Abstract
Background
Identification of the genes underlying psychiatric illness remains a thorny problem. Previously, Quantitative Trait Loci (QTL) for anxiety-like behaviors and beta-carboline-induced seizure vulnerability have been mapped to the distal portion of mouse chromosome 10, using crosses of A/J and C57BL6 mice.
Methods
An interval specific congenic strain for this chromosomal 10 region facilitated the genetic dissection of novelty-induced exploratory behaviors.
Results
By microarray studies, an unsuspected E3 Ubiquitin Ligase, Ring Finger 41 (Rnf41) was differentially expressed in the region of interest, being upregulated in the hippocampi of B6 compared to A/J as well as congenic A.B6chr10 vs. A/J. By qRT-PCR, Rnf41 expression levels were significantly increased 1.5 and 1.3-fold in the hippocampi of C57BL6/J and A.B6chr10 mice compared to A/J mice, respectively. Protein levels of Rnf41 were increased in hippocampi of B6 mice compared to A/J mice across postnatal development with a 5.5-fold difference at P56. Yeast two hybrid studies searching for Rnf41 binding partners in fetal hippocampus identified several potential targets. An interaction between Rnf41 and NogoA was validated by GST-Rnf41 pulldown experiments. Re-analyzing a microarray database of human post-mortem prefrontal cortex (Brodmann’s Area 46/10), RNF41 mRNA expression levels were reduced significantly in patients with major depression and bipolar disorder compared to unaffected controls and confirmed by qRT-PCR.
Conclusion
Overall, Rnf41 is nominated as a candidate gene for anxiety-like behaviors, depression, and vulnerability to seizures. RNF41 and its binding partners suggest molecular pathways underlying behavior, highlighting a potential role for the ubiquitin proteasome system in psychiatric illness.
Keywords: Stress, epilepsy, genetics, mapping, ubiquitin proteasome system, mice
The genetic dissection of psychiatric illness and complex traits remains thorny due to genetic heterogeneity, diagnostic uncertainties, phenocopies, and the small relative magnitude of the underlying genes. From meta-analyses of genetic association studies (excluding HLA associations) and the Wellcome Trust Whole Genome association studies, an inherited risk allele confers modest increased genotypic relative risk (GRR) for illness compared to the alternative wild type (1–3), with some notable exceptions such as Apo E4 (14.9 OR) for Alzheimer’s. Specifically, in 168 larger association studies with more than 500 subjects, the median odds ratio for a given risk or vulnerability gene was 1.15 (2). A recent genome-wide association study for the Neuroticism personality trait studied the phenotypic extremes from a cohort of 88,142 people and failed to find any loci accounting for more than 1% of the variance(4). Overall, for complex human traits, these studies suggest odds ratios for genetic variants will be approximately 1.1–1.5. Hence, mouse models of psychiatric illness facilitate genetic dissection with the key advantages of high homology of mouse genes to human, a controlled environment, access to brain tissue at any stage of development, the ability to perform planned crosses, and the availability of sophisticated transgenic and knockdown techniques to explore function. Quantitative trait loci (QTL) analysis has provided an unbiased approach to understanding the genetic architecture of animal behaviors, demonstrating that most QTL explain less than 10% of the phenotypic variance(5–7). Although many QTLs for behaviors have been defined, the field has struggled to find the underlying quantitative trait genes for behaviors with some notable exceptions such as Rgs2(8) and Mpdz(9). The A/J strain is behaviorally inhibited in locomotor activity to the initial novelty of an brightly lit open field compared to the C57BL6 (B6) strain(10). This inhibited behavior in an open field can be viewed as an anxiety-like behavior(11). Likewise, the A/J strain makes fewer transitions than the B6 strain in the Light Dark Transition Test, an anxiety paradigm having pharmacologic validation with benzodiazepinenes(10). Finally, the A/J strain is more susceptible to a beta-carboline (a GABAA receptor inverse agonist) induced seizure than B6 mice(10). In this study, the polygenic traits of anxiety-like and seizure behaviors were simplified to a monogenic model, focusing on a previously mapped QTL in a distal region of chromosome 10 (12). This QTL from the A/J strain was genetically bred onto the C57BL6/J line, creating a congenic line differing from B6 just in this chr 10 region of interest, namely an interval specific congenic strain (A.B6chr10). This A.B6chr10 line provides a cleaner genetic background and less “noise” for gene expression profiling of brain tissue to develop a candidate gene underlying this QTL. Using a convergent functional genomic approach(13), one positional candidate gene emerged, namely an E3 Ubiquitin Ligase and was characterized.
Materials and methods
Animals, congenics, breeding
Male A/J (A) and C57BL/6J (B6) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Interval-specific congenic strain of Chromosome 10 (hereafter referred to as the A.B6chr10 congenic line) mice and the consomic B6.Ac10tel lines were generated as described previously (12) and in supplemental methods, respectively. All experiments followed the NIH Guide for the Care and Use of Laboratory Animals and received Institutional Animal Care and Use approval.
Behavioral testing
All mice were initially tested on the Open Field Test at 7–8wks of age and then tested in 1–2 subsequent behavioral tests. For microarray studies and protein levels, mice were sacrificed for brain tissue 1 week after open field testing without more stressful testing.
Open Field Test
The automated open field test (Digiscan, Accuscan, Columbus, OH) measured initial novelty locomotor responses, performing it as previously described(12, 14).
Seizure testing
Mice were tested for vulnerability to seizures induced by 1 mg/ml (5 mg/kg) β̃CCM (methyl -β̃ carboline -3-carboxylate; a GABAA receptor inverse agonist; Sigma-Aldrich, St. Louis, Mo.) i.p. at 9–12 weeks of age as previously described (15).
Light-Dark transition test
The light-dark test is a naturalistic conflict test, where an animal is initially placed in the bright side of a two compartment box. There is a conflict between the natural drive to explore vs. the avoidance of bright light. LD testing was performed in a single 10 minute session. The dependent variable is the number of transitions between the two chambers, being a measure of fear-like behavior. This version of the Light-Dark paradigm was selected because of the rigorous pharmacological validation of the paradigm (10, 16).
Other behavioral tests are described in the Supplemental methods in detail.
Brain tissue dissection and RNA preparation (see supplemental methods)
Microarray methods via the Affymetrix and Illumina platforms (see Supplemental Methods) qRT-PCR validation in mice Standard qRT-PCR was performed with SYBR green as previously described(17, 18) with primers and detailed methods described in supplemental methods.
Western blot analysis
For comparison of Rnf41 protein levels, standard methods of immunoblotting were used as described(18) with antibodies, chemiluminescent kits, and quantitative imaging methods detailed in supplemental methods.
Statistical analyses
Behavioral Studies
All experiments with normally distributed, continuous dependent measures were compared by one and two way ANOVAs and ANCOVAs with planned post-hoc comparisons. Gender differences were tested and when no differences were found, the groups were pooled. For non-normally distributed data, the data were transformed to normal distributions or analyzed with non-parametric statistics (Statview 5.1, SuperAnova, and JMP 5.1, SAS Institute).
Microarray analyses, qRT-PCR, and Re-analysis of Post-mortem human brain microarray data and validation samples (see Supplemental Methods)
Identification of proteins that interact with RNF41 by yeast two-hybrid (Y2H) screening
As bait, the C-terminal fragment (residues 173-317) of a Mus Rnf41 cDNA was subcloned into a Hybrigenics Yeast Two-Hybrid (Y2H) vector, checked, and transformed into yeast. Interactors were selected by screening an optimized Hybrigenics yeast two hybrid embryonic mouse brain random-primed cDNA library, testing 70 million interactions using Hybrigenics’ (Paris, France) established procedures ((19–21); see details in supplemental methods).
Glutathionine-S-Transferase (GST) pulldown experiments to validate interactions
The Mus Rnf41 c-terminal region (residues 193-317) was cloned into the vector pGEX-2T (GE Healthcare) upstream of GST, creating a RNF-C-terminus GST fusion as described(22). Pulldown with this GST fusion protein and controls were performed on P0.5 NMRI mouse brain lysate and extracts using standard methods as described (23)(see supplemental methods for details), immunoblotting to detect NogoA followed by the chemiluminescent methods as described in supplemental methods.
Results
Behavioral differences between congenic and consomic lines vs. controls
The rederived A.B6chr10 mice were studied and compared to littermate controls in a variety of behavioral paradigms to determine phenotypes that markedly differed, searching for the range of behaviors affected by this heterozygous, telomeric B6 region of interest (chr. 10 64cM to telomere; 13cM; 12.4 Mbp; estimated 233 genes) on the A/J background. The A/J strain is the strain more behaviorally inhibited to initial novelty and significantly more susceptible to seizures than the B6 strain as measured an established drug induced (β-CCM, a GABAA receptor inverse agonist; 5mg/kg, i.p.) seizure paradigm (10, 15). Using this β-CCM paradigm, A/J mice have a greater susceptibility to seizures and a shorter latency to seizure than B6 mice (i.e., A/J 248s latency vs. B6 510 s latency). The A.B6chr10 congenic showed a significantly reduced susceptibility to seizures (50% congenic; 80% for control littermate; Chi sq. = 13.9; p=.003) and a longer latency to β-carboline-induced seizure than littermate control (A.B6chr10 338 s vs. A control 180 s; U(164)=2290, p=.0005; Table 1). Also, parental B6 mice are much more active (less behavioral inhibition) in an open field than A/J mice(14) and show increased temperature rise after stress(24). The congenic A.B6chr10 mice showed significantly greater novelty-induced exploratory activity in an open field, (Table 1). The A.B6chr10 congenic mice showed a non-significant trend for increased hyperthermia in the Stress Induced Hyperthermia (SIH) task compared to littermate controls (F1,25= 3.0, p=0.095; Table 1) using a smaller sample size. However, no significant differences between A.B6chr10 and littermate controls were detected in the following tests: Passive Avoidance test ( U(26)=74, p=.59), Marble burying ( F1,22 = 1.95, p=.18), and a modified latency to eat a highly palatable food test in an unfamiliar cage(U(45)=121, p=.24).
Table 1.
Behavioral Phenotypes comparing A.B6chr10 congenic mice to littermate control A mice
| Trait | N | A mice Mean(± SD) | N | A.B6chr10 Mean(± SD) | Significance Test | p-value | Effect size Cohen d |
|---|---|---|---|---|---|---|---|
| β-CCM Seizure Latency (sec) | 86 | 180.3 (228.2) | 78 | 337.6 (267.5) | U(164)=2290 | 0.0005 | 0.64 |
| OF Vertical Movement 15min | 152 | 16.6 (16.1) | 129 | 31.1 (25.4) | F1,279=33.7 | <.0001 | 0.69 |
| OF Total Distance 15min | 152 | 672 (401) | 129 | 1044 (521) | F1,279=45.6 | <.0001 | 0.81 |
| Stress Induced Hyperthermia ΔT(°C) | 12 | 1.19 (.59) | 15 | 1.55 (.49) | F1,25= 3.0 | 0.095 | 0.67 |
| Passive Avoidance (sec; 2nd trial) | 13 | 255 (84) | 13 | 220 (120) | U(26)=74 | 0.59 | |
| Marble Burying (marbles buried) | 13 | 13.9 (4.3) | 12 | 15.8 (1.9) | F1,22=1.95 | 0.76 | |
| Latency to Eat (seconds) | 36 | 53.4 (109.0) | 9 | 113.8 (158.6) | U(45)=121, | 0.24 |
A QTL mapping study for anxiety-like traits (including the light-dark paradigm) phenotyped chromosome substitution (consomic) mouse lines and failed to map QTL to chromosome 10 (25), contradicting our findings. The breeding and selection process for this B6.Ac10 consomic strain lacked a telomeric marker in our region of interest. Hence, this B6.Ac10 consomic line’s telomeric region was genotyped with our telomeric markers and found to be heterozygous, namely still segregating for A and B genotypes. Through selective breeding for 5 generations, we derived a B6.Ac10tel mouse line, capturing 3.2 Mbp of the most telomeric region with A/A homozygosity. The B6 strain was significantly higher than this selectively bred B6.Ac10tel line for open field TD15 (F1,42=11.2, p= .0017) and Light-Dark transitions (F2,39=9.25, p=.005; using gender as a covariate), but not Vertical movements (F1,42=0.125, p=.72) (Table 2), consistent with an A/A QTL causing inhibition.
Table 2.
Behavioral Phenotypes comparing B6.Ac10tel consomic mice to B6 mice
| Strain | N | OF Vert. Mov. (15min) Mean (±SD) | OF Tot. Dist. (15min) Mean (± SD) | N (M; F) | Light-Dark Transitions Mean (± SD)a |
|---|---|---|---|---|---|
| B6.Ac10tel | 22 | 91.4 (33.7) | 1894 (751) | 13 (6M; 7F) | 23.8 (8.9) |
| C57Bl6/J | 22 | 94.9 (31.9) | 2612 (693) | 20 (12M; 8F) | 34.9 (8.9) |
| Significance Test | |||||
| F1,42=.125 | F1,42= 11.2 | F2,39= 9.2 | |||
| p-value | 0.73 | 0.0017 | 0.005 | ||
Means were sex adjusted
Gene expression studies for candidate genes of anxiety and β-CCM-induced seizure in the distal region of chromosome 10
Hippocampal tissue was targeted for gene expression studies based on prior theory of anxiety and the behavioral inhibition system (reviewed (26)). Gene expression profiling was performed on adult hippocampi (P56) and whole brains of P0.5 (first day postnatal) to identify candidate genes for these behaviors in the distal chromosome 10 region. Differentially expressed genes were defined using two comparisons, namely between 1) A/J mice vs. B6 mice and 2) A/J mice vs. A.B6chr10 congenic mice, yielding two hippocampal gene lists and two P0.5 gene lists (Suppl. Tables 1–2; Table 3–4). The intersection of these two hippocampal gene lists provided 11 differentially expressed genes as candidates (Suppl. Table 5) and 39 genes in common for the P0.5 whole brain comparisons (Suppl. Table 6). The RING (Really Interesting New Gene) finger E3 ubiquitin ligase gene, Rnf41, was the only candidate gene located in the distal chromosome 10 region of interest found repeatedly in both lists.
Validation of the differential expression of Rnf41 at the mRNA level and protein level
To further validate the differential expression of Rnf41, adult hippocampi of congenic mice and littermate controls as well as a developmental time course for the parental strains were examined using qRT-PCR with gene specific primers. Expression of Rnf41 was increased by 1.30-fold in hippocampi of the congenic line A.B6c10 compared with those of A/J (t(12) = 3.49, p = .004) (Fig. 1A). For the developmental time course, the following A/J and B6 tissues and ages were tested by qRT-PCR: whole brain at P0.5 and hippocampi at P21 and P56(Fig. 1B). Comparing B6 levels to A/J, Rnf41 levels were significantly increased by 1.18-fold in P0.5 whole brains (t(6) = 3.17, p = .019); and increased by 1.30-fold at P21 (t(9)=2.690, p=.023) and by 1.50-fold at P56 (t(8)=2.81, p=.023) in hippocampal tissue. Overall, these qRT-PCR results confirm both sets of microarray data showing higher expression of Rnf41 in the adult hippocampi in B6 mice and A.B6c10 congenic mice compared to A/J mice.
Figure 1.
Quantitative RT-PCR measurements of Rnf41 mRNA levels in A) adult hippocampi of A.B6chr10 congenic mice (line Q) and littermate controls (t(12) = 3.49, p = .004) and B) in postnatal hippocampal tissues of A/J vs B6 parental strains across development (P0.5, P21 and P56), validating microarray results. Expression levels of Rnf41 were normalized to cyclophilin (Ppia) levels. Hipp, Hippocampus. Data expressed as mean ± SEM; N = 4–10/group. * p<0.05.
Western blot analysis showed that the transcriptional changes in Rnf41 were present at the protein level(Fig. 2). Specifically, comparing B6 levels relative to A/J, Rnf41 protein levels were increased by 1.6-fold in the whole brain of P0.5 (t(15)= 2.02, p= .06), by 2.0-fold (t(15)= 3.18, p= 0.006) in P21 hippocampi, and by 5.5-fold (t(12)= 2.77, p= 0.017) in P56 hippocampi .
Figure 2.
Quantitative Western blotting analysis of Rnf41 protein levels in the hippocampus comparing A/J vs. B6 strains across three time points (P0.5, P21 and P56). Twenty micrograms of total protein per lane was used. Estimated amounts of Rnf41 protein were normalized with β-tubulin as the control band. Data expressed as mean ± SEM; N= 8 mice/group. *p<0.05 **p<0.01
Rnf41 expression levels correlate with Open Field behavior among Inbred Strains and LXS RI mice
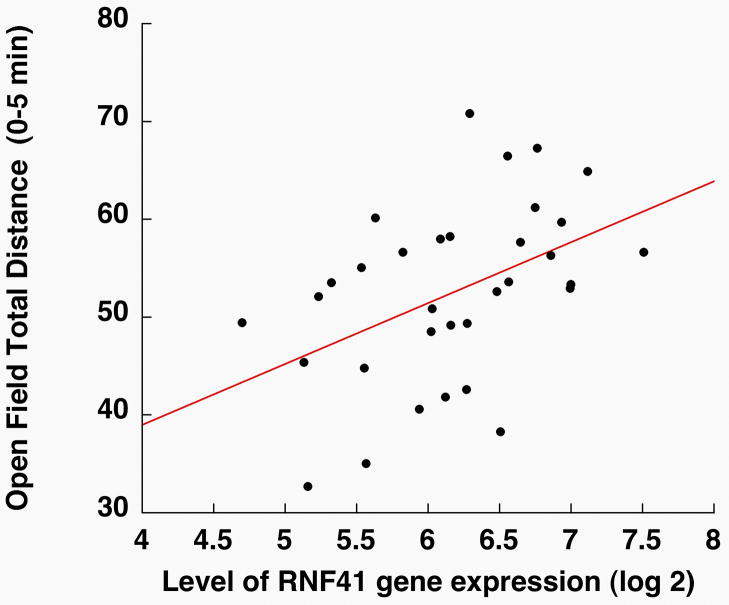
Initially, a limited sample of 6 inbred strains (where hippocampal Rnf41 mRNA levels were available as well as our own phenotypic Open Field data(27)) showed that hippocampal Rnf41 transcript levels correlated with Open Field Vertical Movements over 15 min (VM15; r= .810, P= .051; N=6). Using an independent mouse population with a larger sample size, the Long Sleep × Short Sleep Recombinant Inbred (LxS RI; N= 75 lines) panel was analyzed via the online GeneNetworks algorithms. This database contains hippocampal gene expression data estimated from Illumina microarrays on each line and numerous behavioral phenotypes, including Open Field locomotor activity(28, 29). In the LXS RI panel, hippocampal RNF41 gene expression levels varied over a 11.31-fold range, providing wide variation in levels. By a correlation analysis, this hippocampal RNF41 gene expression level significantly correlated with novelty-induced open field locomotor activity during first 5 min.(r= .454, p=.0073, N=33)(Fig. 3). Using Rnf41 mRNA levels as a trait, no QTL were mapped controlling the hippocampal expression levels of Rnf41. Neither genotyped SNPs nor haplotypes correlated with Rnf41 levels, but the genotyped SNPs were distant from Rnf41 (data not shown).
Figure 3.
Correlation of hippocampal Rnf41 gene expression levels with novelty-induced open field locomotor activity during the first 5 min epoch in LXS RI mice (r = 0.454, p= .0073; r2= .115; N=33 LXS lines). Rnf41 mRNA expression levels were estimated by re-analyzing the Illumina microarrays of hippocampal tissue of LXS RI mice in the GeneNetwork database (accessed 01/07). Each point represents the mean value.
Analysis of post-mortem human microarray data
From twin studies, the genetic vulnerability to anxiety and depressive disorders is thought to share a common set of genes (30). RNF41 provides a plausible candidate gene for stress vulnerability to illness. Using the Neuropathology Consortium Brain Collection of gene expression data from the SMRI, RNF41 expression levels were analyzed among the four available diagnostic groups, namely patients with Major Depression, Bipolar Disorder, Schizophrenia, and matched unaffected controls. These postmortem human frontal cortex samples (Brodmann’s Area 8/9 and 46/10) were examined for all confounding factors by multiple regression, finding brain pH and PMI as the only significant confounding factors. In a model adjusting for brain pH and PMI as covariates, the mean RNF41 expression levels of depression and bipolar disorder patients were significantly decreased compared to unaffected controls and schizophrenic patients (Fig. 4A, p<.001). These microarray findings were confirmed in two sets of independent samples by qRT-PCR, showing downregulation of RNF41 gene expression in depression and in bipolar disorder groups (fig. 4B and 4C).
Figure 4.
Decreased RNF41 gene expression levels in post-mortem human frontal cortex tissue (BA 8/9 and 46/10) in Bipolar Disorder (BD) and Major Depressive Disorder(MDD) compared to controls. A) RNF41 gene expression levels were estimated by reanalyzing an Affymetrix microarray database (Stanley Consortium database) and mean values were obtained after adjustment for brain pH and PMI as covariates among four patient populations, namely Bipolar Disorder (BD; N= 14), Major Depressive Disorder (MDD; N= 14), Schizophrenia (Schiz; N= 15) and normal controls (Control; N= 15). F(3,54) = 4.9, p= .004; Scheffe post-hoc comparison ***p<0.001. By qRT-PCR in independent samples, decreased RNF41 levels were validated as shown in B) MDD samples (N= 14) compared to control samples (N= 33); *p<0.05 and C) BD (N= 27) and Schiz (N= 28) samples compared to controls (N=20), *p<0.05. All means are expressed as standardized residuals ± SEM.
Yeast Two Hybrid Studies Identifying Rnf41 targets
Using a genome-wide screen for potential Rnf41 E3 ligase C-terminal domain (the E3 ubiquitin ligase ring finger domain that binds substrate) binders, six potential E3 ligase substrates were identified as statistically very high confidence interactors, namely ADP ribosylation factor-ike protein 6 interacting protein 4 (Arl6ip4), Jumonji domain containing histone demethylation protein 2B (Jmjd1b), Reticulon 4 (Rtn4; Human Nogo A; Neurite outgrowth inhibitor), Mouse 4932416N17Rik (RIKEN cDNA 4932416N17 gene), and Expressed seq C79267. Six additional proteins were identified as high confidence interactors, including mouse Smarce1 (Suppl. Table 7). Due to the unavailability or limitations of commercially available antibodies for these interactants, only the interaction between Rnf41 and Rtn4/NogoA could be further validated. The interaction of Rnf41 was confirmed using fetal brain extracts mixed with either immobilized recombinant GST-Rnf41 fusion protein or control GST protein. The protein pulled down by the GST-Rnf41 fusion protein and the GST control were eluted, analyzed by SDS-PAGE, and immunoblotted with NogoA antibody, detecting specific pulldown of Nogo A by the GST-Rnf41 fusion only (suppl. Fig 2).
Discussion
The most important finding of this study is the nomination and characterization of the E3 Ubiquitin ligase, Rnf41, as a candidate gene for vulnerability to fear-like behaviors, seizures, and depression. The evidence for this provisional candidate gene includes the following: localization to the region of genomic interest, differential gene expression at the protein and mRNA level (reduced levels in the brain tissues of A/J mice (behaviorally inhibited in novelty responses in the open field, fewer Light dark (LD) transitions/anxious, and greater seizure sensitivity to β-CCM) compared to those of B6 mice (less inhibited/more active, less anxious with more LD transitions, and more seizure resistant)), independent correlation of mRNA levels with Open Field behavior in the LxS panel of mice, and downregulation in RNF41 mRNA levels in frontal cortex of Major Depressive and Bipolar patients. Disappointingly, genomic sequencing data found no coding SNPs correlating with mRNA levels, suggesting that non-coding regulatory SNPs may regulate differential expression. Finally, other literature in the field is consistent with Rnf41 as a candidate gene. Specifically, in a large population of genetically heterogeneous stock mice (N= 2504), a whole genome association study (15K SNP) fine mapped QTL for open field behavior (7), mapping a highly significant QTL (log P = 9.27) on distal chromosome 10 containing a 1 Mbp interval harboring Rnf41 along with 39 other genes (see results online at http://gscan.well.ox.ac.uk/; and suppl. Figure 3).
The second most important finding of this study is the identification of potential Rnf41 substrates via a yeast two-hybrid screen. The Ubiquitin Proteasome System (UPS) has been recognized as regulator of intracellular protein degradation and an important pathway in human disease (reviewed (31). Rnf41, a Ring Finger E3 Ubiquitin Ligase, provides substrate specificity in the UPS, covalently attaching Ubiquitin to a select set of targeted proteins for degradation. The UPS system is built hierarchically with a few E1’s, several E2’s, and almost a thousand E3’s (reviewed (32)). Briefly, Rnf41 is highly conserved protein across evolution being found in flies and humans. In mouse and humans, RNF41 is a 35.8kd protein composed of 317 amino acids with 100% identity. The human RNF41 gene is located on 12q13.13 and has a more complicated splicing pattern with multiple isoforms (~10) due to alternative splicing, including a naturally occurring dominant negative isoforms (transcript variant-2 Genebank NM_194358; (33)). Recently, TRIM23 and other ubiquitin cycle related genes have been identified as down-regulated in bipolar patients (34). The C-terminal portion of the RING finger E3 ligases (Rnf41; aa 179–317) selectively binds the substrate for ubiquitination (22, 35–37). Surprisingly, none of the previously established Rnf41 interacting molecules (i.e., ErbB3-ErbB4 (36, 37), BRUCE/appollon (38, 39), Parkin (40), USP8 (41), and Mark4 (42)) were found in our screen, emphasizing perhaps the role of a specific cell’s microenvironment. Rnf41 binding to Rtn4/Nogo-A was confirmed by pulldown experiments. Reticulon 4 (Rtn4; also named Human Nogo A) is a neurite outgrowth inhibitor, expressed in oligodendrocytes, functioning as an inhibitor of neurite outgrowth in the CNS, but not present in PNS(reviewed (43)). Rnf41’s ubiquitination of Rtn4 may control its trafficking and/or Rtn4’s protein level, leading to functional regulation of neurite outgrowth. If true, then we hypothesize that lentiviral overexpression of an altered Rnf41 with enhanced specificity to Rtn4 via a directed evolution approach might be an alternative strategy for enhancing neurite outgrowth after injury(44). Two other intriguing potential interactors act as regulators of gene expression and chromatin structure, namely Jmjd1b (Jumonji domain containing histone demethylation protein 2B, which demethylates Lys-9 of Histone H3 and derepresses the silencing of H3K9 methylation in chromatin structure)(45) and Mouse Smarce1/BAF57 (a SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1)(46, 47). Future work will address which, if any, of these Rnf41 substrates are salient in explaining behavioral and CNS effects. Nonetheless, a conceptual framework emerges whereby inherited differences in the level of an E3 ubiquitin ligase could lead to consequential effects. Specifically, we speculate that low levels of Rnf41 would be a risk factor for some inhibitory or anxiety-like traits and for vulnerability to stress.
This study has a number of limitations and cautions. First, the nomination of Rnf41 as a candidate gene underlying a QTL is suggestive and correlational, rather than proven and definitive(48). Independent replication is required for the genetic association of Rnf41 with behavioral phenotypes. Second, the list of potential interacting molecules probably contains false positives. Third, additional neighboring genes besides Rnf41 may underlie the QTL in our region of interest as QTLs often break into several pieces. Fourth, we cannot formally rule out the following possibilities: other candidate genes undetected by our approach, residual passenger genes in the congenic strains, epigenetic effects, copy number variations, or the three microRNAs in our region of interest.
In summary, an unbiased genetic approach in a mouse model has identified an unsuspected E3 Ubiquitin ligase, Rnf41, as a candidate gene for fear-like behavior, seizures, and depressive illness. Rnf41 and its potential targeted substrates, as revealed from a yeast two-hybrid screen (such as Rtn4/Nogo A), provide testable pathways for functional analysis and future work on dissecting Rnf41’s function in the brain and its role in psychiatric illness.
Supplementary Material
Acknowledgments
This work was generously supported in part by NIH (RO1 MH067211) and NARSAD Independent Investigators Award (HG). We are grateful to our UTSW colleagues for helpful conversations. The post-mortem brain collection was developed over many years through the generous permission of the families and the long-term support of the SMRI Brain Collection. We thank the SMRI Brain Collection and its Neuropathology Consortium for making its microarray database publicly available. In particular, we acknowledge The Stanley Medical Research Institute’s Investigators, Drs. E. Fuller Torrey, Robert H. Yolken, Michael B. Knable, and Maree J. Webster along with their many collaborators, who made this work possible. We acknowledge Drs. B. Bennett, J. DeFries, T. Johnson, and colleagues (Univ. of Colorado) for developing and phenotyping the LxS RI strains along with Dr. M. Cook for phenotyping them. We appreciate the work and infrastructure of the INIA program and the GeneNetwork database (supported by NIH) allowing skillful exploration of contributed datasets. We appreciated the help and advice from Drs. T. Wheeler, Xiao-Bo Qiu ( gift of a Rnf41 cDNA clone), and K. Carraway. We thank the following groups for their skillful technical work: the UTSW microarray core for the gene expression profiling, Polymorphic DNA Technologies Inc for SNP genotyping and DNA sequencing, and Hybrigenics for the yeast two hybrid screening.
Footnotes
Financial Disclosures: None of the authors has biomedical financial interest or potential conflict of interests relevant to this work.
Authors’ contributions: Conceived and designed the experiments: HG, SK, and SZ. Performed the experiments: SK SZ RR CD KC AFB HG. Analyzed the data: SK SZ HG. Contributed reagents/materials/analysis tools: KC. Wrote the paper: SK HG. All authors read and approved of the final version of manuscript.
Shared Mouse Models: The A.B6c10 congenic and refined B6.Ac10tel consomic mouse lines have been cryopreserved at the Jackson Labs and are available upon request. The microarray data in this study has been submitted to the Gene Expression Omnibus (GEO) database and assigned the series Accession number: GSE8641 available at the URL: http://www.ncbi.nlm.nih.gov/geo/index.cgi
Databases cited:
Allen Brain Bank http://www.brain-map.org/viewGeneInfo.do?geneId=43431
GeneCard http://www.genecards.org/
Gene Expression Omnibus (GEO) database http://www.ncbi.nlm.nih.gov/geo/index.cgi
GeneNetwork online database http://www.genenetwork.org/
Genepaint database http://www.genepaint.org/
GenomeScan in HS Mice http://gscan.well.ox.ac.uk/
GNF SymAtlas http://symatlas.gnf.org/SymAtlas/
NIH Image J software http://rsb.info.nih.gov/ij/
Stanley Med. Res. Inst. Database (SMRIDB) http://www.stanleygenomics.org/
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 2.Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol. 2006;164:609–614. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 3.Wellcome-Trust-Case-Control-Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shifman S, Bhomra A, Smiley S, Wray NR, James MR, Martin NG, et al. A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry. 2008;13:302–312. doi: 10.1038/sj.mp.4002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 6.Flint J. Analysis of quantitative trait loci that influence animal behavior. J Neurobiol. 2003;54:46–77. doi: 10.1002/neu.10161. [DOI] [PubMed] [Google Scholar]
- 7.Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 8.Yalcin B, Willis-Owen SA, Fullerton J, Meesaq A, Deacon RM, Rawlins JN, et al. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet. 2004;36:1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- 9.Shirley RL, Walter NA, Reilly MT, Fehr C, Buck KJ. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat Neurosci. 2004;7:699–700. doi: 10.1038/nn1271. [DOI] [PubMed] [Google Scholar]
- 10.Mathis C, Paul SM, Crawley JN. Characterization of benzodiazepine-sensitive behaviors in the A/J and C57BL/6J inbred strains of mice. Behavior Genetics. 1994;24:171–180. doi: 10.1007/BF01067821. [DOI] [PubMed] [Google Scholar]
- 11.Prut L, Belzung C. The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: a review. Eur J Pharmacol. 2003;463:3–33. doi: 10.1016/s0014-2999(03)01272-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S, Lou Y, Amstein TM, Anyango M, Mohibullah N, Osoti A, et al. Fine mapping of a major locus on chromosome 10 for exploratory and fear-like behavior in mice. Mamm Genome. 2005;16:306–318. doi: 10.1007/s00335-004-2427-8. [DOI] [PubMed] [Google Scholar]
- 13.Le-Niculescu H, McFarland MJ, Mamidipalli S, Ogden CA, Kuczenski R, Kurian SM, et al. Convergent Functional Genomics of bipolar disorder: from animal model pharmacogenomics to human genetics and biomarkers. Neurosci Biobehav Rev. 2007;31:897–903. doi: 10.1016/j.neubiorev.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gershenfeld HK, Neumann PE, Mathis C, Crawley JN, Li X, Paul SM. Mapping quantitative trait loci for open-field behavior in mice. Behav Genet. 1997;27:201–210. doi: 10.1023/a:1025653812535. [DOI] [PubMed] [Google Scholar]
- 15.Gershenfeld HK, Neumann PE, Li X, St Jean PL, Paul SM. Mapping quantitative trait loci for seizure response to a GABAA receptor inverse agonist in mice. J Neurosci. 1999;19:3731–3738. doi: 10.1523/JNEUROSCI.19-10-03731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawley JN. Neuropharmacologic specificity of a simple animal model for the behavioral actions of benzodiazepines. Pharmacology, Biochemistry & Behavior. 1981;15:695–699. doi: 10.1016/0091-3057(81)90007-1. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Choi KH, Baykiz AF, Gershenfeld HK. Suicide candidate genes associated with bipolar disorder and schizophrenia: an exploratory gene expression profiling analysis of post-mortem prefrontal cortex. BMC Genomics. 2007;8:413. doi: 10.1186/1471-2164-8-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Amstein T, Shen J, Brush FR, Gershenfeld HK. Molecular correlates of emotional learning using genetically selected rat lines. Genes Brain Behav. 2005;4:99–109. doi: 10.1111/j.1601-183X.2004.00099.x. [DOI] [PubMed] [Google Scholar]
- 19.Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 21.Wojcik J, Boneca IG, Legrain P. Prediction, assessment and validation of protein interaction maps in bacteria. J Mol Biol. 2002;323:763–770. doi: 10.1016/s0022-2836(02)01009-4. [DOI] [PubMed] [Google Scholar]
- 22.Bouyain S, Leahy DJ. Structure-based mutagenesis of the substrate-recognition domain of Nrdp1/FLRF identifies the binding site for the receptor tyrosine kinase ErbB3. Protein Sci. 2007;16:654–661. doi: 10.1110/ps.062700307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vikis HG, Guan KL. Glutathione-S-transferase-fusion based assays for studying protein-protein interactions. Methods Mol Biol. 2004;261:175–186. doi: 10.1385/1-59259-762-9:175. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Peprah D, Gershenfeld HK. Tail-suspension induced hyperthermia: a new measure of stress reactivity. J Psychiatr Res. 2003;37:249–259. doi: 10.1016/s0022-3956(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 25.Singer JB, Hill AE, Nadeau JH, Lander ES. Mapping quantitative trait loci for anxiety in chromosome substitution strains of mice. Genetics. 2005;169:855–862. doi: 10.1534/genetics.104.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray JA, McNaughton N. The neuropsychology of anxiety: reprise. Nebr Symp Motiv. 1996;43:61–134. [PubMed] [Google Scholar]
- 27.Liu X, Gershenfeld HK. An exploratory factor analysis of the Tail Suspension Test in 12 inbred strains of mice and an F2 intercross. Brain Res Bull. 2003;60:223–231. doi: 10.1016/s0361-9230(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 28.Williams RW, Bennett B, Lu L, Gu J, DeFries JC, Carosone-Link PJ, et al. Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mamm Genome. 2004;15:637–647. doi: 10.1007/s00335-004-2380-6. [DOI] [PubMed] [Google Scholar]
- 29.Downing C, Carosone-Link P, Bennett B, Johnson T. QTL mapping for low-dose ethanol activation in the LXS recombinant inbred strains. Alcohol Clin Exp Res. 2006;30:1111–1120. doi: 10.1111/j.1530-0277.2006.00137.x. [DOI] [PubMed] [Google Scholar]
- 30.Kendler KS, Neale MC, Kessler RC, Heath AC, Eaves LJ. Major depression and generalized anxiety disorder. Same genes, (partly) different environments? Arch Gen Psychiatry. 1992;49:716–722. doi: 10.1001/archpsyc.1992.01820090044008. [DOI] [PubMed] [Google Scholar]
- 31.Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med. 2006;145:676–684. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- 32.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 33.Yen L, Cao Z, Wu X, Ingalla ER, Baron C, Young LJ, et al. Loss of Nrdp1 enhances ErbB2/ErbB3-dependent breast tumor cell growth. Cancer Res. 2006;66:11279–11286. doi: 10.1158/0008-5472.CAN-06-2319. [DOI] [PubMed] [Google Scholar]
- 34.Ryan MM, Lockstone HE, Huffaker SJ, Wayland MT, Webster MJ, Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol Psychiatry. 2006;11:965–978. doi: 10.1038/sj.mp.4001875. [DOI] [PubMed] [Google Scholar]
- 35.Avvakumov GV, Walker JR, Xue S, Finerty PJ, Jr, Mackenzie F, Newman EM, et al. Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8) J Biol Chem. 2006;281:38061–38070. doi: 10.1074/jbc.M606704200. [DOI] [PubMed] [Google Scholar]
- 36.Diamonti AJ, Guy PM, Ivanof C, Wong K, Sweeney C, Carraway KL., 3rd An RBCC protein implicated in maintenance of steady-state neuregulin receptor levels. Proc Natl Acad Sci U S A. 2002;99:2866–2871. doi: 10.1073/pnas.052709799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu XB, Goldberg AL. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc Natl Acad Sci U S A. 2002;99:14843–14848. doi: 10.1073/pnas.232580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lotz K, Pyrowolakis G, Jentsch S. BRUCE, a giant E2/E3 ubiquitin ligase and inhibitor of apoptosis protein of the trans-Golgi network, is required for normal placenta development and mouse survival. Mol Cell Biol. 2004;24:9339–9350. doi: 10.1128/MCB.24.21.9339-9350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu XB, Markant SL, Yuan J, Goldberg AL. Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. Embo J. 2004;23:800–810. doi: 10.1038/sj.emboj.7600075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong L, Tan Y, Zhou A, Yu Q, Zhou J. RING finger ubiquitin-protein isopeptide ligase Nrdp1/FLRF regulates parkin stability and activity. J Biol Chem. 2005;280:9425–9430. doi: 10.1074/jbc.M408955200. [DOI] [PubMed] [Google Scholar]
- 41.Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol Cell Biol. 2004;24:7748–7757. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brajenovic M, Joberty G, Kuster B, Bouwmeester T, Drewes G. Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J Biol Chem. 2004;279:12804–12811. doi: 10.1074/jbc.M312171200. [DOI] [PubMed] [Google Scholar]
- 43.Schwab ME. Nogo and axon regeneration. Curr Opin Neurobiol. 2004;14:118–124. doi: 10.1016/j.conb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Buchli AD, Rouiller E, Mueller R, Dietz V, Schwab ME. Repair of the injured spinal cord. A joint approach of basic and clinical research. Neurodegener Dis. 2007;4:51–56. doi: 10.1159/000100359. [DOI] [PubMed] [Google Scholar]
- 45.Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- 46.Battaglioli E, Andres ME, Rose DW, Chenoweth JG, Rosenfeld MG, Anderson ME, et al. REST repression of neuronal genes requires components of the hSWI.SNF complex. J Biol Chem. 2002;277:41038–41045. doi: 10.1074/jbc.M205691200. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Archer TK. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol Cell Biol. 2005;25:9016–9027. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, et al. The nature and identification of quantitative trait loci: a community’s view. Nat Rev Genet. 2003;4:911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.