Abstract
Helix-helix association within a membrane environment represents one of the fundamental processes in membrane protein folding. However, studying the kinetics of such processes has been difficult because most membrane proteins are insoluble in aqueous solution. Here we present a stopped-flow fluorescence study of the membrane interaction kinetics of a designed, water-soluble transmembrane (TM) peptide, anti-αIIb, which is known to dimerize in phospholipid bilayers. We show that by using two fluorescent amino acids, i.e., tryptophan and p-cyano-phenylalanine, we are able to kinetically dissect distinct phases in the peptide-membrane interaction, representing membrane binding, membrane insertion, and TM helix-helix association. Our results further show that the latter process occurs on a time scale of seconds, indicating that the association of two TM helices is an intrinsically slow event.
While the study of how globular proteins fold has reached an advanced stage, there have been far fewer studies focused on the folding dynamics of membrane proteins.1–4 This is due in part to the fact that most membrane proteins are insoluble in aqueous solution and thus, making kinetic studies (e.g., via mixing) difficult. Here, we show that by using a water-soluble transmembrane (TM) peptide, anti-αIIb,5,6 and two fluorescent probes, i.e., tryptophan (Trp) and p-cyano-phenylalanine (PheCN), it is possible to kinetically dissect distinct phases in the peptide-membrane interaction, representing binding, insertion and dimerization, thus providing new mechanistic and kinetic information on several processes that are fundamentally important to membrane protein folding.
Anti-αIIb (sequence: KKAYV MLLPF FIGLL LGLIF GGAFW GPARH LKK) inserts spontaneously into lipid bilayers and has similar dimerization affinity to that of GpA which is known to form tight TM homodimers.5–7 In addition, the lysine residues appended to both the N- and C-termini greatly enhance the solubility of anti-αIIb in water, allowing kinetic studies of membrane peptide folding starting from aqueous phase. More importantly, the TM anti-αIIb homodimers do not further aggregate to form high order oligomers or pores, nor induce membrane lysis at a lipid to peptide ratio up to 10:1.6 Thus, using this peptide and an appropriate spectroscopic probe it is possible to directly assess the time scales during which several important kinetic steps occur, including peptide binding to membrane from aqueous solution, insertion into the membrane as well as helix-helix association inside the membrane.
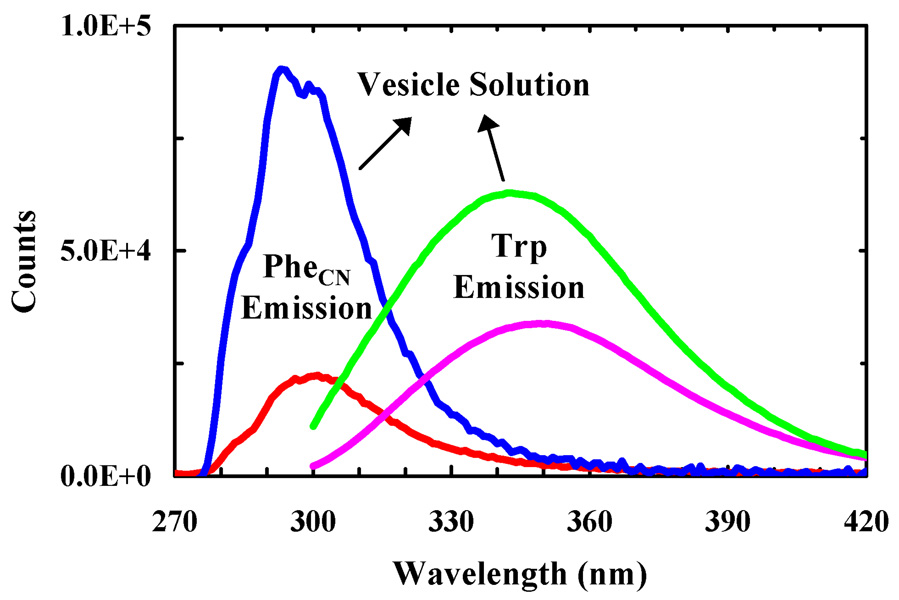
The peptide-membrane interaction was probed using either the native Trp fluorescence in anti-αIIb or that arising from PheCN in anti-αIIb-PheCN, an anti-αIIb mutant wherein the Trp residue is replaced by PheCN. It has been shown earlier that the PheCN fluorescence is sensitive to its environment and thus, can be used as a protein conformational probe.8 As shown (Figure 1), upon association of the respective peptides with POPC/G (3/1, wt/wt) vesicles, both the Trp fluorescence and PheCN fluorescence increase, indicative of the usefulness of these probes.
Figure 1.
Fluorescence spectra of anti-αIIb (pink and green) and anti-αIIb-PheCN (red and blue) in buffer and POPC/G vesicle solutions, as indicated. The peptide and lipid concentrations were 2.5 µM and 0.86 mg/mL, respectively. The excitation wavelengths were 290 and 240 nm for anti-αIIb and anti-αIIb-PheCN, respectively.
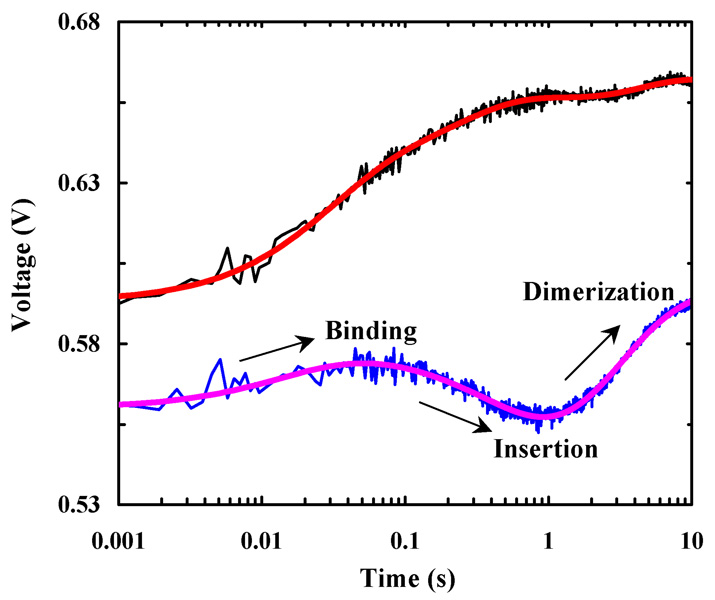
The peptide-membrane association kinetics were measured using a stopped-flow fluorescence techqnique.9 Consistent with the equilibrium results (Figure 1), rapidly mixing anti-αIIb and POPC/G vesicle solutions induces a time-dependent, net increase in the Trp fluorescence although some minor, wavy features occur in the late stages of the stopped-flow kinetics (Figure 2). Similarly, rapidly mixing anti-αIIb-PheCN with POPC/G vesicles also leads to an overall increase in the PheCN fluorescence. However, the resultant stopped-flow kinetics clearly show three distinct phases of alternating signs (Figure 2). Thus, these results not only underscore the importance of employing multiple probes in the study of complex ‘reaction’ kinetics, but also indicate that in the current case the PheCN fluorescence is more sensitive to those molecular events occurring during the time course leading to the formation of TM anti-αIIb homodimer.
Figure 2.
Stopped-flow kinetics of anti-αIIb (black) and anti-αIIb-PheCN (blue) upon association with POPC/G vesicles (0.86 mg/mL). In both cases, the final peptide concentration was 2.5 µM. The smooth lines are fits of these data to the model discussed in the text.
Under current experimental conditions it is reasonable to assume that the peptides form TM homodimers as the reaction reaches equilibrium.5,6 Therefore, similar to that observed for many antimicrobial peptides,9,10 the increase in Trp fluorescence at the early stages of the stopped-flow kinetics of anti-αIIb most likely arises from the binding and insertion of the peptide into the membrane. However, a more insightful assessment of the entire kinetics reported by Trp fluorescence is less straightforward. On the other hand, the distinct features in the stopped-flow kinetics of anti-αIIb-PheCN make them relatively easier to interpret. It has been shown that upon burial in a hydrophobic environment the PheCN fluorescence decreases.8 Hence, it is reasonable to attribute the middle or negative kinetic phase to peptide insertion into the membrane because such a process will bring the PheCN residue to the hydrophobic interior of the bilayer; and attribute the initial, minor positive phase to membrane binding as it will bring the peptide to the polar headgroup region of the membrane. Following these assignments and also the two-stage model11 of membrane protein folding, it is logical to further attribute the third kinetic phase, in which the PheCN fluorescence increases, to the formation of TM homodimers.
To provide quantitative information regarding the kinetics of membrane binding, insertion, and dimerization of anti-αIIb, we further fit all of the stopped-flow traces to the following kinetic scheme, which is a minimally expanded version of the two-stage model by including peptide dimer formation in solution,
where P and V represent the peptide and vesicle, respectively, and D represents the TM peptide dimer. In addition, the subscripts ‘m’ and ‘d’ refer to peptide monomer and dimer, respectively, while the superscripts ‘s’ and ‘i’ represent the surface-bound and membrane-inserted states of the respective peptide species, respectively. As shown (Figure 2), this simple model, despite its lack of consideration of (a) the dynamic equilibria between the peptide monomer and dimer in solution and on the membrane surface and (b) the possibility that anti-αIIb may form higher order oligmers in solution, yields not only satisfactory fits to the anti-αIIb and anti-αIIb-PheCN stopped-flow kinetics, but also self-consistent kinetic and fluorescent parameters (Tables S1 and Table S2, Supporting Information). In addition, kinetic data obtained at other peptide concentrations (5 µM for anti-αIIb and 1, 5 and 15 µM for anti-αIIb-PheCN) can also be fit by this model, yielding comparable microscopic rate constants but different percentages of Pd in solution (Figure S1 to Figure S5 and Table S1 and Table S2, Supporting Information), further substantiating the validity of the model.
Furthermore, and perhaps more importantly, the fitting parameters recovered also make physical sense. For example, all the backward rate constants are negligible compared to the corresponding forward rate constants, indicating that all of the kinetic steps are essentially irreversible. This is consistent with previous thermodynamic measurements.5 In addition, the recovered membrane binding rate constant is also in good agreement with those measured for other peptides,3,8,9 and the membrane insertion rate constant of the anti-αIIb monomer, 2.4 ± 0.7 s−1 (the average of k2 values in Table S1, Supporting Information), is very similar to that (1.9 s−1) of the membrane pH (low) insertion peptide (pHLIP).12 On the other hand, the membrane insertion rate (0.5 ± 0.1 s−1) of the anti-αIIb dimer, as expected, is close to that (~0.24 s−1) of a helical membrane protein, diacylglycerol kinase.3
Moreover, our results indicate that the event of helix-helix association inside a membrane environment takes place on a time scale of a few seconds, which is not only much slower than the folding rate of the coiled-coil motif in solution,13 but also several orders of magnitude slower than that expected for a diffusion-limited association process. The latter is estimated to be on the order of a few hundreds of microseconds using the diffusion constant (3 × 10−9 cm2 s−1) of a model TM peptide14 and a two-dimensional random walk model.15 Taken together, these results thus indicate that the rate of TM helix-helix association is not diffusion-limited, but rather is determined by the actual assembly process of the TM helical dimer, the folding of which requires proper backbone orientations and well-defined intermolecular sidechain-sidechain interactions.16
In summary, using two fluorescent amino acids we are able to dissect the major kinetic events associated with the interaction of a designed TM peptide, anti-αIIb, with a POPC/G model membrane, including membrane binding, insertion, and TM helix-helix association. While the kinetics of the latter process have been studied earlier either in a micelle environment or via vesicle fusion,17 the current study allows for the direct assessment of the kinetics of this fundamental membrane folding event starting from the aqueous phase.
Supplementary Material
Materials, methods, stopped-flow kinetics at other peptide concentrations, and fitting parameters.
Acknowledgement
We thank the NIH (GM-065978, RR-01348) and the NSF (DMR05-20020) for funding.
References
- 1.(a) Bogdanov M, Dowhan W. J. Biol. Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]; (b) MacKenzie KR. Chem. Rev. 2006;106:1931–1977. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]; (c) Stanley AM, Fleming KG. Arch. Biochem. Biophys. 2008;469:46–66. doi: 10.1016/j.abb.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 2.(a) Nagy JK, Lonzer WL, Sanders CR. Biochemistry. 2001;40:8971–8980. doi: 10.1021/bi010202n. [DOI] [PubMed] [Google Scholar]; (b) Otzen DE. J. Mol. Biol. 2003;330:641–649. doi: 10.1016/s0022-2836(03)00624-7. [DOI] [PubMed] [Google Scholar]; (c) Allen SJ, Curran AR, Templer RH, Meijberg W, Booth PJ. J. Mol. Biol. 2004;342:1279–1291. doi: 10.1016/j.jmb.2004.07.040. [DOI] [PubMed] [Google Scholar]
- 3.Lorch M, Booth PJ. J. Mol. Biol. 2004;344:1109–1121. doi: 10.1016/j.jmb.2004.09.090. [DOI] [PubMed] [Google Scholar]
- 4.(a) Nymeyer H, Woolf TB, Garcia AE. Proteins. 2005;59:783–790. doi: 10.1002/prot.20460. [DOI] [PubMed] [Google Scholar]; (b) Bond PJ, Sansom MSP. J. Am. Chem. Soc. 2006;128:2697–2704. doi: 10.1021/ja0569104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Lopez CF, Nielsen SO, Srinivas G, DeGrado WF, Klein ML. J. Chem. Theory and Comput. 2006;2:649–655. doi: 10.1021/ct050298p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Bu L, Im W, Brooks CL. Biophys. J. 2007;92:854–863. doi: 10.1529/biophysj.106.095216. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Chu JW, Voth GA. Biophys. J. 2007;93:3860–3871. doi: 10.1529/biophysj.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Lee J, Im W. J. Am. Chem. Soc. 2008;130:6456–6462. doi: 10.1021/ja711239h. [DOI] [PubMed] [Google Scholar]
- 5.Yin H, Slusky JS, Berger BW, Walters RS, Vilaire G, Rustem LI, Lear JD, Caputo GA, Bennett JS, DeGrado WF. Science. 2007;315:1817–1822. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 6.Caputo GA, Litvinov RI, Li W, Bennett JS, DeGrado WF, Yin H. Biochemistry. 2008;47:8600–8606. doi: 10.1021/bi800687h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Russ WP, Engelman DM. Proc. Natl. Acad. Sci. U.S.A. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Adair BD, Engelman DM. Biochemistry. 1994;33:5539–5544. doi: 10.1021/bi00184a024. [DOI] [PubMed] [Google Scholar]
- 8.(a) Tucker MJ, Oyola R, Gai F. Biopolymers. 2006;83:571–576. doi: 10.1002/bip.20587. [DOI] [PubMed] [Google Scholar]; (b) Aprilakis KN, Taskent H, Raleigh DP. Biochemistry. 2007;46:12308–12313. doi: 10.1021/bi7010674. [DOI] [PubMed] [Google Scholar]
- 9.(a) Tucker MJ, Tang J, Gai F. J. Phys. Chem. B. 2006;110:8105–8109. doi: 10.1021/jp060900n. [DOI] [PubMed] [Google Scholar]; (b) Tang J, Signarvic RS, DeGrado WF, Gai F. Biochemistry. 2007;46:13856–13863. doi: 10.1021/bi7018404. [DOI] [PubMed] [Google Scholar]
- 10.Constantinescu I, Lafleur M. Biochim. Biophys. Acta. 2004;1676:26–37. doi: 10.1016/j.bbamem.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Popot JL, Engelman DM. Biochemistry. 1990;19:4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- 12.Tang J, Gai F. Biochemistry. 2008;47:8250–8252. doi: 10.1021/bi801103x. [DOI] [PubMed] [Google Scholar]
- 13.(a) Mo JM, Holtzer ME, Holtzer A. Proc. Natl. Acad. Sci. U.S.A. 1991;88:916–920. doi: 10.1073/pnas.88.3.916. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wendt H, Berger C, Baici A, Thomas RM, Bosshard HR. Biochemistry. 1995;34:4097–4107. doi: 10.1021/bi00012a028. [DOI] [PubMed] [Google Scholar]; (c) Zitzewitz JA, Bilsel O, Luo J, Jones BE, Matthews CR. Biochemistry. 1995;34:12812–12819. doi: 10.1021/bi00039a042. [DOI] [PubMed] [Google Scholar]
- 14.Gambin Y, Lopez-Esparza R, Reffay M, Sierecki E, Gov NS, Genest M, Hodges RS, Urbach W. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2098–2102. doi: 10.1073/pnas.0511026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tonegawa Y, Umeda N, Hayakawa T, Ishibashi T. Biomed. Res. Tokyo. 2005;26:207–212. doi: 10.2220/biomedres.26.207. [DOI] [PubMed] [Google Scholar]
- 16.(a) DeGrado WF, Gratkowski H, Lear JD. Protein Sci. 2003;12:647–665. doi: 10.1110/ps.0236503. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bowie JU. Nature. 2005;438:581–589. doi: 10.1038/nature04395. [DOI] [PubMed] [Google Scholar]; (c) Slivka PF, Wong J, Caputo GA, Yin H. ACS Chem. Biol. 2008;3:402–411. doi: 10.1021/cb800049w. [DOI] [PubMed] [Google Scholar]
- 17.(a) Popot JL, Gerchman SE, Engelman DM. J. Mol. Biol. 1987;198:655–676. doi: 10.1016/0022-2836(87)90208-7. [DOI] [PubMed] [Google Scholar]; (b) Nannepaga SJ, Gawalapu R, Velasquez D, Renthal R. Biochemistry. 2004;43:550–559. doi: 10.1021/bi034875c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials, methods, stopped-flow kinetics at other peptide concentrations, and fitting parameters.