Abstract
An extensive protein–protein interaction network has been identified between proteins implicated in inherited ataxias. The protein sacsin, which is mutated in the early-onset neurodegenerative disease autosomal recessive spastic ataxia of Charlevoix-Saguenay, is a node in this interactome. Here, we have established the neuronal expression of sacsin and functionally characterized domains of the 4579 amino acid protein. Sacsin is most highly expressed in large neurons, particularly within brain motor systems, including cerebellar Purkinje cells. Its subcellular localization in SH-SY5Y neuroblastoma cells was predominantly cytoplasmic with a mitochondrial component. We identified a putative ubiquitin-like (UbL) domain at the N-terminus of sacsin and demonstrated an interaction with the proteasome. Furthermore, sacsin contains a predicted J-domain, the defining feature of DnaJ/Hsp40 proteins. Using a bacterial complementation assay, the sacsin J-domain was demonstrated to be functional. The presence of both UbL and J-domains in sacsin suggests that it may integrate the ubiquitin–proteasome system and Hsp70 function to a specific cellular role. The Hsp70 chaperone machinery is an important component of the cellular response towards aggregation prone mutant proteins that are associated with neurodegenerative diseases. We therefore investigated the effects of siRNA-mediated sacsin knockdown on polyglutamine-expanded ataxin-1. Importantly, SACS siRNA did not affect cell viability with GFP-ataxin-1[30Q], but enhanced the toxicity of GFP-ataxin-1[82Q], suggesting that sacsin is protective against mutant ataxin-1. Thus, sacsin is an ataxia protein and a regulator of the Hsp70 chaperone machinery that is implicated in the processing of other ataxia-linked proteins.
INTRODUCTION
Causative genes have been identified for at least 45 inherited ataxias. These neurodegenerative disorders include autosomal dominant ataxias, such as the polyglutamine-mediated spinocerebellar ataxias, and the recessive diseases Friedreich's ataxia and autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) (1).
Many of the proteins involved in inherited ataxias share common interacting partners indicating that they are functionally linked (2). Furthermore, although the clinical features of these conditions are variable there is substantial evidence for overlapping pathways of molecular pathogenesis in some forms of ataxia. These include perturbations in normal protein homeostasis that invoke cellular responses from both molecular chaperone and ubiquitin–proteasome systems (UPS).
Hsp70 molecular chaperone networks are associated with neurodegenerative diseases where protein conformational changes and subsequent aberrant protein folding are a feature (3). The pathogenesis of these conditions usually includes the formation of deposits of misfolded ubiquitylated protein with which Hsp70 and Hsp40/DnaJ proteins are frequently associated. Hsp70 and its co-chaperones, particularly Hsp40 proteins, have also been recognized as potent modulators of protein aggregation and survival in cellular and animal models of neurodegenerative disease (3).
The Hsp70 molecular chaperone machine also functions in targeting proteins to the UPS for degradation. For example, the co-chaperone CHIP (C-terminus of Hsc70-interacting protein), which regulates Hsp70 chaperone activity and acts as an ubiquitin ligase for Hsp70 clients (4), has been shown to interact with ataxia proteins such as ataxin-1, thereby modulating their solubility and degradation.
ARSACS (OMIM: 270550) is characterized by early-onset spastic ataxia. Pathological features include atrophy of the upper cerebellar vermis and absence of Purkinje cells (5). Mutations in the SACS gene were identified as causing ARSACS in a Canadian population (6). Subsequent genetic studies indicate that this form of spastic ataxia may be far more common than originally presumed and have a global distribution (7). Initially, the SACS gene was reported to consist of one 12 794 bp exon (6), in which homozygous missense, nonsense and frameshift mutations have been found. Recently, further mutations have been reported in newly identified 5′ exons (7,8).
The SACS gene is predicted to encode a large multidomain protein, sacsin, which has regions of sequence similarity to several proteins. The strongest similarity identified was between a region near the C-terminus of sacsin and the J-domain of Hsp40 proteins (approximately 60% identity over 30 residues compared with Hsp40/Hdj1). The highly conserved J-domain is the defining feature of the Hsp40 family of Hsp70 co-chaperones (9). Therefore, sacsin may represent a direct link between ataxia and the Hsp70 machinery.
This study represents the first characterization of sacsin. We have defined the localization of the sacsin protein in brain. The data show that sacsin has nine coding exons. We identified a ubiquitin-like (UbL) domain at the N-terminus of sacsin that can interact with the proteasome. Furthermore, we used an in vivo assay to demonstrate that the J-domain of sacsin is functional. Finally, sacsin knockdown resulted in a reduction in cells expressing polyglutamine-expanded ataxin-1, which correlated with a loss of cells with large nuclear ataxin-1 inclusions. Together these data suggest that the large multi-domain sacsin protein is able to recruit Hsp70 chaperone action and has the potential to regulate the effects of other ataxia proteins.
RESULTS
Sacsin is expressed in the cell bodies and processes of neurons
The human SACS gene was initially reported to contain a single massive exon encoding a 11.5 kb open reading frame (6). Subsequently eight additional 5′ putative coding exons have been annotated. We confirmed that the human gene transcribed a 9 exon transcript, which is predicted to code a 4579 amino acid protein (NM_014363.4), by reverse transcriptase-polymerase chain reaction (RT–PCR) analysis of human brain mRNA (Supplementary Material, Fig. S1). In mouse, a sacsin isoform of 4579 amino acids, with a molecular weight of 520 kDa, is also predicted (UniProt accession number: Q9JC8).
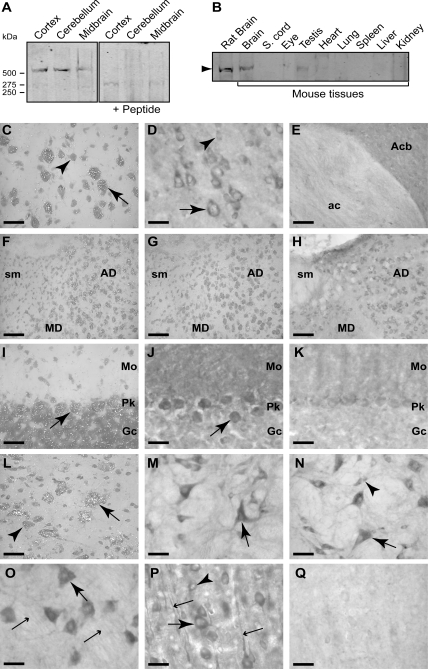
To analyse the expression and localization of sacsin we developed an antiserum (r-sacsin1–17) to the N-terminus of sacsin and an antiserum (r-sacsin4489–4503) to a peptide sequence, which is located near the C-terminus. Western analysis of mouse and rat tissues with r-sacsin4489–4503 detected a specific band of approximately 520 kDa in brain which was competed with immunizing peptide (Fig. 1A and B). r-Sacsin1–17 was unsuitable for western blotting (not shown).
Figure 1.
Sacsin is predominantly expressed in the cell bodies and processes of neurons. Western analysis of mouse and rat tissues was performed using an affinity-purified polyclonal antiserum (r-sacsin4489–4503). (A) r-Sacsin4489–4503 detected a band of >500 kDa in neuronal tissues. The band was not detected when r-sacsin4489–4503 was pre-incubated with immunizing peptide. (B) Sacsin was detected in brain tissue and in testis. (C–Q) In situ hybridization with emulsion autoradiography for SACS mRNA and immunohistochemistry for sacsin protein was performed in rat brain. Sacsin was detected with r-sacsin4489–4503 in D–E, H, J, K, M–O, and with r-sacsin1–17 in P and Q. Layer 5 of the cerebral cortex labelled for SACS mRNA (C) and protein immunoreactivity (D). Larger pyramidal neurons (arrows) had higher expression of both SACS mRNA and sacsin protein than smaller neuronal profiles (arrowhead). (E) Immunohistochemistry showing axonal labelling in the anterior commissure (ac) and heavy neuropil labelling in the nucleus accumbens (Acb). (F) SACS mRNA signal was higher in the anterodorsal thalamus (AD) than the mediodorsal thalamus (MD). The stria medullaris (sm) had no detectable SACS mRNA. (G) Competition control resulted in the loss of specific signal. (H) Immunohistochemistry of the same region showed an identical labelling pattern. (I) Cerebellum in situ hybridization showed high levels of signal over Purkinje cells (Pk), with cells of the granule cell layer (Gc) expressing moderate levels. (J) Immunoreactivity for sacsin was high in Purkinje cells and lower in granule cells. The molecular layer (Mo) had dense immunoreactivity. (L) The magnocellular preoptic nucleus neurons had relatively high levels of autoradiographic signal (arrows) with smaller neurons considerably less labelled (arrowhead). (M) Large neurons (arrow) of the gigantocellular reticular nucleus had high levels of immunoreactivity. Primary dendrites and surrounding fibrous neuropil also labelled. (N) In the medial vestibular nucleus, both large and small neurons and the surrounding fibrous neuropil prominently stained. (O) Large motorneurons in the motor trigeminal nucleus (large arrow) had intense immunoreactivity. Fine, labelled axons (small arrows) in the adjacent motor root of the trigeminal nerve were observed. (P) In layer 5 of the cortex, the N-terminal sacsin antibody stained pyramidal neurons intensely as did the C-terminal antibody (D) but labelled dendritic processes as well. (K and Q) In peptide absorption controls of analogous sections in J and P, respectively, immunoreactivity was significantly reduced. Scale bar = 40 µm in (C and D) and (I–Q); 90 µm in (E–H).
We performed immunohistochemistry and in situ hybridization on rat brain sections using r-sacsin4489–4503 and SACS probes. In Supplementary Material, Figure S2, film autoradiograms of in situ hybridization signal are presented to show the overall distribution of expression. There was a strong correlation between the localization of immunoreactivity and SACS mRNA at the cellular level (Fig. 1). Expression appeared predominantly, if not exclusively neuronal, with sacsin protein localized to the cytoplasm of neuronal cell bodies and processes including both dendrites and axons, the latter as evidenced by axonal tract labelling. Examination of sections with dual-label immunofluorescence histochemistry for r-sacsin4489–4503 and NeuN, a neuron-specific marker that labels most neuronal cell populations (10), confirmed that all NeuN-positive neurons expressed detectable levels of sacsin protein (data not shown), therefore, there were no clear examples of sacsin-negative neurons.
In the forebrain, cerebral cortex expression of SACS mRNA and sacsin protein was abundant with large layer 5 corticospinal neurons expressing the highest levels (Fig. 1C and D). Neuropil labelling for sacsin immunoreactivity was prominent in the striatum and fibre tracts, such as the anterior commissure, exhibited fine labelling (Fig. 1E). In the thalamus expression levels for both mRNA and protein varied between nuclei with the anterodorsal nucleus showing highest expression (Fig. 1F–H).
In the cerebellum, Purkinje cells had the highest levels of expression of both SACS mRNA and sacsin protein. The molecular layer with Purkinje cell dendrites and contacting afferents was clearly stained for sacsin protein (Fig. 1I–K).
Between the forebrain and brainstem, an almost continuous band of large neurons expressing high levels of SACS mRNA and sacsin protein extended from the basal forebrain magnocellular system through the zona incerta and pontine and medullary reticular formation (Fig. 1L and M). Neurons of precerebellar nuclei including the red, pontine, vestibular and lateral reticular nuclei as well as cranial motor nuclei had relatively high levels of expression (Fig. 1M–O). Brainstem sensory nuclei including the nucleus of the trapezoid body, cochlear nuclei and sensory trigeminal nerve nuclei had moderate expression. Large neurons of the mesencephalic nucleus of the trigeminal nerve expressed very high levels of the mRNA (Supplementary Material, Fig. S2).
The N-terminal antiserum r-sacsin1–17 in immunohistochemistry produced a very similar staining pattern to that observed with r-sacsin4489–4503, although the staining of dendritic processes was more intense using r-sacsin1–17 as shown in layer 5 cerebral cortex (Fig. 1P).
Controls for specificity of in situ hybridization (Fig. 1G) and immunohistochemistry (Fig. 1K and Q), using excess oligonucleotide or immunizing peptides respectively, eliminated or dramatically reduced labelling.
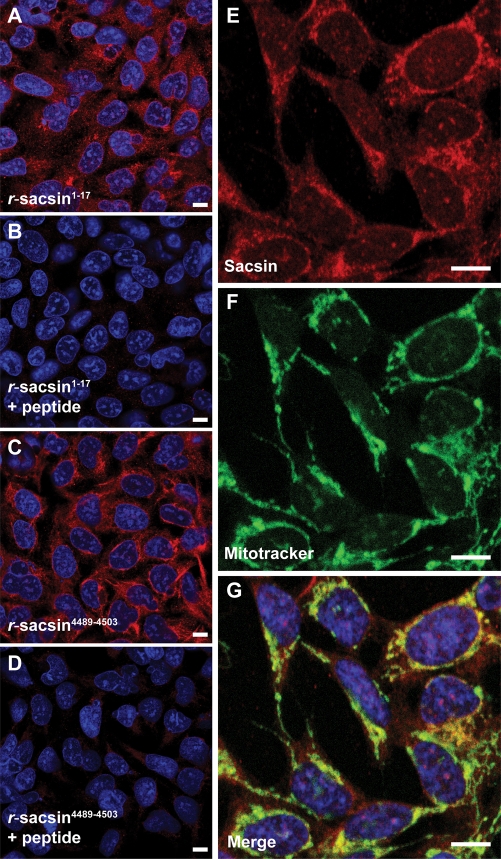
Sacsin has a predominantly cytoplasmic distribution with a mitochondrial component
SACS is expressed in the human neuroblastoma derived cell line SH-SY5Y (Supplementary Material, Fig. S5). We examined the subcellular localization of endogenous sacsin in SH-SY5Y cells by staining with r-sacsin1–17 and r-sacsin4489–4503. Both N- and C-terminal sacsin antibodies produced the same staining pattern (Fig. 2A–D). The protein had a predominantly cytoplasmic localization with some punctate staining also visible. A small proportion of sacsin immunoreactivity appeared nuclear (Supplementary Material, Fig. S3). Interestingly, bioinformatics analysis revealed that sacsin contains multiple putative nuclear localization signals (Supplementary Material, Table S1). To further investigate the subcellular distribution of sacsin we tested for co-localization between sacsin and specific organelle markers. Sacsin immunoreactivity partially overlapped with the distribution of MitoTracker (Fig. 2E–G) a mitochondrial fluorescent probe, and the mitochondrial protein Hsp60 (Supplementary Material, Fig. S3). The overlap between sacsin localization and MitoTracker was quantified using MetaMorph 7.5 image analysis software. About 30% ± 3% of sacsin staining showed co-localization with MitoTracker fluorescence. Significantly, 40% ± 2% of MitoTracker staining did not overlap with sacsin immunoreactivity (similar values were obtained for the overlap between sacsin and Hsp60 immunoreactivity). Analyses of the sacsin protein sequence did not reveal the presence of a mitochondrial leader sequence. Together, these observations suggest that a proportion of sacsin localizes at or near the cytoplasmic face of the mitochondria.
Figure 2.
Sacsin has a predominantly cytoplasmic localization with partial mitochondrial overlap. Immunofluorescent detection of sacsin in SH-SY5Y cells using r-sacsin1–17 (A, B) and r-sacsin4489–4503 (C, D). The antibodies were competed with immunizing peptide in (B and D). (E–G) Sacsin distribution (red) partially overlapped with a fluorescent mitochondria marker (green). Cells were imaged by laser scanning confocal microscopy. Microscope settings were constant for acquisition in the peptide competition experiments. Scale bar = 10 µm.
The N-terminus of sacsin contains an ubiquitin-like domain
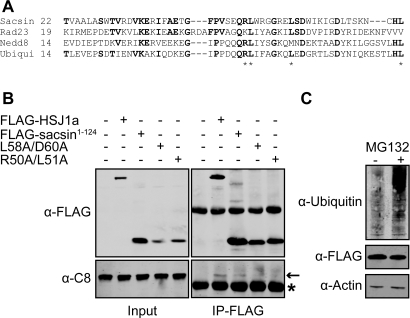
Bioinformatic analyses of the predicted amino acid sequence of the full-length nine exon-encoded sacsin protein revealed a previously unreported, putative UbL domain at the N-terminus (Fig. 3A). UbL domains are present in ubiquitin-like proteins (such as NEDD8 and SUMO-1) and in a variety of other multidomain proteins of diverse biological function (11). Many of these proteins have been shown to bind components of the proteasome via their UbL domain (11). These include the DNA repair protein Rad23 (12), which has an N-terminal UbL domain with 43% identity over 65 residues to the putative sacsin UbL. Within the UbL domain, a potential consensus sequence for proteasome interaction has been identified (13). The sacsin UbL domain contains part of this motif (Fig. 3A). Therefore, we tested the interaction between the sacsin UbL domain and the proteasome. The N-terminus of sacsin (residues 1–124), including the putative UbL domain, was expressed in CHO-1 cells as a chimeric protein with a FLAG epitope tag at its C-terminus. The sacsin-UbL-FLAG was immunoprecipitated and western blot analysis performed (Fig. 3B). The proteasome-associated chaperone HSJ1, which functions as a neuronal shuttling factor for the sorting of chaperone clients to the proteasome (14), was used as a positive control. Importantly, the 20S proteasomal alpha subunit C8 was co-immunoprecipitated with the sacsin UbL domain.
Figure 3.
The N-terminus of sacsin contains a ubiquitin-like (UbL) domain and interacts with the proteasome. (A) Alignment of sacsin's putative UbL domain with human UbL domain proteins and ubiquitin (Ubiqui). Residues conserved between sacsin and the UbL proteins and/or ubiquitin are shown in bold. Sacsin residues that match key hydrophobic amino acids which may play a role in proteasomal targeting are indicated by asterisks. (B) The proteasomal subunit C-8 co-precipitated with the N-terminus of sacsin, containing the UbL domain. Lysates from CHO-1 cells transfected with wild-type sacsin1–124-FLAG, or sacsin1–124-FLAG containing double mutations predicted to inhibit proteasome interaction, L58A/D60A and R50A/L51A, were precipitated with anti-FLAG M2-conjugated agarose beads. HSJ1a-FLAG was used as a positive control for proteasome co-precipitation. Western blot analysis for FLAG-tagged proteins and the C8 subunit were performed. Asterisk highlights immunoglobulin chains. (C) To control for the possibility that the sacsin1–124-FLAG may interact with the proteasome through being targeted for degradation, we investigated the effect of MG132 inhibition on cellular levels of the chimeric protein. Western blot analysis was used to compare cellular levels of sacsin1–124-FLAG in cells treated with MG132 and control cells. Proteasome inhibition was confirmed by blotting cell lysates for ubiquitylated proteins. Protein loading was monitored by blotting for actin.
To confirm that this interaction was dependent on the UbL domain, we mutated conserved UbL protein residues to alanine. The sacsin-UbL domain double mutants L58/D60A and R50A/L51A showed reduced co-immunoprecipitation with C8 (Fig. 3B). It was possible that the proteasome interaction may have been because of the N-terminus of sacsin being ubiquitylated and targeted for degradation. To control for this we compared levels of the sacsin-UbL-FLAG in CHO-1 cells where the proteasome had been inhibited by MG132 and untreated cells (Fig. 3C). There was no increase in levels of the chimeric protein after 16 h of MG132 treatment. However, we did observe an increase in levels of ubiquitylated protein species confirming efficient proteasome inhibition. Together, these data indicate that sacsin has a functional N-terminal UbL domain capable of interaction with the proteasome.
The sacsin J-domain is functional
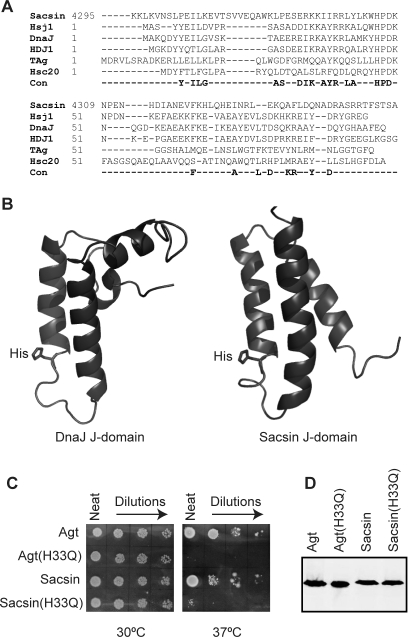
Sacsin was suggested to be a type III Hsp40 family protein based upon the presence of a putative J-domain near the C-terminus of its predicted amino acid sequence. Alignment of the human sacsin sequence with that of Hsp40/HDJ1 revealed approximately 60% identity over 30 residues (Fig. 4A) exhibiting more divergence than most mammalian J-domains. The structure of several J-domains has revealed the presence of four α-helices and a loop region between helices II and III (9), which contains the highly conserved histidine-proline-aspartic acid (HPD) motif. To further investigate if the sacsin J-domain was likely to be functional, we compared its structure (PDB:1IUR) to the structure of the Escherichia coli DnaJ J-domain (PDB: 1XBL) (15) (Fig. 4B). Helix II of the Sacsin J-domain is similar in size and structure to that of the J-domain of E. coli DnaJ with the histidine residue of the sacsin HPD motif projecting into the J-domain core in a comparable orientation (Fig. 4B). To test if the sacsin J-domain was able to function in the Hsp70 chaperone system, we utilized a bacterial in vivo complementation assay. E. coli strain OD259 contains disrupted genes for DnaJ and CbpA such that it will not grow at temperatures of 37°C and above (16) (Fig. 4C). Complementation with Agrobacterium tumefaciens (Agt) DnaJ and chimeric proteins containing J-domains from a wide range of organisms have been shown to compensate for the lack of chromosomal-encoded E. coli DnaJ and CbpA allowing growth at ≥37°C (17,18). To analyse the effect of sacsin's J-domain in this experimental system, the J-domain of Agt DnaJ was substituted with the J-domain of human sacsin to make a chimeric protein. The Agt DnaJ-sacsin was able to confer growth at ≥37°C (Fig. 4C). To confirm that the complementation was dependent on a DnaK (E. coli Hsp70)/sacsin J-domain interaction, the histidine residue in the HPD motif was mutated to glutamine. The modified Agt DnaJ-sacsin chimeric protein was unable to reverse the thermosensitivity of E. coli OD259 (Fig. 4C), consistent with previous studies that have also shown that the histidine to glutamine substitution in the HPD motif disrupted the ability of J-domains to functionally interact with Hsp70 (17). Western blot analysis confirmed that the chimeric proteins were produced at similar levels (Fig. 4D).
Figure 4.
The sacsin J-domain is functional. (A) Structure-aided alignment of the sacsin J-domain to four other J-domains of known structure and the HSJ1 J-domain. Comparison with a consensus of highly conserved residues derived from over 200 J-domain sequences is also shown. The J domains in the alignment are derived from Homo sapiens Sacsin, H. sapiens HSJ1, Escherichia coli DnaJ, H. sapiens HDJ1, the murine polyoma virus T antigen (TAg) and E. coli Hsc20. Those residues conserved in greater than 70% of all known J domains are indicated below the alignment as a consensus (Con). The position of each sequence is indicated at the start of each sequence. (B) Secondary structure of the sacsin J-domain (PDB code 1IUR) compared with the E. coli J-domain (PDB code 1XBL). The ribbon representations of the structures were rendered using PyMol. (C) The J-domain of sacsin was found to be functional using an in vivo complementation assay. Agt DnaJ (Agt) and Agt DnaJ with its J-domain swapped for that of sacsin (Sacsin) were able to replace the lack of chromosomally expressed DnaJ and CbpA in E. coli OD259 allowing growth at 37°C. Agt J-domain (H33Q) (Agt(H33Q)) and the Agt DnaJ-sacsin J-domain chimera with the H33Q substitution (Sacsin(H33Q)) were unable to replace DnaJ and CbpA in E. coli OD259, resulting in a lack of growth at 37°C. (D) Expression of Agt DnaJ and its chimeric derivatives was confirmed by western blot analysis.
Recently an ARSACS-causing compound heterozygous mutation, Lys1715X/Arg4331Gln, was reported (7). The arginine at position 4331 lies within helix II of the sacsin J-domain and is partially conserved. However, a glutamine is occasionally found at this position (equivalent to residue 27 in E. coli DnaJ) in J-domains, for example in the murine polyoma virus T antigen. Interestingly, we have previously shown that in other J-domains substitution of an adjacent helix II arginine residue (Arg26) for an alanine disrupted function (18). Therefore, to investigate if the Arg4331Gln mutation affected the function of the sacsin J-domain, we introduced it into Agt DnaJ-sacsin chimeric protein and tested the effect of this change in the bacterial complementation assay (Supplementary Material, Fig. S4). The Arg4331Gln change did not affect bacterial growth at 37°C (or more stringent temperatures up to 44°C) indicating that this change does not significantly impair sacsin J-domain function.
Sacsin is recruited to mutant ataxin-1 inclusions and modulates the incidence of nuclei with large inclusions
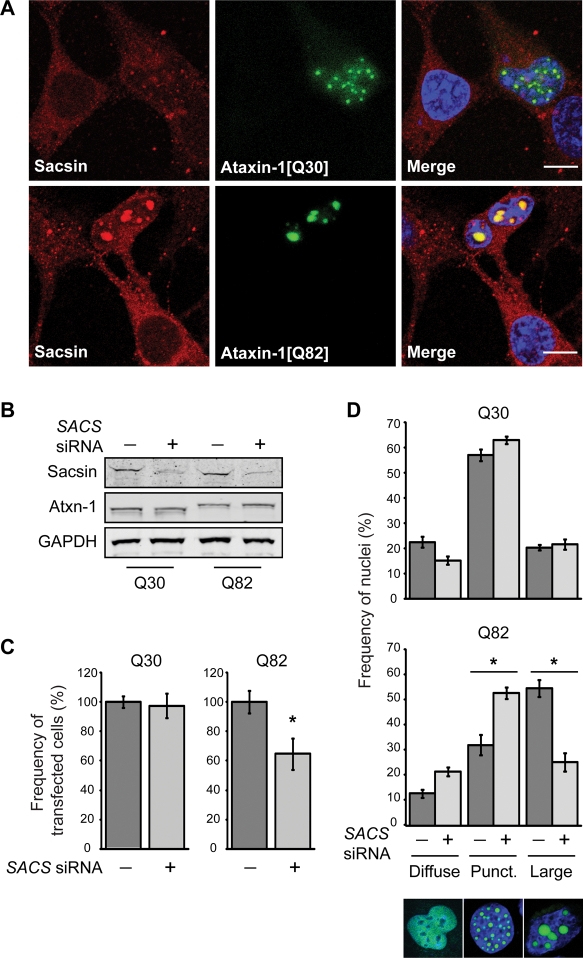
Sacsin has previously been reported to be part of an ataxia protein interactome (2). Moreover, mutant proteins linked to many ataxic diseases form intracellular inclusions of ubiquitylated protein that contain interacting proteins, as well as molecular chaperones and components of the UPS. We therefore tested if sacsin was recruited to nuclear ataxin-1 inclusions. Constructs for expression of wild-type and pathogenic ataxin-1, with polyglutamine tracts of 30 and 82 glutamine residues, respectively, were transfected into SH-SY5Y cells, and the cells subsequently stained for endogenous sacsin (Fig. 5A). GFP-ataxin-1 has previously been reported to localize to the nucleus where the GFP-ataxin-1 nuclear staining is either diffuse or in multiple small punctate foci, with an increased incidence of large nuclear inclusions in cells expressing polyglutamine-expanded ataxin-1 (19). Importantly, we observed that sacsin was localized to intranuclear ataxin-1 inclusions in cells expressing GFP-ataxin-1[82Q].
Figure 5.
Sacsin knockdown reduces the incidence of cells with large nuclear inclusions of polyglutamine-expanded ataxin-1. (A) Confocal immunofluorescence microscopy showing recruitment of sacsin to ataxin-1 nuclear inclusions and in particular large inclusions of GFP-ataxin-1[82Q]. The scale bar represents 10 µm. (B) Western blot analysis confirming sacsin knockdown. Total cellular levels of ataxin-1 were not significantly affected by sacsin knockdown. (C) The total number of cells expressing GFP-ataxin-1 was compared in cells co-transfected with SACS siRNA, or a scrambled control siRNA. Sacsin knockdown did not affect the number of cells expressing GFP-ataxin-1[30Q], but did cause a reduction in the number of cells expressing GFP-ataxin-1[82Q] after 48 h. The frequency of ataxin-1-expressing cells with SACS siRNA is shown as a percentage of the remaining ataxin-1-expressing cells in the control siRNA treatment. (D) The distribution of ataxin-1 in the nuclei of SH-SY5Y cells co-transfected with GFP-ataxin-1[30Q] or GFP-ataxin-1[82Q] and SACS siRNAs, or a scrambled control siRNA, was quantified. Confocal images of nuclei with representative ataxin-1 distributions are shown. Ataxin-1 was either diffusely localized within the nucleus, localized to multiple small punctate foci, or present in fewer larger inclusions. Sacsin knockdown reduced the number of cells with large inclusions in cells expressing GFP-ataxin-1[82Q]. A minimum of 50 transfected cells were scored for each treatment from five separate transfections. Scoring was conducted blind to experimental status 48 h post-transfection. P < 0.05 are indicated with an asterisk.
Hsp70 proteins and their co-chaperones have been shown to modulate aggregation in models of neurodegenerative diseases. Polyglutamine-expanded ataxin-1 has been suggested to favour a toxic and aggregation-prone conformation of the protein that the wild-type protein backbone already has a tendency to adopt (20). Furthermore, ataxin-1 has been shown to interact with Hsp70 and its co-chaperones (20,21). As sacsin was also recruited to GFP-ataxin-1[82Q] inclusions, we investigated the effects of siRNA-mediated sacsin knockdown on ataxin-1 inclusion formation in SH-SY5Y cells. Western blot analysis confirmed effective knockdown of sacsin and suggested a slight increase in GFP-ataxin-1[82Q] levels in cells where sacsin was knocked-down. However, densitometric analyses revealed no significant differences in the level of GFP-ataxin-1 in cell lysates expressing wild-type [Q30] and mutant [Q82] protein or between cells treated with scrambled control and SACS siRNAs (Fig. 5B). We quantified the percentage of cells expressing GFP-ataxin-1, when co-transfected with control or SACS siRNA after 48 h (Fig. 5C). We did not observe any difference in the number of cells expressing GFP-ataxin-1[30Q] upon sacsin knockdown. Therefore, SACS siRNA is not inherently toxic to SH-SY5Y cells. The expression of GFP-ataxin-1[82Q] leads to cell death (22,23). Importantly, there was a significant (P < 0.05) reduction in the number of cells expressing GFP-ataxin-1[82Q] upon sacsin knockdown after 48 h. To control for the possibility that the observed reduction was because of less GFP-ataxin-1[82Q] expression as opposed to cell death, we repeated the experiment using DsRedII to monitor transfected cell survival. In cells co-transfected with DsRedII and GFP-ataxin-1[82Q], the number of DsRedII-expressing cells was reduced to a similar extent when treated with SACS siRNA (not shown), confirming that there was enhanced toxicity of the GFP-ataxin-1[82Q] when sacsin was knocked down.
We next examined the phenotype of cells expressing GFP-ataxin-1 more closely. In SH-SY5Y cells transfected with scrambled control siRNA and GFP-ataxin-1[30Q], we observed a diffuse nuclear ataxin-1 staining in 23 ± 2% of cells. In 57 ± 2% of nuclei, the diffuse staining was accompanied by multiple small punctate inclusions of <1.5 µm in diameter. In 20 ± 1% of cells fewer, large (≥1.5 µm), ataxin-1 inclusions were observed in the nucleus (Fig. 5D). When pathogenic GFP-ataxin-1[82Q] was expressed, the percentage of cell nuclei harbouring large inclusions increased dramatically (55 ± 3% had large inclusions). SACS siRNA on GFP-ataxin-1[30Q] did not significantly alter the incidence of small punctate or large inclusions, compared with control siRNA. However, in cells expressing GFP-ataxin-1[82Q] downregulation of sacsin resulted in a significant (P < 0.0001) reduction in the percentage of cells with large nuclear inclusions, compared with cells treated with the control siRNA, and a commensurate increase in the percentage of cells with the diffuse and punctate nuclear phenotypes (Fig. 5C). The reduction in cells with large nuclear inclusions correlated with the overall reduction in cell number (approximately 35%), suggesting that this population of cells was the most affected by SACS siRNA. The frequency of nuclei with inclusions was not significantly different in cells transfected with control siRNA and cells where siRNA was omitted from the transfections (not shown).
DISCUSSION
The ARSACS gene product, sacsin, was originally reported to be encoded by a single large exon. However, data presented in this study are consistent with a SACS mRNA, derived from a nine-exon gene, expressing a 4579 amino acid sacsin isoform, with a molecular weight of 520 kDa. This is in agreement with the recent identification of ARSACS mutations in exons 6, 7 and 8 of SACS (7,8).
Our data indicate that sacsin is widely expressed in the brain. SACS message and sacsin protein were expressed at high levels in Purkinje cells. Consistent with this, cerebellar Purkinje cell degeneration is a common pathology of ataxias, including ARSACS. Precerebellar nuclei which project afferent processes to the cerebellum also had significant expression of sacsin suggesting that input to the cerebellum could also be affected by altered sacsin function. Corticospinal tract and peripheral nerve axonal degeneration and changes in myelination and conductance are reported in ARSACS patients (5). Interestingly, neurons contributing to the corticospinal tract and cranial motoneurones express high levels of sacsin and axons contain sacsin immunoreactivity.
Very large proteins, such as sacsin, are extremely difficult to express heterologously. Therefore, to elucidate the cellular role of sacsin we have adopted a strategy of analysing the function of specific domains and siRNA. Bioinformatic analysis revealed that the N-terminus of human sacsin contains a putative UbL domain. This domain co-immunoprecipitated with the 20S proteasomal subunit and thus can occur in a complex with the proteasome, possibly interacting with it directly. Interestingly, a number of proteins that function in the UPS are linked to ataxias. These include ataxin-3, which contains a ubiqutin interaction motif (UIM) and functions as a de-ubiquitinating enzyme (24). Furthermore, a number of protein–protein interactions have been observed between UbL and UIM domain proteins. For example, the UbL domain protein PLIC-1, which is a component of the quality control machinery that regulates protein aggregation, is able to interact with both the proteasome and some UIM domain proteins, including ataxin-3 and HSJ1 (25). Thus, it is possible that sacsin may also interact with UIM-containing proteins.
The human proteome contains 12 Hsp70 family members, the activity of which is regulated by numerous co-chaperones, including nearly 50 Hsp40 family members (26). Hsp70 switches from a low substrate affinity when bound to ATP, to a high affinity state upon the hydrolysis of ATP to ADP (27). This cycle is regulated by Hsp70 co-chaperones and in particular Hsp40 proteins, which interact with Hsp70 via the J-domain, stimulating ATP hydrolysis and altering substrate-binding (27). Importantly, Hsp40 proteins act to recruit the Hsp70 machinery to specific cellular locales and functions. We demonstrated that the sacsin J-domain is able to function with the E. coli Hsp70, DnaK, thus confirming that sacsin is a type III Hsp40 protein and supporting an alternative Hsp40 family member designation of DNAJC29 (26). As a component of the sacsin staining was observed to be associated with mitochondrial markers, sacsin could potentially recruit a Hsp70 to specific function at the surface of mitochondria. It will be important to establish with which Hsp70 partners sacsin functions in vivo to further understand its cellular role. Particularly, as although the Arg4331Gln change, which was reported as a compound heterozygous sacsin mutation, did not affect the ability of the sacsin J-domain to function with the E. coli Hsp70, DnaK, it may inhibit the ability of the sacsin J-domain to stimulate the ATPase activity of other Hsp70 proteins. Interestingly, in addition to its J-domain, sacsin also contains multiple regions of homology to Hsp90 (6), further suggesting a possible chaperone function.
We observed a significant enhancement in the toxicity of GFP-ataxin-1[82Q] when sacsin levels were reduced by siRNA. This reduction in cell number correlated with the loss of cells with the large nuclear inclusion phenotype from the population. These data suggest that sacsin is protective against polyglutamine-expanded ataxin-1 toxicity. Interestingly, the overall level of GFP-ataxin-1[82Q], detected by western blot analysis of cell lysates, was not significantly altered after sacsin knockdown, even though there was a reduction in the number of cells with detectable GFP signal. This implies that the remaining cells have higher levels of GFP-ataxin-1[82Q] and may indicate a role for sacsin in the degradation of aberrantly folded ataxin-1.
The dynamics of ataxin-1 inclusion formation is controversial. Ataxin-1 nuclear inclusion formation has been shown to be dependent on RNA-binding. Wild-type ataxin-1, has been reported to shuttle between the nucleus and cytoplasm, whereas the polyglutamine-expanded mutant protein cannot shuttle (28). This suggests a gain of function of the mutant protein such as retention of associated RNAs or proteins within the nucleus. Conversely, it has also been reported that polyglutamine expansion does not affect nuclear shuttling, but enhances the intracellular kinetics of ataxin-1 (29). Inhibition of the proteasome leads to the apparent fusion of ataxin-1 nuclear inclusions and an increase in their size (28), while the type I Hsp40 protein HDJ-2/DnaJA1 has been reported to suppress ataxin-1 aggregation and is localized to inclusions (30). It is unknown if sacsin's localization to ataxin-1-[Q82] inclusions, and effect on their incidence, is dependent on its J-domain, or UBL domain, or if it requires sacsin to bind ataxin-1 as a chaperone.
A recent study undertook a yeast two-hybrid-based screen to develop a protein–protein interaction network for proteins involved in inherited ataxias and disorders of Purkinje cell degeneration (2). The C-terminal region of sacsin was found to share four interacting partners with other ataxia proteins (2). The best studied of these is PICK1 (protein interacting with C-kinase), which was identified as binding ataxin-7, PRKCG, aprataxin and frataxin. Interestingly, frataxin, mutations in which cause the autosomal recessive disease Friedreich's ataxia, is also targeted to mitochondria. The recruitment of sacsin to polyglutamine-expanded ataxin-1 intranuclear inclusions and the disappearance of cells with these inclusions, when sacsin levels are reduced by siRNA, further points to sacsin being in common cellular pathways with other ataxia proteins.
Type I and II Hsp40 proteins predominantly act to stimulate Hsp70 general protein folding function, while type III Hsp40 proteins recruit Hsp70s to specialized roles. Sacsin's large size and multiple domains suggest that it may have additional cellular roles to protecting against mutant ataxin-1 expression. We hypothesize that sacsin may act as a molecular scaffold for assembly of a specific protein complex and that regulation of this complex requires integration of molecular chaperone machinery and the UPS.
METHODS
Sacsin antibody generation and characterization
Two rabbit antisera were raised against the human sacsin peptides DYAVRGKSDKDVKPT (amino acid residues 4489–4503) and METKENRWVPVTVLPGC (amino acid residues 1–17) conjugated to KLH. The antisera were affinity-purified against immunizing peptides. For western blotting of sacsin, in tissue and SH-SY5Y cells, samples were separated on a 4–12% NuPAGE gel (Invitrogen).
Analysis of sacsin localization
Adult male Wistar rats were used for in situ and immunohistochemical analyses of SACS mRNA and sacsin protein distribution in the brain. In situ hybridization was performed using radioactive oligonucleotide probes and emulsion autoradiography as described previously (31). Oligonucleotide probes complementary to rat SACS mRNA (accession number XM_001063390) at nucleotides 569–602 (aagccggtctcagcgaagatgcgctccttgaggt), 13881–13914 (aaggtagcacttgaagcacacccactcattggca) and 14062–14095 (acctttgagagtatgtctgtccagcagagggagg) were tested and found to give an identical cellular distribution for sacsin. Free-floating sections of 40 µm were processed for immunohistochemistry with standard methodologies previously described (32). Immunofluorescent processing and subsequent confocal microscopy were as described previously (14). The overlap between sacsin staining and mitochondrial markers was quantified from 10 representative images using the co-localization function of the MetaMorph 7.5 image analysis software (Molecular Devices).
Co-immunoprecipitation of the N-terminus of sacsin with the proteasome
The 5′ end of the SACS open reading frame was sub-cloned into p3xFLAG-CMV-14, such that the N-terminal 124 amino acids of sacsin, encompassing the UbL domain, was expressed with a FLAG-tag fused at the C-terminus. UbL domain mutations L58/D60A and R50A/L51A were introduced to this construct by site-directed mutagenesis. CHO-1 cells transiently transfected with constructs expressing sacsin1–124-FLAG or HSJ1-FLAG, which was used as a positive control (14), were harvested in 50 mm Tris HCl pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, protease inhibitors. Supernatant from a 17 000g 30 min centrifugation were incubated at 4°C with anti-FLAG (M2)-conjugated agarose beads (Sigma). After 16 h, beads were washed and proteins eluted in sodium dodecyl sulphate–polyacrylamide gel electrophoresis sample buffer and subjected to western blot analyses. Co-immunoprecipitated proteasome subunit was detected with anti-human C8 (Affiniti). For proteasome inhibition, cells were incubated in media containing 10 nm MG132 for 16 h.
Bacterial complementation assay for J-domain function
Complementation assays were performed in E. coli strain OD259 (16) as described previously (17). Briefly, the pQE30-derived pRJ30 vector containing the A. tumefaciens DnaJ coding sequence served as a base vector for J-domain swapping using the strategy we have outlined before (18). The coding region for the sacsin J-domain (residues 4296–4384) was amplified by PCR from BAC clone RP11-40O20 (http://bacpac.chori.org). The histidine to glutamine mutations of the sacsin HPD motif was produced by site-directed mutagenesis. E. coli OD259 cells exogenously producing Agt DnaJ and Agt DnaJ(H33Q) served as controls. Western blot analysis using an anti-His antibody was performed on whole cell lysates of E. coli OD259 and its transformants producing 6xHis-tagged Agt DnaJ and chimeric proteins to confirm equivalent levels of expression.
SACS siRNA-mediated knockdown in ataxin-1-expressing cells
Target sequences in exons 6 (GGATGATCCTCGAAGGTC), 7 (GCGGCCGAATTCTATAAAG) and 9 (CGTAAGATTTCTAGATGAC) of SACS were identified and used to generate siRNA molecules (Ambion). siRNAs were validated individually by comparison of levels of SACS mRNA in SH-SY5Y-transfected cells with the siRNA or a scrambled control by RT-PCR (Supplementary Material, Fig. S5). A mixture of the three siRNAs, at a concentration of 20 µm was used in subsequent experiments. siRNAs were co-transfected with ataxin-1 constructs and cells processed for immunofluorescence or western blot analysis after 48 h. DR-RedII (Clontech) was used at a ratio of 1:3, to GFP-ataxin-1, to monitor transfection efficiency. Quantification of transfected cells and scoring of cell nuclei containing ataxin-1 inclusions was performed blinded to experimental status. A Mann–Whitney U test was used for statistical analysis.
SUPPLEMENTARY MATERIAL
FUNDING
The Medical Research Council (MRC Grant ID: 81373). The Barts and the London Charity. The National Research Foundation (South Africa to G.L.B.) and the Ernest Oppenheimer Memorial Trust. Funding to pay the Open Access Charge came from the MRC grant ID: 81373.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Huda Zoghbi for the ataxin-1 expression plasmids.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Breedveld G.J., van Wetten B., te Raa G.D., Brusse E., van Swieten J.C., Oostra B.A., Maat-Kievit J.A. A new locus for a childhood onset, slowly progressive autosomal recessive spinocerebellar ataxia maps to chromosome 11p15. J. Med. Genet. 2004;41:858–866. doi: 10.1136/jmg.2004.019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim J., Hao T., Shaw C., Patel A.J., Szabo G., Rual J.F., Fisk C.J., Li N., Smolyar A., Hill D.E., et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 3.Muchowski P.J., Wacker J.L. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- 4.Connell P., Ballinger C.A., Jiang J., Wu Y., Thompson L.J., Hohfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard J.P., Richter A., Mathieu J., Brunet D., Hudson T.J., Morgan K., Melancon S.B. Autosomal recessive spastic ataxia of Charlevoix-Saguenay. Neuromuscul. Disord. 1998;8:474–479. doi: 10.1016/s0960-8966(98)00055-8. [DOI] [PubMed] [Google Scholar]
- 6.Engert J.C., Berube P., Mercier J., Dore C., Lepage P., Ge B., Bouchard J.P., Mathieu J., Melancon S.B., Schalling M., et al. ARSACS, a spastic ataxia common in northeastern Quebec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat. Genet. 2000;24:120–125. doi: 10.1038/72769. [DOI] [PubMed] [Google Scholar]
- 7.Vermeer S., Meijer R.P., Pijl B.J., Timmermans J., Cruysberg J.R., Bos M.M., Schelhaas H.J., van de Warrenburg B.P., Knoers N.V., Scheffer H., Kremer B. ARSACS in the Dutch population: a frequent cause of early-onset cerebellar ataxia. Neurogenetics. 2008;9:207–214. doi: 10.1007/s10048-008-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang Y., Takiyama Y., Sakoe K., Shimazaki H., Ogawa T., Nagano S., Yamamoto Y., Nakano I. Sacsin-related ataxia (ARSACS): expanding the genotype upstream from the gigantic exon. Neurology. 2006;66:1103–1104. doi: 10.1212/01.wnl.0000204300.94261.ea. [DOI] [PubMed] [Google Scholar]
- 9.Cheetham M.E., Caplan A.J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullen R.J., Buck C.R., Smith A.M. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann-Petersen R., Gordon C. Integral UBL domain proteins: a family of proteasome interacting proteins. Semin. Cell Dev. Biol. 2004;15:247–259. doi: 10.1016/j.semcdb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Kim I., Mi K., Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol. Biol. Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Upadhya S.C., Hegde A.N. A potential proteasome-interacting motif within the ubiquitin-like domain of parkin and other proteins. Trends Biochem. Sci. 2003;28:280–283. doi: 10.1016/S0968-0004(03)00092-6. [DOI] [PubMed] [Google Scholar]
- 14.Westhoff B., Chapple J.P., van der S.J., Hohfeld J., Cheetham M.E. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr. Biol. 2005;15:1058–1064. doi: 10.1016/j.cub.2005.04.058. [DOI] [PubMed] [Google Scholar]
- 15.Pellecchia M., Szyperski T., Wall D., Georgopoulos C., Wuthrich K. NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J. Mol. Biol. 1996;260:236–250. doi: 10.1006/jmbi.1996.0395. [DOI] [PubMed] [Google Scholar]
- 16.Deloche O., Kelley W.L., Georgopoulos C. Structure-function analyses of the Ssc1p, Mdj1p, and Mge1p Saccharomyces cerevisiae mitochondrial proteins in Escherichia coli. J. Bacteriol. 1997;179:6066–6075. doi: 10.1128/jb.179.19.6066-6075.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennessy F., Boshoff A., Blatch G.L. Rational mutagenesis of a 40 kDa heat shock protein from Agrobacterium tumefaciens identifies amino acid residues critical to its in vivo function. Int. J. Biochem. Cell Biol. 2005;37:177–191. doi: 10.1016/j.biocel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Nicoll W.S., Botha M., McNamara C., Schlange M., Pesce E.R., Boshoff A., Ludewig M.H., Zimmermann R., Cheetham M.E., Chapple J.P., Blatch G.L. Cytosolic and ER J-domains of mammalian and parasitic origin can functionally interact with DnaK. Int. J. Biochem. Cell Biol. 2007;39:736–751. doi: 10.1016/j.biocel.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skinner P.J., Koshy B.T., Cummings C.J., Klement I.A., Helin K., Servadio A., Zoghbi H.Y., Orr H.T. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 20.Al Ramahi I., Lam Y.C., Chen H.K., de Gouyon B., Zhang M., Perez A.M., Branco J., de Haro M., Patterson C., Zoghbi H.Y., Botas J. CHIP protects from the neurotoxicity of expanded and wild-type ataxin-1 and promotes their ubiquitination and degradation. J. Biol. Chem. 2006;281:26714–26724. doi: 10.1074/jbc.M601603200. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen N.D., Andresen J.M., Pitt J.E., Swenson M.A., Zoghbi H.Y., Orr H.T. Hsp70/Hsc70 regulates the effect phosphorylation has on stabilizing ataxin-1. J. Neurochem. 2007;102:2040–2048. doi: 10.1111/j.1471-4159.2007.04678.x. [DOI] [PubMed] [Google Scholar]
- 22.Bolger T.A., Zhao X., Cohen T.J., Tsai C.C., Yao T.P. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J. Biol. Chem. 2007;282:29186–29192. doi: 10.1074/jbc.M704182200. [DOI] [PubMed] [Google Scholar]
- 23.Rich T., Varadaraj A. Ataxin-1 fusion partners alter polyQ lethality and aggregation. PLoS One. 2007;2:e1014. doi: 10.1371/journal.pone.0001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burnett B., Li F., Pittman R.N. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- 25.Heir R., Ablasou C., Dumontier E., Elliott M., Fagotto-Kaufmann C., Bedford F.K. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006;7:1252–1258. doi: 10.1038/sj.embor.7400823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kampinga H.H., Hageman J., Vos M.J., Kubota H., Tanguay R.M., Bruford E.A., Cheetham M.E., Chen B., Hightower L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartl F.U., Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 28.Irwin S., Vandelft M., Pinchev D., Howell J.L., Graczyk J., Orr H.T., Truant R. RNA association and nucleocytoplasmic shuttling by ataxin-1. J. Cell Sci. 2005;118:233–242. doi: 10.1242/jcs.01611. [DOI] [PubMed] [Google Scholar]
- 29.Krol H.A., Krawczyk P.M., Bosch K.S., Aten J.A., Hol E.M., Reits E.A. Polyglutamine expansion accelerates the dynamics of ataxin-1 and does not result in aggregate formation. PLoS One. 2008;3:e1503. doi: 10.1371/journal.pone.0001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cummings C.J., Mancini M.A., Antalffy B., DeFranco D.B., Orr H.T., Zoghbi H.Y. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 31.Storr H.L., Clark A.J., Priestley J.V., Michael G.J. Identification of the sites of expression of triple A syndrome mRNA in the rat using in situ hybridisation. Neuroscience. 2005;131:113–123. doi: 10.1016/j.neuroscience.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 32.Dyall S.C., Michael G.J., Whelpton R., Scott A.G., Michael-Titus A.T. Dietary enrichment with omega-3 polyunsaturated fatty acids reverses age-related decreases in the GluR2 and NR2B glutamate receptor subunits in rat forebrain. Neurobiol. Aging. 2007;28:424–439. doi: 10.1016/j.neurobiolaging.2006.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.