Abstract
Actinomyces spp., predominant members of human oral biofilms, may use extracellular sialidase to promote adhesion, deglycosylate immunoglobulins and liberation of nutrients. Partial nanH gene sequences (1077 bp) from Actinomyces oris (n =74), Actinomyces naeslundii (n =30), Actinomyces viscosus (n =1) and Actinomyces johnsonii (n =2) which included the active-site region and the bacterial neuraminidase repeats (BNRs) were compared. The sequences were aligned and each species formed a distinct cluster with five isolates having intermediate positions. These five isolates (two A. oris and three A. naeslundii) exhibited interspecies recombination. The nonsynonymous/synonymous ratio was <1 for both A. oris and A. naeslundii indicating that nanH in both species is under stabilizing selective pressure; nonsynonymous mutations are not selected. However, for A. oris significant negative values in tests for neutral selection suggested the rate of mutation in A. oris was greater than in A. naeslundii but with selection against nonsynonymous mutations. This was supported by the observation that the frequency of polymorphic sites in A. oris, which were monomorphic in A. naeslundii was significantly greater than the frequency of polymorphic sites in A. naeslundii which were monomorphic in A. oris (χ2=7.011; P =0.00081). The higher proportions of A. oris in the oral biofilm might be explained by the higher mutation rate facilitating an increased ability to respond successfully to environmental stress.
Keywords: Actinomyces, sialidase, dental plaque
Introduction
Sialidase activity is produced by many commensal oral bacteria including Actinomyces spp. (Moncla & Braham, 1989; Beighton & Whiley, 1990; Beighton et al., 1991; Braham & Moncla, 1992). The activity produced by Actinomyces spp. desialylates IgA by the removal of the terminal sialic acid residues rendering the molecule more susceptible to proteolysis (Reinholdt et al., 1990; Frandsen, 1994). Sialidase activity also appears important in the nutrition of the oral biofilm because sialidase activity increases in the absence of host diet, and withdrawal of the normal diet increases the proportions of sialidase-producing bacteria (Smith & Beighton, 1986; Lucas et al., 1997; Sheehy et al., 2000). Sialic acid is utilized by various oral streptococci, Actinomyces naeslundii and by different mixed populations of subgingival plaque bacteria (ter Steeg et al., 1987; Frandsen, 1994; Byers et al., 1996, 1999; Homer et al., 1996). Sialidase also exposes galactose residues of O- and N-linked glycans which mediate the adherence of human A. naeslundii strains described previously as genospecies 1 and 2 to human glycans including those of the salivary pellicle (Costello et al., 1979; Gibbons et al., 1990).
The complete genome of ‘A. naeslundii’ MG1 (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi/) contains ORFs for two sialidase genes (ANA2709, nanH, a sialidase and ANA1493 an exosialidase). The sequence of the nanH gene (ANA2709) of MG1 has >97% similarity with the sequences of the two previously reported Actinomyces sialidase gene sequences (Henningsen et al., 1991; Yeung, 1993) while the ANA1493 exhibits <5% sequence similarity with the nanH genes.
Two of the strains from which the nanH was sequenced were described as Actinomyces viscosus but such human strains were subsequently described as A. naeslundii genospecies 2 (Johnson et al., 1990). However, on the basis of a phylogenetic analysis of partial gene sequences of housekeeping genes, we (Henssge et al., in press) have reported that strains previously identified as A. naeslundii genospecies 2, including the sequenced strain MG1, should be reclassified as Actinomyces oris. The species A. naeslundii has been amended to include only strains previously identified as A. naeslundii genospecies 1. Therefore, we have nanH sequence data on three A. oris strains but no information of the nanH genes of A. naeslundii, Actinomyces johnsonii (previously A. naeslundii genospecies WVA 963) or animal strains of A. viscosus.
In this report, we have compared the nucleotide and amino acid sequences of the region of the nanH gene containing the active site (Crennell et al., 1993) and the five Asp boxes or bacterial neuraminidase repeats (BNRs, Henningsen et al., 1991) of these Actinomyces species. In the mouth A. naeslundii and A. oris occupy the same sites but A. oris is the predominant species (Bowden et al., 1999) while A. johnsonii was isolated from the gingival crevice (Johnson et al., 1990). We present a phylogenetic analysis of a partial sequence of nanH in these four species and present evidence of interspecies recombination between nanH genes.
Materials and methods
Bacterial strains
The isolates have all been reported and identified in a previous taxonomic study (Henssge et al., in press) describing the new species A. oris (previously A. naeslundii genospecies 2) and A. johnsonii (previously A. naeslundii genospecies WVA 963) and reporting an emended description of A. naeslundii (previously A. naeslundii genospecies 1). The strains included in this study were 30 A. naeslundii (CCUG 33521, CCUG 33522, CCUG 33519, CCUG 33523, ATCC 12104, CCUG 34725, CCUG 35334 and CCUG 37599 and 22 human clinical and oral isolates); 71 A. oris (P2G, P5K, P6K, P7K, P8K, P9K, Pn4D, Pn5D, CCUG 33915, CCUG 33919, CCUG 33920, CCUG 33914, CCUG 34285, CCUG 34286 and ATCC 27044 and 56 human clinical and oral isolates); two A. johnsonii (CCUG 33932 and CCUG 34287) isolates and one A. viscosus (NCTC 10951). The oral and clinical isolates were given study numbers from 1 to 94. All isolates were cultured anaerobically at 37 °C on Fastidious Anaerobe Agar (LabM) supplemented with 5% (v/v) defibrinated horse blood and stored at −80 °C in brain–heart infusion (Oxoid) containing 50% glycerol.
PCR conditions and nanH sequencing
DNA was extracted from cells as described previously (Henssge et al., in press). Owing to the length of the gene fragment which was likely to contain the sites of interest, two pairs of primers were used to obtain a 1077-bp fragment of the nanH gene in all species: Sial-F1 5′-ACACGATCACGCAAGCCGA-3′ and Sial-R1 5′-CGACCTTGTTCTCATCCA-3′ and Sial-F2 5′-AACCACATCGTCCA-3′ and Sial-R2 5′-GAGCCAGTTCATCGTGAA-3′. The PCRs were performed using Reddymix (Abgene, Epsom, Surrey, UK) and the PCR conditions for each pair of primers were an initial denaturation for 10 min at 94 °C followed by 30 cycles of 94 °C for 30 s, 49 °C for 30 s and 72 °C for 90 s. A final extension was carried out for 5 min at 72 °C. The PCR products were visualized on a 1% agarose gel stained with Gel Red (Biotium Inc.). The amplicons were cleaned using a 50:50 mixture of 40% polyethylene glycol and 3 M NaCl, washed twice with 70% ethanol and rehydrated in sterile water. The same primers were used for the sequencing reactions and all amplicons were sequenced in both directions using the BigDye Terminator Sequencing kit (Applied Biosystems) and reaction products were run on a 3730xl sequencer (Applied Biosystems).
nanH sequence analysis
The DNA sequences were aligned using bioedit (Wide-field images were captured using an Olympus BX51 upright wide-field microscope with a ×40/1.00 UPlan Apo objective and a Coolsnap ES camera (Photometrics) through MetaVuesoftware (Molecular Devices). All wide-field images were captured using the same exposure and image scaling settings, and image scaling was adjusted to exclude background immunostaining. Offline image analysis used ImageJsoftware (http://www.mbio.ncsu.edu/BioEdit/bioedit.html/). The phylogenetic relationships between partial nanH nucleotide sequences of the reference strains, the human oral and clinical isolates and the two ‘A. viscosus’nanH sequences in GenBank (L06898 and X62276) and the nanH of the sequenced strain MG1 (ANA2709) were analysed using mega 4 (Tamura et al., 2007). Distances were calculated using the Kimura two-parameter model and for clustering the neighbour-joining method of Saitou & Nei (1987) using bootstrap values based on 500 replicates was used. The amino acid sequences were clustered using clustalw2 (http://www.ebi.ac.uk/).
dnasp (Rozas et al., 2003) was used to investigate the nanH sequences of A. naeslundii and A. oris. The G+C content of the partial genes sequences, the number of discrete sequences, the number of polymorphic sites, average nonsynonymous/synonymous ratios (dN/dS) were calculated and nucleotide diversity was estimated by determining π (nucleotide diversity) and θ (the total number of mutations). To test for neutral molecular evolution three tests were used; Tajima's D, based on the differences between the number of segregating sites and the average number of nucleotide differences, Fu and Li's D*, based on the differences between the number of singletons (mutations appearing only once among the sequences) and the total number of mutations and Fu and Li's F*, based on the differences between the number of singletons, and the average number of nucleotide differences between pairs of sequences. The extent of the DNA sequence polymorphism between A. naeslundii and A. oris was calculated and compared using a χ2 test.
Split decomposition trees were constructed with 1000 bootstrap replicates based on parsimony splits as implemented in the splitstree 4.0 (Hudson & Bryant, 2006) and the statistic phi was calculated. To identify strains with evidence of recombination the Recombination Detection Package (Martin et al., 2005) was used with the default settings.
Identification of BNR sequences and active site residues
The BNRs were identified using the InterProScan Sequence Search tool (http://www.ebi.ac.uk/Tools/InterProScan/). The amino acid polymorphisms in the BNRs were determined by visual inspection. The partial nanH amino acid sequences of the four type strains were aligned with the sequence of a Salmonella typhimurium sialidase (M55342) using Parallel Protein Information Analysis System (http://www.cbrc.jp/papia/papia.html/) and the 12 putative active site residues of the S. typhimurium sialidase (Crennell et al., 1993) were identified.
Detection of nanH transcripts
Each type strain was grown on FAA supplemented with 5% (v/v) defibrinated horse blood and total RNA was extracted using the UltraClean Microbial RNA Isolation Kit (MO BIO Laboratories Inc.). Reverse transcription was performed using the Omniscript RT kit (Qiagen Ltd) with primers, Sial-F1 and Sial-R1, for nanH and AtpA-F (CCCTGGAGTACACCACCAT) and AtpA-R (CGCCAGGGTGATCTTGAG), to amplify the housekeeping gene, atpA (ATP synthase F1, α subunit, ANA_0169). The thermal cycling was as follows: cDNA synthesis at 37 °C for 30 min, 94 °C for 1 min, followed by amplification with 40 cycles of denaturation at 94 °C for 20 s, annealing at 55 °C for 30 s, extension at 72 °C for 1 min, and then the final extension at 72 °C for 5 min. The products were run on a 1% agarose gel, with Gel Red incorporated, for 10 min at 100 V. The identity of the amplicons was confirmed by sequencing as described above.
Results
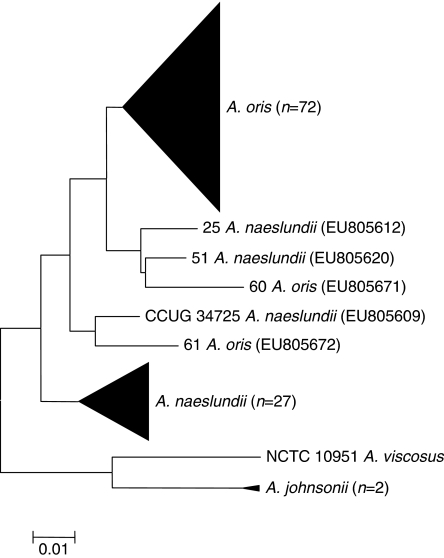
The construction of a phylogenetic tree using the nanH sequences indicated that the four species were separated from each other except that two of A. oris strains (strains 60 and 61) were separate from the major A. oris cluster and three A. naeslundii strains (strains 25, 51 and CCUG 34725) were distinct from the major A. naeslundii cluster (Fig. 1). Essentially, the same tree topography was found for the analysis of the derived amino acid sequences (data not shown) with the same isolates being found on the periphery of the clusters composed of the majority of the A. oris and A. naeslundii isolates and clearly distinct from the A. johnsonii and A. viscosus sequences.
Fig. 1.
Neighbour-joining tree showing relationships between type and reference strains of Actinomyces naeslundii, Actinomyces oris, Actinomyces johnsonii and Actinomyces viscosus and oral and clinical isolates determined by partial nanH gene sequence analysis. Actinomyces naeslundii strains 25, 51 and CCUG 34725 and A. oris strains 60 and 61 (each shown with the nanH sequence accession number) did not cluster with the majority of strains of the same species and were identified as strains with significant evidence of interspecies recombination. Scale bar=0.01 substitutions per site.
The dN/dS values for A. oris and A. naeslundii were 0.1466 and 0.2134, respectively. Tajima's D, Fu and Li's D* and F* were all significantly negative for A. oris but did not achieve significance for A. naeslundii (Table 1). The test for DNA divergence between populations implemented in dnasp indicated that the number of polymorphic sites in A. naeslundii and monomorphic in A. oris was 59 but the number of polymorphic sites in A. oris and monomorphic in A. naeslundii was 287 (χ2=7.011; P =0.00081).
Table 1.
Genetic variation of partial nanH sequences of type and reference strains and clinical isolates of Actinomyces oris and Actinomyces naeslundii
| A. naeslundii (n =30) | A. oris (n =74) | |
|---|---|---|
| G+C content | 0.687 | 0.693 |
| No. of segregating sites, S | 147 | 319 |
| No. of discrete sequences | 29 | 71 |
| dN/dS | 0.2134 | 0.1466 |
| π | 0.0210 | 0.0296 |
| θ | 0.0361 | 0.0728 |
| Tajima's D | −1.6094 | −2.0658* |
| Fu and Li's D* | −1.9615 | −3.6456** |
| Fu and Li's F* | −1.6815 | −3.5907** |
π and θ, nucleotide diversity.
All analyses were performed using dnasp version 4.5 (http://www.ub.es/dnasp/index.html/).
P <0.05;
P <0.02.
Analysis of the amino acid sequences of the BNRs of the four species is shown in supporting Table S1. There was overall great consistency in the pattern SXDXGXTW within each of the BNRs for all of the species with only single exceptions in the first for BNRs. In the fifth BNR the pattern was SXDXGXSW in all of the isolates except for one A. johnsonii isolate which had the sequence SCGNGASW.
The alignment of the amino acid sequences inferred from the partial nanH sequences from the four species with that of S. typhimurium LT2 demonstrated that the active site identified in the S. typhimurium sialidase was recognized in each of the Actinomyces sequences. Nine of the 12 amino acids in the S. typhimurium active site were identical in all the Actinomyces sequences (supporting Fig. S1). The exceptions among the sequences were the met-99 and trp-121 in S. typhimurium which were both replaced by serine and leu-175 which was replaced by phenylalanine.
To test for the statistical evidence of recombination in the first instance the Splitstrees method was used and the phi test provided evidence of significant recombination when all strains, except the A. viscosus and A. johnsonii strains, were included in the analysis (P =<10−20). Consideration of A. oris or A. naeslundii strains alone yielded phi values with P =3.43 × 10−6 and P =1.37 × 10−5, respectively, indicating statistically significant evidence of recombination in each of the species. To identify the strains with evidence of recombination we analysed the data of these two species together using the seven programs within the RDP suite and found that only A. naeslundii strains 25, 51 and CCUG 34725 and A. oris strains 60 and 61 gave significant evidence of recombination. The recombination events are summarized in Fig. 2. In the A. naeslundii strains 25, 51 and CCUG 34725 and A. oris strain 60 the recombination event extended beyond the available partial sequences.
Fig. 2.
Recombination events found in nanH of Actinomyces oris strains 60 (EU805671) and 61 (EU805672) and Actinomyces naeslundii strains 51 (EU805620), 25 (EU805612) and CUG 34725 (EU805609). Solid line indicates portion of sequence derived from nanH of A. naeslundii and broken line indicates portion of sequence derived from nanH of A. oris. Insertion in strain 25 between 365 and 1038, in strain 51 between 629 and 1038, in strain 60 between 1 and 477, in strain 61 between 432 and 1038 and between 1 and 655 in CUG 34725. Breakpoints determined using RDP suite of programs with significant evidence (P <0.001) for recombination obtained with ≥5 recombination tests in all cases.
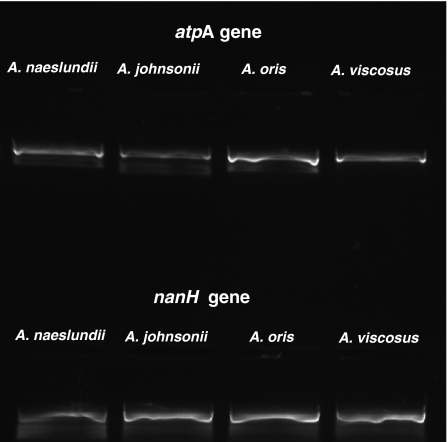
The type strains of each of the four species examined expressed the nanH gene when grown on a complex medium containing defibrinated horse blood and 1% (w/v) glucose (Fig. 3) and sequence analysis confirmed that the individual products were derived from the nanH genes of the appropriate species.
Fig. 3.
Reverse transcriptase (RT)-PCR assays showing expression of nanH and the housekeeping gene atpA in the type strains of Actinomyces oris (CCUG 34288; EU805602), and Actinomyces johnsonii (CCUG 34287; EU805600), Actinomyces naeslundii (CCUG 2238; EU805601) and Actinomyces viscosus (NCTC 10951; EU805603). Negative controls, omitting the RT in the reaction mix, yielded no amplicons from any strain.
Discussion
The complete functions of sialidase in the oral cavity are certainly not fully understood but dietary restriction in macaque monkeys resulted in the increase in levels of sialidase activity in the oral biofilm suggesting a role in bacterial nutrition (Beighton & Smith, 1986). The levels of sialidase-producing bacteria also increase in the absence of host's diet, again supporting the hypothesis that it plays a role in providing nutrient, both C and N, for microbial growth and proliferation (Beighton & Smith, 1986; Lucas et al., 1997; Sheehy et al., 2000). Bacterial sialidases, including those of the oral Actinomyces, also expose galactose residues of glycoprotein glycans located on mucosal surfaces or adsorbed to the tooth as part of the enamel pellicle. The exposed galactose residues may then be used as attachment sites by members of the oral biofilm promoting dental plaque accumulation (Costello et al., 1979; Gibbons et al., 1990).
All sialidase enzymes have a highly conserved array of residues, which in the tertiary structure of the enzyme, form the active and binding sites of the molecule (Crennell et al., 1993). In addition to these residues all sialidases also have a number of BNRs structural entities for which a function cannot be definitely ascribed because Asp boxes are found in many protein families (Henningsen et al., 1991; Copley et al., 2001). In determining the sequence diversity of nanH in these Actinomyces species, we selected a portion of the gene which contained the 12 residues involved in substrate interactions with, and stabilization of, the active site and the BNRs originally described in ‘A. viscosus’ strain DSM 43798, now identified as A. oris. The gene fragment of 1077 bp represents c. 30% of the entire nanH sequence in A. oris. The determination of the partial sequences of the nanH genes from multiple isolates of A. oris and A. naeslundii has enabled the amino acid conservation within each species and nucleotide sequence variation between the multiple independent strains of A. oris and A. naeslundii to be determined. The phylogenetic analysis clearly demonstrated that these two species have nanH genes with different nucleotide sequences. It was also apparent that the nucleotide sequences of A. johnsonii and A. viscosus were characteristic and that, despite considerable site-specific conservation of amino acid residues, the amino acid sequences of all four species were distinct and characteristic. The BNRs of the four Actinomyces species showed a very high degree of conservation with respect to the amino acid sequence as would be expected (Vimr, 1994). However, in the fifth BNR the threonine residue was replaced by a serine in all the 107 sequences determined; such a structurally synonymous substitution would not be expected to significantly modify the activity of the enzyme. The same variations observed here in the conserved amino acids involved in the active-site region have also been reported in other high G+C organisms (Sakurada et al., 1992; Jost et al., 2001).
The dN/dS values were <1 for both A. oris and A. naeslundii indicating that nanH in both species is under stabilizing selective pressure; nonsynonymous mutations are not selected. However, for A. oris significant negative values Tajima's D and Fu and Li's D* and F* were calculated suggesting that, as all nucleotides of any codon are assumed to be equally mutatable, the rate of mutation in A. oris must be greater than in A. naeslundii but with selection against nonsynonymous mutations. The proposed higher rate of mutation in the nanH of A. oris was supported by the observation that the frequency of polymorphic sites in A. oris which were monomorphic in A. naeslundii was significantly greater than the frequency of polymorphic sites in A. naeslundii which were monomorphic in A. oris. Consideration of our previously published sequence data on six housekeeping genes (Henssge et al., in press) indicated that the frequency of polymorphic sites in A. oris which were monomorphic in A.naeslundii was also significantly (P <0.05) greater in gltA, metG, pgi and rpoB, while in gyrA and atpA no significant difference was found. The reasons for the apparent greater frequency of mutations in A. oris is not known but may suggest that A. oris is subject to greater stress than A. naeslundii in the oral biofilm because environmental stress may increase the rate of mutation of organisms in biofilms (Bjedov et al., 2003; Tenaillon et al., 2004). However, given the structural requirements of a functioning sialidase, nonsynonymous mutations may not be beneficial and will, therefore, not be selected. Actinomyces oris (as A. naeslundii genospecies 2) is numerically more successful in the oral biofilm than A. naeslundii (as A. naeslundii genospecies 1) (Bowden et al., 1999) which might in part be explained by its higher mutation rate facilitating genome plasticity and an increased ability to respond successfully to environmental stress.
The methods used to interrogate data for the presence of recombinational events provided evidence for significant but limited recombination between the A. oris and A. naeslundii. This is the first evidence of recombination between these two species although recombination between sialidase genes of other oro-pharyngeal organisms, Streptococcus oralis and Streptococcus pneumoniae, has been reported (King et al., 2005). We found evidence of recombination in three of 30 A. naeslundii strains and in two of 74 A. oris isolates suggesting that recombination between these species is either not uncommon or these may be rare events and the recombinants may be more successful than the parent strains. However, the growth or survival benefit acquired by these recombinant strains is not known but because they have persisted and proliferated, at least in the mouths of the individuals from whom they were isolated, this suggests benefit has been accrued. The survival and growth benefits may not be due to recombinational events in nanH but to other unknown events, congruent with possession of a recombinant nanH gene, which might be responsible for the proliferation of these particular strains.
Acknowledgments
This project was supported by a grant from the Wellcome Trust (GR076381MA). The authors would like to thank Professor Nicklas Stromberg, Umeå University, Sweden, for gift of many of the Actinomyces strains used in this study; other strains were purchased from the Culture Collection, University of Göteborg, Sweden, the National Collection of Type Cultures, HPA, Colindale, England and the human clinical isolates were purchased from Dr Val Hall, Anaerobe Reference Unit, Department of Medical Microbiology and Public Health Laboratory, University Hospital of Wales, Cardiff.
Note
nanH partial gene sequences have been deposited in GenBank with accession numbers EU805600–EU805702.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Comparison of the amino acid sequence of bacterial neuraminidase repeats [BNRs] and frequency of detection amongst A. naeslundii, A. oris, A. johnsonni and A. viscosus strains.
Fig. S1. Alignment of partial sequences of the nanH gene of the type strains of A. naeslundii (accession number EU805601), A. oris (EU805602), A. johnsonii (EU805600) and A. viscosus (EU805603) with the nanH gene sequence of S. typhimurium (M55342).
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Beighton D, Smith K. The modulation of exoglycosidic enzymes in the supragingival plaque of macaque monkeys. Arch Oral Biol. 1986;34:319–322. doi: 10.1016/0003-9969(86)90137-8. [DOI] [PubMed] [Google Scholar]
- Beighton D, Whiley RA. Sialidase activity of the “Streptococcus milleri group” and other viridans group streptococci. J Clin Microbiol. 1990;28:1431–1433. doi: 10.1128/jcm.28.6.1431-1433.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beighton D, Hardie JM, Whiley RA. A scheme for the identification of viridans streptococci. J Med Microbiol. 1991;35:367–372. doi: 10.1099/00222615-35-6-367. [DOI] [PubMed] [Google Scholar]
- Bjedov I, Tenaillon O, Gérard B, Souza V, Denamur E, Radman M, Taddei F, Matic I. Stress-induced mutagenesis in bacteria. Science. 2003;300:1404–1409. doi: 10.1126/science.1082240. [DOI] [PubMed] [Google Scholar]
- Bowden GH, Nolette N, Ryding H, Cleghorn BM. The diversity and distribution of the predominant ribotypes of Actinomyces naeslundii genospecies 1 and 2 in samples from enamel and from healthy and carious root surfaces of teeth. J Dent Res. 1999;78:1800–1809. doi: 10.1177/00220345990780120601. [DOI] [PubMed] [Google Scholar]
- Braham PH, Moncla BJ. Rapid presumptive identification and further characterization of Bacteroides forsythus. J Clin Microbiol. 1992;30:649–654. doi: 10.1128/jcm.30.3.649-654.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers HL, Homer KA, Beighton D. Utilization of sialic acid by viridans streptococci. J Dent Res. 1996;75:1564–1571. doi: 10.1177/00220345960750080701. [DOI] [PubMed] [Google Scholar]
- Byers HL, Tarelli E, Homer KA, Hambley H, Beighton D. Growth of viridans streptococci on human serum alpha1-acid glycoprotein. J Dent Res. 1999;78:1370–1380. doi: 10.1177/00220345990780071201. [DOI] [PubMed] [Google Scholar]
- Copley RR, Russell RB, Ponting CP. Sialidase-like Asp-boxes: sequence-similar structures within different protein folds. Protein Sci. 2001;10:285–292. doi: 10.1110/ps.31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello AH, Cisar JO, Kolenbrander PE, Gabriel O. Neuraminidase-dependent hamagglutination of human erythrocytes by human strains of Actinomyces viscosus and Actinomyces naeslundii. Infect Immun. 1979;26:563–572. doi: 10.1128/iai.26.2.563-572.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crennell SJ, Garman EF, Laver WG, Vimr ER, Taylor GL. Crystal structure of a bacterial sialidase (from Salmonella typhimurium LT2) shows the same fold as an influenza virus neuraminidase. Proc Natl Acad Sci USA. 1993;90:9852–9856. doi: 10.1073/pnas.90.21.9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen EV. Carbohydrate depletion of immunoglobulin A1 by oral species of gram-positive rods. Oral Microbiol Immunol. 1994;9:352–358. doi: 10.1111/j.1399-302x.1994.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Hay DI, Childs WC, III, Davis G. Role of cryptic receptors (cryptitopes) in bacterial adhesion to oral surfaces. Arch Oral Biol. 1990;35(suppl):107S–114S. doi: 10.1016/0003-9969(90)90139-2. [DOI] [PubMed] [Google Scholar]
- Henningsen M, Roggentin P, Schauer R. Cloning, sequencing and expression of the sialidase gene from Actinomyces viscosus DSM 43798. Biol Chem Hoppe Seyler. 1991;372:1065–1072. doi: 10.1515/bchm3.1991.372.2.1065. [DOI] [PubMed] [Google Scholar]
- Henssge U, Do T, Radford DR, Gilbert SC, Clark D, Beighton D. Emended description of Actinomyces naeslundii and descriptions of Actinomyces oris sp. nov. and Actinomyces johnsonii sp. nov. previously identified as Actinomyces naeslundii genospecies 1, 2 and WVA 963. Int J Syst Evol Microbiol. 2008 doi: 10.1099/ijs.0.000950-0. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer KA, Kelley S, Hawkes J, Beighton D, Grootveld MC. Metabolism of glycoprotein-derived sialic acid and N-acetylglucosamine by Streptococcus oralis. Microbiology. 1996;142:1221–1230. doi: 10.1099/13500872-142-5-1221. [DOI] [PubMed] [Google Scholar]
- Hudson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Moore LV, Kaneko B, Moore WE. Actinomyces georgiae sp. nov., Actinomyces gerencseriae sp. nov., designation of two genospecies of Actinomyces naeslundii, and inclusion of A. naeslundii serotypes II and III and Actinomyces viscosus serotype II in A. naeslundii genospecies 2. Int J Syst Bacteriol. 1990;40:273–286. doi: 10.1099/00207713-40-3-273. [DOI] [PubMed] [Google Scholar]
- Jost BH, Songer JG, Billington SJ. Cloning, expression, and characterization of a neuraminidase gene from Arcanobacterium pyogenes. Infect Immun. 2001;69:4430–4437. doi: 10.1128/IAI.69.7.4430-4437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Whatmore AM, Dowson CG. NanA, a neuraminidase from Streptococcus pneumoniae, shows high levels of sequence diversity, at least in part through recombination with Streptococcus oralis. J Bacteriol. 2005;187:5376–5386. doi: 10.1128/JB.187.15.5376-5386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas VS, Beighton D, Roberts GJ, Challacombe SJ. Changes in the oral streptococcal flora of children undergoing allogeneic bone marrow transplantation. J Infect. 1997;35:135–141. doi: 10.1016/s0163-4453(97)91545-0. [DOI] [PubMed] [Google Scholar]
- Martin DP, Williamson C, Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Moncla BJ, Braham P. Detection of sialidase (neuraminidase) activity in Actinomyces species by using 2′-(4-methylumbelliferyl)alpha-d-N-acetylneuraminic acid in a filter paper spot test. J Clin Microbiol. 1989;27:182–184. doi: 10.1128/jcm.27.1.182-184.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinholdt J, Tomana M, Mortensen SB, Kilian M. Molecular aspects of immunoglobulin A1 degradation by oral streptococci. Infect Immun. 1990;58:1186–1194. doi: 10.1128/iai.58.5.1186-1194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messegyer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakurada K, Ohta T, Hasegawa M. Cloning, expression, and characterization of the Micromonospora viridifaciens neuraminidase gene in Streptomyces lividans. J Bacteriol. 1992;174:6896–6903. doi: 10.1128/jb.174.21.6896-6903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy EC, Beighton D, Roberts GJ. The oral microbiota of children undergoing liver transplantation. Oral Microbiol Immunol. 2000;15:203–210. doi: 10.1034/j.1399-302x.2000.150309.x. [DOI] [PubMed] [Google Scholar]
- Smith K, Beighton D. The effects of the availability of diet on the levels of exoglycosidases in the supragingival plaque of macaque monkeys. J Dent Res. 1986;65:1349–1352. doi: 10.1177/00220345860650111401. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Denamur E, Matic I. Evolutionary significance of stress-induced mutagenesis in bacteria. Trends Microbiol. 2004;12:264–270. doi: 10.1016/j.tim.2004.04.002. [DOI] [PubMed] [Google Scholar]
- ter Steeg PF, Van der Hoeven JS, de Jong MH, van Munster PJ, Jansen MJ. Enrichment of subgingival microflora on human serum leading to accumulation of Bacteroides species, peptostreptococci and fusobacteria. Antonie Van Leeuwenhoek. 1987;53:261–272. doi: 10.1007/BF00393933. [DOI] [PubMed] [Google Scholar]
- Vimr ER. Microbial sialidases: does bigger always mean better? Trends Microbiol. 1994;12:271–277. doi: 10.1016/0966-842x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Yeung MK. Complete nucleotide sequence of the Actinomyces viscosus T14V sialidase gene: presence of a conserved repeating sequence among strains of Actinomyces spp. Infect Immun. 1993;61:109–116. doi: 10.1128/iai.61.1.109-116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.