated by arrows. c. ChIP assays to show binding of TLX to the consensus TLX binding sites downstream of miR-9-1 gene. d. ChIP assays to show binding of HDAC5 to TLX binding sites TLX-1/2 and TLX-3. The association of these sites with H3K9me3, but not with AcH3 and H3K4me3, was shown in the same assays. e. TLX represses miR-9-1 reporter activity. Luciferase activity of wild type (WT) or mutant (MT) miR-9-1 reporter gene was measured in the absence (-) or presence (+) of transfected TLX expression vector in mouse neural stem cells. Error bars are s.d. of the mean. * p=0.009 by Student's t-test.
Sequence analysis revealed several consensus TLX binding sites in the flanking regions of miR-9-1 (Fig. 6b) and miR-9-2 genes (data not shown). To further explore whether TLX regulates miR-9 expression by direct binding to miR-9 genomic sequences, chromatin immunoprecipitation (ChIP) analysis was performed. MiR-9-1 was chosen for this analysis due to its more substantial induction in TLX-null brains (Fig. 6a). Three pairs of primers were designed for ChIP analysis, with one pair of primers covering both TLX binding sites 1 and 2 (TLX-1/2). The other two pairs of primers were designed for TLX binding site 3 (TLX-3) and site 4 (TLX-4) individually. ChIP assays revealed that TLX binds to the consensus TLX binding sites, TLX-1/2 and TLX-3, that are downstream of miR-9-1 genes and upstream of a previously characterized REST binding site32,33, whereas no binding was detected on TLX-4 that is downstream of the REST binding site (Fig. 6c).
Consistent with TLX binding to miR-9-1 genomic sequences, the TLX-interacting transcriptional corepressor HDAC5 was also detected on the TLX binding sites, TLX-1/2 and TLX-3, in the miR-9-1 genomic locus (Fig. 6d). Furthermore, these sites are associated with the repressive chromatin marker, trimethylated histone H3 lysine 9 (H3K9me3), but are not associated with active chromatin markers, acetylated histone H3 (AcH3) and trimethylated histone H3 lysine 4 (H3K4me3)34,35 (Fig. 6d).
To validate the regulation of miR-9-1 gene expression by TLX, we cloned the 1.2 kb miR-9-1 downstream genomic sequence that contains the consensus TLX binding sites, TLX-1/2 and TLX-3, and inserted it downstream of a Renilla luciferase reporter gene in the siCHECK vector. Co-transfection of TLX with the reporter gene in neural stem cells led to 2.2-fold repression of the reporter activity (Fig. 6e). Mutation of the TLX binding sites relieved the repression mediated by TLX substantially (Fig. 6e). These results together suggest that TLX represses miR-9 expression by binding to the consensus TLX binding sites in the 3′ genomic sequence of miR-9-1.
Discussion
We show here that microRNA miR-9 and nuclear receptor TLX forms a feedback regulatory loop to regulate neural stem cell proliferation and differentiation. TLX is highly expressed in neural stem cells but is repressed upon differentiation16, in contrast, the level of the miR-9 mature form is increased upon differentiation13,24,36. The temporal relationship between miR-9 and TLX expression resembles that between miR-124 and its target genes lamc1, itgb1, and REST32,37,38. In both instances, miRNA targets are preferentially expressed at high levels when the targeting miRNA expression is low. Conversely, the expression of these targets is down-regulated as their targeting miRNAs accumulate. These data support the hypothesis that miRNAs induced during differentiation function to ensure proper cell fate transitions by suppressing leftover stem cell maintenance transcripts in stem cells39.
This study demonstrates that miR-9 plays an important role in neural stem cell proliferation and differentiation and that TLX is a key target of miR-9 in neural stem cells. Every miRNA could have multiple target genes23,26, and indeed, several target genes have been predicted for miR-9, including transcription factors REST, FoxG1 and senseless32,33,40,41. One of the questions addressed here is whether the cell proliferation and differentiation effect mediated by miR-9 in neural stem cells is directly related to repression of TLX expression. Transfection of miR-9 into neural stem cells that are stably transduced with a TLX-expressing vector lacking the miR-9 target site showed that such ectopically expressed TLX rescued the proliferative deficiency induced by overexpression of miR-9, and compromised miR-9-induced precocious differentiation. The result of this rescue experiment suggests that miR-9 regulates neural stem cell proliferation and differentiation through repression of TLX expression. While TLX is an important target gene of miR-9, additional targets may also play a role in miR-9 function in neural stem cells.
Recent evidence suggests that miRNAs often act as fine-tuning devices rather than as primary gene regulators42. Consistent with this concept, we failed to induce neural differentiation by overexpressing miR-9 alone in cultured neural stem cells. Instead, an accelerated differentiation program was detected when differentiation of neural stem cells was induced in culture or in E13.5 brains, where active endogenous neurogenesis occurs. Furthermore, it has been suggested that inhibiting a miRNA may not generate a strong or even detectable phenotype, as expression of its target genes are already repressed at the transcriptional level, whereas overexpressing a miRNA in cells where its target genes are highly expressed, may render the action of the miRNA more detectable37. In accordance with this theory, we failed to detect a change in cell differentiation in miR-9 antisense RNA-treated neural stem cells (data not shown). However, we were able to detect a precocious differentiation program upon miR-9 overexpression in neural stem cells that were primed for differentiation.
In addition to being a direct target of miR-9, TLX also transcriptionally inhibits miR-9 genes, suggesting a negative feedback loop between TLX and miR-9 for a rapid transition from neural stem cells to differentiated cells. In neural stem cells, TLX is expressed at relatively high levels. During differentiation, as TLX levels decrease, miR-9 expression accumulates. MiR-9, in turn, post-transcriptionally suppresses TLX expression to further promote neural differentiation. This regulatory loop may represent a key mechanism to sense the intricate balance between cell proliferation and differentiation and to confer cell fate determination in a timely manner. Overall, our study suggests that the brain-specific miRNA miR-9 plays a key role in vertebrate brain development. MiR-9 provides a novel strategy to control neural stem cell fate determination by forming a feedback regulatory loop with TLX.
Methods
Neural stem cell culture and differentiation
We isolated mouse neural stem cells from adult mouse forebrains using percoll gradient as described16 and cultured in DMEM F12 medium with 1 mM L-glutamine, N2 supplement (Gibco-BRL), EGF (20 ng ml− 1), FGF2 (20 ng ml− 1), and heparin (50 ng ml− 1) for proliferation. For differentiation, we exposed neural stem cells to DMEM F12 media with N2 supplement, 5 μM forskolin and 0.5% (v/v) FBS or 1 μM retinoic acid and 0.5% (v/v) FBS.
miR-9 expression vector and reporter construct
We amplified miR-9-1 gene by PCR from genomic locus of mouse miR-9-1, which contains the 89 nt hairpin sequences and 200 nt of genomic sequences flanking each side of it. We inserted the 489 nt DNA fragment into a miRNA expression vector, MDH1-PGK-GFP2, to generate miR-9 expression construct. For reporter construct, we subcloned DNA fragments encoding mouse TLX 3′ UTR (1813-3232bp) into psiCHECK 2 (Promega) to make the TLX3′ UTR-siCHECK construct. We created mutant miR-9 target site by site-directed mutagenesis in TLX 3′ UTR-siCHECK vector. We mutated the wild type miR-9 binding site AACCAAAG into TTGGTTTC. To make miR-9-1 reporter construct, we inserted mouse miR-9-1 downstream genomic sequence (1340 bp to 2546 bp downstream of miR-9 hairpin structure), which contains the consensus TLX binding sites, TLX-1/2 and TLX-3, downstream of a Renilla luciferase reporter gene in psiCHECK2 vector. The miR-9-1 mutant reporter construct has the consensus TLX binding site AAGTCA mutated to AGATCA at TLX-1/2 and TLX-3 sites by sequential site-directed mutagenesis.
BrdU labeling and immunostaining analysis
We seeded mouse neural stem cells at 1 × 105 cells per ml in 4-well chamber slides. We added BrdU to cells 48 hrs after seeding and pulsed for 1 to 2 hrs. The BrdU-treated cells were fixed and acid-treated, followed by immunostaining analysis with BrdU-specific antibody16. We performed immunostaining using antibodies for BrdU (Accurate; diluted 1:1,000), Ki67 (Thermo Scientific, 1:400), nestin (Pharmingen; 1:1000), Tuj1 (Covance, 1:6,000), DCX (Santa Cruz, 1:300), and GFAP (Advance Immuno; 1:500).
Transfection, Western blot, and reporter assay
We transfected plasmid DNA or DNA/RNA mix using Transfectin (Bio-Rad). We transfected RNA duplexes using SilentFect™ (Bio-Rad). For 50 nM, 100 nM or 200 nM final concentration of miR-9, 0.5 μl, 1 μl or 2 μl of 50 μM RNA duplexes and 1 μl SilentFect™ were mixed in 50 μl media, incubated at room temperature for 20 min, and added dropwise to cells in a 24-well plate with 450 μl medium to a total volume of 500 μl. The transfected cells were harvested 48 hrs after transfection and subjected to subsequent analyses. The wild type miR-9 RNA duplex sense sequence is UCU UUG GUU AUC UAG CUG UAU GA. The mutant miR-9 RNA duplex sense sequence is UGA AAC CAA AUC UAG CUG UAU GA. Western blot of TLX and GAPDH was performed using rabbit anti-TLX antibody (1:1,000) and rabbit anti-GAPDH antibody (Santa Cruz, 1:1,000). We measured Renilla luciferase activity 48 hrs after transfection, normalized it with firefly luciferase or β-galactosidase internal control and expressed it as relative luciferase activity.
miRNA Northern blotting
We extracted total RNA from tissues or cultured cells by Trizol. We separated 8 μg of RNA on a 10% polyacrylamide gel containing 8M urea and transferred the RNA to Nylon membranes electrophorectically. Membranes were crosslinked by UV irradiation and hybridized overnight with 32P-labeled oligonucleotide probes. We quantified miRNA signals using Phosphor Imager (Molecular Dynamics). DNA probes for Northern blotting include miR-9-antisense probe (as): CAT ACA GCT AGA TAA CCA AAG A and U6-as: TAT GGA ACG CTT CTC GAA TT.
RT-PCR analysis
We prepared cDNA from total RNA using Omniscript® Reverse Transcription kit (Qiagen) for RT-PCR analyses. Primers for RT-PCR include TLX forward primer: GTC TTT ACA AGA TCA GCT GAT G, reverse primer: ATG TCA CTG GAT TTG TAC ATA TC; GFAP forward primer: GCT ACA TCG AGA AGG TCC GC, reverse primer: GTC TCT TGC ATG TTA CTG GTG; Tuj-1 forward primer: CTG GAG CGC ATC AGC GTA TAC, reverse primer: ATC TGC TGC GTG AGC TCA GG; p21 forward primer: ATG TCC AAT CCT GGT GAT GTC CG, reverse primer: TCA GGG TTT TCT CTT GCA GAA GA; GAPDH forward primer: CAT CAC CAT CTT CCA GGA GC, reverse primer: GCT GTA GCC GTA TTC ATT GTC; actin forward primer: ACC TGG CCG TCA GGC AGC TC, reverse primer: CCG AGC GTG GCT ACA GCT TC.
In utero electroporation
We performed all animal experiments in accordance with City of Hope and NIH guidelines and regulations. We carried out in utero electroporation as described21. We injected 37.5 pmol of miR-9 or control RNA duplex into the lateral ventricles of embryos along with 0.625 μg of pActin-EGFP reporter plasmid using electroporator CUY-21 (Protech International). The electroporated mice were allowed to survive for 2 days. Brains of embryos were collected and analyzed as described 21.
Chromatin immunoprecipitation (ChIP) assays
We performed ChIP assays using EZ-ChIP kit (Upstate) with precleaned chromatin from 2×106 mouse neural stem cells and 5 μg antibody for each ChIP assay. Antibodies used include antibodies for TLX, tri-methyl Histone H3 lysine 4 (H3K4me3, Cell Signaling Technology), acetylated histone H3 (AcH3, Cell Signaling Technology), histone deacetylase 5 (HDAC5, Santa Cruz technology), and tri-methyl Histone H3 lysine 9 (H3K9me3, Abcam). Primers for ChIP assays include miR-9-1 TLX-1/2 forward primer: GGT AGG GGT GGT GGG GAT GAA, reverse primer: TCT AGG ATG CCC AAG AAC TTG CT; miR-9-1 TLX-3 forward primer: GCT GGG ACA CTG GGG ATG CTA GA, reverse primer: AGG AGA GAT CCA TGG AGA TAT C; miR-9-1 TLX-4 forward primer: TCC AGG CAG ACA TCC TGC ACT AC, reverse primer: CCT GGT TCT TAG GGA TAC TTC AC.
Supplementary Material
Acknowledgments
We thank Drs. John Rossi and John Zaia for their critical comments on the manuscript, Dr. Chang-Zheng Chen (Stanford University), Drs Harvey F. Lodish and David P. Bartel (Massachusetts Institute of Technology) for providing the micro RNA expression vector, MDH1-PGK-GFP, and Dr. Qiang Lu (City of Hope) for providing the pEF-pUb-EGFP plasmid. This work was supported by NIH NINDS R01 NS059546 (to Y.S.).
Footnotes
Author Contributions: Y.S., C.Z, and G.S. designed the project; C.Z., G.S., and S.L. performed the experiments; Y.S., and C.Z. analyzed the data and wrote the manuscript.
References
- 1.McKay R. Stem cells in the central nervous system. Science. 1997;276:66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez-Buylla A, Temple S. Stem cells in the developing and adult nervous system. J Neurobiol. 1998;36:105–10. [PubMed] [Google Scholar]
- 3.Gage FH, Kempermann G, Palmer TD, Peterson DA, Ray J. Multipotent progenitor cells in the adult dentate gyrus. J Neurobiol. 1998;36:249–66. doi: 10.1002/(sici)1097-4695(199808)36:2<249::aid-neu11>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 4.Weiss S, van der Kooy D. CNS stem cells: where's the biology (a.k.a. beef)? J Neurobiol. 1998;36:307–14. doi: 10.1002/(sici)1097-4695(199808)36:2<307::aid-neu14>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 8.Suh MR, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 10.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–8. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 11.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–42. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 13.Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–81. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235:2538–48. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- 15.Kapsimali M, et al. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y, et al. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 17.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci U S A. 2007;104:15282–7. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang CL, Zou Y, He W, Gage FH, Evans RM. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004–7. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- 19.Roy K, et al. The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci. 2004;24:8333–45. doi: 10.1523/JNEUROSCI.1148-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenman JM, Wang B, Campbell K. Tlx controls proliferation and patterning of lateral telencephalic progenitor domains. J Neurosci. 2003;23:10568–76. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, et al. Nuclear Receptor TLX Regulates Cell Cycle Progression in Neural Stem Cells of the Developing Brain. Mol Endocrinol. 2008;22:56–64. doi: 10.1210/me.2007-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 24.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 26.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 27.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. Rna. 2004;10:544–50. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian X, et al. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 35.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–64. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–6. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–9. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 40.Bredenkamp N, Seoighe C, Illing N. Comparative evolutionary analysis of the FoxG1 transcription factor from diverse vertebrates identifies conserved recognition sites for microRNA regulation. Dev Genes Evol. 2007;217:227–33. doi: 10.1007/s00427-006-0128-x. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–4. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.