Abstract
Duarte galactosemia is a mild to asymptomatic condition that results from partial impairment of galactose-1-phosphate uridylyltransferase (GALT). Patients with Duarte galactosemia demonstrate reduced GALT activity and carry one profoundly impaired GALT allele (G) along with a second, partially impaired GALT allele (Duarte-2, D2). Molecular studies reveal at least five sequence changes on D2 alleles: a p.N314D missense substitution, three intronic base changes and a 4 bp deletion in the 5′ proximal sequence. The four non-coding sequence changes are unique to D2. The p.N314D substitution, however, is not; it is found together with a silent polymorphism, p.L218(TTA), on functionally normal Duarte-1 alleles (D1, also called Los Angeles or LA alleles). The HapMap database reveals that p.N314D is a common human variant, and cross-species comparisons implicate D314 as the ancestral allele. The p.N314D substitution is also functionally neutral in mammalian cell and yeast expression studies. In contrast, the 4 bp 5′ deletion characteristic of D2 alleles appears to be functionally impaired in reporter gene transfection studies. Here we present allele-specific qRT–PCR evidence that D2 alleles express less mRNA in vivo than their wild-type counterparts; the difference is small but statistically significant. Furthermore, we characterize the prevalence of the 4 bp deletion in GG, NN and DG populations; the deletion appears exclusive to D2 alleles. Combined, these data strongly implicate the 4 bp 5′ deletion as a causal mutation in Duarte galactosemia and suggest that direct tests for this deletion, as proposed here, could enhance or supplant current tests, which define D2 alleles on the basis of the presence and absence of linked coding sequence polymorphisms.
INTRODUCTION
Duarte galactosemia is a mild to asymptomatic condition that results from partial impairment of galactose-1-phosphate uridylyltransferase (GALT) [reviewed in (1)]. Individuals with Duarte galactosemia are identified by newborn screening at an incidence as high as one in 4000 live births (2,3), which is close to 10 times the detection rate of classic galactosemia (4). Patients with Duarte galactosemia carry one GALT allele (G) that is profoundly impaired, and a second GALT allele (Duarte-2, D2; sometimes called D) that is partially impaired. Hemolysates from patients with Duarte galactosemia demonstrate, on average, ∼25% normal GALT activity, although there is a broad range in individual values (3). Duarte hemolysates also have a characteristic pattern of altered GALT isozyme mobility on native or isoelectric focusing gels (5–7).
Prior studies have shown that D2 alleles carry the amino acid substitution p.N314D (c.940A>G) (8–11), which fully accounts for the altered isozyme mobility (12), but this amino acid substitution does not cause the partial impairment of activity (11,12). Of note, p.N314D is also found on Duarte-1 alleles (D1, also called Los Angeles or LA alleles), which exhibit normal or even above normal activity (13–15). On the basis of HapMap comparative sequence studies (http://www.hapmap.org/index.html.en), p.N314D is now considered a common variant or polymorphism (16) with an allele frequency of ∼11% in European populations (CEPH, or Centre d’Etude du Polymorphisme Humain) and lower frequencies in other populations, for a ‘pan-ethnic’ frequency near 8% (4).
A number of different approaches have been taken to the question of why D2 alleles are functionally compromised, whereas D1 alleles are not. Andersen et al. (17) used immunochemistry to reveal that the different activities attributed to D1 and D2 alleles reflect differential GALT protein abundance, rather than differential specific activity. Later studies of the coding and non-coding nucleotide sequences of D1 and D2 alleles revealed that p.N314D exists on both alleles in linkage disequilibrium with other sequence variants, and that these variants differ between D1 and D2 alleles. Specifically, D1 alleles carry a c.652C>T nucleotide change that results in a silent substitution at codon 218 (p.L218) (CTA to TTA, p.L218, sometimes called L218L) (14,15,18,19), whereas D2 alleles carry a 4 bp 5′ deletion (c.-119_-116delGTCA) (20) along with three intronic base changes [c.378-27G>C or IVS4-27G>C, c.508-24G>A or IVS5-24G>A, and c.507 + 62G>A or IVS5-62G>A (20–22)]. Both D1 and D2 alleles also carry an extended sequence of adenine nucleotides in intron 10 (22). Large-scale studies of GALT alleles tested for the presence or absence of the p.D314 and p.L218(TTA) sequence variants estimated the pan-ethnic frequencies of D1 and D2 alleles at 2.7 and 5.1%, respectively (4).
The question of which reported base changes account for up to 50% reduction in hemolysate D2 GALT protein levels has remained a point of controversy for many years. One group proposed that the intronic base changes in D2 alleles might compromise processing or expression of the encoded message (15). Another group (14,23) compared the GALT mRNA and protein levels in NN versus DD human cell lines, using RNase protection and western blots, and concluded that the D2 GALT message levels were normal, but that the p.N314D GALT protein was destabilized. These authors proposed that D1 alleles, which also carry p.D314, might fail to manifest destabilization due to improved translation of the p.L218(TTA) codon (14,23). However, mammalian and yeast expression studies found no evidence of compromised p.D314 GALT protein expression or function (11,12).
The plot thickened when Kozak and Francova (20) speculated that the 5′ 4 bp deletion specific to D2 alleles might be functionally significant, because it disrupted the predicted binding sites of two transcriptional activators (AP1Q2 and AP1Q4). Two years later, GALT promoter–luciferase reporter gene studies confirmed that the D2 promoter was indeed less active than the wild-type promoter in transient transfection experiments (24,25). One study (25) also reported diminished total GALT message in lymphoblasts cultured from a Duarte carrier (DN) and a Duarte patient (DG) compared with lymphoblasts cultured from five (NN) controls. These results implicated the 4 bp 5′ deletion and perhaps the other D2 non-coding sequence changes as functionally significant. Nonetheless, variations in GALT message levels between the individual samples in the study combined with the inability to distinguish p.N314 from p.D314 GALT messages in individual samples meant that altered D2 message abundance could not be confirmed as the basis for diminished D2 GALT expression.
Here we present studies that address the origin, distribution and expression of the D2 GALT allele. Specifically, we compared GALT sequences among humans, non-human hominids, non-hominid primates and non-primate placental mammals, and these comparisons clearly implicate p.D314 as the ancestral allele and p.N314 as a recent sequence variant that may be unique to humans. We also report that the 4 bp 5′ deletion in D2 alleles represents a one-unit contraction of a GTCA tetranucleotide repeat whose repeat number has fluctuated through evolution. To track the distribution of this 4 bp deletion within cohorts of patient and control samples, we developed and applied a robust PCR-based assay that confirmed complete linkage of the 4 bp deletion with GALT alleles that also carry p.D314 + L218(CTA), and complete repulsion of the 4 bp deletion with GALT alleles that carry p.D314 + p.L218(TTA). Finally, we applied an allele-specific qRT–PCR approach to quantify the relative abundance of ‘normal’ versus p.D314 GALT messages in four separate DN individuals. Our results confirm a small, but statistically significant, under-representation of p.D314 GALT message in each of these carriers. These data revealed that under-expression, at the mRNA level, contributes to the compromised function of the D2 GALT allele.
RESULTS
Origins of the p.N314D and p.L218(TTA) polymorphisms and the 4 bp 5′ deletion in human GALT
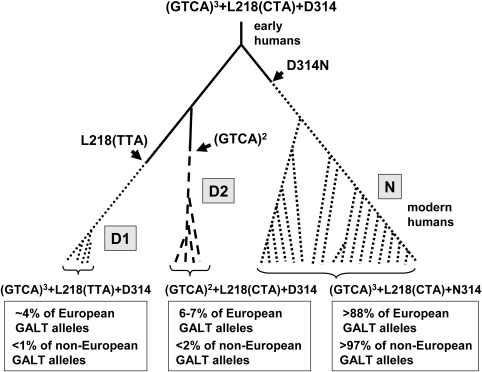
As a first step toward understanding the origin and relationship of the p.N314D (rs2070074 at position 34,639,442 of chromosome 9 in NCBI Build 36.1) and 5′ variations in human GALT, we performed cross-species comparisons of the appropriate coding and non-coding sequences from representative human, non-human hominid (e.g. chimpanzee), non-hominid primate (e.g. macaque) and non-primate placental mammalian (e.g. mouse) species. All species examined, other than human, encoded D rather than N at position 314 (Table 1), strongly implicating D314 as the ancestral GALT allele; the variant that is predominant among modern humans is therefore most appropriately termed p.D314 N (Fig. 1).
Table 1.
Cross-species sequence comparisons suggest origins of the p.N314D and p.L218(TTA) polymorphisms and 5′ promoter deletion characteristic of the D1 and D2 alleles of GALT
| Sequence | Humans (European)a | Other hominids | Other primates | Other placental mammals |
|---|---|---|---|---|
| Amino acid at residue 314 | Asparagine (N) ∼89% | Aspartate (D) | Aspartate (D) | Aspartate (D) |
| Aspartate (D) ∼11% | ||||
| Sequence at residue 218 | CTA (Leu) ∼96% | CTA (Leu) | CTA (Leu) | Some CTA (Leu) |
| TTA (Leu) ∼4% | Some TTA (Leu) | |||
| GTCA repeats | (GTCA)3 in control and D1 alleles | (GTCA)3 | (GTCA)2 | (GTCA)1 |
| (GTCA)2 in D2 alleles |
Species examined that encode D at residue 314 included: chimp, gibbon, gorilla, orangutan, rhesus, galago, tree shrew, mouse, rat, guinea pig, shrew, dog, cat, horse, cow and armadillo. Species examined encoding N at residue 314 included human only. Species examined with p.L218(CTA) included: chimp, gibbon, gorilla, orangutan, rhesus, galago, tree shrew, dog, cat, horse and armadillo. Species examined with p.L218(TTA) included: mouse and rat. Of note, cow GALT encodes p.L218(CTT), whereas guinea pig and rabbit GALT encode p.L218(CTG). Species examined that carry a 5′ (GTCA)3 included: chimp, gibbon, gorilla and orangutan. Species examined that carry a 5′ (GTCA)2 included: rhesus and macaque. Species examined that carry a 5′ (GTCA)1 included: tree shrew, mouse, rat, guinea pig, shrew, hedgehog, dog, cat, cow, elephant and armadillo. Notably, horses carry one copy of ATCA in place of the GTCA sequence found in other species.
Figure 1.
Proposed origins and relationship of the p.N314, D1 and D2 alleles of human GALT. The predicted ancestral human GALT allele carries the (GTCA)3 + p.L218(CTA) + p.D314 sequences found in other hominid species. The p.D314N substitution occurs early in human evolution, and the 4 bp 5′ deletion and p.L218(TTA) silent substitution occur later, on distinct branches of the tree. The three intronic base changes reported to exist in cis with D2 alleles [c.378-27G>C or IVS4-27G > C, c.508-24G > A or IVS5-24G > A and c.507 + 62G > A or IVS5-62G > A (20–22), not illustrated here] presumably occurred subsequent to, or concurrently with, the 4 bp 5′ deletion on the D2 branch of the tree.
Within the CEPH population, the frequency of the ancestral p.D314 allele is ∼11.3% (http://hapmap.org), which is unusually high compared with other human populations; Yoruba, Chinese and Japanese populations each exhibit a frequency of p.D314 well under 3%. Thus, it seems that the common allele p.D314 is not only the derived state, but also that the ancestral allele is nearly absent among non-European populations.
We also explored the origins of the codon 218 sequence; a silent substitution in this codon constitutes the p.L218 variation (CTA to TTA, rs2070075 at position 34 638 418 of chromosome 9 in NCBI Build 36.1) found on D1, but not on D2, alleles. In non-human hominids and non-hominid primates, the CTA codon also predominates. Among non-primate placental mammals, some published sequences carry CTA (e.g. dog, cat, horse), whereas others carry TTA (e.g. mouse, rat). These data implicate CTA as the ancestral allele in humans, but also suggest that the TTA silent substitution may have arisen more than once through the course of evolution.
As summarized in Table 1, the ancestral CTA (Leu) codon accounts for ∼95.5% of alleles in the CEPH population, while the derived TTA (Leu) codon accounts for ∼4.5% of alleles. The derived TTA allele is even rarer in non-European populations, with an observed frequency of ∼1% in the HapMap Chinese sample and a complete absence in both the Yoruba and Japanese samples (http://hapmap.org).
Cross-species comparisons of the GALT proximal 5′ sequence also reveal an interesting pattern (Table 1). GALT alleles that are predominant in humans and non-human hominids carry three tandem GTCA repeats (GTCA)3, but GALT alleles in non-hominid primates carry only two repeats (GTCA)2 and GALT alleles in non-primate placental mammals carry only one repeat unit (GTCA)1. Hence, this repeat sequence appears to have expanded through the course of evolution, and the 4 bp deletion seen in human D2 alleles represents a contraction by one repeat unit.
A PCR-based assay to detect the 4 bp 5′ deletion associated with D2 GALT
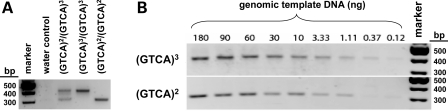
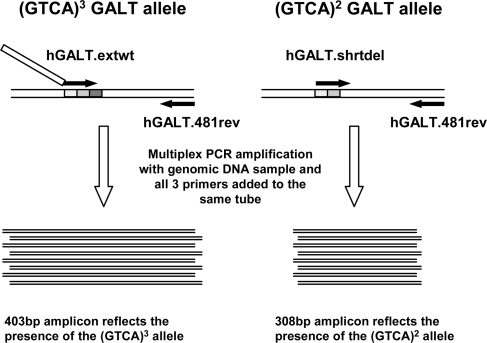
To facilitate rapid and direct detection of the 4 bp 5′ deletion in human genomic DNA, we designed a multiplex allele-specific PCR-based assay (Fig. 2); allele-specific amplicons are generated using forward primers specific for either the wild-type (GTCA)3 or mutant (GTCA)2 GALT sequences, together with a shared reverse primer. As illustrated in Figure 2, the (GTCA)3-specific primer includes a 91 bp ‘tail’ of non-human (Saccharomyces cerevisiae) sequence that extends the length of the amplicon without compromising annealing specificity. The wild-type amplicon is 403 bp, whereas the corresponding (GTCA)2-specific amplicon, which lacks the 91 bp tail, is 308 bp. This size difference makes for easy visual distinction between the two amplified fragments following agarose gel electrophoresis. Figure 3A illustrates the application of this method to a set of controls, including one sample homozygous for the (GTCA)3 wild-type allele, one homozygous for the (GTCA)2 D2 allele and one heterozygous for the (GTCA)2 and (GTCA)3 alleles.
Figure 2.
Strategy for allele-specific amplification of (GTCA)2 versus (GTCA)3 5′ GALT sequences. As illustrated, the forward primer specific for the (GTCA)3 allele includes a 91 bp ‘tail’ of S. cerevisiae sequence that increases the size of the corresponding amplicon.
Figure 3.
Multiplex allele-specific amplification of (GTCA)2 and (GTCA)3 5′ GALT sequences. (A) Amplification of (GTCA)2/(GTCA)2 and (GTCA)3/(GTCA)3 templates demonstrates the specificity of the primer sets. (B) A dilution series of template levels establishes the tolerance of both amplification reactions to a wide range of template concentrations.
To test the sensitivity of this method to template concentration, we performed assays on wild-type and D2 homozygous genomic DNAs ranging in concentration from 1 to 180 ng/reaction. As illustrated in Figure 3B, bands of the expected sizes, and the expected sizes only, were easily detected in all reaction lanes, demonstrating tolerance of the assay for a wide range of template concentrations.
Distribution of the 4 bp 5′ deletion among D1 and D2 GALT alleles
To explore the distribution of the (GTCA)2 variant among D1 and D2 GALT alleles, we applied our assay to 183 genomic samples representing 366 GALT alleles, of which 202 were known to encode p.D314 and 163 were known to encode p.N314 (Table 2). Of these 202 p.D314 alleles, 4 had previously tested positive for the p.L218(TTA) silent base substitution; the remainder were L218(CTA). Our results demonstrated the anticipated one-to-one correspondence between the presence of the 4 bp 5′ deletion and the absence of the p.L218(TTA) variation among the D2 alleles (Table 2). Our results also revealed a perfect one-to-one correspondence between the absence of the 4 bp 5′ deletion and the presence of the p.L218(TTA) variation among D1 alleles (Table 2). As expected, these results were also fully consistent with biochemical data associated with each sample, with D2 alleles being partially impaired, and D1 alleles not impaired at all (data not shown).
Table 2.
Distribution of the 4 bp 5′ deletion among D1 and D2 alleles of GALT
| GALT genotype of patient (biochemically confirmed) | No. of samples | No. of D2 alleles [p.D314 + p.L218(CTA)] | No. of D1 alleles [p.D314 + p.L218L(TTA)] | Other alleles (N or G) | No. of alleles carrying (GTCA)2 | No. of alleles carrying (GTCA)3 |
|---|---|---|---|---|---|---|
| D2/D2 | 18 | 36 | 0 | 0 | 36 | 0 |
| D2/N | 52 | 52 | 0 | 52 N | 52 | 52 |
| D2/G | 109 | 109 | 0 | 109 G | 109 | 108a |
| D2/D1 | 1 | 1 | 1 | 0 | 1 | 1 |
| D1/G | 3 | 0 | 3 | 3 G | 0 | 6 |
| Total | 183 | 198 | 4 | 164 | 198 | 167 |
D1 alleles were identified on the basis of p.D314 + p.L218L(TTA); D2 alleles were identified on the basis of p.D314 + p.L218(CTA). N alleles were identified on the basis of N314, no detected mutations, and normal biochemical activity.
aOne G allele in this cohort was the 5 kb deletion, which removes the 5′ GALT sequence.
Absence of the 4 bp 5′ deletion among N (control) and G (classic galactosemia) GALT alleles that carry N314
To test the distribution of the 4 bp 5′ deletion among control (N) and classic galactosemia (G) GALT alleles that encode p.N314, we applied our assay to 98 control genomic DNAs (196 control alleles) and 48 genomic DNAs derived from GG patients (96 G alleles). Of note, each of these alleles had already been shown to not carry p.D314 or p.L218(TTA). As expected, none of these alleles demonstrated the presence of the 4 bp 5′ deletion (Table 3).
Table 3.
Distribution of the 4 bp promoter deletion among normal (N) and classic (G) galactosemia alleles of GALT
| GALT genotype of volunteer | No. of samples | No. of alleles | No. of alleles carrying (GTCA)2 | No. of alleles carrying (GTCA)3 |
|---|---|---|---|---|
| N/N (normal) [p.N314 + p.L218(CTA)] | 98 | 196 | 0 | 196 |
| G/G (classic) [p.N314 + p.L218(CTA)] | 48 | 96 | 0 | 96 |
Distribution of the 4 bp 5′ deletion among classic galactosemia (G) GALT alleles that also carry D314
From the many classic galactosemia patient DNA samples tested, we identified eight unrelated samples that encoded p.D314 on one or both alleles in cis with a classic (G) mutation. Indeed, of the 16 alleles represented, 11 carried p.D314 (Table 4). The GALT genotypes identified in these samples illustrate two important points. First, classic galactosemia (G) mutations are found on both D1 and D2 genetic backgrounds. Secondly, we found no apparent association of the D1 or D2 background with a specific G mutation.
Table 4.
Distribution of the D1- and D2-related variations among GALT alleles that also carry a classic galactosemia (G) mutation
| Patient | G mutation genotype | D1- or D2-related variations |
|---|---|---|
| A | p.R204X/p.P265A | p.D314 + (GTCA)2/p.D314 + (GTCA)2 |
| B | p.Q207X/p.Q207X | p.D314 + (GTCA)2/p.D314 + (GTCA)2 |
| C | p.H132Q, p.R204X, p.P265A cis/trans relationships unknown | p.D314/p.N314 (GTCA)2/(GTCA)3 |
| D | p.Q188R, p.R259Q, p.D273H cis/trans relationships unknown | p.D314 + p.L218(TTA) + (GTCA)3/p.N314 + (GTCA)3 |
| E | p.Q188R/p.R333W | p.D314 + p.L218(TTA) + (GTCA)3/p.N314 + (GTCA)3 |
| F | p.R204X/unknown | p.D314 + (GTCA)2/p.D314 + (GTCA)2 |
| G | p.Q188R/unknown | p.D314 + p.L218(TTA) + (GTCA)3/p.N314 + (GTCA)3 |
| H | Unknown/unknown | p.D314 + (GTCA)2/p.N314 + (GTCA)3 |
If the codon at 218 is not specified, it is CTA.
D2 GALT mRNA is under-represented in the blood of DN carriers
To test the hypothesis that D2 alleles express a lower level of message in vivo than wild-type alleles, we performed allele-specific quantitative RT–PCR on blood-derived RNA samples from DN (D2 carrier) volunteers, as well as from NN (control) and DD volunteers. Equivalent amounts of RNA derived from each sample were subjected to qRT–PCR with each of two sets of primers: one specific for the p.N314 (normal) transcript and one specific for the p.D314 (D2) transcript. As anticipated, all NN samples amplified well with the N314-specific primers and showed only minimal (∼3.5%) background signal when amplified with the p.D314-specific primers. Similarly, a DD sample amplified well with the p.D314-specific primers and showed only minimal (∼1.5%) background signal when amplified with the N314-specific primers. As indicated (Table 5), the ratio of p.N314:p.D314 signals, corrected for background, detected in each of the four DN samples was close to 54:46%. Though small, the difference between this distribution and the null hypothesis (50:50%) was quite statistically significant (P < 0.002). These data demonstrate that D2 alleles are mildly under-expressed at the level of RNA in vivo relative to normal GALT alleles.
Table 5.
Relative expression levels of the p.N314 and p.D314 GALT messages in DN carriers
| Sample | GALT genotype | Relative expression of p.N314 GALT message | Relative expression of p.D314 GALT message |
|---|---|---|---|
| 1 | p.D314 + (GTCA)2/N | 0.525 ± 0.048 | 0.475 ± 0.048 |
| 2 | p.D314 + (GTCA)2/N | 0.573 ± 0.072 | 0.427 ± 0.072 |
| 3 | p.D314 + (GTCA)2/N | 0.541 ± 0.052 | 0.459 ± 0.052 |
| 4 | p.D314 + (GTCA)2/N | 0.530 ± 0.072 | 0.470 ± 0.072 |
| Total | 0.540 ± 0.011 (n = 4) | 0.460 ± 0.011 (n = 4, P < 0.002) |
These values were corrected for background cross-reactivity of the primers, which was measured at 1.5% for p.D314 signal and 3.5% for p.N314 signal.
GALT activity in hemolysates from DG patients
As a final test of the GALT activities attributable to D alleles, we examined GALT activity levels determined in the Emory Biochemical Genetics Lab for 133 NN samples, 90 DG samples and 27 GG samples. As expected (Table 6), average DG GALT values were ∼21% of those in NN patients. Notably, however, the ranges of both sets were very large. There was a more than 2-fold range in the GALT levels of NN samples, and a more than 6-fold range in the GALT levels of DG samples. Indeed, ‘high-activity’ DG samples had >60% of the GALT activity seen in ‘low-activity’ NN samples. These ranges suggest that there are likely many factors, including GALT coding and non-coding sequences, and also factors beyond GALT sequence, that influence GALT expression or activity, or both, in human cells.
Table 6.
GALT activities detected in hemolysates from GG, DG and NN individuals
| GALT genotype (n) | GALT activity (μmol/h/g Hb) |
|
|---|---|---|
| Average ± SD | Range | |
| NN (133) | 33.5 ± 6.8 | 20.1–47.8 |
| DG (90) | 7.2 ± 2.0 | 2.0–12.5 |
| GG (27) | 0.2 ± 0.6 | 0.0–2.4 |
DISCUSSION
The data presented here both extend and clarify our understanding of the origins, distribution and expression of the D2 GALT allele associated with Duarte galactosemia.
Origins
To explore the origins of the p.D314, p.L218(TTA) and (GTCA)2 variations found on D1 and/or D2 GALT alleles, we used interspecies sequence comparisons to reconstruct the variants most likely to represent the ancestral state. These analyses clearly implicated p.D314 as the ancestral allele and p.N314 as a polymorphism that may be predominant in modern humans, but which also appears unique to humans. Of note, p.D314 is not found with any significant frequency in populations of African descent, although it is found in African Americans, presumably due to admixture (26). This distribution pattern suggests that the p.D314N variant arose in Africa very early in human evolution, and that the persistence of the p.D314 allele in European and, to a lesser extent, Asian populations may reflect a founder effect or other factors acting upon descendants of the early waves of human migration out of Africa.
We similarly noted that the 4 bp 5′ deletion found in D2 GALT alleles is a one-unit contraction of a tetranucleotide repeat sequence that has otherwise expanded through evolution from placental mammals (one repeat unit), to primates (two repeat units), to hominids, including humans (three repeat units). That the repeat number of this tetranucleotide sequence has varied through evolution is consistent with the instability observed in other repeat tracts in the mammalian genome.
Distribution of the 4 bp 5′ deletion on human GALT alleles
To explore the distribution of the (GTCA)2 sequence variant in human GALT alleles, we developed and applied a simple and robust PCR-based assay to identify and distinguish wild-type (GTCA)3 from D2-associated (GTCA)2 5′ sequences. Using this assay we confirmed the previously described strict linkage of the (GTCA)2 sequence with p.D314 + L218(CTA) coding sequence alleles, and the strict repulsion of the (GTCA)2 5′ sequence with the p.N314 and p.D314 + p.L218(TTA) coding sequence alleles. Although each of these relationships had been reported previously (20,24,25), to our knowledge ours is the largest pan-ethnic collection of GG, DG and control samples genotyped with regard to the 4 bp 5′ deletion. It is also noteworthy that, though the HapMap database cites allele frequencies for the p.D314 and p.L218(TTA) polymorphisms, that study, which restricted itself to SNPs, did not assay the (GTCA)2 promoter allele in human GALT. Applying the apparently tight linkage of the (GTCA)2 5′ variation with GALT alleles that carry p.D314 but lack p.L218(TTA), we can estimate the likely allele frequency of this non-coding variation in the HapMap-tested European population at near 6–7% [the overall frequency of p.D314 (11%) minus the frequency of the p.L218(TTA) allele (4–5%)], with lower frequencies in other human populations.
It is also interesting to note that the p.D314 sequence, with or without p.L218(TTA), is also found in cis with many different G (classic galactosemia) mutations, suggesting that through the course of human history, although most G mutations have arisen on the predominant p.N314 GALT genetic background, others have arisen on p.D314 + p.L218(TTA) backgrounds or on p.D314 + (GTCA)2 backgrounds. In short, with the caveat of a small p.D314-G data set, we see no clear evidence of preferential association between specific classic galactosemia mutations and any given GALT genetic background. Assuming that all classic galactosemia GALT mutations are younger than any of the GALT polymorphisms discussed, and assuming G mutations arise independently of genetic background, we therefore predict that in European populations ∼89% of GALT mutations will be found on a p.N314 background, 6–7% on a D2 [p.D314 + (GTCA)2] background and 4–5% on a D1 [p.D314 + p.L218(TTA)] background. In non-European populations, nearly all GALT mutations will be found on a p.N314 background.
Expression
Another goal of this work was to clarify the basis of the ∼50% decrease in GALT expression and activity typically associated with D2 alleles. As explained in the Introduction, earlier reports addressing this point were contradictory. Some studies claimed normal D2 GALT mRNA levels and attributed the decrease to altered p.N314D protein stability (14,23); others claimed decreased D2 GALT mRNA levels ostensibly resulting from intronic mutations (15) or from impaired D2 promoter function (24,25). All of these studies were limited by extremely small sample sizes and by analyses of cultured transformed cell lines as opposed to primary tissues. Given the more than 6-fold difference in GALT activity attributed to Duarte alleles in different DG patients (Table 7), it is difficult to generalize from the less than 2-fold differences in results obtained using samples from individual study volunteers, much less individual cell lines derived from those individual volunteers. The decreased p.N314D protein stability hypothesis needed to account for the apparently normal or above normal levels of GALT protein and activity associated with D1 alleles, which also encode p.N314D. The explanation posed (but not tested)—namely, improved translation efficiency of D1 messages due presence of the silent p.L218(TTA) variant (14,23)—is also problematic. Both the D1-associated TTA (Leu) and D2-associated CTA (Leu) codons occur at other places in N, D1 and D2 GALT alleles. Indeed, N and D2 alleles each carry a total of six CTA codons and one TTA codon, whereas D1 alleles carry a total of five CTA codons and two TTA codons.
Table 7.
Primers used in this study
| Primer name | Sequence (5′–3′) | Comments |
|---|---|---|
| hGALT.extwt (primer 1) | TACAAAGTGAAAGTACTTCTAAAATTGTTTTGGTTACAGGTGGTGCTGGATACATTGGTTCACACACTGTGGTAGAGCTAATTGAGAATGGggcagcccagtcagtcagt | Anneals only to alleles that do not carry the 4 bp 5′ deletion |
| hGALT.shrtdel (primer 2) | ggcagcccagtcagtcacg | Anneals only to alleles that carry the 4 bp 5′ deletion |
| hGALT.481rev (primer 3) | gagcgttccaaccttcggaggg | Reverse primer anneals to both alleles |
| hGALT.N314.f3 | gaggctggggccaactgga | anneals to p.N314 encoding message |
| hGALT.D314.f3 | gaggctggggccaactggg | anneals to p.D314 encoding message |
| hGALT.end.r2 | aggtggtaatgaacctcaggaagtgc | reverse primer anneals to both p.N314 and p.D314 encoding messages |
Uppercase letters represent non-human sequence (S. cerevisiae) used to extend the length of the wild-type amplicon. Lowercase letters represent human GALT sequence.
To test whether D2 GALT alleles are under-expressed relative to N alleles in human blood, we applied allele-specific qRT–PCR to RNA samples derived from each of the four individuals heterozygous for the D2 GALT allele. This study design had the advantages of primary human tissue samples and the inclusion of an internal control within each sample. Our results demonstrated a small but consistent and statistically significant under-representation of p.D314 encoding message relative to ‘normal’ p.N314 encoding message in each sample tested. Specifically, the mRNA distribution detected was 54% p.N314 to 46% p.D314 encoding messages. While statistically distinct from the 50:50% distribution predicted if there were no decrease in D2 GALT mRNA, this 54:46% distribution also falls short of the 67% p.N314 to 33% p.D314 distribution predicted if mRNA deficits could account fully for the average 50% loss of expression/activity typically attributed to D2 alleles.
The reason for this ‘middle ground’ result remains unclear, though there may be a small protein expression or stability effect. It is possible that the full effect is RNA mediated, but that the primers and qRT–PCR technology applied here were unable to confirm it. Another possibility is that individual-to-individual naturally occurring variation between samples prevented unambiguous demonstration of the predicted 67:33% p.N314:p.D314 message distribution.
When considering our results, it is also important to note that, unlike prior reports, our studies used mRNA samples isolated directly from whole blood, and not from cultured transformed, cell lines. These RNAs therefore derived predominantly from circulating white cells, whereas the GALT activity values listed here (Table 6) or reported previously (3,14) were derived from red blood cells or from cultured transformed cells. To our knowledge, there have been no systematic studies of D2-associated GALT activity or abundance in circulating white cells. It is therefore possible that the 54:46% distribution reported here is an accurate reflection of the relative levels of p.N314 to p.D314 encoding GALT messages in the circulating white blood cells of DN carriers. Regardless, it is now clear that altered mRNA levels can indeed account for at least some of the ‘missing’ GALT activity attributable to D2 GALT alleles.
Testing for the D2 allele
Most clinical diagnostic labs currently test for the presence or absence of a Duarte GALT allele using a combination of molecular and biochemical studies. The molecular studies generally involve a direct assay for p.N314D and/or p.L218(TTA) alleles, or full exon sequencing. The biochemical studies typically involve a GALT activity assay that will show high activity in the case of a D1 allele and low but detectable activity in the case of a D2 allele. Isozyme studies are used to detect the shifted banding pattern characteristic of both D1 and D2 GALT. These combined approaches can correctly identify and distinguish D1 from D2 GALT alleles; however, given the broad range of GALT activity values obtained for the suspected DG patient population, together with the realization that p.N314D and p.L218(TTA) are themselves most likely linked polymorphisms rather than causal mutations, an alternative diagnostic plan might prove beneficial.
We propose that DG patients could be accurately diagnosed through a combination of GALT enzyme assay and molecular studies to query the coding sequence and/or candidate G mutations, with an added test for the presence or absence of the 5′ 4 bp deletion. If low but detectable activity was present, one G mutation was found, and the 4-bp deletion seen, the patient would most likely have DG galactosemia. Defining the presence or absence of the p.N314D substitution or a shifted isozyme banding pattern would be superfluous. Given the prevalence of p.N314D in the general population and its linkage to D1, D2 and G GALT alleles, its utility as a clinically relevant marker is compromised at best. In fact, if the 5′ 4 bp deletion is causative, tests that rely on the p.N314D polymorphism, with or without p.L218(TTA), could lead to faulty conclusions in any individual with a recent recombination between the 5′ deletion and the tested coding sequence variants.
MATERIALS AND METHODS
Study subjects
All DNA samples were obtained either from consenting volunteers from an ongoing IRB-approved study of galactosemia (Emory IRB Protocol # 618-99, PI: Fridovich-Keil) or as anonymized discards from the Emory Genetics Laboratory. All of the samples studied were tested by the Emory Genetics Laboratory to detect the presence of p.N314D, p.L218(TTA) and a set of common GALT gene mutations associated with classic galactosemia.
Quantitative allele-specific RT–PCR
RNA samples were prepared from whole blood collected in Tempus™ Blood RNA Tubes using the PerfectPure™ RNA Purification Kit according to the manufacturer’s instructions (5 Prime Inc., Gaithersburg, MD, USA); RNA was quantified by UV absorbance. Whole blood was collected and stored at −20°C until RNA isolation. RNA was stored at −80°C, until it was used to prepare cDNA for quantitative PCR. SuperScript III First-Strand Synthesis System for RT–PCR using Oligo(dT)20 primers was used to prepare cDNA according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Each cDNA synthesis reaction contained 250 ng of RNA.
Relative abundance of the D2 transcript in four DN individuals, of whom three were unrelated, was determined by comparison of the results of qRT–PCR reactions from each cDNA sample using primers that specifically amplified a 225 bp fragment of the D314 sequence versus primers that specifically amplified the corresponding 225 bp fragment of the N314 sequence. The primers used in these reactions were hGALT.D314.f3, hGALT.N314.f3 and hGALT.end.r2 (Table 7). The specificity/selectivity of each allele-specific primer set was confirmed by qRT–PCR with RNA derived from NN or DD homozygous individuals, and subsequent analyses of DN-mixed message populations were corrected for the low level of background cross-reactivity (1.5% for D314 primers and 3.5% for N314 primers). All qRT–PCR reactions were performed using SYBR Green for the LightCycler® 480 in a 96-well plate format (Roche, Indianapolis, IN, USA). The absolute abundance of the D314 and N314 messages in each DN sample was calculated using titration curves of DN cDNA amplified with both primer sets. The relative abundance of the D314 and N314 messages in each sample was calculated as D/(D + N) and N/(D + N), respectively. Assays were performed in triplicate or more, with results reported as mean ± SD.
Detection of the 4 bp GTCA proximal promoter deletion
The presence or absence of the 4 bp GALT promoter deletion was determined by allele-specific genomic PCR with primers hGALT.extwt (specific for the WT allele), hGALT.shrtdel (specific for the deletion allele) and hGALT481rev (reverse primer that recognizes both alleles). Each 20 μl PCR mixture contained 3.75 pmol of hGALT.extwt (Table 7), 10 pmol of hGALT.shrtdel (Table 7) 20 pmol of hGALT481rev (Table 7), between 1 and 50 ng of genomic DNA, 4 µl of 5× GoTaq™ buffer (Promega), 3.2 µl of 2.5 mm dNTPs, 0.4 µl DMSO and 0.5 U of GoTaq (Promega). The reaction mixture was collected in 0.5 ml tubes on ice and transferred directly to a thermal cycler pre-heated to 95°C. The reactions were melted at 95°C for 5 min, followed by 38 cycles of 94°C for 45 s, 65°C for 45 s, and 74°C for 1 min. In the final cycle, the reactions were extended at 72°C for 5 min, and then held at 16°C for at least 10 min. Following amplification of each sample, 2 µl of PCR product were diluted 1:5 in ddH2O and run at 90 V for 55 min on a 2% agarose/TAE gel in conjunction with an appropriate marker (100 bp ladder, New England Biolabs); bands were visualized by staining with ethidium bromide.
FUNDING
This work was supported by funds from the National Institutes of Health, grant numbers DK046403, DK059904 to J.L.F.K. and DK074297, to K.R.G. R.D.S. was supported in part by funds from NIH Training, grant number 2T32GM008367-16, and A.E.C. was supported in part by funds from the Howard Hughes Medical Institute under grant number 52003727. Funding to pay the Open Access charge was provided by National Institutes of Health grant number HG003461 to D.J.C.
ACKNOWLEDGEMENTS
Our profound thanks to the many study volunteers and their families who participated in this project, and to Dr. Rani Singh and Ms. Mary Jane Kennedy for their help with volunteer recruitment. We also thank Dr. Miao He and Ms. Tisa Harper for sharing anonymized GALT activity values from their clinical lab. We are grateful to the National Institutes of Health and the Howard Hughes Medical Institute for supporting this research.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Fridovich-Keil J., Walter J. Galactosemia. In: Valle D., Beaudet A.L., Vogelstein B., Kinzler K.W., Antonarakis S.E., Ballabio A., editors. The Online Metabolic and Molecular Bases of Inherited Disease—OMMBID. Chapter 72. New York: McGraw-Hill; 2008. [Google Scholar]
- 2.Ficicioglu C., Yager C., Segal S. Galactitol and galactonate in red blood cells of children with the Duarte/galactosemia genotype. Mol. Genet. Metab. 2005;84:152–159. doi: 10.1016/j.ymgme.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Ficicioglu C., Thomas N., Yager C., Gallagher P., Hussa C., Mattie A., Day-Salvatore D., Forbes B. Duarte (DG) galactosemia: a pilot study of biochemical and neurodevelopmental assessment in children detected by newborn screening. Mol. Genet. Metab. 2008;95:206–212. doi: 10.1016/j.ymgme.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki M., West C., Beutler E. Large-scale molecular screening for galactosemia alleles in a pan-ethnic population. Hum. Genet. 2001;109:210–215. doi: 10.1007/s004390100552. [DOI] [PubMed] [Google Scholar]
- 5.Beutler E., Baluda M.L., Sturgeon P., Day R.W. A new genetic abnormality resulting in galactose-1-phosphate uridyltransferase deficiency. Lancet. 1965;1965–1961:353–354. doi: 10.1016/s0140-6736(65)91782-4. [DOI] [PubMed] [Google Scholar]
- 6.Kelley R.I., Harris H., Mellman W.J. Characterization of normal and abnormal variants of galactose-1-phosphate uridyltransferase (EC 2.7.7.12) by isoelectric focusing. Hum. Genet. 1983;63:274–279. doi: 10.1007/BF00284663. [DOI] [PubMed] [Google Scholar]
- 7.Shin Y.S., Niedermeier H.P., Endres W., Schaub J., Weidinger S. Agarose gel isoelectric focusing of UDP-Gal pyrophorylase and galactose-1-phosphate uridyl transferase: developmental aspects of UDP-galactose pyrophorylase. Clin. Chim. Acta. 1987;166:27–35. doi: 10.1016/0009-8981(87)90191-4. [DOI] [PubMed] [Google Scholar]
- 8.Elsas L.J., Dembure P.P., Langley S.D., Paulk E.M., Hjelm L.N., Fridovich-Keil J.L. A common mutation associated with the Duarte galactosemia allele. Am. J. Hum. Genet. 1994;54:1030–1036. [PMC free article] [PubMed] [Google Scholar]
- 9.Leslie N.D., Immerman E.B., Flach J.E., Florez M., Fridovich-Keil J.L., Elsas L.J. The human galactose-1-phosphate uridyl transferase gene. Genomics. 1992;14:474–480. doi: 10.1016/s0888-7543(05)80244-7. [DOI] [PubMed] [Google Scholar]
- 10.Lin H.-C., Kirby L.T., Ng W.G., Reichardt J.K.V. On the molecular nature of the Duarte variant of galactose-1-phosphate uridyl transferase (GALT) Hum. Genet. 1994;93:167–169. doi: 10.1007/BF00210604. [DOI] [PubMed] [Google Scholar]
- 11.Reichardt J.K.V., Woo S.L.C. Molecular basis of galactosemia: mutations and polymorphisms in the gene encoding human galactose-1-phosphate uridylyltransferase. Proc. Natl Acad. Sci. USA. 1991;88:2633–2637. doi: 10.1073/pnas.88.7.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridovich-Keil J.L., Quimby B.B., Wells L., Mazur L.A., Elsevier J.P. Characterization of the N314D allele of human galactose-1-phosphate uridylyltransferase using a yeast expression system. Biochem. Mol. Med. 1995;56:121–130. doi: 10.1006/bmme.1995.1067. [DOI] [PubMed] [Google Scholar]
- 13.Bergren W., Donnell G. A new variant of galactose-1-phosphate uridyltransferase in man: the Los Angeles variant. Ann. Hum. Genet. 1973;37:1–8. doi: 10.1111/j.1469-1809.1973.tb01808.x. [DOI] [PubMed] [Google Scholar]
- 14.Langley S.D., Lai K., Dembure P.P., Hjelm L.N., Elsas L.J. Molecular basis for Duarte and Los Angeles variant galactosemia. Am. J. Hum. Genet. 1997;60:366–372. [PMC free article] [PubMed] [Google Scholar]
- 15.Podskarbi T., Kohlmetz T., Gathof B.S., Kleinlein B., Bieger W.P., Gresser U., Shin Y.S. Molecular characterization of Duarte-1 and Duarte-2 variants of galactose-1-phosphate uridyltransferase. J. Inherit. Metab. Dis. 1996;19:638–644. doi: 10.1007/BF01799840. [DOI] [PubMed] [Google Scholar]
- 16.Calderon F., Phansalkar A., Crockett D., Miller M., Mao R. Mutation database for the galactose-1-phosphate uridyltransferase (GALT) gene. Hum. Mutat. 2007;28:939–943. doi: 10.1002/humu.20544. [DOI] [PubMed] [Google Scholar]
- 17.Andersen M.W., Williams V.P., Sparkes M.C., Sparkes R.S. Transferase-deficiency galactosemia: immunochemical studies of the Duarte and Los Angeles variants. Hum. Genet. 1984;65:287–290. doi: 10.1007/BF00286519. [DOI] [PubMed] [Google Scholar]
- 18.Gathof B., Sommer M., Podskarbi T., Reichardt J., Braun A., Gresser U., Shin Y. Characterization of two stop codon mutations in the galactose-1-phosphate uridyltransferase gene of three male galactosemic patients with severe clinical manifestation. Hum. Genet. 1995;96:721–725. doi: 10.1007/BF00210306. [DOI] [PubMed] [Google Scholar]
- 19.Greber S., Guldberg P., Scheibenreiter S., Strobl W. Mutations in classical and Duarte2 galactosemia (abstract) Pediatr. Res. 1995;38:434. [Google Scholar]
- 20.Kozak L., Francova H. Presence of a deletion in the 5′ upstream region of the GALT gene in Duarte (D2) alleles. J. Med. Genet. 1999;36:576–578. [PMC free article] [PubMed] [Google Scholar]
- 21.Lin H., Reichardt J. Linkage disequilibrium between a SacI restriction fragment length polymorphism and two galactosemia mutations. Hum. Genet. 1995;95:353–355. doi: 10.1007/BF00225208. [DOI] [PubMed] [Google Scholar]
- 22.Shin Y.S., Koch H.G., Kohler M., Hoffmann G., Patsoura A., Podskarbi T. Duarte-1 (Los Angeles) and Duarte-2 (Duarte) variants in Germany: two new mutations in the GALT gene which cause a GALT activity decrease by 40–50% of normal in red cells. J. Inherit. Metab. Dis. 1998;21:232–235. doi: 10.1023/a:1005303818858. [DOI] [PubMed] [Google Scholar]
- 23.Lai K., Langley S., Dembure P., Hjelm L., Elsas L.I. The Duarte allele impairs biostability of galactose-1-phosphate uridyltransferase in human lymphoblasts. Hum. Mutat. 1998;11:28–38. doi: 10.1002/(SICI)1098-1004(1998)11:1<28::AID-HUMU5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 24.Trbusek M., Francova H., Kozak L. Galactosemia: deletion in the 5′ upstream region of the GALT gene reduces promoter efficiency. Hum. Genet. 2001;109:117–120. doi: 10.1007/s004390100540. [DOI] [PubMed] [Google Scholar]
- 25.Elsas L.J., Lai K., Saunders C.J., Langley S.D. Functional analysis of the human galactose-1-phosphate uridyltransferase promoter in Duarte and LA variant galactosemia. Mol. Genet. Metab. 2001;72:297–305. doi: 10.1006/mgme.2001.3157. [DOI] [PubMed] [Google Scholar]
- 26.Ashino J., Okano Y., Suyama I., Yamazaki T., Yoshino M., Furuyama J.-I., Lin H.-C., Reichardt J.K.V., Isshiki G. Molecular characterization of galactosemia (Type 1) Mutations in Japanese. Hum. Mutat. 1995;6:36–43. doi: 10.1002/humu.1380060108. [DOI] [PubMed] [Google Scholar]