Abstract
Objective
Sequence variation in gene promoters is often associated with disease risk. In this study, we tested the hypothesis that common promoter variation in the APOH gene (encoding for β2-glycoprotein I) is associated with systemic lupus erythematosus (SLE) risk and SLE-related clinical phenotypes in a Caucasian cohort.
Methods
We used a case-control design and genotyped 345 SLE women and 454 healthy control women for 8 APOH promoter single nucleotide polymorphisms (SNPs) (−1284C>G, −1219G>A, −1190G>C, −759 A>G, − 700C>A, −643T>C, −38G>A, and −32C>A). Association analyses were performed on single SNPs and haplotypes. Haplotype analyses were performed using EH (Estimate Haplotype-frequencies) and Haploview programs. In vitro reporter gene assay was performed in COS-1 cells. Electrophoretic mobility shift assay (EMSA) was performed using HepG2 nuclear cells.
Results
Overall haplotype distribution of the APOH promoter SNPs was significantly different between cases and controls (P = 0.009). The −643C allele was found to be protective against carotid plaque formation (adjusted OR = 0.37, P = 0.013) among SLE patients. The −643C allele was associated with a ~ 2-fold decrease in promoter activity as compared to wild-type −643T allele (mean ± standard deviation: 3.94 ± 0.05 vs. 6.99 ± 0.68, P = 0.016). EMSA showed that the −643T>C SNP harbors a binding site for a nuclear factor. The −1219G>A SNP showed a significant association with the risk of lupus nephritis (age-adjusted OR = 0.36, P = 0.016).
Conclusion
Our data indicate that APOH promoter variants may be involved in the etiology of SLE, especially the risk for autoimmune-mediated cardiovascular disease.
Keywords: APOH, β2-glycoprotein I, promoter, SLE, lupus, polymorphism
INTRODUCTION
Systemic lupus erythematosus (SLE) is a prototypic autoimmune inflammatory disease that can affect virtually any organ system. It primarily affects premenopausal women (female:male ratio = 9:1) and is characterized by a plethora of clinical manifestations, including glomerulonephritis, arthritis, pleuritis, pericarditis and vasculitis 1. Nephritis is a predictor of poor prognosis and a major cause of death in patients with active SLE 2. The serological hallmark of SLE is the production of autoantibodies directed against components of the cellular nucleus. Antiphospholipid antibodies (APA) represent a group of heterogeneous antibodies [anticardiolipin antibodies (aCL), lupus anticoagulant (LAC), and anti-β2-glycoprotein I (anti-β2GPI)] which are detected in a variety of conditions, including SLE. APA are found in ~ 1–10% of a general Caucasian US population versus (vs.) 30–70% in patients with SLE and antiphospholipid syndrome (APS) 3–5. APS manifests clinically with arterial and venous thrombosis, recurrent pregnancy loss, and thrombocytopenia 6. Premature coronary heart disease is a major cause of morbidity and mortality in SLE patients; more pronounced in younger women (35–44 years) for whom the estimated risk is more than 50 fold 7. Genetic factors play an important role in SLE susceptibility, as reflected by heritability of up to 66% 8 and a 20–80 times greater risk for family members 9.
Human apolipoprotein H (APOH), also known as β2-glycoprotein I (β2GPI), is a 50-kDa single chain plasma glycoprotein (in this paper, we will use APOH to refer to the gene as used in human genome databases and β2GPI to refer to the protein as commonly used in the rheumatology literature). APOH spans 18 kilobases (kb) on chromosome 17q23–24 and is comprised of 8 exons that encode for a protein of 345 amino acid (aa) residues including 19 aa in the signal peptide 10–12. Complete DNA sequence analysis of APOH 13 has identified ~ 150 single nucleotide polymorphisms (SNPs), including four in the coding region (Ser88Asn, Val247Leu, Cys306Gly, and Trp316Ser). Significant structural homology of β2GPI across several mammalian species indicates evolutionary conserved regions in the protein that may be critical for its function 14, 15.
Several functions have been implicated for β2GPI, including a role in lipid metabolism 16 and blood coagulation 17, binding with oxidized low-density lipoproteins (oxLDL) to form oxLDL/β2GPI complexes 18 and being a cofactor necessary for the binding of certain APA to anionic phospholipids in patients with autoimmune disorders 19–21. Previously, two coding variants (Trp316Ser and Val247Leu) of APOH have shown association with the occurrence of APA in different studies 4, 22–24. The presence of autoantibodies to phospholipid-free β2GPI has also been reported in patients with primary APS, indicating that there may be two epitopes on β2GPI targeted by autoantibodies 25–27.
The association of genetic variation in the coding region of APOH with SLE risk has been examined by us 23; however, a comprehensive analysis of APOH promoter SNPs has not been reported yet. Since variation in promoter region could affect gene expression and disease risk, the objective of this study was to examine the association of APOH promoter SNPs with SLE risk and/or SLE-related clinical phenotypes in Caucasian SLE cases and controls. For this purpose, all eight APOH promoter SNPs previously reported to be present in Caucasians (−1284C>G, −1219G>A, −1190G>C, −759 A>G, − 700C>A, −643T>C, −38G>A, and −32C>A) 13 were examined.
PATIENTS AND METHODS
Subjects
Peripheral blood samples were obtained from 345 Caucasian women with SLE and 454 Caucasian healthy control women. SLE subjects were derived from the Pittsburgh Lupus Registry and were 18 years of age or older (mean age ± standard deviation (SD) = 43.45 ± 11.40), and met the 1982 28 and the 1997 revised 29 American College of Rheumatology (ACR) classification criteria for definite or probable SLE. The spectrum of ACR criteria was as follows in our Caucasian patient population: skin (malar rash and discoid rash) 55%, photosensitivity 59%, oral ulcers 54 %, arthritis 91%, serositis 45%, renal involvement 30%, neurologic involvement 9%, hematologic involvement 52%, immunologic involvement 72%, antinuclear antibody 98%. Blood samples from Caucasian control subjects (mean age ± SD = 45.28 ± 13.14) with no history of SLE were obtained from Central Blood Bank of Pittsburgh. A second cohort of Caucasian female SLE patients (n = 109) and controls (n = 81) from the Chicago SOLVABLE (Study of Lupus Vascular and Bone Longterm Endpoints) study was used to replicate the most significant SLE disease association and the most significant carotid plaque association (cases only) detected in the Pittsburgh cohort. Detailed description of the Chicago study population is reported elsewhere 30. This study was approved by the University of Pittsburgh Institutional Review Board and all participants provided written informed consent.
Diagnosis criteria for lupus nephritis among SLE patients was based on the presence of either 1) renal biopsy showing lupus nephritis, 2) at least two readings of proteinuria > 0.5 gm/24 hours or 3 + protein by dipstick, or 3) red blood cell casts. Two main clinical parameters that were used to define subclinical cardiovascular disease (CVD) in a subgroup of SLE patients (n = 245) were the measurements of carotid intima-media thickness (IMT) and carotid plaque index. B-mode carotid ultrasound was performed using a Toshiba SSA-270 A scanner (Tustin, CA) equipped with a 5-mHz linear array imaging probe as described previously 31. Sonographers scanned the right and left common carotid artery, carotid bulb, and the first 1.5 cm of the internal and external carotid arteries. Plaque was defined as a distinct area protruding into the vessel lumen that was at least 50% thicker than the surrounding areas. The average carotid IMT was measured across 1-cm segments of both the right and left sides of the near and far walls of the distal common carotid artery and the far wall of the carotid bulb and internal carotid artery. Values from each location were then averaged to produce an overall measure of carotid IMT 31.
Measurement of Antiphospholipid antibodies (APA)
Serum samples were screened in duplicate for the presence of aCL (IgG aCL positivity defined as >15 IgG phospholipid units, IgM aCL positivity defined as >10 IgM phospholipid units; kits obtained from Incstar, Stillwater, MN), LAC (by partial thromboplastin time or Russell’s viper venom time with mix), and anti-β2GPI (Quantalite β2GPI screen, INOVA Diagnostics, San Diego, CA). Additional information regarding the methods that were used to measure these three APA is provided elsewhere 14, 23.
Genotyping
QIAamp kit (Qiagen, Chatsworth, CA) was used to isolate genomic DNA from buffy coat. The eight APOH promoter SNPs numbered in relation to the translation start site (−1284C>G, −1219G>A, −1190G>C, −759 A>G, −700C>A, −643T>C, −38G>A, and −32C>A) were amplified as three separate PCR fragments. Two of the amplicons, each containing three APOH promoter SNPs, were genotyped by Pyrosequencing using the universal biotinylated primer tag (5' – Biotin – GCTGCTCCGGTTCATAGATT - 3'). The PCR primers used to amplify these two fragments were: Fragment I (−1284C>G, −1219G>A, −1190G>C): Forward (F) = 5' TCTCCCTGACAGATGGAGATT 3' and Reverse (R) = 5' CACACCTGAAGCCTTTCC 3', Fragment II (−759 A>G, −700C>A, −643T>C): F = 5' CGAACCC TCTCAAGCA ACA 3' and R = 5' TTGCAAGCTCCTATAGCTCCA 3'. Fragment III contained two promoter SNPs (−38G>A, and −32C>A) and the primers used for PCR were F = 5'/BioGTGGGTCTCAGAGTT CCATTCAA 3' and R = 5'CCTGACATATACGAAGGGGTTGGA 3'.
Transient DNA transfection and Dual-luciferase assay
The wild-type APOH plasmid construct containing the ~1.4 kb 5' region of the putative APOH promoter was cloned into the luciferase reporter vector (pGL3-Basic). This wild-type construct contained the major alleles at all of the APOH promoter SNPs (wild-type haplotype). Site-directed mutagenesis of this wild-type clone was performed using Stratagene's QuikChange® Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, U.S.A.) as per the manufacturer's protocol. We prepared APOH promoter-pGL3-Basic luciferase constructs bearing minor allele only at SNP −643T>C (H3 haplotype) and minor alleles at SNPs −643T>C, −1190G>C and −759A>G (H7 haplotype). For the −643 mutation, the forward and reverse mutagenic primers, 5' GACAGATCCAAGACATACTAAGAATGGATGAGGAGG 3' and 5' CCTCCTCATCCATTCTTAGTATGTCTTGGATCTGTC 3', were designed to incorporate the desired base pair change.The −1190G>C and −759A>G mutations were introduced using the forward and reverse mutagenic primers, 5' GAGCTTGCTATAGCAAGGGAGGCAGC 3', 5' CTGCCTCCC TTGCTATA GCAAGCTC 3', 5' CAGCACTGGCCCATTGTCTTATCCTACTCAAG 3', and 5' CTTGAGTAGGATAAGACAATGGGCCAGTGCTG 3', respectively. Transfection of the COS-1 cells was carried out with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. The wild-type and mutant H3 and H7 haplotype pGL3-basic firefly luciferase reporter vectors (Promega, Madison, WI) harboring the ~1.5 kb region of the putative APOH promoter were used to transiently co-transfect COS-1 cells along with the Renilla luciferase control vector (pRL-TK) (Promega). Transfected cells were lysed with the Dual-luciferase lysis buffer 48 hours after the transfection, and luciferase light outputs were measured through the TD-20/20 Luminometer (Turner Design, Sunnyvale, CA). In each well, the ratio of firefly luciferase to Renilla luciferase activity was calculated after normalization against the transfection reference vector (promoter-less pGL3-Basic) to yield data reflecting fold-activity increase over baseline levels. Each experiment was performed in triplicate (intra-experiment variation) and was repeated at least three times (inter-experiment variation).
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed for the −643T>C site. Two double-stranded 30-mer oligonucleotides corresponding to the wild-type (5′-GACAGATCCAAGACATATTAAGAATGG-3′ and 3′-CCATTCTTAATATGTCTTGGATCTGTC-5′) and mutant allele (5′-GACAGATCCAAGACATACTAAGAATGG-3′ and 3′-CCATTCTTAGTATGTCTTGGATCTGTC-5′) were prepared as described elsewhere32. The wild-type T oligonucleotide was 5'-end-labeled with α-32P ATP and purified by means of the QIAquick Purification kit (Qiagen). Equally concentrated, nonradioactive competitor DNA was added in 1x, 5x, 20x, 50x, 100x, 200x, and 400x excess volumes of the labeled probe. The mixture of unlabeled and labeled oligos were incubated with 2 µL (5µg) of human HepG2 cell nuclear extracts for 20 minutes at room temperature in 5x gel shift binding buffer (1 mmol/L MgCl2, 0.5 mmol/L EDTA, 0.5 mmol/L dithiothreitol, 50 mmol/L NaCl, 10 mmol/L Tris-HCl [pH = 7.5], 20% glycerol). The DNA-protein complexes were then separated on 5% nondenaturing polyacrylamide gel at 120 volts for 2 hours, the gel was dried and autoradiographed overnight.
Statistical Analysis
Allele frequencies were determined by direct allele counting. Concordance of the genotype distribution to Hardy-Weinberg equilibrium was tested using χ2 goodness-of-fit test for each polymorphism. Linkage disequilibrium (LD) pattern was determined using the Haploview version 3.32 (http://www.broad.mit.edu/mpg/haploview/). To control for multiple testing concerns, we performed two types of “gate-keeper” analyses to determine whether there was evidence that any of these SNPs influenced risk of SLE: (1) multiple regression incorporating all SNPs and (2) haplotype analyses. The multiple regression analysis was performed under additive model for genotypic effects, that is, major allele homozygote = 0, heterozygote = 1, minor allele homozygote = 2. Subsequent to observing associations with SLE risk, we performed additional analyses to determine if any of the promoter SNPs also influenced specific clinical manifestations of SLE to gain insights into possible mechanisms of action. Again, to control for multiple testing concerns, we initially assessed possible associations of all SNPs with clinical variables assessed in patients only (the cases who were negative for these variables were treated as controls) using a multiple regression analysis. In these analyses, the genotypic effects were modeled as dominant (i.e., major homozygote vs. heterozygote + minor homozygote) because of the small sample size. Only for those SNPs that showed significant association in multiple regression analysis, we performed follow-up single-site analysis of the genotype (using Fisher’s exact test or analysis of deviance when adjusting for covariates) and allele frequencies (standard Z-test of 2 binomial proportions). Carotid plaque was categorized as plaque positive (degree of plaque equal to or greater than 1) and plaque negative (degree of plaque equal to zero). The carotid IMT data was transformed to reduce the effects of non-normality. The covariates considered for analysis of variance (ANOVA), multiple regression analysis, and odds ratio (OR) calculation were age, body mass index (BMI), ever smoking, lipid profile (high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total cholesterol and triglycerides) and C-reactive protein (CRP) levels whenever they were significant for a given phenotype. All computations were performed using the R statistical software package (version 2.3.1, http://www.r-project.org). The haplotype analysis was performed using both EH (Estimate Haplotype-frequencies) (version 1.2, http://linkage.rockefeller.edu/ott/eh.htm) and Haploview programs to check for overall haplotype distribution differences and individual haplotype associations, respectively. A P value of less than 0.05 was considered as suggestive evidence of association. Student’s t-test was used to determine statistical significance of the expression difference between the wild-type and mutant constructs. Power analysis was performed using the QUANTO software (http://hydra.usc.edu/GxE).
RESULTS
APOH promoter polymorphisms and SLE
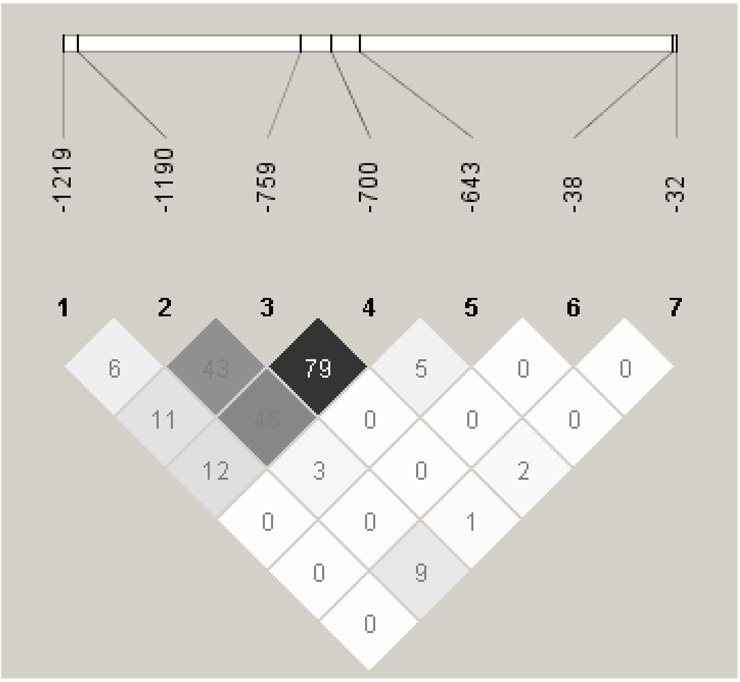
Minor allele frequencies (MAFs) of the APOH promoter SNPs that we observed in our control population (−1284G: 0.1%, −1219A: 9.9%, −1190C: 37.6%, −759G: 24.1%, −700A: 24.4%, −643C: 13.8%, −38A: 2.5%, −32A:6.2%) were similar to those previously reported in the literature 13 and/or in public databases. Due to its rare presence (MAF < 0.01), −1284C>G SNP was excluded from further analyses. Pairwise LD analysis using seven SNPs showed similar patterns in cases and controls (data not shown), therefore only the results from combined case + control cohort are presented in Figure 1. There was a strong LD with high correlation (D' = 0.92, r2 = 0.79) between −759A>G and −700C>A SNPs, hence one of these SNPs (−700C>A) was excluded from subsequent multiple regression and haplotype analyses.
Figure 1.
Pairwise Linkage disequilibrium analysis of APOH promoter SNPs showing r 2 (x 100) values.The intensity of gray is proportional to r2, with the darkest gray being the highest r2 value.
Multiple regression analysis (Table 1) of the remaining 6 SNPs showed significant association of the >1219G>A and −643T>C SNPs with SLE risk after adjusting for age (P = 0.016 for each). However, the follow-up single-site analysis for the difference in genotype distribution and allele frequencies showed a significant association for only −643T>C; the genotype distribution of −643T>C in cases/controls was: 68.8%/74.6% for TT, 26.2%/23.2% for TC and 5.0%/2.2% for CC (age-adjusted P = 0.020); the −643C allele frequency was 18.1% in SLE cases vs. 13.8% in controls. The age-adjusted OR for the −643C allele under the additive model was 1.41 (95% CI: 1.07–1.86, P = 0.014). In order to replicate the −643 finding, we genotyped an independent sample from Chicago (109 SLE cases, 81 controls). No association/trend for association was observed in the Chicago (P = 0.919) ; the genotype distribution of −643T>C in cases/controls was: 73.3%/73.8% for TT and 26.7%/26.3% for TC). When the data from both cohorts was combined and analyzed by adjusting for the recruitment site, the association became less significant despite the increase in sample size and power (combined adjusted OR was 1.33 (95% CI: 1.04–1.70, P = 0.025). This suggests that the association of −643 with SLE risk may not be real.
Table 1.
Multiple Regression * analysis of APOH promoter variants for SLE disease status, Nephritis and subclinical CVD (carotid plaque and IMT)
| P Value for ** | ||||
|---|---|---|---|---|
| SLE | Nephritis | Carotid Plaque | Carotid IMT | |
| rs8178819 (−1219G>A) | 0.016 | 0.014 | 0.344 | 0.887 |
| rs3760290 (−1190G>C) | 0.982 | 0.790 | 0.070 | 0.322 |
| rs8178820 (−759A>G) | 0.182 | 0.185 | 0.884 | 0.903 |
| rs3760292 (−643T>C) | 0.016 | 0.944 | 0.003 | 0.036 |
| (−38G>A) | 0.295 | 0.200 | 0.022 | 0.836 |
| rs8178822 (−32C>A) | 0.646 | 0.344 | 0.033 | 0.753 |
SLE risk was analyzed under additive model while lupus nephritis and carotid plaque and IMT under dominant model
Adjusted for age for SLE and nephritis risk; adjusted for age, HDL-C, LDL-C,cholesterol and triglycerides for carotid plaque and IMT
Six-site haplotype analysis identified a total of nine haplotypes with a frequency of > 1% (Table 2). Using EH program, the overall haplotype distribution was found to differ significantly between cases and controls (P = 0.009). Using Haploview program, one particular haplotype (H7) was found to be present predominantly in cases (χ2 = 19.151, P = 1.21 × 10−5). This haplotype harbored the minor alleles for the −1190G>C, −759A>G and −643T>C SNPs. When we carried out a permutation test for haplotypes in Haploview (permutation number = 1000), none of the permutations exceeded the highest observed χ2, confirming that the observed association of H7 was not just by chance.
Table 2.
Six-site haplotype analysis* of APOH promoter polymorphisms in SLE cases and controls
| Haplotype frequencies | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype | rs8178819 (−1219G>A) |
rs3760290 (−1190G>C) |
rs8178820 (−759A>G) |
rs3760292 (−643T>C) |
(−38G>A) | rs8178822 (−32C>A) |
Total (n=783) |
Cases (n=339) |
Controls (n=444) |
|
| H1 | G | G | A | T | G | C | 0.441 | 0.424 | 0.455 | |
| H2 | G | C | G | T | G | C | 0.156 | 0.138 | 0.168 | |
| H3 | G | G | A | C | G | C | 0.119 | 0.104 | 0.129 | |
| H4 | G | C | A | T | G | C | 0.083 | 0.099 | 0.071 | |
| H5 | A | C | G | T | G | C | 0.057 | 0.043 | 0.069 | |
| H6 | G | C | A | T | G | A | 0.053 | 0.054 | 0.052 | |
| H7 | G | C | G | C | G | C | 0.018 | 0.045 | 0.000 | |
| H8 | G | G | A | T | A | C | 0.017 | 0.014 | 0.019 | |
| H9 | A | G | A | T | G | C | 0.012 | 0.018 | 0.007 | P** = 0.009 |
Using program EH version 1.2 (excluding individuals with missing genotypes)
P value for overall haplotype distribution difference between cases and controls
APOH promoter polymorphisms and lupus nephritis
Because we obtained significant evidence that APOH promoter SNPs influenced SLE risk, we next investigated whether these SNPs influenced specific clinical manifestations of SLE. Multiple regression analysis in lupus patients stratified by the presence (n = 103) or absence (n = 241) of renal disease showed significant association with −1219G>A (P = 0.014) after adjusting for age (Table 1). The −1219A allele frequency was 4.4% in SLE cases with nephritis vs. 9.0% in SLE cases without nephritis. The age-adjusted OR for the −1219A allele under dominant model was 0.36 (95% CI: 0.16–0.83, P = 0.016).
APOH promoter polymorphisms and subclinical cardiovascular disease (carotid plaque and carotid IMT)
Multiple regression analysis of 6 SNPs in lupus patients stratified by the presence (n = 81) or absence (n = 164) of carotid plaque revealed significant associations with −643T>C (P = 0.003), −38G>A (P = 0.022) and −32C>A (P = 0.033) SNPs after adjusting for age and lipid profile (Table 1). Subsequent single-site analyses (adjusted for age and lipid profile) revealed significant genotype differences between the two groups for −643T>C and −32C>A SNPs but not for −38G>A. The −643C allele frequency was 8.6% in SLE cases with carotid plaque vs. 18.4% in patients without plaque; the age- and lipid profile-adjusted OR for the −643C allele carriers (TT vs. TC + CC genotypes) was 0.37 (95% CI: 0.17–0.81, P = 0.013). The −32A allele frequency was 8.6% in SLE cases with carotid plaque vs. 5.8% in patients without plaque; the adjusted OR for the −32A allele carriers (CC vs. AA+CA genotypes) was 2.63 (95% CI: 1.09–6.35, P = 0.031). Multiple linear regression analysis of carotid IMT using the same model as for plaque demonstrated a significant association for only −643T>C after adjusting for age and lipid profile (P = 0.036) (Table 1) which was not observed in the single-site analysis.
Because −643T>C SNP was found to be associated with both carotid plaque and IMT in multiple regression analysis, we also genotyped this SNP in our Chicago SLE sample. The association of the −643T>C SNP with carotid plaque was confirmed in this independent sample of SLE patients from Chicago (37 with carotid plaque vs. 72 without carotid plaque). The age and lipid profile adjusted OR for the >643C allele carriers (TC + CC vs. TT genotypes) was 0.22 (95% CI: 0.06–0.78, P = 0.019). When the data from both Pittsburgh and Chicago cohorts were combined and analyzed by adjusting for the recruitment site, the adjusted OR was 0.35 (95% CI: 0.18–0.67, P = 0.002) suggesting a protective effect of −643C allele against developing carotid plaque.
APOH promoter polymorphisms and the occurrence of antiphospholipid antibodies
We categorized subjects into “APA-positive” and “APA-negative” groups. APA-positive group included participants positive for any of the following 3 antibodies; aCL (IgG and/or IgM), anti-β2GPI (IgA and/or IgG and/or IgM) and LAC. APA-negative group contained individuals negative for all APA. None of the APOH promoter SNPs was found to have a significant association with the presence of APA in controls in multiple regression. However, in cases, the distribution of the −38G>A SNP differed significantly between APA-positive and APA-negative groups in both multiple regression (P = 4.57 × 10−4) and single-site analyses (P = 0.004). It is important to note that −38G>A is a relatively rare variant and was only detected in APA-positive patients (MAF = 0.027) but not in APA-negative patients. The significant difference in −38G>A genotype distribution was also observed when the comparison was subcategorized between aCL-positive vs. APA-negative (P = 0.018), LAC-positive vs. APA-negative (P = 0.001) and anti-β2GPI-positive vs. APA-negative (P = 0.002) cases using multiple regression under dominant model.
Reporter gene expression and Electrophoretic mobility shift assay
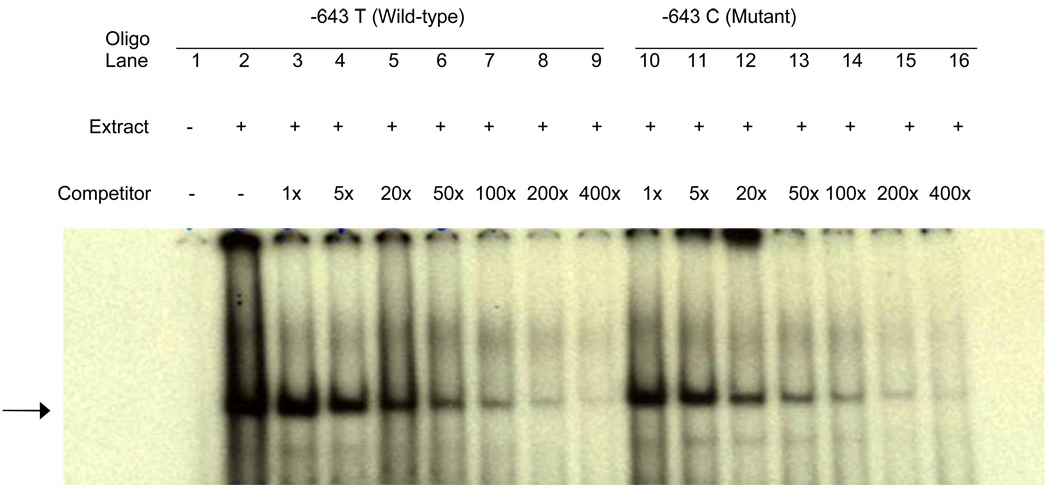
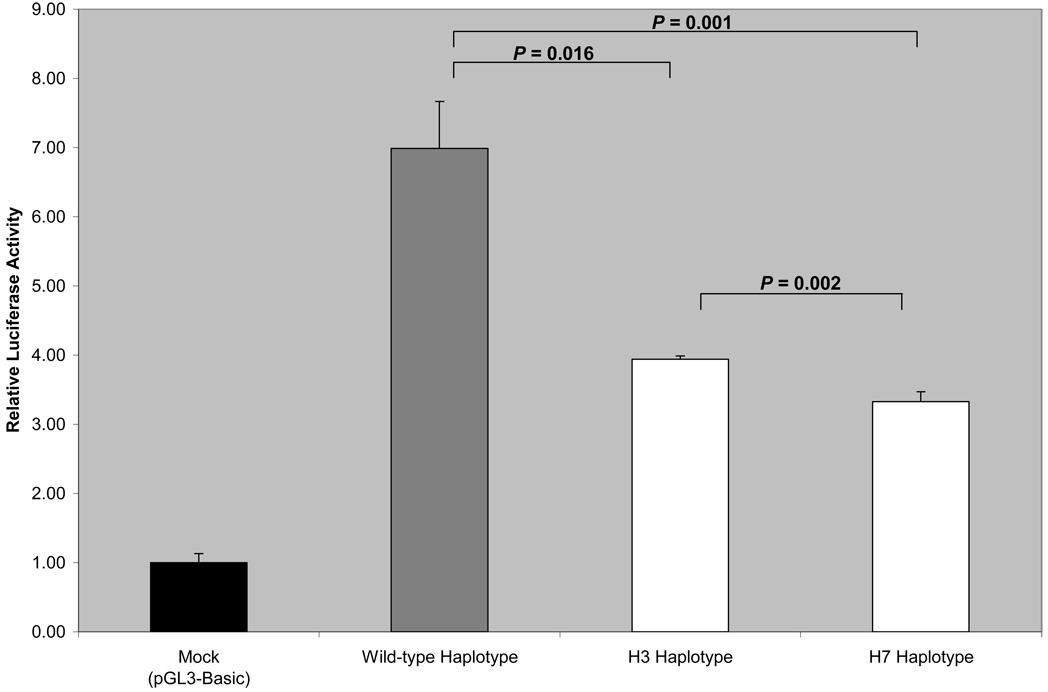
Since the −643T>C SNP was found to be consistently associated with carotid plaque in two independent samples, we hypothesized that it might harbor a binding site for transcription factor and also affect APOH expression. In order to determine whether −643T>C polymorphism affects the binding of putative transcription factors, EMSA was performed using allele-specific consensus oligonucleotide probes. Upon incubation of radiolabeled oligonucleotides specific for −643T and −643C alleles with HepG2 nuclear extracts, both oligonucleotides formed a DNA-protein complex (Figure 2), indicating that a nuclear factor(s) binds to this sequence. However, competition assays using increasing amounts of unlabeled wild-type (T allele) or mutant (C allele) oligonucleotides showed no allele-specific differences.To determine if the wild-type (−643T) and the mutant (−643C) alleles had different promoter activities, we created wild-type and mutant APOH promoter-firefly luciferase reporter constructs. As shown in Figure 3, our results of reporter-gene assays showed a significant decrease in promoter activity associated with the mutant allele (−643C or H3 haplotype) as compared to the wild-type allele (mean ± SD: 3.94 ± 0.05 vs. 6.99 ± 0.68, P = 0.016). Although the −643C allele appears to be functional, the two haplotypes (H3 and H7 in Table 2) that carry −643C, only H7 showed significant association with SLE risk. This suggests that other variants in the H7 haplotype might also be important in affecting APOH expression. To test this hypothesis, we mutated the defining alleles in H7 haplotype (−1190G>C, −759A>G, −643T>C) and compared its gene expression with the wild-type and H3 haplotypes. As shown in Figure 3, the H7 haplotype had the lowest luciferase activity as compared to the H3 (mean ± SD: 3.33 ± 0.14 vs. 3.94 ± 0.05, P = 0.002) and the wild-type haplotype (mean ± SD: 3.33 ± 0.14 vs. 6.99 ± 0.68, P = 0.001).
Figure 2.
EMSA result for −643T>C polymorphism: Each sample contains a mixture of 5 µg of nuclear extract derived from human HepG2 cell nuclear extract and 30xmer 32P-labeled wild-type oligonucleotide containing T allele. Arrowhead indicates specific DNA-protein complex associated with the −643T>C polymorphic site. Lane 1, labeled oligonucleotide without nuclear extract from HepG2 cells; 2, labeled oligonucleotide with nuclear extracts. Competition assay was performed by adding excess cold oligonucleotides containing either the −643 T allele (lanes 3 to 9) or the C allele (lanes 10 to 16). Lanes 3 to 9 have increasing amounts of T oligo competitor (1x, 5x, 20x, 50x, 100x, 200x, and 400x, respectively); lanes 10 to 16 have increasing amounts of C oligo competitor (1x, 5x, 20x, 50x, 100x, 200x, and 400x, respectively)
Figure 3.
Dual-luciferase reporter gene expression: The effect of wild-type and mutants were measured as the mean of the firefly luciferase levels, which were normalized by the Renilla luciferase activity, which served as the reference for the transfection efficiency. The results presented are from one out of two independent experiments. The wild-type haplotype contains the major alleles at all sites. While the H3 haplotype is defined by minor allele at SNP −643T>C, the H7 haplotype is defined by minor alleles at SNPs −643T>C, −1190G>C and −759A>G.
DISCUSSION
Because of the important role of β2GPI in the production of APA and the observation that β2GPI-mediated immune response in patients with autoimmune diseases may lead to atherosclerosis 18, it is important to understand the role of APOH genetic variation in relation to autoimmune diseases and associated premature CVD. A priori, one might expect that genetic variation in elements which control APOH expression (promoter region) can be associated with the disease risk. Promoter sequences are potential sources of polymorphisms affecting gene expression and phenotypic variation 33. Promoter variants may potentially alter the affinities of existing protein-DNA interactions or recruit new proteins to bind to the DNA, altering the specificity and kinetics of the transcription process. To our knowledge, this is the first study to evaluate the role of APOH promoter SNPs in relation to SLE and related phenotypes, including lupus nephritis and subclinical CVD.
Our study revealed a significant association for the −643T>C SNP with SLE risk in the Pittsburgh sample (P = 0.014). However, this association was not confirmed in a second relatively small sample from Chicago (P = 0.919) and furthermore, the significance level decreased in the combined Pittsburgh + Chicago sample, (P = 0.025) despite increase in the sample size and power. This suggests that either the effect of the −643T>C SNP on SLE risk is small which is difficult to reproduce in all samples or this association is due to chance. The latter assumption was confirmed in the haplotype analysis where one haplotype that carried the defining −643C allele (H3 haplotype in Table 2) was not associated with SLE risk. On the other hand, our six-site haplotype analysis yielded a significant difference in overall haplotype distribution between SLE cases and controls (P = 0.009). The haplotype (H7 haplotype) with the most striking difference between cases and controls (P = 1.21 × 10−5) had a frequency of less than 2% in the total sample suggesting that only a small number of individuals were predicted to carry this haplotype. Notably, while no example of this haplotype was observed in 888 control chromosomes based on EH program, 4.5% of the patients carried this haplotype. H7 haplotype was not defined by a particular allele, although it carried the minor alleles for three APOH promoter SNPs (−643T>C, −1190G>C and −759A>G). The minor alleles at these three SNPs do not appear to be causative, as the presence of the −643C allele in H3 haplotype alone or the presence of the −1190C and −759G alleles together in H2 and H5 haplotypes did not demonstrate significant association. It appears that the simultaneous presence of these three alleles affect the risk of SLE in an additive fashion as demonstrated by the H7 haplotype. The apparently additive effects of these alleles were also obvious in the gene expression assay where the H7 haplotype was associated with greatest effect on luciferase activity. Alternatively, the unique H7 haplotype is a marker for a functional variant present in APOH or a nearby gene. Sequencing of the individuals who carry the haplotype H7 may help to identify putative functional variant(s).
The −643T>C SNP showed convincing association with the presence of carotid plaque among SLE patients as it was found to be associated in two independent samples from Pittsburgh and Chicago. In the combined Pittsburgh + Chicago SLE sample the association was more significant than observed in individual samples from each site (adjusted OR = 0.35, P = 0.002). The accumulation of oxLDL is believed to initiate the process of atherosclerosis and β2GPI is known to inhibit the uptake of oxLDL by macrophages in vitro while it promotes the influx of oxLDL in the presence of APA33. Several lines of evidence suggest that autoimmune vascular inflammation and oxidative stress may promote the formation of oxLDL/ β2GPI complexes in the arterial wall as seen in SLE and APS patients 18. Since many SLE patients are positive for APA, it is likely that low β2GPI expression associated with the −643C allele may retard the influx of oxLDL to macrophages in the presence of APA and thus provide protection against plaque formation. Alternatively, there may be another mechanism underlying the effect of −643C allele on carotid plaque formation or it may simply be in strong LD with another variant, especially given that another APOH promoter variant (−32A allele), which is also associated with low APOH expression 24, was associated with increased risk for carotid plaque after adjusting for covariates (OR = 2.63, P = 0.031). Additional studies in large data sets may help to delineate the role of APOH promoter SNPs in relation to carotid plaque formation.
Renal disease is a major cause of morbidity in SLE patients. Some studies reported an increased production of APA in patients with Lupus nephritis , including anti-β2GPI and anti-oxLDL 34–36. We found that −1219G>A SNP may potentially affect the lupus nephritis risk (age-adjusted OR = 0.36, P = 0.016) and this effect seems to be independent from APA occurrence given that this SNP did not show significant association with any of the APA examined in our sample. Rather, we found significant association of the −38G>A SNP with the occurrence of APA (P = 4.57 × 10−4), aCL (P = 0.018), LAC (P = 0.001) and anti-β2GPI (P = 0.002) in SLE cases but not in controls using multiple regression under dominant model. It is possible that the −38G>A SNP may be interacting with other factors that are involved in SLE etiopathogenesis to show its effect on APA in an autoimmune background. It is reasonable to expect that some factors would specifically show their effects on APA in association with disease status, thus contributing to the increased prevalence of APA in patients with autoimmune diseases as compared to the general population. Alternatively, the APOH variants may have modest effects on the occurrence of APA which may be difficult to reproduce in all samples. Finally, given the very low MAF of −38G>A SNP, we cannot exclude the possibility of its spurious association.
The power for most of the examined SNPs in this study was adequate to detect SLE disease - related risk. For four SNPs (−1190G>C, −759 A>G, − 700C>A, −643T>C), we had 80% power to detect ORs between 1.50 and 1.55. For the −1219G>A and −32C>A SNPs, we had 80% power to detect an OR of 1.61 and 1.75, respectively. The power for the less common SNPs (−1284C>G and −38G>A) with MAF < 0.05 was low.
In summary, we present evidence that individual APOH promoter SNPs as well as haplotypes may be involved in the etiology of SLE and especially the risk for autoimmune-mediated CVD. Our results merit further investigation of APOH in independent large SLE case-control samples with subclinical phenotype information.
ACKNOWLEDGEMENTS
We thank Bonnie Kane, BS and Beverly Smulevitz, BS for their technical assistance.
Grant support: This study was supported by the National Heart, Lung, and Blood Institute Grant HL 54900; the grants from the National Institutes of Health F32-AR51681, K23-AR054418, and Mary Kirkland Center for Lupus Research and Rheuminations, Inc.; K24-AR02318, P60 -AR30692, P60-AR48098, NCRR/GCRC M01-RR00048.
Footnotes
Dedication as a footnote: This paper is dedicated to Professor Robert E. Ferrell on his 65th birthday.
REFERENCES
- 1.Frostegard J. SLE, atherosclerosis and cardiovascular disease. J Intern Med. 2005;257(6):485–495. doi: 10.1111/j.1365-2796.2005.01502.x. [DOI] [PubMed] [Google Scholar]
- 2.Trager J, Ward MM. Mortality and causes of death in systemic lupus erythematosus. Curr Opin Rheumatol. 2001;13(5):345–351. doi: 10.1097/00002281-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Atsumi T, Tsutsumi A, Amengual O, et al. Correlation between beta2-glycoprotein I valine/leucine247 polymorphism and anti-beta2-glycoprotein I antibodies in patients with primary antiphospholipid syndrome. Rheumatology (Oxford) 1999;38(8):721–723. doi: 10.1093/rheumatology/38.8.721. [DOI] [PubMed] [Google Scholar]
- 4.Hirose N, Williams R, Alberts AR, et al. A role for the polymorphism at position 247 of the beta2-glycoprotein I gene in the generation of anti-beta2-glycoprotein I antibodies in the antiphospholipid syndrome. Arthritis Rheum. 1999;42(8):1655–1661. doi: 10.1002/1529-0131(199908)42:8<1655::AID-ANR14>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 5.Prieto GA, Cabral AR, Zapata-Zuniga M, et al. Valine/valine genotype at position 247 of the beta2-glycoprotein I gene in Mexican patients with primary antiphospholipid syndrome: association with anti-beta2-glycoprotein I antibodies. Arthritis Rheum. 2003;48(2):471–474. doi: 10.1002/art.10771. [DOI] [PubMed] [Google Scholar]
- 6.Greaves M. Antiphospholipid antibodies and thrombosis. Lancet. 1999;354(9183):1031. doi: 10.1016/S0140-6736(05)76636-8. [DOI] [PubMed] [Google Scholar]
- 7.Manzi S, Meilahn EN, Rairie JE, et al. Age-specific incidence rates of myocardial infarction and angina in women with systemic lupus erythematosus: comparison with the Framingham Study. Am J Epidemiol. 1997;145(5):408–415. doi: 10.1093/oxfordjournals.aje.a009122. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence JS, Martins CL, Drake GL. A family survey of lupus erythematosus. 1. Heritability. J Rheumatol. 1987;14(5):913–921. [PubMed] [Google Scholar]
- 9.Wong M, Tsao BP. Current topics in human SLE genetics. Springer Semin Immunopathol. 2006;28(2):97–107. doi: 10.1007/s00281-006-0031-6. [DOI] [PubMed] [Google Scholar]
- 10.Lozier J, Takahashi N, Putnam FW. Complete amino acid sequence of human plasma beta 2-glycoprotein I. Proc Natl Acad Sci U S A. 1984;81(12):3640–3644. doi: 10.1073/pnas.81.12.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehdi H, Nunn M, Steel DM, et al. Nucleotide sequence and expression of the human gene encoding apolipoprotein H (beta 2-glycoprotein I) Gene. 1991;108(2):293–298. doi: 10.1016/0378-1119(91)90449-l. [DOI] [PubMed] [Google Scholar]
- 12.Kristensen T, Schousboe I, Boel E, et al. Molecular cloning and mammalian expression of human beta 2-glycoprotein I cDNA. FEBS Lett. 1991;289(2):183–186. doi: 10.1016/0014-5793(91)81065-g. [DOI] [PubMed] [Google Scholar]
- 13.Chen Q, Kamboh MI. Complete DNA sequence variation in the apolipoprotein H (beta-glycoprotein I) gene and identification of informative SNPs. Ann Hum Genet. 2006;70(Pt 1):1–11. doi: 10.1111/j.1529-8817.2005.00211.x. [DOI] [PubMed] [Google Scholar]
- 14.Sanghera DK, Nestlerode CS, Ferrell RE, Kamboh MI. Chimpanzee apolipoprotein H (beta2-glycoprotein I): report on the gene structure, a common polymorphism, and a high prevalence of antiphospholipid antibodies. Hum Genet. 2001;109(1):63–72. doi: 10.1007/s004390100549. [DOI] [PubMed] [Google Scholar]
- 15.Kamboh MI, Sanghera DK, Mehdi H, et al. Single nucleotide polymorphisms in the coding region of the apolipoprotein H (beta2-glycoprotein I) gene and their correlation with the protein polymorphism, anti-beta2glycoprotein I antibodies and cardiolipin binding: description of novel haplotypes and their evolution. Ann Hum Genet. 2004;68(Pt 4):285–299. doi: 10.1046/j.1529-8817.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- 16.Kamboh MI, Ferrell RE. Apolipoprotein H polymorphism and its role in lipid metabolism. Advances in Lipids Research. 1991;1:9–18. [Google Scholar]
- 17.Schousboe I. beta 2-Glycoprotein I: a plasma inhibitor of the contact activation of the intrinsic blood coagulation pathway. Blood. 1985;66(5):1086–1091. [PubMed] [Google Scholar]
- 18.Matsuura E, Kobayashi K, Hurley BL, Lopez LR. Atherogenic oxidized low-density lipoprotein/beta2-glycoprotein I (oxLDL/beta2GPI) complexes in patients with systemic lupus erythematosus and antiphospholipid syndrome. Lupus. 2006;15(7):478–483. doi: 10.1191/0961203306lu2337oa. [DOI] [PubMed] [Google Scholar]
- 19.McNeil HP, Simpson RJ, Chesterman CN, Krilis SA. Anti-phospholipid antibodies are directed against a complex antigen that includes a lipid-binding inhibitor of coagulation: beta 2-glycoprotein I (apolipoprotein H) Proc Natl Acad Sci U S A. 1990;87(11):4120–4124. doi: 10.1073/pnas.87.11.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galli M, Comfurius P, Maassen C, et al. Anticardiolipin antibodies (ACA) directed not to cardiolipin but to a plasma protein cofactor. Lancet. 1990;335(8705):1544–1547. doi: 10.1016/0140-6736(90)91374-j. [DOI] [PubMed] [Google Scholar]
- 21.Jones JV, James H, Tan MH, Mansour M. Antiphospholipid antibodies require beta 2-glycoprotein I (apolipoprotein H) as cofactor. J Rheumatol. 1992;19(9):1397–1402. [PubMed] [Google Scholar]
- 22.Yasuda S, Atsumi T, Matsuura E, et al. Significance of valine/leucine247 polymorphism of beta2-glycoprotein I in antiphospholipid syndrome: increased reactivity of anti-beta2-glycoprotein I autoantibodies to the valine247 beta2-glycoprotein I variant. Arthritis Rheum. 2005;52(1):212–218. doi: 10.1002/art.20741. [DOI] [PubMed] [Google Scholar]
- 23.Kamboh MI, Manzi S, Mehdi H, et al. Genetic variation in apolipoprotein H (beta2-glycoprotein I) affects the occurrence of antiphospholipid antibodies and apolipoprotein H concentrations in systemic lupus erythematosus. Lupus. 1999;8(9):742–750. doi: 10.1191/096120399678840909. [DOI] [PubMed] [Google Scholar]
- 24.Mehdi H, Manzi S, Desai P, et al. A functional polymorphism at the transcriptional initiation site in beta2-glycoprotein I (apolipoprotein H) associated with reduced gene expression and lower plasma levels of beta2-glycoprotein I. Eur J Biochem. 2003;270(2):230–238. doi: 10.1046/j.1432-1033.2003.03379.x. [DOI] [PubMed] [Google Scholar]
- 25.Arvieux J, Roussel B, Ponard D, Colomb MG. IgG2 subclass restriction of anti-beta 2 glycoprotein 1 antibodies in autoimmune patients. Clin Exp Immunol. 1994;95(2):310–315. doi: 10.1111/j.1365-2249.1994.tb06529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabiedes J, Cabral AR, Alarcon-Segovia D. Clinical manifestations of the antiphospholipid syndrome in patients with systemic lupus erythematosus associate more strongly with anti-beta 2-glycoprotein-I than with antiphospholipid antibodies. J Rheumatol. 1995;22(10):1899–1906. [PubMed] [Google Scholar]
- 27.Cabral AR, Cabiedes J, Alarcon-Segovia D. Antibodies to phospholipid-free beta 2-glycoprotein-I in patients with primary antiphospholipid syndrome. J Rheumatol. 1995;22(10):1894–1898. [PubMed] [Google Scholar]
- 28.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 29.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 30.Rhew EY, Manzi SM, McPherson D, et al. Differences in Subclinical Cardiovascular Disease in African-American and Caucasian Women with SLE: A Combined Analysis with SOLVABLE (Chicago) and HEARTS (Pittsburgh) Arthritis Rheum. 2007;56:S822. [Google Scholar]
- 31.Selzer F, Sutton-Tyrrell K, Fitzgerald SG, et al. Comparison of risk factors for vascular disease in the carotid artery and aorta in women with systemic lupus erythematosus. Arthritis Rheum. 2004;50(1):151–159. doi: 10.1002/art.11418. [DOI] [PubMed] [Google Scholar]
- 32.Chen Q, Reis SE, Kammerer C, et al. Genetic variation in lectin-like oxidized low-density lipoprotein receptor 1 (LOX1) gene and the risk of coronary artery disease. Circulation. 2003;107(25):3146–3151. doi: 10.1161/01.CIR.0000074207.85796.36. [DOI] [PubMed] [Google Scholar]
- 33.Hoogendoorn B, Coleman SL, Guy CA, et al. Functional analysis of human promoter polymorphisms. Hum Mol Genet. 2003;12(18):2249–2254. doi: 10.1093/hmg/ddg246. [DOI] [PubMed] [Google Scholar]
- 34.Loizou S, Samarkos M, Norsworthy PJ, Cazabon JK, Walport MJ, Davies KA. Significance of anticardiolipin and anti-beta(2)-glycoprotein I antibodies in lupus nephritis. Rheumatology (Oxford) 2000;39(9):962–968. doi: 10.1093/rheumatology/39.9.962. [DOI] [PubMed] [Google Scholar]
- 35.Natejumnong C, Ruangkanchanasetr P, Aimpun P, Supaporn T. Significance of antiphospholipid antibodies in lupus nephritis. J Med Assoc Thai. 2006;89 Suppl 2:S121–S128. [PubMed] [Google Scholar]
- 36.Fialova L, Zima T, Tesar V, et al. Antiphospholipid antibodies in patients with lupus nephritis. Ren Fail. 2003;25(5):747–758. doi: 10.1081/jdi-120024290. [DOI] [PubMed] [Google Scholar]