Abstract
Large case–control genome-wide association studies primarily expose common variants contributing to disease pathogenesis with modest effects. Thus, alternative strategies are needed to tackle rare, possibly more penetrant alleles. One strategy is to use special populations with a founder effect and isolation, resulting in allelic enrichment. For multiple sclerosis such a unique setting is reported in Southern Ostrobothnia in Finland, where the prevalence and familial occurrence of multiple sclerosis (MS) are exceptionally high. Here, we have studied one of the best replicated MS loci, 5p, and monitored for haplotypes shared among 72 regional MS cases, the majority of which are genealogically distantly related. The haplotype analysis over the 45 Mb region, covering the linkage peak identified in Finnish MS families, revealed only modest association at IL7R (P = 0.04), recently implicated in MS, whereas most significant association was found with one haplotype covering the C7-FLJ40243 locus (P = 0.0001), 5.1 Mb centromeric of IL7R. The finding was validated in an independent sample from the isolate and resulted in an odds ratio of 2.73 (P = 0.000003) in the combined data set. The identified relatively rare risk haplotype contains C7 (complement component 7), an important player of the innate immune system. Suggestive association with alleles of the region was seen also in more heterogeneous populations. Interestingly, also the complement activity correlated with the identified risk haplotype. These results suggest that the MS predisposing locus on 5p is more complex than assumed and exemplify power of population isolates in the identification of rare disease alleles.
INTRODUCTION
Multiple sclerosis [MS (MIM #126200)] is a chronic inflammatory disease of the central nervous system (CNS) characterized by multifocal demyelination. MS is a multifactorial trait with complex inheritance and a putative autoimmune pathogenesis (1–5), presenting a quite uniform prevalence of 100/105 in populations of Northern European origin (6). The strongest and most consistent evidence of linkage and association with MS is found with the HLA class II region on 6p, which has been estimated to explain 15–40% of the genetic fraction of MS etiology (7–9). Recently, the first MS genome-wide association (GWA) study revealed two additional loci, IL2RA (MIM *147730) and IL7R (MIM *146661), associated with an increased risk of MS (10), the latter being simultaneously identified also through a candidate gene approach in independent and overlapping data sets (11,12). Interestingly, the IL7R gene is located in chromosome 5p region, to which also linkage has been reported in MS families from several populations, including Finns (13–18), and which is syntenic to the experimental autoimmune encephalomyelitis (EAE) locus on mouse chromosome 15 (19).
The new loci identified in the recent International MS Genetics Consortium GWA scan revealed important new information about the pathogenesis of MS but were estimated to explain only 0.2% of the variance in the risk of the development of multiple sclerosis (10). Thus, it is evident that other loci remain to be identified for a more comprehensive understanding of the genetic susceptibility of MS. Since large GWA studies using common SNPs of HapMap identify common variants of disease loci, alternative strategies are needed to identify less common, possibly more penetrant variants contributing to the molecular background of MS. These relatively rare variants with at least moderate penetrance most probably give rise to a familial concentration of MS cases. One approach to tackle the discovery of relatively rare alleles is to use special populations with a founder effect and isolation, where one could hypothesize enrichment of the disease alleles (20). For MS, such a setting is provided by the MS high-risk region of Southern Ostrobothnia in Western Finland, where exceptionally high incidence (12/105) and prevalence (200/105) rates, as well as increased familial occurrence, have been observed in epidemiological studies over three decades (21–26).
As we previously have detected linkage to chromosome 5p in Finnish MS families, enriched for cases from the high-risk internal isolate, we first wanted to use the Finnish GWA data to study whether IL7R alone or possibly together with other loci in this chromosomal region could be identified as a susceptibility locus for MS in the isolate. A set of 72 MS cases, originating from Southern Ostrobothnia and majority of the samples being genealogically traced to two founding couples living in this geographical area in the 15th and 16th centuries, was monitored for enriched haplotypes using the HumanHap300 Illumina panel.
RESULTS
The impact of IL7R in Finnish MS patients
We initially wanted to study whether the variants of the IL7R gene on 5p, one of the main findings in the recent international GWA study (10), contribute to MS susceptibility in Finland. The non-synonymous SNP rs6897932, which is the likely causative variant (10,12), and three additional SNPs (rs11567701, rs3194051 and rs6871748) from previous association studies (11,27) were genotyped in 922 Finnish MS cases, of which 197 originate from the Southern Ostrobothnian MS high-risk region, and 1392 population controls (Table 1, FIN-isolate1 + 2, FIN-OUT). There was a slight difference in SNP allele frequencies between cases and controls both in the study set from the high-risk isolate (FIN-isolate1 + 2) and in the study set from the rest of Finland (FIN-OUT), providing modest evidence for association in the combined Finnish study sample [Table 2, rs6897932 and rs6871748, P = 0.002, odds ratio (OR) 1.24 (95% CI: 1.09–1.41), the SNPs are in full linkage disequilibrium (LD) (r2 = 1) with each other]. A test for heterogeneity between the data sets from the high-risk isolate and rest of Finland showed no evidence for stratification at this locus, permitting the combined analysis (Mantel–Haenszel corrected P = 0.0001). The OR corresponds to that observed in other studies. However, the associated allele C of the likely causal SNP rs6897932 is less common among Finnish MS cases (0.69) than among cases of other populations reported (Sweden: 0.74; UK and Belgium: 0.76 and USA: 0.78) (11,12).
Table 1.
Description of the study material
| Study Set | Origin | MS cases | Controls |
|---|---|---|---|
| FIN-isolate1 | Finland (Southern Ostrobothnia isolate) | 72 | 68 |
| FIN-isolate2 | Finland (Southern Ostrobothnia isolate) | 125 | 365 |
| FIN-OUT | Finland (other parts of the country) | 725 | 959 |
| SWE | Sweden | 651 | 651 |
| NOR | Norway | 359 | 471 |
| US | USA | 920 | 177 |
Table 2.
Association analysis of the critical IL7R SNPs in Finnish case–control samples
| dbSNP | Location | Allelesa | FIN-isolate1 + 2 |
FIN-OUT |
FIN (isolate1 + 2 + OUT) |
P-valuec | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Freq MSb | Freq Ctrl | Freq MSb | Freq Ctrl | Freq MSb | Freq Ctrl | |||||
| rs11567701 | Intron 1 | G/T | 0.26 | 0.25 | 0.30 | 0.29 | 0.29 | 0.28 | 0.282 | — |
| rs6897932 | Exon 6 | T/C | 0.68 | 0.63 | 0.70 | 0.65 | 0.69 | 0.65 | 0.002 | 1.24 (1.09–1.41) |
| rs3194051 | Exon 8 | A/G | 0.26 | 0.24 | 0.30 | 0.30 | 0.29 | 0.28 | 0.489 | — |
| rs6871748 | Intergenic | C/T | 0.69 | 0.63 | 0.69 | 0.65 | 0.69 | 0.64 | 0.002 | — |
aAssociated alleles underlined.
bFrequency of the associated allele.
cHaploview custom association test.
Screen of the linked region on 5p15-q11
As the single SNP association observed with markers in the IL7R gene was quite modest in the Finnish MS samples, we wanted to systematically monitor if a haplotype in the whole 45 Mb linked locus on 5p (between markers D5S667 and D5S407, 11.1–56.0 Mb, UCSC Genome Browser, hg18 assembly, March 2006) (Fig. 1A) would be enriched in the internal high-risk isolate and potentially contribute more substantially to MS susceptibility.
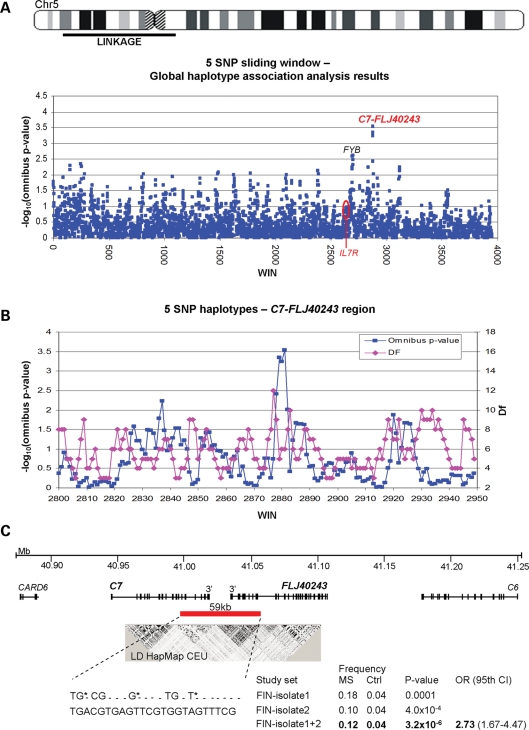
Figure 1.
Screen of the linked region on 5p15-q11 utilizing the SNP data of the Finnish MS GWA study and validation of the finding. (A) A 45 Mb region showing linkage in Finnish MS families was screened with a five SNP haplotype association analysis from the Illumina HumanHap 300 chip genotypes using the sliding window option of the PLINK program. The –log(P-values) for the haplotypes are shown as blue dots. The analysis revealed only one region with omnibus P-values 10−4. These haplotype windows map to the C7 and FLJ40243 genes. (B) Five SNP sliding window results zoomed to the C7-FLJ40243 region. Number of degrees of freedom (df) is given to illustrate the recombination spots, which flank the four associated consecutive five SNP windows. The haplotype showing strongest evidence for association was extended with PLINK to both orientations until reaching sites with increased df, resulting to an eight SNP haplotype covering one haplotype block in the Finnish isolate. (C) The critical region, extending from 3′ end of C7 to 3′ end of FLJ40243, was further covered with 24 HapMap tagging SNPs, which were genotyped in an independent case–control set from the high-risk region. The original finding was validated. To estimate the effect size of the 59 kb risk haplotype identified, an OR was calculated for the combined study set from the high-risk region. Alleles of the original eight SNP haplotype (rs324058, rs8264 and rs10512754), which correspond (r2 = 1) to the tagging SNPs (rs324061, rs7732104 and rs3817324), are marked with asterisks. LD between the SNPs flanking the C7 and FLJ40243 genes was measured using Haploview and publicly available genotype data of CEPH individuals of European origin (CEU). Positions are obtained from the UCSC Genome Browser, hg18 assembly, March 2006.
Haplotypes generated from the Illumina HumanHap300 SNP panel genotypes were monitored for association with MS in the genealogically linked sample from the internal isolate. Specifically, 72 MS cases (Table 1, FIN-isolate1), having either both parents born within the high-risk region of Southern Ostrobothnia (n = 64) or one Southern Ostrobothnia born parent as well as a family history of MS (n = 8), have been genotyped for the Finnish GWA study (E. Jakkula et al., manuscript in preparation), and this data were first utilized to screen the chromosome 5p-linked region. Forty-one of the 72 MS cases belonged to either one or both of the two large interconnected mega-pedigrees, which we were able to construct via genealogical studies. However, none of the MS cases included in the study were first-degree relatives with each other. We hypothesized that the relatively short history, with common ancestors only 14–16 generations ago (ancestors born in 1490 and 1594), might expose shared haplotypes between the distantly related MS cases. Thus, the linked interval on 5p was screened with a five SNP sliding window haplotype association analysis using PLINK (28). Sixty-eight identity-by-state (IBS) matched Finns were used as controls (Table 1, FIN-isolate1). We used genome-wide SNP data and identity-by-sharing, multidimensional scaling and identity-by-descent analyses to select these controls so that their genetic background would be similar with the cases, as parental birthplace information was not available for all the controls (Supplementary Material, Fig. S1). The genomic inflation factor (λ), calculated using genome-wide single SNP data in PLINK, was 1.0758 for our GWA data set, suggesting that cases and controls are well-matched and there is no large-scale population stratification within our final study set.
In the sliding window analysis, only very modest association was observed with the IL7R haplotypes (single haplotype P = 0.043 and omnibus haplotype P = 0.084) (Supplementary Material, Tables S1 and S2). Instead, the haplotype analysis revealed one region (5p13.1 at 41.0 Mb region, P = 0.00029) with omnibus P-values ≤10−4 when all the haplotypes were compared between cases and controls (Fig. 1A; Supplementary Material, Table S1). This region is located at the C7-FLJ40243 gene locus, 5.1 Mb centromeric from IL7R. The same C7-FLJ40243 haplotype also provided the strongest evidence for association in a single haplotype analysis (P = 0.00017, flanked by SNPs rs1901167–rs10512754), whereas only two other single haplotypes within the MS-linked 5p locus provided a P-value of <0.001: an intergenic region in 5p15.2 at 13.6 Mb (P = 0.00085, flanked by SNPs rs3868287–rs10491318) and the FYB region (MIM *602731) in 5p13.1 at 39.2 Mb (P = 0.00088, flanked by SNPs rs1549690–rs66782) (Supplementary Material, Table S2).
The C7-FLJ40243 haplotype was further extended with PLINK to both orientations until reaching sites where the number of observed haplotypes increased corresponding to potential recombination spots (shown in Fig. 1B as an increase in the degrees of freedom), resulting to an eight SNP haplotype. This haplotype had a frequency of 0.183 in MS cases compared with 0.037 in controls in the GWA study sample (P = 0.00013) and it covers the 3′ ends of the C7 (complement component 7 precursor) and the FLJ40243 (hypothetical protein LOC133558) genes (Fig. 1C).
Most promising C7-FLJ40243 haplotype region in an independent study sample from the isolate
To confirm the initial finding and to study the genetic variation of the C7-FLJ40243 region in more detail, the region with strongest original association was tagged with 24 HapMap SNPs and analyzed in an independent set of 125 Finnish MS cases having at least one parent born within the Southern Ostrobothnian isolate and 365 population controls from the same geographical region (Table 1, FIN-isolate2). The initial C7-FLJ40243 haplotype association was validated in this larger independent sample from the internal isolate with a P-value of 4.0 × 10−4 (Fig. 1C, FIN-isolate2). To further estimate the effect size of the 59 kb risk haplotype in the MS high-risk region, the two study sets from the isolate were combined. The frequency of the risk haplotype in this combined study set was 0.12 among MS cases and 0.04 among controls, providing a P-value of 3.2 × 10−6 (pperm = 5.0 × 10−5, P = 0.012 after Bonferroni correction for the 3976 haplotype windows tested) and an allelic OR of 2.73 (95% CI: 1.67–4.47) (Fig. 1C, FIN-isolate1 + 2). The main haplotypes of the region (freq ≥ 0.05) and their frequencies in the combined study set from the high-risk region are shown in Table 3.
Table 3.
Frequencies and P-values for the C7-FLJ40243 haplotypes in different study sets
| Haplotype (freq ≥ 0.05)a | FIN-isolate1 + 2 |
FIN-OUT |
NOR |
SWE |
US |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq MS | Freq Ctrl | P-valueb | Freq MS | Freq Ctrl | P-valueb | Freq MS | Freq Ctrl | P-valueb | Freq MS | Freq Ctrl | P-valueb | Freq MS | Freq Ctrl | P-valueb | |
| TAACAGGCGCTTGTGGTAGTTTCG | 0.16 | 0.18 | ns | 0.20 | 0.16 | 0.003 | 0.18 | 0.18 | ns | 0.18 | 0.15 | 0.098 | 0.17 | 0.14 | ns |
| TGCTAGTCGTTCGTAAAGGTCTCG | 0.07 | 0.09 | ns | 0.10 | 0.10 | ns | 0.15 | 0.13 | ns | 0.15 | 0.13 | ns | 0.18 | 0.22 | ns |
| TGACAGTCACTTGTGAAAATTCGA | 0.02 | 0.05 | 0.060 | 0.06 | 0.05 | ns | 0.10 | 0.09 | ns | 0.08 | 0.09 | ns | 0.06 | 0.07 | ns |
| CGACAGGCGCGTGTGGTAGTTTCG | 0.06 | 0.05 | ns | 0.05 | 0.05 | ns | 0.01 | 0.02 | ns | 0.03 | 0.03 | ns | 0.01 | 0.02 | ns |
| TGACGTGAGTTCGTGGTAGTTTCG | 0.12 | 0.04 | 3 × 10−6 | 0.05 | 0.05 | ns | 0.04 | 0.05 | ns | 0.04 | 0.03 | ns | 0.03 | 0.06 | ns |
| CGACAGGCGCGCATGAAAGGCTCG | 0.04 | 0.04 | ns | 0.02 | 0.04 | 0.004 | 0.03 | 0.05 | 0.085 | 0.04 | 0.04 | ns | 0.03 | 0.03 | ns |
| TGACAGTCACTTGTGAAAATTCGG | 0.05 | 0.04 | ns | 0.04 | 0.03 | ns | 0.03 | 0.03 | ns | 0.03 | 0.03 | ns | 0.02 | 0.03 | ns |
| TGCTAGTCGTTCGTGGTAGTTTCG | 0.02 | 0.02 | ns | 0.03 | 0.04 | ns | 0.05 | 0.03 | 0.012 | 0.04 | 0.04 | ns | 0.04 | 0.04 | ns |
| CGACAGGCGCGCGTGGTAGTTTCG | 0.02 | 0.02 | ns | 0.04 | 0.04 | ns | 0.04 | 0.05 | ns | 0.04 | 0.05 | ns | 0.04 | 0.06 | ns |
aSNPs 6, 7, 10, 11, 13, 16, 18, 19, 20 and 23, respectively, have been genotyped only in the FIN-isolate study set.
bHaploview custom association test. P-values >0.1 are marked as ‘ns’.
Additional family members were available for 156 MS patients from the MS high-risk region (Table 1, FIN-isolate1 and FIN-isolate2). The 24 SNPs over the critical C7-FLJ40243 region were further analyzed in these families using the gamete competition test (29) to perform family based association analysis that avoids any potential population stratification. Convincingly, also family based evidence for association with MS (P < 0.05) was observed with eight SNPs, SNP rs6860438 in C7 providing the strongest P-value of 0.006 (Supplementary Material, Table S3).
C7-FLJ40243 risk region in more heterogeneous populations
We next monitored the frequency of the identified C7-FLJ40243 risk haplotype in various populations utilizing the Human Diversity Panel SNP data publicly available. The MS risk haplotype, enriched among the MS cases of the Finnish isolate, was observed to be relatively rare globally, found at the ∼4% of alleles in the general European population, being almost absent in the Africans, Southern Americans and Oceanians, and having the highest frequency of ∼6% in the Eastern populations (Supplementary Material, Table S4).
To evaluate the role of the identified C7-FLJ40243 risk region in MS susceptibility in more heterogeneous populations, 14 of the 24 tagging SNPs (see Materials and Methods) were genotyped in four independent study sets: Finland outside the MS high-risk region (725 MS cases and 959 controls), Sweden (651 MS cases and 651 controls), Norway (359 MS cases and 471 controls) and USA (920 MS cases and 177 controls) (Table 1, FIN-OUT, SWE, NOR, US). Association of the main haplotypes (alleles with freq ≥ 0.05) was tested in each population sample. Frequencies and P-values for the haplotypes are shown in Table 3.
The relatively rare allele, which was observed to be enriched among MS cases from the high-risk region, had a comparable frequency in control samples of the isolate (0.04) and the other populations of Northern European origin (0.05 in Finland and Norway; 0.03 in Sweden and 0.06 in USA). No evidence for association was seen with this haplotype allele in other study sets. However, the Finnish MS cases were observed to carry another risk allele (freq MS 0.20, controls 0.16, P = 0.003), which was also marginally over-represented in the MS cases from Sweden and USA (Sweden: freq MS 0.18, controls 0.15; USA: freq MS 0.17, controls 0.14). A third allele was discerned to be over-represented among the Norwegian cases (freq MS 0.05, controls 0.03, P = 0.012). Further, single SNPs also provided marginal evidence for association with MS (best P-values: FIN-OUT P = 0.006; SWE P = 0.012; NOR P = 0.013; US P = 0.018), but again, there seems to be allelic heterogeneity within the region in more heterogeneous populations, and strongest association was detected with different SNPs in different populations (Supplementary Material, Table S5).
Notably, the identified 59 kb risk region can be divided into a low LD region (first-half) and a more clear block structure (end-half) in heterogeneous populations according to the HapMap CEU data (Fig. 1C), whereas it seems to cover only one haplotype block in the Finnish MS isolate (based on degrees of freedom in the GWA data, Fig. 1B).
Sequencing of the coding regions of the C7 and FLJ40243 genes
To exclude structural alterations in the coding regions of the C7 and FLJ40243 genes in MS, the C7 promoter, all exons of C7 and the 3′ exons 25–42 of FLJ40243 (covered by the risk haplotype) were sequenced in 16 Finnish individuals from the high-risk region, of which four MS cases were known to carry two copies, four MS cases one copy and eight controls no copies of the C7-FLJ40243 risk haplotype.
Altogether 11 variants were observed, 2 of which were located in the promoter of C7, 4 in the coding sequence of C7 and 5 in the coding sequence of FLJ40243. The conserved alleles of the non-synonymous SNPs (rs2271708 in C7 exon 5, rs1063499 in C7 exon 10, rs13157656 and a novel SNP in C7 exon 14, rs10054110 in FLJ40243 exon 27 and rs2271704 in FLJ40243 exon 33) are present both in the MS risk haplotype and in most of the other common haplotypes. Thus, these SNPs are not likely candidates for the causative variant. Importantly, none of the variants identified was observed to be in tight LD with the risk haplotype identified. All the sequenced variants are shown in Table 4.
Table 4.
Sequencing of the coding regions of the C7-FLJ40243 risk region
| Rs# | Positiona | Location | Function | Allelesb | Freq CEUc | Allele of the risk haplotype |
|---|---|---|---|---|---|---|
| Novel | 40944608 | C7 promoter | — | T/C | Unknown | T |
| rs1376178 | 40944658 | C7 promoter | — | A/C | 0.61 | A/C |
| rs2271708 | 40972298 | C7 exon 5 | Non-synonymous (Cys-Arg) | T/C | 0.99 | T |
| rs1063499 | 40991318 | C7 exon 10 | Non-synonymous (Ser-Thr) | G/C | 0.40 | G/C |
| rs13157656 | 41000609 | C7 exon 14 | Non-synonymous (Thr-Pro) | A/C | 0.76 | A |
| Novel | 41000642 | C7 exon 14 | Non-synonymous (Thr-Ser) | A/T | Unknown | A |
| rs722575 | 41033992 | FLJ40243 exon 42 | Synonymous | C/T | 0.58 | T |
| rs704040 | 41033953 | FLJ40243 exon 42 | Synonymous | T/C | 0.33 | C |
| rs1001420 | 41036100 | FLJ40243 exon 39 | Synonymous | G/A | 0.29 | A |
| rs2271704 | 41044537 | FLJ40243 exon 33 | Non-synonymous (Leu-Pro) | A/G | 0.17 | G |
| rs10054110 | 41054209 | FLJ40243 exon 27 | Non-synonymous (Asn-Lys) | A/T | 0.64 | A |
aPosition on chromosome 5 according to UCSC Genome Browser, March 2006 freeze.
bReference allele underlined.
cFrequency of the reference allele for the Centre d’Etude du Polymorphisme Humain (CEPH) individuals of European (CEU) origin (HapMap).
Tissue-specific expression of FLJ40243
The function of FLJ40243 is unknown, but it is known to encode for a protein. Based on the human GNF Expression Atlas 2 Data (UCSC Genome Browser, hg18 assembly, March 2006), FLJ40243 is expressed at an extremely low level at least in ovary, testis, adrenal gland, pancreas, kidney, cerebellum, caudate nucleus, globus pallidus, olfactory bulb, ciliary ganglion and dorsal root ganglia.
No expression of FLJ40243 was observed in peripheral blood mononuclear cells (PBMCs) of which we had RNA available (data not shown). Thus, we could not test whether the identified risk haplotype has an effect on FLJ40243 expression in MS. We further monitored the expression of FLJ40243 in several tissues using human tissue cDNA panels (Clontech, Mountain View, CA, USA) and FLJ40243-specific PCR probes. The fetal (fetal brain, lung, liver, kidney, heart, spleen, thymus and skeletal muscle) and immune (adult spleen, lymph node, thymus, tonsil, bone marrow and peripheral blood leukocyte) cDNA panels were tested. The gene was observed to be expressed at very low level only in spleen, lymph node, fetal liver and fetal skeletal muscle (data not shown).
Identified risk haplotype correlates with complement levels
The C7 gene is an excellent functional candidate for an inflammatory disease like MS, for it encodes the seventh component of the complement system. C7 is mainly expressed in cell types impractical for RNA quantification studies (for example, hepatocytes and endothelial cells). No expression of C7 was observed in PBMCs (data not shown).
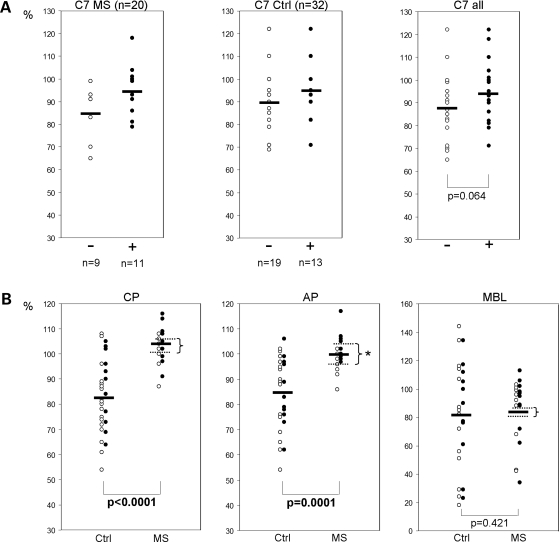
To evaluate, whether the identified risk haplotype has an effect on complement levels, we collected serum and plasma samples from 20 Finnish MS cases and 32 unaffected controls. Of the MS cases 11 and of the controls 13 carried the C7-FLJ40243 risk allele, either as heterozygote (MS n = 9 and Ctrl n = 11) or as homozygote (MS n = 2 and Ctrl n = 2). Most of the plasma C7 protein levels were within the reference range. However, we observed a distinct trend, the C7 protein levels being higher in the C7-FLJ40243 risk allele carriers than in non-carriers (Fig. 2A), although the difference did not reach a statistical significance in this small set of samples (P = 0.064). The difference was seen both in MS cases (mean no risk allele = 85% and mean risk allele = 94%) and in unaffected controls (mean no risk allele = 90% and mean risk allele = 95%).
Figure 2.
C7 protein levels and the amount of formed TCC following complement activation C7 protein levels and total complement activity were measured in 20 Finnish MS cases (MS) and in 32 unaffected Finnish controls (Ctrl). Eleven of the cases and 13 of the controls were known to carry the C7-FLJ40243 risk haplotype, whereas 9 of the MS cases and 19 of the controls did not carry the risk allele. The dots indicate individual values, whereas the horizontal lines indicate the mean value of each group. P-values were calculated with the non-parametric Mann–Whitney U test. (A) Plasma complement component 7 protein levels (C7, measured with radial immunodiffusion) were noticed to be consistently higher in carriers of the risk haplotype (+, black dots) compared with non-carriers (−, open dots). However, the difference was not statistically significant in this relatively small study sample. Reference range: 80–120%. (B) Amount of serum terminal complement complexes (TCC, C5b–C9, measures with ELISA), formed as a consequence of the activation of each (classical = CP, alternative = AP, lectin = MBL) pathway, was observed to be higher in MS patients compared with controls. Reference values: CP > 60%, AP > 40%, MBL > 10%. Carriers of the risk allele are indicated with black dots and the non-carriers with open dots. The complement system was most active in MS cases carrying the identified C7-FLJ40243 risk allele. The star (*) indicates the statistically significant difference between the mean values of the carriers (upper dashed line) and non-carriers (lower dashed line) of the risk haplotype identified to be enriched in the MS high-risk region.
C7 is a component of the terminal complement complex (TCC, C5b-9), which, when assembled on a cell membrane, forms the cytolytic membrane attack complex (MAC). Thus, also the total complement activity was studied in the same set of MS cases and controls by measuring the number of TCC formed as a consequence of activation of each pathway: classical (CP), alternative (AP) and lectin (MBL). The MS cases were observed to have a significantly increased number of functional TCC generated via the classical and the alternative pathway when the complement cascade was activated compared with the controls: CP mean Ctrl = 83%, mean MS = 104%, P < 0.0001; AP mean Ctrl = 85%, mean MS = 100%, P = 0.0001 (Fig. 2B). Importantly, the complement system was most active in MS cases carrying the identified C7-FLJ40243 risk allele (MS cases, AP: no risk allele = 96%, risk allele = 104%, P = 0.0049; MS cases, CP: no risk allele = 101%, risk allele = 106%, P> 0.05) (Fig. 2B).
The effect of the two associated loci on 5p, C7-FLJ40243 and IL7R
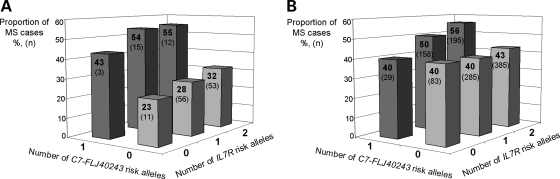
The two reported MS associated loci, C7-FLJ40243 and IL7R, are 5.0 Mb apart from each other and there is no LD between the two loci. We looked for a potential sum effect of the risk alleles to show how these two independent MS risk loci factors contribute to the probability of developing the disease in an additive way (Fig. 3). Individuals carrying the C7-FLJ40243 risk allele seem to be in higher risk to get the disease compared with non-carriers, and the risk is even higher when an additional IL7R risk allele is present. This was observed both in the study set from the Southern Ostrobothnia high-risk region (Fig. 3A) and in the combined study set from more heterogeneous populations, namely Finland outside the high-risk region and Sweden (Fig. 3B).
Figure 3.
Effect of the C7-FLJ40243 and IL7R risk alleles on MS predisposition. Y-axis refers to the proportion (%) of the MS cases among all the individuals of the study set (cases and controls) carrying a certain number of the IL7R (0, 1 or 2 copies) and C7-FLJ40243 (0 or 1 copies) risk alleles. The population-specific C7-FLJ40243 risk allele was selected according to the association results in Table 3. The absolute number of the MS cases in each risk group is shown within the bars in parenthesis. Groups with two copies of the C7-FLJ40243 risk alleles were excluded from the analysis due to the small number of individuals. IL7R genotypes were not available for the study sets from Norway and USA, thus, these samples were not included in the analysis. (A) Southern Ostrobothnia high-risk region. (B) Two study sets from more heterogeneous population, Finland outside the high-risk region and Sweden, combined.
DISCUSSION
Identification of genes predisposing to MS has proven to be a daunting task. A large number of genome-wide linkage scans have been conducted, but the strong linkage and association with the HLA locus remained to be the only systematically replicated finding with a genome-wide significance until 2007 (8). Finally, the first MS GWA study (10), together with other studies (11,12), revealed two additional loci, IL7R and IL2RA, associated with an increased risk of MS. These loci have now been replicated by independent groups of investigators (30–32) and are associated with MS susceptibility at a genome-wide level of significance. Despite of initial disappointment often faced with HLA-associated diseases, we are beginning to uncover the genetic architecture of this disease. However, the risk alleles in the IL7R and IL2RA are estimated to explain only a small proportion (0.2%) of the genetic risk of MS (10,33). Many more such low penetrance alleles remain to be discovered using conventional GWA approaches, but a change of tack will also be needed to discover relatively rare alleles, enriched in some populations or familial study samples, and potentially having a stronger effect on disease susceptibility. Here, we have attempted to pinpoint a rare MS predisposing gene on the 5p linkage region, for which most of the linkage information emerges from an internal founder population. An effort was previously made to narrow the wide locus (34), but only the most ‘telomeric’ 20 cM of the original wide 40 cM-linked region was analyzed in the restriction step, as the LODs were highest (maximum LOD 3.4) in that area. Importantly, no gene in the restricted region was found to explain the linkage observed (data not shown). Here, the complete ∼40 cM-linked region was scanned using the Illumina HumanHap300 panel.
In this linkage-guided association study, we initially used the haplotype association analysis results for ranking the most interesting regions for follow-up. Scanning of the complete linkage region of 45 Mb revealed strong association only with one haplotype flanking the C7 and FLJ40243 genes, and this finding was validated in an independent case–control set originating from the same high-risk geographical region. The same markers also provided nominal evidence for association in a family based analysis. The association analysis combining all the samples from the high-risk region resulted in a P-value of 3.2 × 10−6, which remained significant even after the conservative Bonferroni correction.
Eighty-nine percentage of the MS cases used in the initial screen had both parents born within the MS high-risk region, and genealogical studies showed that majority of the patients were distantly related. Thus, our study design was specifically aimed to enrich for relatively rare, potentially more penetrant variants. Indeed, the identified risk allele was found to be present only in ∼4% of alleles in the general European population, whereas it has a slightly higher frequency of ∼6% in the Asian populations and it is almost absent among Africans, Southern Americans and Oceanians. Importantly, it has obviously become enriched in the Southern Ostrobothnian high-risk MS region due to the founder effect and isolation, and is found in 12% of cases. Within the high-risk region, carriers of the C7-FLJ40243 risk allele have higher risk to get the disease compared with non-carriers (OR: 2.7), whereas the previously identified MS risk alleles in IL7R and IL2RA have been reported to have OR of 1.2 (32). Suggestive evidence for association with C7-FLJ40243 alleles and MS was seen also in more heterogeneous populations, but as expected, there seems to be more allelic heterogeneity within the region, and the causative variant is most probably carried in diverse allelic backgrounds.
The advantages of the Finnish population and, especially, its subisolates are that most of the affected individuals typically share the same major risk allele and that the relatively rare variants can be exposed by the common HapMap markers due to the wide LD intervals (35). However, it is worth noting that these relatively rare variants identified using population isolates are much more challenging to detect with common HapMap SNPs in more heterogeneous populations having distinct LD patterns. Moreover, it is also possible that there are various causative mutations within the same gene in different populations. Thus, replication of the original allelic association in a more heterogeneous population might not be straightforward. This holds true, for example, for familial hypercholesterolemia [FH (MIM #143890)], an autosomal dominant disorder, where majority of mutations occur in the gene encoding low-density lipoprotein receptor (LDLR) and in two other genes (36). There is notable allelic heterogeneity in FH patients worldwide. Over 1000 mutations have been reported (37), and allelic heterogeneity is seen even within Finland in LDLR: there is a founder mutation (FH-North Karelia) found as the major mutation in a limited region in the Northeastern part of Finland and other mutations are found elsewhere in Finland (38).
C7 has previously been studied as an MS candidate gene (39). Three SNPs, C7-327 corresponding to rs7713884 in intron 13, C7-363 corresponding to rs13157656 in exon 14 and a SNP called C7-396, were genotyped in 227 MS families (including only 59 whole trio families) and 72 controls originating from UK. Suggestive evidence for linkage was observed (P = 0.0061). Further, SNP rs7713884 provided nominal evidence for association in the case–control analysis (P = 0.009), but family based association with the transmission disequilibrium test (TDT) was not seen with any of the SNPs, and the authors concluded that C7 does not confer susceptibility to MS. Notably the family based study was underpowered.
Both human C7 (C7) and FLJ40243 (RIKEN cDNA 4930455B06 gene) exist also in the corresponding location of the mouse chromosome 15 (Mouse Genome Informatics database, http://www.informatics.jax.org/). The genes coding for the sixth and ninth components of the complement cascade [C6 (MIM +217050), C9 (MIM +120940)] are also located in the chromosome 5-linked region, but no signal of association with MS was seen with those in the Finnish GWA study sample.
Based on our results, neither C7 nor FLJ40243 was expressed in PBMCs. Thus, expression levels of these genes could not be measured in RNA samples obtained from mononuclear cells of MS patients. However, we tested expression of FLJ40243 in human tissue cDNA panels. FLJ40243 was observed to be expressed in spleen, lymph node, fetal liver and fetal skeletal muscle, but no evidence for expression was seen in fetal brain, adult thymus, tonsil, bone marrow or peripheral blood leukocytes.
C7 is synthesized not only by hepatocytes, but also by endothelial cells, polymorphonuclear cells, macrophages, platelets, fibroblasts, synovial tissue and even in the CNS by astrocytes and oligodendrocytes (40–44), cells relevant for MS. We measured circulating C7 protein levels and total complement activity in MS patients and in unaffected controls. The causative variant of the C7-FLJ40243 risk region is yet unknown, but based on our results, it may affect protein levels of the complement component 7, which is an excellent functional candidate for MS. Importantly, the complement system was observed to be significantly more active among MS cases compared with unaffected population controls; the MS patients carrying the identified C7-FLJ40243 risk haplotype producing most functional TCCs when the complement cascade gets activated.
The complement system is a biochemical cascade of the innate immune system that helps to clear pathogens from the organism. In addition, the first components (C1–C4) of the classical and the lectin pathway are important in the clearance of apoptotic cells and cellular debris (45). Activation of C3 by the classical, lectin or alternative pathway leads to activation of the terminal components (C5–C9), which then form the TCC (C5b–C9), also called as the MAC when assembled on a cell membrane. The MAC forms a transmembrane channel on the cell membrane of the target cell, resulting in the osmotic lysis of the target. The classical complement pathway requires antibodies for activation, whereas the alternative pathway can be activated directly by microbial surfaces and the lectin pathway by certain carbohydrates, such as mannan. The end result of each pathway is in any case the formation of the MAC.
C7 is a single-chain plasma glycoprotein involved in the cytolytic phase of the complement activation cascade. C7 deficient individuals (MIM #610102) are prone to Neisseria meningitidis infections, because the MAC complex, which is important in destroying gram-negative bacteria by cytolysis, cannot be formed correctly. Importantly, C7 is a critical limiting factor of complement activation: only when the local expression of C7 is sufficient, C7 binds to preformed C5b6 and the resulting C5b–C7 complex is able to insert into the phospholipid membrane to start the formation of the MAC (46). The oligodendrocytes are, especially, sensitive to complement-mediated injury because of the relative deficiency of protective complement regulatory proteins, which normally protect host cells from complement-mediated lysis (47).
Several publications support the hypothesis that complement activation and especially the MAC may play a key role in the pathogenesis of autoimmune demyelination. Increased levels of terminal complement complexes (C5b–9) have been detected in the cerebrospinal fluid of MS patients during relapses, and their levels have been shown to correlate with the Expanded Standard Disability Status Scale score, measuring the neurological disability (48). C6-deficient rats, unable to form the MAC, exhibit no demyelination and show a significantly reduced clinical score in the Ab-mediated demyelinating form of EAE, the animal model of MS (49). Moreover, C9 deposition (corresponding to the number of MAC complexes), P-selectin expression and cellular infiltrates were observed to be reduced in the spinal cords of these rats compared with rats capable of forming the MAC (50). On the other hand, mice lacking the complement regulatory protein CD59, which normally protects host cells from complement-mediated damage by blocking the formation of the MAC, have more severe EAE and increased demyelination, inflammatory cell infiltration and axonal injury (51).
It is tempting to speculate that the causative variant of the C7-FLJ40243 region may lead to increased complement activation in the CNS and further to demyelination by damaging the defenceless myelin forming oligodendrocytes. MS can be classified into four different pathological patterns based on the composition of active lesions (52). Pattern II, accounting for over 50% of patients, is characterized by immunoglobulins and MAC deposition at the areas of myelin destruction. Thus, the C7-FLJ40243 risk allele identified here may predispose, especially, to this type of disease.
To summarize, the C7-FLJ40243 risk region, here shown to contain a risk allele enriched in a population isolate, is an excellent candidate for the susceptibility of autoimmune demyelination and seems to be a second MS locus in this region on 5p, initially identified by linkage in multiple study samples. Our observations highlight the value of population isolates in the identification of rare disease alleles in common diseases and further demonstrate the complexity of the genome regions initially identified as potential risk loci.
MATERIALS AND METHODS
Study sample
We selected 72 MS patients, having either both parents born within the Southern Ostrobothnia high-risk region for MS (n = 64) or one high-risk region born parent as well as a family history of MS (n = 8), for our Finnish GWA study. In addition, 68 IBS-matched population controls from Finnish genome-wide studies (from a total of 227 control individuals with GWA data) were used as the control set (Table 1, FIN-isolate1). Fourteen of the controls were known to have both parents born in Southern Ostrobothnia (Supplementary Material, Fig. S1, SOB Ctrls), while we did not have the information of the place of birth of the parents for rest of the controls. However, 13 of the controls were known to live in Southern Ostrobothnia (Supplementary Material, Fig. S1, SOB living Ctrls), and 41 controls were part of the Health 2000 project (Supplementary Material, Fig. S1, H2000 selected Ctrls) and have been used as controls in a genome-wide study of schizophrenia (O. Pietiläinen et al., manuscript in preparation). Genealogical studies enabled us to construct two large interconnected mega-pedigrees, and 41 MS cases out of 72 were noticed to belong to either one (n = 14) or both (n = 27) of them.
An independent set of 125 MS cases, having at least one parent born within the Southern Ostrobothnia high-risk region (including 74 MS cases with both parents born in the high-risk region), and 365 population controls, collected from the same region in Southern Ostrobothnia, were used to validate the initially observed association (Table 1, FIN-isolate2). Nine first-degree relatives of the cases of the GWA study were included in this validation sample set.
Additional family members (33 MS patients and 392 parents or sibs) were available for 156 MS patients included either in the FIN-isolate1 or FIN-isolate2 sets. Forty-one of the MS cases in these case–control sets had no healthy family members available and were not included in the family based analysis. We could construct 22 multiplex (two to six affected individuals per each pedigree) and 134 nuclear (one affected individual per family) MS families originating from Southern Ostrobothnia high-risk region to be used in the family based association analysis. Of the nuclear families, 48 contained two parents, 57 contained one parent and 76 had one or more healthy siblings.
A total of 725 unrelated MS cases and 959 population controls from Finland outside the high-risk region were analyzed as one case–control study set from more heterogeneous populations (Table 1, FIN-OUT). In addition, 651 MS cases and 651 controls from Sweden, 359 MS cases and 471 controls from Norway and 920 MS cases and 177 controls from United States (Table 1, SWE, NOR, US) were obtained from the collaborators of the Nordic MS Genetics Network and the Partners Multiple Sclerosis Center in Boston, MA, USA.
The diagnosis of MS has strictly followed Poser’s or McDonald’s diagnostic criteria (53,54). All the samples were of Northern European descent. All individuals have given their informed consent, and the study has been approved by the Ethics Committee for Ophthalmology, Otorhinolaryngology, Neurology and Neurosurgery in the Hospital District of Helsinki and Uusimaa (Decision 46/2002, Dnro 192/E9/02) and the Ethics Committees of the institutions involved.
SNP selection and genotyping
Illumina HumanHap300 SNP chip (Illumina, San Diego, CA, USA) was used to genotype samples of the Finnish MS project. Genotyping was performed according to manufacturer’s instructions at the Broad Institute, Cambridge, MA, USA and at the Finnish Genome Center, Helsinki, Finland. The samples for schizophrenia study were genotyped with Illumina HumanHap300 chip at DeCode Genetics, Reykjavik, Iceland. Alleles were called on TOP-strand orientation for each project. Only samples and markers with success rates >95% were included in the analyses.
SNPs used to screen the original linked region map to 11.1–56.0 Mb according to UCSC Genome Browser (hg18 assembly, March 2006, http://genome.ucsc.edu/). The identified eight SNP haplotype region (40 996 921–41 039 556 according to UCSC Genome Browser, hg18 assembly, March 2006) was covered with tag SNPs using the HapMap tagger tool [r2 = 0.9, minor allele frequency (MAF) > 0.05] (http://hapmap.org/), resulting in 24 SNPs with an average 2.4 kb inter-SNP interval: rs1901167, rs324061, rs1315656, rs3805221, rs3805226, rs3828511, rs6860438, rs1055021, rs7732104, rs4957361, rs13161930, rs2122564, rs1376174, rs7736582, rs896117, rs1530812, rs3817324, rs2167229, rs16870603, rs7712140, rs2305314, rs1450658, rs1450659 and rs10058056. All the 24 SNPs were genotyped in the Finnish study set from the Southern Ostrobothnian isolate (Table 1, FIN-isolate2), whereas only 14 of these SNPs (rs1901167, rs324061, rs13157656, rs3805221, rs3805226, rs1055021, rs7732104, rs2122564, rs7736582, rs896117, rs3817324, rs2305314, rs1450658 and rs10058056) were genotyped in other study materials (Table 1, FIN-OUT, SWE, NOR, US; controls for complement level assay), because they provide sufficiently enough information to estimate the main C7-FLJ40243 haplotypes (freq ≥ 0.05), capture 92% of the common variation with r2> 0.5 and 75% of the variation with r2 > 0.8.
In addition, four SNPs (rs11567701, rs6897932, rs3194051 and rs6871748) mapping to the IL7R gene and the 3′ flanking region were selected based on previous publications (10–12,27).
The SNPs tagging the C7-FLJ40243 risk region and the SNPs mapping to the IL7R gene were genotyped using the Sequenom’s MassARRAY iPLEX system (Sequenom, San Diego, CA, USA). The SNP assays were designed using Assay Designer 3.1 (Sequenom), and the PCR and extension reactions were done as specified by the manufacturer (specific details available upon request). We used 15 ng of genomic DNA/SNP-multiplex. Genotypes were both automatically called with the automatic caller of the software (Sequenom) and manually verified, as described in detail by Silander et al. (55).
Statistical analyses
Genotypes from controls of the schizophrenia project were merged with the MS study set, and genome-wide IBS sharing and multidimensional scaling analyses were performed using PLINK to select the best matching controls (28), resulting in the total number of 68 controls to be used in this study (Supplementary Material, Fig. S1). We then calculated pairwise IBD-sharing estimates for our study set and no first-degree relative pairs were found in any pair combination (case–case, case–control and control–control). There were three case–case pairs with IBD sharing estimates consisting with second-degree relatives (sharing 0.25 of their genome by IBD) and on third-degree relative level (sharing 0.125 of their genome by IBD), there were four case–case, two case–control and one control–control pairs. All the other relationships between all pair combinations were more distant. As the cases and controls were genotyped in different centers, and we had more genealogical data for our cases, we calculated genomic inflation factor (λ) using genome-wide single SNP data in PLINK (28). The genomic inflation factor, λ, was 1.0758 for our data set, which suggests that cases and controls are well-matched and there is no large-scale population stratification within our final study set.
SNPs mapping to 11.1–56.0 Mb according to UCSC Genome Browser (hg18 assembly, March 2006) and passing the quality control criteria in the genome-wide study (minimum of 95% call rate per individual and per SNP) were used in the sliding window five SNP haplotype analysis. All the analyses were performed with the PLINK program (version 1.00). Altogether 3976 consecutive windows were analyzed both for global haplotype association (omnibus P-value) and for the degrees of freedom (number of haplotypes − 1). In addition to global haplotype association, also single haplotype association P-values for each haplotype with a frequency of over 1.0% was calculated. All the results are available as Supplementary Material, Tables S1 and S2 and are represented as uncorrected P-values.
SNP genotypes in the C7-FLJ40243 validation and IL7R analyses were checked for Hardy–Weinberg equilibrium using Pearson’s χ2 test. Allele and genotype frequencies were determined from the data. Only markers with MAF over 5%, overall genotyping success rate >95% and validity of Hardy–Weinberg equilibrium in control samples were accepted for statistical analyses. Five SNPs used to form the C7-FLJ40243 haplotype were genotyped using both the Illumina HumanHap300 SNP chip and Sequenom system in 77 individuals. No discrepancies were observed between the two genotyping methods. There were no discrepancies between the 100 duplicate samples, which were added to the sample plates and genotyped using Sequenom system.
The haplotype frequencies for the validation sample set (FIN-isolate2), the combined Southern Ostrobothnian study set (FIN-isolate1 + 2) and the more heterogeneous populations (FIN-OUT, SWE, NOR, US) were estimated using Haploview (version 4.0beta6) (56), and the case–control P-values were obtained from the Haploview custom association test. P-value for the combined Southern Ostrobothnian study set (FIN-isolate1 + 2) was corrected for multiple testing using the permutation option available in Haploview, 1 000 000 permutations. Both the uncorrected and the permutated P-values are shown in the text.
LD between the SNPs flanking the C7 and FLJ40243 genes was measured using Haploview and the publicly available genotype data of Centre d’Etude du Polymorphisme Humain (CEPH) individuals of European origin (Fig. 1C).
To estimate the effect size of the identified risk haplotype in the Finnish isolate, the haplotypes were estimated with the Phase 2.1.1 program (57,58). Haplotypes with probability of >0.6 were accepted for further analyses. Haplotype frequencies estimated by Haploview 4.0beta6 and the Phase 2.1.1 program were almost identical (MS: 0.12 Haploview, 0.12 Phase; Ctrls: 0.04 Haploview, 0.05 Phase). ORs for the C7-FLJ40243 risk haplotype and for the IL7R risk allele were calculated using a Pearson’s χ2 test, comparing allele carriage counts between cases and controls (Fig. 1C and Table 2).
The SNPs in MS families were analyzed using the gamete competition option of the Mendel 7.0.0 program (29), which utilizes the genotype data of the whole pedigree and is better adapted to missing data than the classical TDT test.
P-values for the C7 protein and total complement activity differences between carriers and non-carriers of the risk haplotype as well as for the cases and the controls were calculated with the non-parametric Mann–Whitney test, because the number of individuals studied was relatively small.
Risk alleles for both the C7-FLJ40243 region (population-specific risk haplotype, see Table 3) and IL7R (rs6897932 allele C) were available for 150 MS cases and 323 controls from the Southern Ostrobothnia high-risk region and for 1135 MS cases and 1402 controls of the combined study set from Finland outside the high-risk region and Sweden, and these individuals were analyzed to estimate the co-effect of the C7-FLJ40243 and IL7R risk alleles on MS predisposition (Fig. 3). IL7R genotypes for the Swedish individuals were obtained from the collaborators of the MS Nordic Genetics Network. IL7R genotypes were not available for the study sets from Norway and USA. Groups with two copies of the C7-FLJ40243 risk alleles were excluded from the analysis due to the small number of individuals (two C7-FLJ40243 risk alleles and zero IL7R risk alleles: Southern Ostrobothnia MS n = 0; other parts of Finland and Sweden MS n = 4).
A test of heterogeneity between the study sets from the high-risk isolate and rest of Finland was performed to allow for a combined analysis for IL7R. Mantel–Haenszel corrected P-values were calculated using PLINK (28).
Sequencing
In 890 bases of the C7 promoter region, all the exons of the C7 gene and exons 25–42 of FLJ40243 (3′ end, covered by the risk haplotype) were sequenced in four MS cases with two copies, four MS cases with one copy and eight controls with no copies of the C7-FLJ402437 risk haplotype of the high-risk region. Four of the controls had only the head or the tail of the C7-FLJ402437 risk haplotype, and these controls enabled us to sort out the variants, which were in LD with only the head or the tail, but were not inherited with the actual risk haplotype (head and tail). Coverage of the sequencing was 100%. All the individuals sequenced originated from the Finnish high-risk region. Sequencing was done both in the Broad Institute Center for Genotyping and Analysis (C7) and in the National Public Health Institute of Finland (FLJ40243). Briefly, the primer design was done using Primer3 (http://primer3.sourceforge.net/), 10 µl PCRs were cleaned up using a SAP/Exo mix (Amersham Biosciences, Piscataway, NJ, USA) or SOPE Resin (Edge Biosystems Gaithersburg, MD, USA), and detection was done using ABI 3730xl’s (Applied Biosystems, Foster City, CA, USA).
Expression of FLJ40243 and C7
The primer design was done using Primer3. Primers (C7: 5′-GGCCAGATGTGGAGAAGATT-3′ and 5′-ATGCCATCCATCAGTACAGG-3′; FLJ40243: 5′-TGATTTCATTCCTTCTGCACCT-3′ and 5′-CACAGAATTGCCTGTAGAAATCC-3′) mapped to exons within the risk haplotype region.
Tissue expression of FLJ40243 was tested by PCR analysis using the Human Fetal and Immune cDNA panels (Clontech). Tissue-specific products were detected by agarose gel electrophoresis.
Expression of FLJ40243 and C7 was further tested in mononuclear cell samples using the same method. These mononuclear cells are collected from 10 Finnish MS patients. All donors have given their informed consent and the study has been approved by the Ethics Committee for Ophthalmology, Otorhinolaryngology, Neurology and Neurosurgery in the Hospital District of Helsinki and Uusimaa (Decision 46/2002, Dnro 192/E9/02). The PBMCs were collected using Vacutainer CPT tubes (Becton-Dickinson, Franklin Lakes, NJ, USA). The total cellular RNA was extracted using RNAeasy Plus Mini Kit columns (Qiagen, Valencia, CA, USA) according to the manufacturers’ instructions. The quality of the RNA was assessed using the RNA 6000 Nano assay in the Bioanalyzer (Agilent, Foster City, CA, USA) monitoring for ribosomal S28/S18 RNA ratio (acceptable 1.5–2.5), signs of degradation and DNA contamination. The concentration and the A260/A280 ratio of the samples were measured using a spectrophotometer, the acceptable ratio being 1.8–2.2. Oligo(dT) primer and the TaqMan Gold RT–PCR kit (Applied Biosystems) were used to perform first-strand cDNA synthesis starting from 1 µg of total RNA, according to manufacturer’s instructions.
Complement level quantification
Serum and plasma samples of 20 Finnish MS cases from the Southern Ostrobothnia high-risk region were collected, and the total complement activity as well as the C7 protein levels were measured at the Department of Medical Microbiology and Immunology, University of Turku, Finland. All donors have given their informed consent and the study has been approved by the Ethics Committee for Ophthalmology, Otorhinolaryngology, Neurology and Neurosurgery in the Hospital District of Helsinki and Uusimaa (Decision 46/2002, Dnro 192/E9/02). Two of the MS cases had two C7-FLJ40243 risk alleles of the isolate, nine had one risk allele and nine had no risk alleles. There was approximately the same number of females (no risk alleles 63% and risk allele 55%) and users of immunomodulation drugs (cortisone during 6 months, interferon β or glatirameracetate; no risk alleles 43% and risk allele 33%) in different risk groups. Only one patient had a primary progressive form of MS (no risk allele). A Wielisa total complement system screen kit (COMPL 300, Wieslab, Lund, Sweden) was used for measuring functional activity of the classical (CP), alternative (AP) and lectin (MBL) pathways of the complement system in serum (59). In brief, dilutions of sera were incubated at 37°C in microtiter wells precoated with IgM, LPS or mannan for activation of the CP, AP and MBL pathways, respectively. After washing, alkaline phosphatase-conjugated antihuman C5b-9 was added before incubation for 30 min at room temperature. After additional washings and incubation with a substrate, the absorbance was measured at 405 nm. Reference values are CP > 60%, AP > 40%, MBL > 10% (MBL deficiency < 10%, decreased 10–40%). Plasma C7 protein levels were measured by radial immunodiffusion (Mancini technique) using goat anti-human-C7 antiserum (A308, Quidel, San Diego, CA, USA). Reference values are 80–120%. The results are quantitative.
A set of 150 healthy Finnish individuals was obtained as described in more detail by Seppänen et al. (60). These control samples have been collected from Helsinki and represent mixed Finnish population. Thirteen of these controls were noticed to carry the C7-FLJ40243 risk allele of the FIN-OUT. In addition to these 13 risk allele carriers, 19 non-carriers of the risk allele were selected for the complement level assays. There was approximately the same number of females in the control group compared with the MS group (no risk alleles 68% and risk allele 54%). The total complement activity and the C7 protein levels were measured at the Department of Medical Microbiology and Immunology, University of Turku, Finland, using the same methods described above.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health (grant RO1 NS 43559), Center of Excellence for Disease Genetics of the Academy of Finland, the Sigrid Juselius Foundation, the Biocentrum Helsinki Foundation, Helsinki University Central Hospital Research Foundation, the Multiple Sclerosis Foundation of USA, a Harry Weaver Neuroscience Scholar Award from National Multiple Sclerosis Society (P.L.D.) and the Neuropromise EU project (grant LSHM-CT-2005-018637). The Broad Institute Center for Genotyping and Analysis is supported by the National Center for Research Resources (grant U54 RR020278). The genotyping of the Health 2000 controls was funded by the SGENE EU project (LSHM-CT-2006-037761) and Simons Foundation (R01MH71425-01A1).
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank all participating MS patients and families, the Finnish Genome Center, Dr Robert Onofrio and Dr Daniel Mirel for their help with sequencing, Mrs Anne Nyberg for her help in genotyping and Mrs Seija Laanti for expert laboratory help in complement determinations. The Norwegian Bone Marrow Donor Registry is acknowledged for collaboration in establishment of the Norwegian control material.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Sadovnick A.D., Armstrong H., Rice G.P., Bulman D., Hashimoto L., Paty D.W., Hashimoto S.A., Warren S., Hader W., Murray T.J., et al. A population-based study of multiple sclerosis in twins: update. Ann. Neurol. 1993;33:281–285. doi: 10.1002/ana.410330309. [DOI] [PubMed] [Google Scholar]
- 2.Ebers G.C., Sadovnick A.D., Risch N.J. A genetic basis for familial aggregation in multiple sclerosis. Canadian Collaborative Study Group. Nature. 1995;377:150–151. doi: 10.1038/377150a0. [DOI] [PubMed] [Google Scholar]
- 3.Oksenberg J.R., Barcellos L.F. Multiple sclerosis genetics: leaving no stone unturned. Genes Immun. 2005;6:375–387. doi: 10.1038/sj.gene.6364237. [DOI] [PubMed] [Google Scholar]
- 4.Mumford C.J., Wood N.W., Kellar-Wood H., Thorpe J.W., Miller D.H., Compston D.A. The British Isles survey of multiple sclerosis in twins. Neurology. 1994;44:11–15. doi: 10.1212/wnl.44.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Hansen T., Skytthe A., Stenager E., Petersen H.C., Brønnum-Hansen H., Kyvik K.O. Concordance for multiple sclerosis in Danish twins: an update of a nationwide study. Mult. Scler. 2005;11:504–510. doi: 10.1191/1352458505ms1220oa. [DOI] [PubMed] [Google Scholar]
- 6.Compston A. Genetic epidemiology of multiple sclerosis. J. Neurol. Neurosurg. Psychiatry. 1997;62:553–561. doi: 10.1136/jnnp.62.6.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jersild C., Svejgaard A., Fog T. HL-A antigens and multiple sclerosis. Lancet. 1972;1:1240–1241. doi: 10.1016/s0140-6736(72)90962-2. [DOI] [PubMed] [Google Scholar]
- 8.Dyment D.A., Ebers G.C., Sadovnick A.D. Genetics of multiple sclerosis. Lancet Neurol. 2004;3:104–110. doi: 10.1016/s1474-4422(03)00663-x. [DOI] [PubMed] [Google Scholar]
- 9.Hafler D.A., Slavik J.M., Anderson D.E., O’Connor K.C., De Jager P., Baecher-Allan C. Multiple Sclerosis. Immunol. Rev. 2005;204:208–231. doi: 10.1111/j.0105-2896.2005.00240.x. [DOI] [PubMed] [Google Scholar]
- 10.International Multiple Sclerosis Genetics Consortium (IMSGC) Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 11.Lundmark F., Duvefelt K., Iacobaeus E., Kockum I., Wallstrom E., Khademi M., Oturai A., Ryder L.P., Saarela J., Harbo H.F., et al. Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat. Genet. 2007;39:1108–1113. doi: 10.1038/ng2106. [DOI] [PubMed] [Google Scholar]
- 12.Gregory S.G., Schmidt S., Seth P., Oksenberg J.R., Hart J., Prokop A., Caillier S.J., Ban M., Goris A., Barcellos L.F., et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007;39:1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 13.Kuokkanen S., Sundvall M., Terwilliger J.D., Tienari P.J., Wikstrom J., Holmdahl R., Pettersson U., Peltonen L. A putative vulnerability locus to multiple sclerosis maps to 5p14-p12 in a region syntenic to the murine locus Eae2. Nat. Genet. 1996;13:477–480. doi: 10.1038/ng0896-477. [DOI] [PubMed] [Google Scholar]
- 14.Kuokkanen S., Gschwend M., Rioux J.D., Daly M.J., Terwilliger J.D., Tienari P.J., Wikström J., Palo J., Stein L.D., Hudson T.J., et al. Genomewide scan of multiple sclerosis in Finnish multiplex families. Am. J. Hum. Genet. 1997;61:1379–1387. doi: 10.1086/301637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebers G.C., Kukay K., Bulman D.E., Sadovnick A.D., Rice G., Anderson C., Armstrong H., Cousin K., Bell R.B., Hader W., et al. A full genome search in multiple sclerosis. Nat. Genet. 1996;13:472–476. doi: 10.1038/ng0896-472. [DOI] [PubMed] [Google Scholar]
- 16.Oturai A., Larsen F., Ryder L.P., Madsen H.O., Hillert J., Fredrikson S., Sandberg-Wollheim M., Laaksonen M., Koch-Henriksen N., Sawcer S., et al. Linkage and association analysis of susceptibility regions on chromosomes 5 and 6 in 106 Scandinavian sibling pair families with multiple sclerosis. Ann. Neurol. 1999;46:612–616. [PubMed] [Google Scholar]
- 17.Eraksoy M., Kurtuncu M., Akman-Demir G., Kilic M., Gedizlioglu M., Mirza M., Anlar O., Kutlu C., Demirkiran M., Idrisoglu H.A., et al. A whole genome screen for linkage in Turkish multiple sclerosis. J. Neuroimmunol. 2003;a 143:17–24. doi: 10.1016/j.jneuroim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Eraksoy M., Hensiek A., Kurtuncu M., Akman-Demir G., Kilic M., Gedizlioglu M., Petek-Balci B., Anlar O., Kutlu C., Saruhan-Direskeneli G., et al. A genome screen for linkage disequilibrium in Turkish multiple sclerosis. J. Neuroimmunol. 2003;b 143:129–132. doi: 10.1016/j.jneuroim.2003.08.027. [DOI] [PubMed] [Google Scholar]
- 19.Sundvall M., Jirholt J., Yang H.T., Jansson L., Engström A., Pettersson U., Holmdahl R. Identification of murine loci associated with susceptibility to chronic experimental autoimmune encephalomyelitis. Nat. Genet. 1995;10:313–317. doi: 10.1038/ng0795-313. [DOI] [PubMed] [Google Scholar]
- 20.Varilo T., Peltonen L. Isolates and their potential use in complex gene mapping efforts. Curr. Opin. Genet. Dev. 2004;14:316–323. doi: 10.1016/j.gde.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Sumelahti M.L., Tienari P.J., Wikstrom J., Palo J., Hakama M. Regional and temporal variation in the incidence of multiple sclerosis in Finland 1979–1993. Neuroepidemiology. 2000;19:67–75. doi: 10.1159/000026241. [DOI] [PubMed] [Google Scholar]
- 22.Sumelahti M.L., Tienari P.J., Wikstrom J., Palo J., Hakama M. Increasing prevalence of multiple sclerosis in Finland. Acta Neurol. Scand. 2001;103:153–158. doi: 10.1034/j.1600-0404.2001.103003153.x. [DOI] [PubMed] [Google Scholar]
- 23.Rinne U.K., Panelius M., Kivalo E., Hokkanen E., Palo J. Distribution of multiple sclerosis in Finland, with special reference to some geological factors. Acta Neurol. Scand. 1966;42:385–399. doi: 10.1111/j.1600-0404.1966.tb01191.x. [DOI] [PubMed] [Google Scholar]
- 24.Kinnunen E., Wikström J., Porras J., Palo J. The epidemiology of multiple sclerosis in Finland: increase of prevalence and stability of foci in high-risk areas. Acta Neurol. Scand. 1983;67:255–262. doi: 10.1111/j.1600-0404.1983.tb04574.x. [DOI] [PubMed] [Google Scholar]
- 25.Wikström J., Palo J. Studies on the clustering of multiple sclerosis in Finland I: comparison between the domiciles and places of birth in selected subpopulations. Acta Neurol. Scand. 1975;51:85–98. doi: 10.1111/j.1600-0404.1975.tb01362.x. [DOI] [PubMed] [Google Scholar]
- 26.Tienari P.J., Sumelahti M.L., Rantamäki T., Wikström J. Multiple sclerosis in western Finland: evidence for a founder effect. Clin. Neurol. Neurosurg. 2004;106:175–179. doi: 10.1016/j.clineuro.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z., Duvefelt K., Svensson F., Masterman T., Jonasdottir G., Salter H., Emahazion T., Hellgren D., Falk G., Olsson T., et al. Two genes encoding immune-regulatory molecules (LAG3 and IL7R) confer susceptibility to multiple sclerosis. Genes Immun. 2005;6:145–152. doi: 10.1038/sj.gene.6364171. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lange K., Cantor R., Horvath S., Perola M., Sabatti C., Sinsheimer J., Sobel E. Mendel version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am. J. Hum. Genet. 2001;69:A504. [Google Scholar]
- 30.Ramagopalan S.V., Anderson C., Sadovnick A.D., Ebers G.C. Genomewide study of multiple sclerosis. N. Engl. J. Med. 2007;357:2199–2200. doi: 10.1056/NEJMc072836. [DOI] [PubMed] [Google Scholar]
- 31.O’Doherty C., Kantarci O., Vandenbroeck K. IL7RA polymorphisms and susceptibility to multiple sclerosis. N. Engl. J. Med. 2008;358:753–754. doi: 10.1056/NEJMc0707553. [DOI] [PubMed] [Google Scholar]
- 32.International Multiple Sclerosis Genetics Consortium (IMSGC) Refining genetic associations in multiple sclerosis. Lancet Neurol. 2008;7:567–569. doi: 10.1016/S1474-4422(08)70122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peltonen L. Old suspects found guilty—the first genome profile of multiple sclerosis. N. Engl. J. Med. 2007;357:927–929. doi: 10.1056/NEJMe078147. [DOI] [PubMed] [Google Scholar]
- 34.Riise Stensland H.M., Saarela J., Bronnikov D.O., Parkkonen M., Jokiaho A.J., Palotie A., Tienari P.J., Sumelahti M.L., Elovaara I., Koivisto K., et al. Fine mapping of the multiple sclerosis susceptibility locus on 5p14-p12. J. Neuroimmunol. 2005;170:122–133. doi: 10.1016/j.jneuroim.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Service S., DeYoung J., Karayiorgou M., Roos J.L., Pretorious H., Bedoya G., Ospina J., Ruiz-Linares A., Macedo A., Palha J.A., et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat. Genet. 2006;38:556–560. doi: 10.1038/ng1770. [DOI] [PubMed] [Google Scholar]
- 36.Dedoussis G.V., Schmidt H., Genschel J. LDL-receptor mutations in Europe. Hum. Mutat. 2004;24:443–459. doi: 10.1002/humu.20105. [DOI] [PubMed] [Google Scholar]
- 37.Leigh S.E., Foster A.H., Whittall R.A., Hubbart C.S., Humphries S.E. Update and analysis of the University College London low density lipoprotein receptor familial hypercholesterolemia database. Ann. Hum. Genet. 2008;72:485–498. doi: 10.1111/j.1469-1809.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- 38.Vuorio A.F., Aalto-Setälä K., Koivisto U.M., Turtola H., Nissen H., Kovanen P.T., Miettinen T.A., Gylling H., Oksanen H., Kontula K. Familial hypercholesterolaemia in Finland: common, rare and mild mutations of the LDL receptor and their clinical consequences. Ann. Med. 2001;33:410–421. doi: 10.3109/07853890108995954. [DOI] [PubMed] [Google Scholar]
- 39.Chataway J., Sawcer S., Sherman D., Hobart M., Fernie B., Coraddu F., Feakes R., Broadley S., Gray J., Jones H.B., et al. No evidence for association of multiple sclerosis with the complement factors C6 and C7. J. Neuroimmunol. 1999;99:150–156. doi: 10.1016/s0165-5728(99)00054-5. [DOI] [PubMed] [Google Scholar]
- 40.Gasque P., Fontaine M., Morgan B.P. Complement expression in human brain. Biosynthesis of terminal pathway components and regulators in human glial cells and cell lines. J. Immunol. 1995;154:4726–4733. [PubMed] [Google Scholar]
- 41.Hogasen A.K., Wurzner R., Abrahamsen T.G., Dierich M.P. Human polymorphonuclear leukocytes store large amounts of terminal complement components C7 and C6, which may be released on stimulation. J. Immunol. 1995;154:4734–4740. [PubMed] [Google Scholar]
- 42.Langeggen H., Pausa M., Johnson E., Casarsa C., Tedesco F. The endothelium is an extrahepatic site of synthesis of the seventh component of the complement system. Clin. Exp. Immunol. 2000;121:69–76. doi: 10.1046/j.1365-2249.2000.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hosokawa M., Klegeris A., Maguire J., McGeer P.L. Expression of complement messenger RNAs and proteins by human oligodendroglial cells. Glia. 2003;42:417–423. doi: 10.1002/glia.10234. [DOI] [PubMed] [Google Scholar]
- 44.Morgan B.P., Gasque P. Extrahepatic complement biosynthesis: where, when and why? Clin. Exp. Immunol. 1997;107:1–7. doi: 10.1046/j.1365-2249.1997.d01-890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meri S. Loss of self-control in the complement system and innate autoreactivity. Ann. N. Y. Acad. Sci. 2007;1109:93–105. doi: 10.1196/annals.1398.011. [DOI] [PubMed] [Google Scholar]
- 46.Thompson R.A., Lachmann P.J. Reactive lysis: the complement-mediated lysis of unsensitized cells. I. The characterization of the indicator factor and its identification as C7. J. Exp. Med. 1970;131:629–641. doi: 10.1084/jem.131.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scolding N.J., Morgan B.P., Compston D.A. The expression of complement regulatory proteins by adult human oligodendrocytes. J. Neuroimmunol. 1998;84:69–75. doi: 10.1016/s0165-5728(97)00241-5. [DOI] [PubMed] [Google Scholar]
- 48.Sellebjerg F., Jaliashvili I., Christiansen M., Garred P. Intrathecal activation of the complement system and disability in multiple sclerosis. J. Neurol. Sci. 1998;157:168–174. doi: 10.1016/s0022-510x(98)00086-0. [DOI] [PubMed] [Google Scholar]
- 49.Mead R.J., Singhrao S.K., Neal J.W., Lassmann H., Morgan B.P. The membrane attack complex of complement causes severe demyelination associated with acute axonal injury. J. Immunol. 2002;168:458–465. doi: 10.4049/jimmunol.168.1.458. [DOI] [PubMed] [Google Scholar]
- 50.Tran G.T., Hodgkinson S.J., Carter N., Killingsworth M., Spicer S.T., Hall B.M. Attenuation of experimental allergic encephalomyelitis in complement component 6-deficient rats is associated with reduced complement C9 deposition, P-selectin expression, and cellular infiltrate in spinal cords. J. Immunol. 2002;168:4293–4300. doi: 10.4049/jimmunol.168.9.4293. [DOI] [PubMed] [Google Scholar]
- 51.Mead R.J., Neal J.W., Griffiths M.R., Linington C., Botto M., Lassmann H., Morgan B.P. Deficiency of the complement regulator CD59a enhances disease severity, demyelination and axonal injury in murine acute experimental allergic encephalomyelitis. Lab. Invest. 2004;84:21–28. doi: 10.1038/labinvest.3700015. [DOI] [PubMed] [Google Scholar]
- 52.Lucchinnetti C., Bruck W., Parisi J., Scheithauer B., Rodriguez M., Lassmann H. Heterogeneity of multiple sclerosis lesions: implications for the pathogenesis of demyelination. Ann. Neurol. 2000;47:707–717. doi: 10.1002/1531-8249(200006)47:6<707::aid-ana3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 53.Poser C.M., Paty D.W., Scheinberg L., McDonald W.I., Davis F.A., Ebers G.C., Johnson K.P., Sibley W.A., Silberberg D.H., Tourtellotte W.W. New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann. Neurol. 1983;13:227–231. doi: 10.1002/ana.410130302. [DOI] [PubMed] [Google Scholar]
- 54.McDonald W.I., Compston A., Edan G., Goodkin D., Hartung H.P., Lublin F.D., McFarland H.F., Paty D.W., Polman C.H., Reingold S.C., et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann. Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 55.Silander K., Komulainen K., Ellonen P., Jussila M., Alanne M., Levander M., Tainola P., Kuulasmaa K., Salomaa V., Perola M., et al. Evaluating whole genome amplification via multiply-primed rolling circle amplification for SNP genotyping of samples with low DNA yield. Twin Res. Hum. Genet. 2005;8:368–375. doi: 10.1375/1832427054936664. [DOI] [PubMed] [Google Scholar]
- 56.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 57.Stephens M., Smith N., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stephens M., Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seelen M.A., Roos A., Wieslander J., Mollnes T.E., Sjoholm A.G., Wurzner R., Loos M., Tedesco F., Sim R.B., Garred P., et al. Functional analysis of the classical, alternative, and MBL pathways of the complement system: standardization and validation of a simple ELISA. J. Immunol. Methods. 2005;296:187–198. doi: 10.1016/j.jim.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 60.Seppänen M., Suvilehto J., Lokki M.L., Notkola I.L., Järvinen A., Jarva H., Seppälä I., Tahkokallio O., Malmberg H., Meri S., et al. Immunoglobulins and complement factor C4 in adult rhinosinusitis. Clin. Exp. Immunol. 2006;145:219–227. doi: 10.1111/j.1365-2249.2006.03134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.