Abstract
We report comprehensive structure activity relationship studies on a novel series of c-Jun N-terminal kinase (JNK) inhibitors. The compounds are substrate competitive inhibitors that bind to the docking site of the kinase. The reported medicinal chemistry and structure-based optimizations studies resulted in the discovery of selective and potent thiadiazole JNK inhibitors that displays promising in vivo activity in mouse models of insulin insensitivity.
Introduction
Protein kinases comprise 2% of the mammalian genome and catalyze the transfer of γ-phosphoryl group of ATP to specific protein substrates.1 The c-Jun N-terminal kinases (JNKs) are a series of serine/threonine protein kinases of the mitogen activated protein kinase (MAPK) family. Three distinct genes encoding JNKs have been identified, JNK-1, JNK-2, and JNK-3, and at least 10 different splicing isoforms exist in mammalian cells.2–4 The three JNK isoforms share more than 90% amino acid sequence identity and the ATP pocket is >98% homologous. These proteins are activated in response to cellular stresses such as heat shock, irradiation, hypoxia, chemotoxins, and peroxides. They are also activated in response to various cytokines and participate in the onset of apoptosis.5,6 It is reported that up-regulation of JNK activity is associated with a number of disease states such as type- 2 diabetes, obesity, cancer, inflammation, and stroke.1–3 Therefore, JNK inhibitors are expected to be effective therapeutic agents against a variety of diseases.
JNKs bind to substrates and scaffold proteins, such as JIP-1, that contain a D-domain, as defined by the consensus sequence R/KXXXXLXL.7,8 A peptide corresponding to the D-domain of JIP-1 (aa 153–163; pep-JIP1), inhibits JNK activity in vitro and displays remarkable selectivity with little inhibition of the closely related Erk and p38 MAPKs.9–12 Recent in vivo data, generated for studies focusing on pep-JIP1 fused to the cell permeable HIV-TAT peptide, show that its administration in various mouse models of insulin resistance and type-2 diabetes restores normoglycemia without causing hypoglycemia in lean mice.13 The peptide was further improved by the synthesis of an all-D retro-inverso peptide, D-JNK1 containing a cell-penetrating sequence. However, peptide’s instability in vivo, its short half-life, and the costly and inefficient large-scale manufacturing and purification processes all hamper the development on novel therapies for diabetes based on D-JNK1 alone. Nevertheless, despite some skepticism surrounding this peptide, the efficacy of such substrate competitive peptides is such that several recent studies report preliminary success on the use of D-JNK1 in early pre-clinical and clinical studies. 9–13
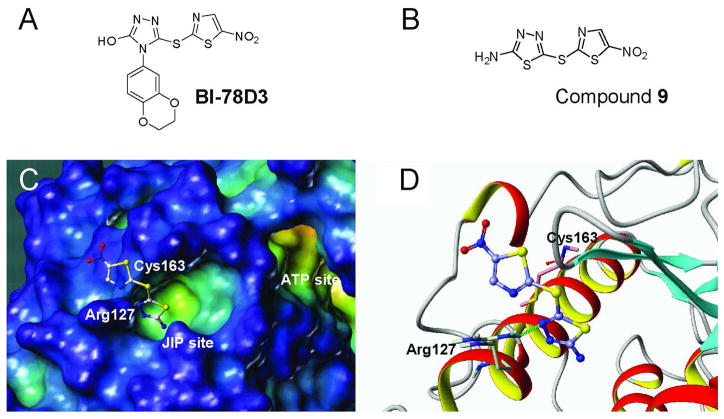
There has been considerable effort to identify JNK inhibitors over the past several years.14–20 A drug discovery program in our laboratory was initiated with the aim of identifying and characterizing small–molecule JNK inhibitors as novel chemical entities targeting the JIP docking site. 21,22 As a result of these efforts, we have very reported on the identification of compound 12 (BI-78D3) (Figure 1A) (4-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol) from our HTS libraries, as the first potent and selective JNK inhibitor of this class with demonstrated in vivo activity in mice model of insulin resistance. 21
Figure 1.
Chemical structures and docked geometry. (A) Chemical structure of the previously reported compound 12 (BI-78D3) 21; (B) Chemical structure of compound 9; (C) and (D) Docked structure of compound 9 in the JIP site of JNK1.
As a continuation of our work21,22 we now report a comprehensive structure activity relationship studies describing the discovery of novel JNK inhibitors that target the JIP-JNK interaction site. We developed a triazole series followed by a thiadiazole series based on structure–activity relationship (SAR) studies carried out on the initial hit compound 12 (Figure 1A)21 which ultimately led to the discovery of compound 9 (Figure 1B). We describe here the pharmacological properties, design, and SAR studies that have lead to its identification.
Results and discussion
Screening of our internal compound collection for JNK inhibitors resulted in the identification of compounds belonging to the triazole series.21 The 4-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (compound 12, Figure 1A)21 was qualified as a hit and became the starting point of our medicinal chemistry efforts.
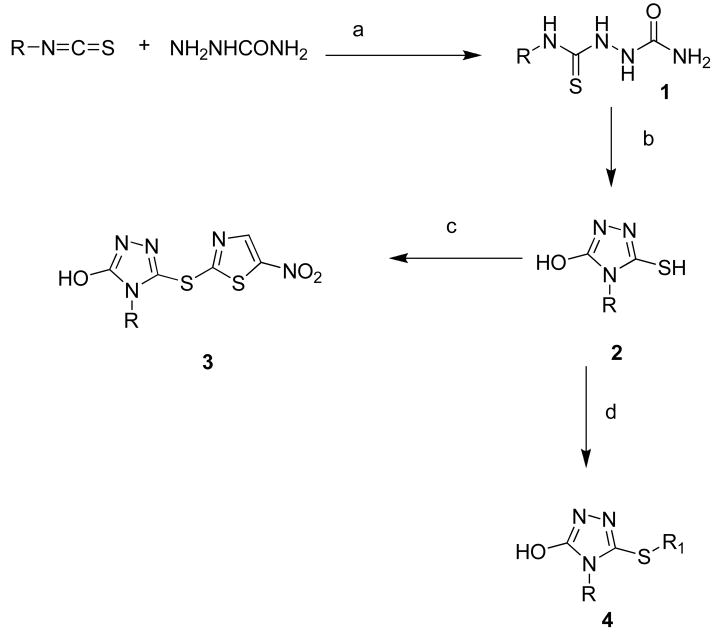
To investigate the effects on potency induced by small changes in the structure of 12, we developed the general synthetic route for the preparation of this series. Compound 1, thiobiurea was synthesized according to the reported procedure (Scheme 1).23 A variety of commercially available arylisothiocyanates were treated with semicarbazide in the presence of sodium acetate in acetonitrile at room temperature to give 1a–1q and 4a–e in good yields (Scheme 1). The thiobiureas (1a–e) and 4a–e were heated at 100 °C in the presence of 2 M NaOH aqueous solution for 5 to 8 h to afford 3-hydroxy-5-thiol derivatives of triazoles in moderate to good yields. Compounds (3a–q) were synthesized by nucleophilic substitution of 2-bromo-5-nitrothiazole with thiol of triazoles in NaOMe/MeOH solution. We found that the thiol is consistently more reactive than hydroxyl; we never isolated any hydroxyl derivative compounds even using excess amounts of 2-bromo-5-nitrothiazole. Similarly, the compounds (4a–e) were prepared using different alkyl and aryl bromides.
Scheme 1.
Reagents and conditions: (a) sodium acetate, CH3CN, rt; (b) 2 M NaOH solution, 100 °C; (c) sodium methoxide, MeOH, 2-bromo-5-nitrothiazole, rt; (d) aryl or alkyl bromide, sodium methoxide, MeOH, rt.
1–3 (a) R = 4-tert-butylphenyl; (b) R = 3,4-methylenedioxyphenyl; (c) R = 4-fluoropheny; (d) R = 2,4-dimethoxyphenyl; (e) R = 3,5- dimethoxyphenyl; (f) R= 3,4-dimethoxyphenyl; (g) R = naphthyl; (h) R = 2,5-dimethoxyphenyl; (i) R = 4-trifluromethoxyphenyl; (j) R = 4-nitrophenyl; (k) R = 3,4-difluorophenyl; (l) R = 3-fluorophenyl; (m) R = 3-methoxyphenyl,; (n) R = 2-methoxyphenyl; (o) R = 3-trifluoromethylphenyl; (p) R = 4-trifluromethylphenyl; (q) R = cyclohexyl; (r) R = 4-methoxyphenyl.
(4a) R = phenyl, R1 = n-butyl; (4b) R = phenyl, R1 = ethyl; (4c) R = 3,4-benzodioxine, R1 = benzyl; (4d) R= phenyl, R1 = 5-nitrothiophene.
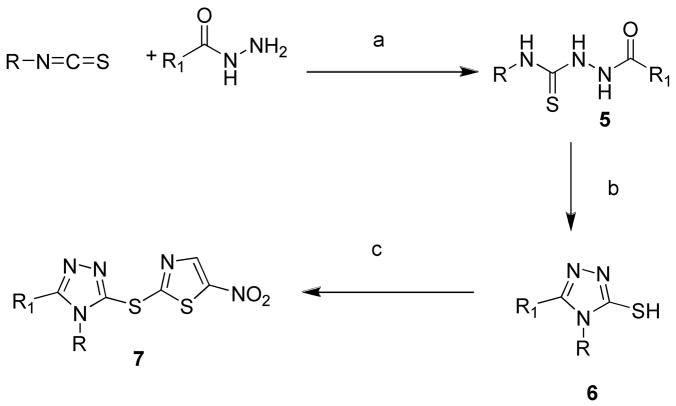
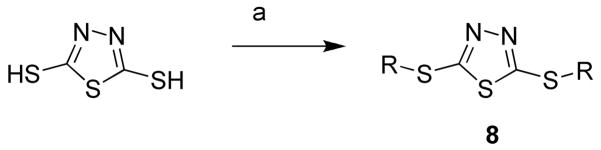
Exploration of triazole C-3 position (replacing hydroxyl to alky or aryl) required a different synthesis of the core scaffold. Arylbithioureas (5a–g) (Scheme 2) were prepared according to published procedures.24 Compounds 6a–g were synthesized following the similar conditions mentioned before in Scheme 1 and a few of them were commercially available (6b, d–g). The final compounds (7a–g) were successfully obtained by displacing bromide with thiols as previously described. The dithioether of thiadiazoles (8a–g) were synthesized from commercially available 1, 3, 4-thiadiazole-2,5-dithiol (Scheme 3). Similarly, we prepared compounds 9 and 10 from 5-amino-1,3,4-thiadiazole-2-thiol to introduce further diversity into the series (Scheme 4).
Scheme 2.
Reagents and conditions: (a) sodium acetate, CH3CN, rt; (b) 2 M NaOH solution, 100 °C; (c) sodium methoxide, MeOH, 2-bromo-5-nitrothiazole, rt.
5–7 (a) R = 3,4-benzodioxine, R1 = phenyl; (b) R = 2,4-dichlorophenyl, R1= 1-methylpyrole; (c) R = 3-methoxyphenyl, R1 = phenyl; (d) R = furfuryl; R1 = naphthyl; (e) R = phenyl, R1 = thiophene; (f) R = phenyl, R1 = 4-pyridyl; (g) R = methyl, R1 = pyrazinyl-oxadiazole.
Scheme 3.
Reagents and conditions (a) alkyl or aryl bromide, sodium methoxide, MeOH, rt. (8a) R = benzyl; (8b) R = 4-fluorobenznyl; (8c) R = 4-methoxybenzyl; (8d) R = 3-nitrobenzyl; (8e) R = 4-nitrobenzyl; (8f) R = 5-nitrothiazole; (8g) R = 5-carboxylthiazole.
Scheme 4.
Reagents and conditions: (a) 2-bromo-5-nitrothiazole, sodium methoxide, MeOH, rt; (b) 4-Fluorobenzoic acid, EDC, HOBt, DIEA, DMF, 50 °C; (c) 4-fluorobenzyl bromide, sodium methoxide, MeOH, rt.
In our work, the basis for the biochemical assay was JNK-1 mediated phosphorylation of ATF-2. High-throughput screening identified compound 12, with an IC50 of 280 nM.21 Competition experiments confirmed that this compound is a competitive inhibitor of the interactions between JNK and pepJIP1.21
We began our investigation of this initial scaffold by synthesizing compounds with variations in the 3-, 4-, and 5-positions of the triazol. As shown in Table 1, among the combinations tested, changes in the 4- and 5-positions are well tolerated. We first synthesized and tested a series of alkyl-thio- ethers but none of these showed any remarkable JNK inhibition (4a, 4b in Table 1). Aryl- and benzyl- thio- ethers lead to a drastic drop in activity. Replacing the thiazole with a thiophene also results in a decrease in activity. We found that the only suitable moiety in the 3-position was a 5-nitrothiazole, and changing from nitro to carboxyl group on the thiazole unit was also not tolerated. Based on our results, we retained the 5-position as a 5-nitro-thiazole and varied the other positions of the parent compound 12 (Figure 1A).
Table 1.
Triazole series
| Compound | Lantha Screen Kinase activity assay, IC50 (μM) or % inhibition at 100 μM | pepJIP1 DELFIA displacement assay, IC50 (nM) or % inhibition at 100 μM |
|---|---|---|
| 4a | 25% | NDa |
| 4b | 16% | ND |
| 4c | 2% | ND |
| 4d | 22% | 55% |
| 3a | 39 | 95 |
| 3b | 3.3 | 102 |
| 3c | 1.8 | 225 |
| 3d | 1.1 | 66 |
| 3e | 4.3 | 105 |
| 3f | 1.0 | 43 |
| 3g | 2.7 | 48 |
| 3h | 2.4 | 85 |
| 3i | 2 | 130 |
| 3j | 0.4 | 86 |
| 3k | 6.6 | 73% |
| 3l | 84 | 626 |
| 3m | 16 | 118 |
| 3n | 2.2 | 53 |
| 3o | 2.9 | 160 |
| 3p | 3 | 75 |
| 3q | 25 | 56 |
| 3r | 17 | 45 |
ND: No displacement at 100 μM.
Modifications investigated in the 4-phenyl moiety of the triazole series are detrimental to JNK potency as shown in Table 1. The 3- and 4-positions of the phenyl group were more active. Compound 3f has a 3,4-methylene-dioxyphenyl group in place of the 3,4-benzodioxin ring in the parent compound, and has comparable activity. However, a 4-tert-butylphenyl group was not tolerated in this position likely due to its bulky size (3a). The most active compound contains 4-nitrophenyl on the 4-position of the triazole, resulting in an IC50 of 0.4 μM (3j); likewise, groups such as fluoro (3c), methoxy (3d, 3e, 3f, 3h, 3m, 3n, and 3r), trifluoromethyl (3o and 3p), trifluoromethoxy (3i) show similar activities. However, a 3-fluoro on phenyl group (3l) does not show any good activity and 3,4- difluorophenyl (3k) has a moderate activity.
We further modified the 4- and 5-positions on the triazole ring while maintaining the 5-nitro-thiazole in the 3-position (Table 2). In this series, phenyl group in the 5-position reduces the activity in the kinase assay as compared to the parent compound 12, but has a lower IC50 (108 nM) in the displacement assay against pepJIP1; whereas replacing the dioxin ring with a 3-methoxypenyl group (7c) results in a 3-fold increase in activity, indicating that two large ring structures in both the 4- and 5-position may constitute too much bulk. We subsequently synthesized compounds contains with smaller aromatic structures in both positions (7b and 7d) and found that 1-methylpyrrole is tolerated while both naphthyl and pyrazinyl-oxadiazole in the 5-position reduce the activity. We next focused on a 1,3,4-thiadiazole-2,5-dithiol as the scaffold ring to further explore the effect of the substituents in the 5-position (Table 3). We had very little success with benzyl groups, but smaller moieties such as a nitrothiazole and a carboxylthiazole were very promising, which led to compound 9 (Table 4). The use of 5-amino-1,3,4-thiadiazole-2-thiol as a scaffold allowed for nonsymmetrical modifications, however, further modifications of the amine group on compound 9 did not increase potency (10). Addition of a methylphenyl group on the thiol resulted in a complete loss of activity (11).
Table 2.
Modification of C-3 position (replacing hydroxyl to alkyl or aryl) on triazoles
| Compound | Lantha Screen Kinase activity assay, IC50 (μM) | pepJIP1 DELFIA displacement assay, IC50 (nM) |
|---|---|---|
| 7a | 27 | 108 |
| 7b | 2 | 83 |
| 7c | 7.9 | 168 |
| 7d | 13.6 | 57 |
| 7e | > 50 | 107 |
| 7f | 3.1 | 122 |
| 7g | 100 | 131 |
Table 3.
Dithioether of 1,3,4-thiadiazoles
| Compound | Lantha Screen kinase activity assay, IC50 (μM) | pepJIP1 DELFIA displacement assay, IC50 (nM) |
|---|---|---|
| 8a | >100 | NDa |
| 8b | >100 | ND |
| 8c | >100 | ND |
| 8d | >100 | ND |
| 8e | >100 | ND |
| 8f | 0.8 | 187 |
| 8g | 84 | 100 |
ND: No displacement at 100 μM.
Table 4.
2-amino-1,3,4- thiadiazole series.
| Compound | Lantha Screen kinase activity assay, IC50 (μM | pepJIP1 DELFIA displacement assay, IC50 (nM) |
|---|---|---|
| 9 | 0.7 | 239 |
| 10 | 84 | NDa |
| 11 | >100 | >1000 |
ND: No displacement at 100 μM.
Modeling studies suggest that compound 9 may bind at the JIP site with the nitrothiazol group crossing the ridge close to residues Arg127 and Cys163 (Figure 1C and 1D). Its thiazole group can form a hydrogen bond with Arg127 and can bind into a sub-pocket nearby, which based on this hypothesis could tolerate groups with the similar size to substitute the thiadiazole group of compound 9.
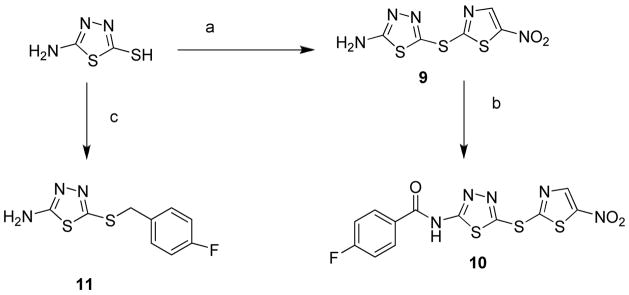
Compound 9 showed an IC50 of 0.7 μM in the kinase assay (Figure 2A) in a substrate competitive manner. In addition, compound 9 also showed an IC50 of 239 nM in displacing pepJIP1, as measured by a DELFIA assay (Figure 2B).21 Compound 9 was found to be 142 fold less active against p38α, a member of the MAPK family with high structural similarity to JNK and completely inactive against Akt kinase (Akt inhibitions at 100 μM for compound 3j, 7b, 8f, and 9 were 20%, 14%, 27%, and 11% respectively). These compounds were also very selective against other protein targets under investigation in our laboratory including the metalloprotease LF (lethal factor) and the proprotein convertase furin (Table 5). This selectivity is in agreement with our previous findings with compound 12 and with the reported data on pepJIP1.10–13, 21
Figure 2.
In vitro assays. (A) Kinase inhibition assay for 9; (B) Dose dependent displacement of biotinylated pepJIP1 from GST-JNK1.
Table 5.
Selectivity profile
| compound | p38α, IC50 (μM) | Akt1, IC50 (μM) | Furin, IC50 (μM) | LF, IC50 (μM) |
|---|---|---|---|---|
| 3j | >100 μM | 20% at 100 μM | > 50 μM | >100 μM |
| 7b | >100 μM | 14% at 100 μM | > 50 μM | >100 μM |
| 8f | >100 μM | 27% at 100 μM | > 50 μM | >100 μM |
| 9 | >100 μM | 11% at 100 μM | > 50 μM | >100 μM |
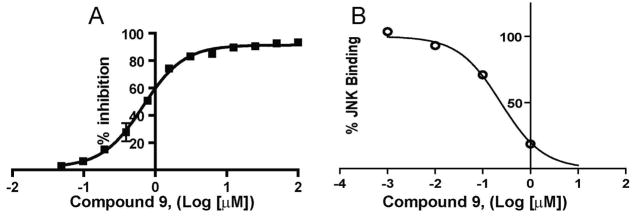
In an attempt to profile the properties of compound 9 in the context of a complex cellular milieu, we employed the cell -based LanthaScreen™ kinase assay. In this assay compound 9 is able to inhibit TNF-α stimulated phosphorylation of c-Jun in cell (EC50 = 6.23 μM; Figure 3A). It should be noted that the cell-based system employed makes use of a GFP-c-Jun stable expression system. As a result, the levels of GFP-c-Jun in these cells are higher than endogenous levels. This could have an inflationary effect on the EC50 values obtained with this assay when testing substrate competitive compounds. Nonetheless, this finding establishes that compound 9 is able to function in a cellular context.
Figure 3.
Cell based and in vivo efficacy studies with compounds 9 and 7b; (A). TR-FRET analysis of c-Jun phosphorylation upon TNF-alpha stimulation of HeLa cells in the presence of increasing concentrations 9; (B) Effects on insulin resistance in 11-week-old BKS.Cg-+Leprdb/+Leprdb/OlaHsd db/db mice (Harlan Sprague Dawley, Inc.; Indianapolis, IN). Diamonds, vehicle control; triangles, 25 mg/kg 9; circles, 25 mg/kg 7b; squares, 25 mg/kg 8f. Data shown as means ± S.D. (n =6). *P = 0.0022, **P = 0.0001.
The link between the JNK pathway and type-2 diabetes has been established previously.10–13 Thus in an attempt to further our bio-analysis of the JNK-inhibitory properties of compound 9, we monitored the ability of compound 9 to restore insulin sensitivity in a mouse model of type-2 diabetes. For this analysis, insulin insensitive mice from Harlan (Harlan Sprague Dawley, Inc.; Indianapolis, IN) were injected once with 25 mg/kg, of compounds 9, 7b, and 8f, 30 minutes prior to insulin injection. The effect of insulin on blood glucose levels was then measured (Figure 3B). Compound 9 resulted in a statistically significant reduction in blood glucose levels as compared to the vehicle control (Figure 3B). Hence, the ability of compound 9 to restore insulin sensitivity is consistent with its proposed function as an effective JNK inhibitor.21 Liquid chromatography/mass spectrometry bio-availability analysis demonstrates that compound 9 has favorable microsomal and plasma stability (T1/2 = 27 min. see supporting information) which support its use in further in vivo experiments.
Conclusion
We successfully developed a new series of JNK inhibitors, many of which are very potent in vitro. Our initial in vivo screens indicate that compound 9 possess the ability to restore insulin sensitivity in mice models of diabetes. Our results indicate that targeting the protein-protein interaction between JNK and JIP with a small molecule is a new and promising avenue for the development of novel pharmacological tools that inactivates the JNK pathway.
Experimental Section
General
Unless otherwise indicated, all anhydrous solvents were commercially obtained and stored in Sure-seal bottles under nitrogen. All other reagents and solvents were purchased as the highest grade available and used without further purification. Thin-layer chromatography (TLC) analysis of reaction mixtures was performed using Merck silica gel 60 F254 TLC plates, and visualized using ultraviolet light. NMR spectra were recorded on Varian 300 or 500 MHz instruments. Chemical shifts (δ) are reported in parts per million (ppm) referenced to 1H (Me4Si at 0.00). Coupling constants (J) are reported in Hz throughout. Mass spectral data were acquired on Shimadzu LCMS-2010EV for low resolution, and on an Agilent ESI-TOF for either high or low resolution. Purity of all compounds was obtained in a HPLC Breeze from Waters Co. using an Atlantis T3 3μm 4.6×150 mm reverse phase column. The eluant was a linear gradient with a flow rate of 1 ml/min from 95% A and 5% B to 5% A and 95% B in 15 min followed by 5 min at 100% B (Solvent A: H2O with 0.1% TFA; Solvent B: ACN with 0.1% TFA). The compounds were detected at λ=254 nm. Purity of key compounds was established by elemental analysis as performed on a Perkin Elmer series II-2400 (see supporting information).
2-(4-tert-butylphenylcarbamothioyl)hydrazinecarboxamide (1a)
To a suspension of sodium acetate (216 mg, 2.63 mmol) and semicarbazide (293 mg, 2.63 mmol) in acetonitrile (10 mL) was added 4-tert-butylphenyl isothiocyanate (506 mg, 2.63 mmol) and the reaction mixture was stirred for 24 h. The solvent was removed by rotary evaporation and the resulting residue was dissolved in 2 M NaOH aqueous solution (30 mL) and filtered through activated charcoal to remove any unreacted isothiocyanate. The filtrate was acidified with 1 N HCl (40 mL). The precipitate was collected and washed with water (3 × 20 mL), and hexanes (2 × 20 mL) to afford 1a as an white solid (601 mg, 86%). 1H NMR (300 MHz, DMSO-d6) δ 1.26 (s, 9 H), 6.03 (s, 2 H, NH2), 7.31 (d, J = 8.4 Hz, 2 H), 7.39 (d, J = 8.1 Hz, 2 H), 7.99 (s, 1 H, NH), 9.32 (s, 1 H), NH), 9.55 (s, 1 H, NH); MS m/z 555(2M+Na)+, 533 (2M+H)+, 289 (M+Na)+, 267 (M+H)+, 250, 224, 158, 88, 74; HRMS calcd for C12H19N4OS (M+H) 267.1274, found 267.1279.
Following above mentioned procedure and the appropriate starting materials and reagents used; compounds (1b–q) were synthesized.
2-(benzo[d][1,3]dioxol-5-ylcarbamothioyl)hydrazinecarboxamide (1b)
Yield: 88%; 1H NMR (300 MHz, DMSO-d6) δ 5.99 (s, 2 H), 6.03 (s, 2 H, NH2), 6.80–6.86 (m, 2 H), 7.09 (s, 1 H), 7.97 (s, 1 H, NH), 9.32 (s, 1 H, NH), 9.54 (s, 1 H, NH); MS m/z 531 (2M+Na)+, 277 (M+Na)+, 255 (M+H)+, 238, 212, 151, 147, 129, 106, 102, 97; HRMS calcd for C9H11N4O3S (M+H) 255.0546, found 255.0546.
2-(4-fluorophenylcarbamothioyl)hydrazinecarboxamide (1c)
Yield: 76%; 1H NMR (300 MHz, DMSO-d6) δ 6.05 (s, 2 H, NH2), 7.10–7.18 (m, 2 H), 7.44–7.50 (m, 2 H), 8.02 (s, 1 H, NH), 9.41 (s, 1 H, NH), 9.69 (s, 1 H, NH); MS m/z 251 (M+Na)+, 229 (M+H)+, 212, 192, 186, 141, 129, 106, 102, 97, 85; HRMS calcd for C8H10FN4OS (M+H) 229.0554, found 229.0560.
2-(2,4-dimethoxyphenylcarbamothioyl)hydrazinecarboxamide (1d)
Yield: 81%; 1H NMR (300 MHz, DMSO-d6) δ 3.74 (s, 3 H, OMe), 3.77 (s, 3 H, OMe), 6.07(s, 2 H, NH2), 6.48 (d, J = 8.1 Hz, 1 H), 6.59 (s, 1 H), 7.84 (d, J = 8.1 Hz, 1 H), 8.08 (s, 1 H, NH), 8.93 (s, 1 H, NH), 9.36 (s, 1 H, NH); HRMS calcd for C10H15N4O3S (M+H) 271.0865, found 271.0866.
2-(3,5-dimethoxyphenylcarbamothioyl)hydrazinecarboxamide (1e)
Yield: 85%; 1H NMR (300 MHz, DMSO-d6) δ 6.06 (s, 2 H, NH2), 6.26 (s, 1 H), 6.87 (s, 2 H), 7.99 (s, 1 H, NH), 9.40 (s, 1 H, NH), 9.56 (s, 1 H, NH); HRMS calcd for C10H15N4O3S (M+H) 271.0865, found 271.0868.
2-(3,4-dimethoxyphenylcarbamothioyl)hydrazinecarboxamide (1f)
Yield: 82%; 1H NMR (300 MHz, DMSO-d6) δ 6.04 (s, 2 H, NH2), 6.86 (d, J = 8.7 Hz, 1 H), 6.99 (d, J = 8.7 Hz, 1 H), 7.25 (s, 1 H), 7.99 (s, 1 H, NH), 9.28 (s, 1 H, NH), 9.51 (s, 1 H, NH); HRMS calcd for C10H15N4O3S (M+H) 271.0865, found 271.0864.
2-(naphthalen-1-ylcarbamothioyl)hydrazinecarboxamide (1g)
Yield: 79%; 1H NMR (300 MHz, DMSO-d6) δ 6.10 (s, 2 H, NH2), 7.38–7.55 (m, 4 H), 7.84 (d, J = 7.8 Hz, 1 H), 7.91–7.97 (m, 2 H), 8.21 (s, 1 H, NH), 9.46 (s, 1 H, NH), 9.86 (s, 1 H, NH); MS m/z 543 (2M+Na)+, 521 (2M+H)+, 283 (M+Na)+, 261 (M+H)+, 244, 218, 159, 144, 85, 74, 56; HRMS calcd C12H13N4OS (M+H) 261.0805, found 261.0809.
2-(2,5-dimethoxyphenylcarbamothioyl)hydrazinecarboxamide (1h)
Yield: 84%; 1H NMR (300 MHz, DMSO-d6) δ 3.68 (s, 3 H, OMe), 3.77 (s, 3 H, OMe), 6.19 (s, 2 H, NH2), 6.65 (dd, J = 9 and 2.7 Hz, 1 H), 6.95 (d, J = 9 Hz, 1 H), 8.20 (s, 1 H), 8.28 (s, 1 H, NH), 9.14 (s, 1 H, NH), 9.62 (s, 1 H, NH); HRMS calcd for C10H15N4O3S (M+H) 271.0865, found 271.0862.
2-(4-(trifluoromethoxy)phenylcarbamothioyl)hydrazinecarboxamide (1i)
Yield: 82%; 1H NMR (300 MHz, DMSO-d6) δ 6.07 (s, 2 H, NH2), 7.29 (d, J = 8.4 Hz, 2 H), 7.62 (d, J = 9 Hz, 2 H), 8.07 (s, 1 H, NH), 9.51 (s, 1 H, NH), 9.83 (s, 1 H, NH); HRMS calcd for C9H10F3N4O2S (M+H) 295.0477, found 295.04778.
2-(4-nitrophenylcarbamothioyl)hydrazinecarboxamide (1j)
Yield: 88%; 1H NMR (300 MHz, DMSO-d6) δ 6.13 (s, 2 H, NH2), 7.96 (d, J = 9.0 Hz, 2 H), 8.13 (s, 1 H, NH), 8.18 (d, J = 9.3 Hz, 2 H), 9.80 (s, 1 H, NH), 10.13 (s, 1 H, NH); MS m/z 278 (M+Na)+, 256 (M+H)+, 253, 239, 169, 146, 107, 104, 87, 76; HRMS calcd for C8H9N5O3S 256.0499 (M+H), found 256.0500.
2-(3,4-difluorophenylcarbamothioyl)hydrazinecarboxamide (1k)
Yield: 72%; 1H NMR (300 MHz, DMSO-d6) δ 6.08 (s, 2 H, NH2), 7.30–7.39 (m, 2 H), 7.70–7.724 (m, 1 H), 8.03 (s, 1 H, NH), 9.55 (s, 1 H, NH), 9.81 (s, 1 H, NH); MS m/z 515 (2M+Na)+, 269 (M+Na)+, 247 (M+H)+, 230, 204, 192, 147, 121, 102, 97; HRMS calcd for C8H9F2N4OS (M+H) 247.0460, found 247.0455.
2-(3-fluorophenylcarbamothioyl)hydrazinecarboxamide (1l)
Yield: 81%; 1H NMR (300 MHz, DMSO-d6) δ 6.08 (s, 2 H, NH2), 6.94 (t, J = 8.4 Hz, 1 H), 7.31–7.36 (m, 2 H), 7.58–7.63 (m, 1 H), 8.03 (s, 1 H, NH), 9.53 (s, 1 H, NH), 9.78 (s, 1 H, NH); MS m/z 251 (M+Na)+, 229 (M+H)+, 212, 186, 147, 118, 106, 85; HRMS calcd for C8H10FN4OS (M+H) 229.0554, found 229.0553.
2-(3-methoxyphenylcarbamothioyl)hydrazinecarboxamide (1m)
Yield: 84%; 1H NMR (300 MHz, DMSO-d6) δ 3.73 (s, 3 H, OMe), 6.05 (s, 2 H, NH2), 6.69 (d, J = 7.8 Hz, 1 H), 7.10 (d, J = 7.8 Hz, 1 H), 7.20 (t, J = 7.8 Hz, 1 H), 7.26 (s, 1 H), 8.01 (s, 1 H, NH), 9.40 (s, 1 H, NH), 9.61 (s, 1 H, NH); MS m/z 503 (2M+Na)+, 481 (2M+H)+, 263 (M+Na)+, 241 (M+H)+, 224, 198, 166, 147, 129, 118, 107, 97; HRMS calcd for C9H12N4O2S (M+H) 241.0754, found 241.0755.
2-(2-methoxyphenylcarbamothioyl)hydrazinecarboxamide (1n)
Yield: 86%; 1H NMR (300 MHz, DMSO-d6) δ 3.81 (s, 3 H, OMe), 6.15 (s, 2 H, NH2), 6.94 (t, J = 6.9 Hz, 1 H), 7.04 (d, J = 7.2 Hz, 1 H), 7.09–7.14 (m, 2 H), 8.33 (s, 1 H, NH), 9.11 (1 H, NH), 9.56 (s, 1 H, NH); HRMS calcd for C9H12N4O2S (M+H) 241.0754, found 241.0757.
2-(3-(trifluoromethyl)phenylcarbamothioyl)hydrazinecarboxamide (1o)
Yield: 69%; 1H NMR (300 MHz, DMSO-d6) δ 6.10 (s, 2 H), 7.45–7.54 (m, 2 H), 7.87 (d, J = 6.6 Hz, 1 H), 8.00 (s, 1 H), 8.07 (s, 1 H), 9.61 (s, 1 H), 9.96 (s, 1 H); MS m/z 579 (2M+Na)+, 557 (2M+H)+, 301 (M+Na)+, 279 (M+H)+, 262, 236, 169, 126, 88, 74; HRMS calcd for C9H9F3N4OS 279.0522 (M+H), found 279.0526.
2-(4-(trifluoromethyl)phenylcarbamothioyl)hydrazinecarboxamide (1p)
Yield: 56%; 1H NMR (300 MHz, DMSO-d6) δ 6.10 (s, 2 H), 7.66 (d, J = 8.7 Hz, 2 H), 7.82 (d, J = 8.4 Hz, 2 H), 8.07 (s, 1 H), 9.63 (s, 1 H), 9.56 (s, 1 H); HRMS calcd for C9H9F3N4OS 279.0522 (M+H), found 279.0524.
2-(4-methoxyphenylcarbamothioyl)hydrazinecarboxamide (1r)
Yield: 89%; 1H NMR (300 MHz, DMSO-d6) δ 3.77 (s, 3 H, OMe), 6.02 (s, 2 H, NH2), 6.87 (d, J = 9 Hz, 2 H), 7.33 (d, J = 9 Hz, 2 H), 7.98 (s, 1 H, NH), 9.28 (s, 1 H, NH), 9.53 (s, 1 H, NH); HRMS calcd for C9H12N4O2S (M+H) 241.0754, found 241.0756.
4-(4-tert-butylphenyl)-5-mercapto-4H-1,2,4-triazol-3-ol (2a)
2-thiobiurea compound 1a (502 mg, 1.88 mmol) in 2 M NaOH aqueous solution (20 mL) was stirred at reflux for 6 h. The reaction mixture was allowed to cool to room temperature and then acidified with 1 N HCl. The resulting precipitate was collected by filtration and washed with water (3 × 20 mL), hexanes (2 × 20 mL), 1:1 hexanes: diethyl ether (20 mL) respectively and dried in vacuo to afford 2a as a white solid (355 mg, 76%). 1H NMR (300 MHz, DMSO-d6) δ 1.31 (s, 9 H), 7.28 (d, J = 8.4 Hz, 2 H), 7.51 (d, J = 8.4 Hz, 2 H); MS m/z 521 (2M+H)+, 497 (2M-H)−, 272 (M+Na)+, 250 (M+H)+, 151, 147, 134, 106, 97, HRMS calcd for C12H16N3OS (M+H) 250.1009, found 250.1005.
Following above mentioned procedure and the appropriate starting materials and reagents used; compounds (2b–q) were prepared.
4-(benzo[D][1,3]dioxol-5-yl)-5-mercapto-4H-1,2,4-triazol-3-ol (2b)
Yield: 71%; 1H NMR (300 MHz, DMSO-d6) δ 6.10 (s, 2 H), 6.80–7.01 (m, 3 H); MS m/z 497 (2M+Na)+, 475 (2M+H)+, 260 (M+Na)+, 238 (M+H)+, 158, 120, 88, 56; HRMS calcd for C9H8N3O3S (M+H) 238.0281, found 238.0278.
4-(4-fluorophenyl)-5-mercapto-4H-1,2,4-triazol-3-ol (2c)
Yield: 68%; 1H NMR (300 MHz, DMSO-d6) δ 7.28–7.37 (m, 2 H), 7.40–7.46 (m, 2 H); MS m/z 445 (2M+ Na)+, 423 (2M+H)+, 234 (M+Na)+, 212 (M+H)+, 158, 88, 56; HRMS calcd for C8H7FN3OS (M+H) 212.0288, found 212.0289.
4-(2,4-dimethoxyphenyl)-5-mercapto-4H-1,2,4-triazol-3-ol (2d)
Yield: 65%; 1H NMR (300 MHz, DMSO-d6) δ3.72 (s, 3 H, OMe), 3.81 (s, 3 H, OMe), 6.58 (d, J = 8.7 Hz, 1 H), 6.69 (s, 1 H), 7.08 (d, J = 8.7 Hz, 1 H); MS m/z 529 (2M+Na)+, 507 (2M+H)+, 276 (M+Na)+, 254 (M+H)+, 212, 158, 146, 141, 97, 88, 74, 56; HRMS calcd for C10H12N3O3S (M+H) 254.0595, found 254.0586.
4-(3,5-dimethoxyphenyl)-5-mercapto-4H-1,2,4-triazol-3-ol (2e)
Yield: 68%; 1H NMR (300 MHz, DMSO-d6) δ 3.75 (s, 6 H, 2 × OMe), 6.54 (d, J = 2.1 Hz, 2 H), 6.57 (d, J = 2.4 Hz, 1 H); HRMS calcd for C10H12N3O3S (M+H) 254.0595, found 254.0588.
4-(3,4-dimethoxyphenyl)-5-mercapto-4H-1,2,4-triazol-3-ol (2f)
Yield: 71%; 1H NMR (300 MHz, DMSO-d6) δ 3.72 (s, 3 H, OMe), 3.79 (s, 3 H, OMe), 6.87 (d, J = 8.4 Hz, 1 H), 6.94 (s, 1 H), 7.02 (d, J = 8.4 Hz, 1 H); HRMS calcd for C10H12N3O3S (M+H) 254.0595, found 254.0588.
5-mercapto-4-(naphthalen-1-yl)-4H-1,2,4-triazol-3-ol (2g)
Yield: 74%; 1H NMR (300 MHz, DMSO-d6) δ 7.48–768 (m, 5 H), 8.01–8.112 (m, 2 H); MS m/z 509 (2M+Na)+, 487 (2M+H)+, 266 (M+Na)+, 244 (M+H)+, 186, 158, 144, 116, 85, 74, 67, 56; HRMS C12H10N3OS (M+H) 244.0539, found 244.0530.
4-(2,5-dimethoxyphenyl)-5-mercapto-4H-1,2,4-triazol-3-ol (2h)
Yield: 68%; 1H NMR (300 MHz, DMSO-d6) δ 3.68 (s, 3 H, OMe), 3.71 (s, 3 H, OMe), 6.83 (d, J = 2.7 Hz, 1 H), 7.03 (dd, J = 8.4 and 2.7 Hz, 1 H), 7.08 (d, J = 8.4 Hz, 1 H); HRMS calcd for C10H12N3O3S (M+H) 254.0595, found 254.0589.
5-mercapto-4-(4-(trifluoromethoxy)phenyl)-4H-1,2,4-triazol-3-ol (2i)
Yield: 72%; 1H NMR (300 MHz, DMSO-d6) δ 7.45–7.58 (m, 4 H); MS m/z 577 (2M+Na)+, 300 (M+Na)+, 278 (M+H)+, 201, 178, 141, 134, 129, 106, 97, 85; HRMS calcd for C9H7F3N3O2S (M+H) 278.0206, found 278.0211.
5-mercapto-4-(4-nitrophenyl)-4H-1,2,4-triazol-3-ol (2j)
Yield: 25%; 1H NMR (300 MHz, DMSO-d6) δ 7.79 (d, J = 9.3 Hz, 2 H), 8.35 (d, J = 9.0 Hz, 2 H); MS m/z 261 (M+Na)+, 242, 239 (M+H)+, 229, 130, 88, 85; HRMS calcd for C8H6N4O3S 239.0233 (M+H), found 239.0229.
4-(3,4-difluorophenyl)-5-mercapto-4H-1,2,4-triazol-3-ol (2k)
Yield: 57%; 1H NMR (300 MHz, CD3OD) δ 7.02–7.08 (m, 1 H), 7.14–7.27 (m, 2 H), MS m/z 252 (M+Na)+, 230 (M+H)+, 184, 169, 107, 95, 85, 56; HRMS calcd for C8H6F2N3OS (M+H) 230.0194, found 230.0191.
4-(3-fluorophenyl)-5-mercapto-4H-1,2,4-triazol-3-ol (2l)
Yield: 56%; 1H NMR (300 MHz, DMSO-d6) δ 7.24–7.35 (m, 3 H), 7.52–7.56 (m, 1 H); HRMS calcd for C8H7FN3OS (M+H) 212.0288, found 212.0286.
5-mercapto-4-(3-methoxyphenyl)-4H-1,2,4-triazol-3-ol (2m)
Yield: 74%; 1H NMR (300 MHz, DMSO-d6) δ 6.94 (d, J = 8.7 Hz, 1 H), 6.96 (s, 1 H), 7.02 (d, J = 8.4 Hz, 1 H), 7.40 (t, J = 7.5 Hz, 1 H); MS m/z 246 (M+Na)+, 224 (M+H)+, 192, 147, 134, 106, 97; HRMS calcd for C9H10N3O2S (M+H) 224.0488, found 224.0488.
5-mercapto-4-(2-methoxyphenyl)-4H-1,2,4-triazol-3-ol (2n)
Yield: 65%; 1H NMR (300 MHz, DMSO-d6) δ 3.74 (s, 3 H, OMe), 7.04 (t, J = 7.2 Hz, 1 H), 7.15–7.24 (m, 2 H), 7.4 (t, J = 7.2 Hz, 1 H); HRMS calcd for C9H10N3O2S (M+H) 224.0488, found 224.0486.
5-mercapto-4-(3-(trifluoromethyl)phenyl)-4H-1,2,4-triazol-3-ol (2o)
Yield: 68%; 1H NMR (300 MHz, DMSO-d6) δ 7.75–7.77 (m, 2 H), 7.82 (d, J = 3.9 Hz, 1 H), 7.86 (s, 1 H); MS m/z 543 (2M+Na)+, 284 (M+Na)+, 262 (M+H)+, 226, 159, 141, 102, 74; HRMS calcd for C9H9F3N4OS 279.0522 (M+H), found 279.0526.
5-mercapto-4-(4-(trifluoromethyl)phenyl)-4H-1,2,4-triazol-3-ol (2p)
Yield: 45%; 1H NMR (300 MHz, DMSO-d6) δ 7.58 (d, J = 9.0 Hz, 2H), 8.06 (d, J = 8.4 Hz, 2H); MS m/z 523 (2M+H)+, 284 (M+Na)+, 262 (M+H)+, 260 (M-H)−, 238, 102, 91, 74; HRMS calcd for C9H6F3N3OS 262.0256 (M+H), found 262.0253.
4-cyclohexyl-5-mercapto-4H-1,2,4-triazol-3-ol (2q) was commercially available.
5-mercapto-4-(4-methoxyphenyl)-4H-1,2,4-triazol-3-ol (2r)
Yield: 69%; 1H NMR (300 MHz, DMSO-d6) δ 3.79 (s, 3 H, OMe), 7.02 (d, J = 7.5 Hz, 2 H), 7.25 (d, J = 7.5 Hz, 2 H); HRMS calcd for C9H10N3O2S (M+H) 224.0488, found 224.0489.
4-(4-tert-butylphenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3a)
To a solution of 2a (94 mg, 0.377 mmol) in MeOH (2 mL) was added MeONa (0.75 mL, 0.5 M solution in MeOH) and stirred. After 5 minutes, 2-bromo-5-nitrothiazole (78 mg, 0.377 mmol) was added to the reaction mixture and stirred until deemed complete by TLC (6 h). The reaction mixture was acidified with 1 N HCl and the resulting precipitate was collected by filtration and washed with water (2 × 30 mL), hexanes (2 × 30 mL), and 10% ethyl acetate in hexanes (2 × 30 mL) to give a white solid. The residue was chromatographed over silica gel (40% ethyl acetate in hexane) to afford the 3a (102 mg, 71%). 1H NMR (300 MHz, DMSO-d6) δ 1.27 (s, 9 H), 7.27 (d, J = 8.4 Hz, 2 H), 7.47 (d, J = 8.4 Hz, 2 H), 8.68 (s, 1 H); MS m/z 777 (2M+Na)+, 755 (2M+H)+, 400 (M+Na)+, 378 (M+H)+, 354, 298, 250, 258, 207, 85, 74; HRMS calcd for C15H16N5O3S2 (M+H) 378.0689, found 378.0690.
Following above mentioned procedure (3a) and the appropriate starting materials and reagents used; compounds (3b–q) were synthesized.
4-(benzo[d][1,3]dioxol-5-yl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3b)
Yield: 60%; 1H NMR (300 MHz, DMSO-d6) δ 6.08 (s, 2 H), 6.84 (d, J = 8.1 Hz, 1 H), 6.96 (s, 1 H), 7.01 (d, J = 6 Hz, 1 H), 8.68 (s, 1 H); MS m/z 387 (M+Na)+, 365 (M+H)+, 354, 349, 190, 158, 125, 102, 97; HRMS calcd for C12H8N5O5S2 (M+H) 365.9961, found 365.9964.
4-(4-fluorophenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3c)
Yield: 62%; 1H NMR (300 DMSO-d6) δ 7.28–7.35 (m, 2 H), 7.43–7.50 (m, 2 H), 8.65 (s, 1 H); 361 (M+Na)+, 339 (M+H)+, 294, 158, 85,74, 67, 60; HRMS calcd for C11H7FN5O3S2 (M+H) 339.9969, found 339.9964.
4-(2,4-dimethoxyphenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3d)
Yield: 62%; 1H NMR (300 MHz, DMSO-d6) δ 3.66 (s, 3 H), 3.79 (s, 3 H), 6.65 (d, J = 8.4 Hz, 2 H), 7.18 (s, 1 H), 8.69 (s, 1 H); MS m/z 785 (2M+Na)+, 763 (2M+H)+, 404 (M+Na)+, 382 (M+H)+, 359, 345, 158, 102, 85, 74, 55; HRMS calcd for C13H12N5O5S2 (M+H) 382.0274, found 382.0281.
4-(3,5-dimethoxyphenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3e)
Yield: 67%; 1H NMR (300 MHz, DMSO-d6) δ 3.70 (s, 6 H, 2 × OMe), 6.57 (s, 3 H), 8.69 (s, 1 H); HRMS calcd for C13H12N5O5S2 (M+H) 382.0274, found 382.0280.
4-(3,4-dimethoxyphenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3f)
Yield: 64%; 1H NMR (300 MHz, DMSO-d6) δ 3.67 (s, 3 H, OMe), 3.76 (s, 3 H, OMe), 6.88 (dd, J = 8.4 and 1.8 Hz, 1 H), 6.98 (s, 1 H), 7.01 (d, J = 8.4 Hz, 1 H), 8.68 (s, 1 H); HRMS calcd for C13H12N5O5S2 (M+H) 382.0274, found 382.0282.
4-(naphthalen-1-yl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3g)
Yield: 61%, 1H NMR (300 MHz, DMSO-d6) δ 7.38–7.76 (m, 5 H), 8.01–8.10 (m, 2 H), 8.53 (s, 1 H); MS m/z 394 (M+Na)+, 372 (M+H)+, 217, 169, 130, 107,85, 74, 67, 45; HRMS calcd for C15H10N5O3S2 (M+H) 372.0220, found 372.0224.
4-(2,5-dimethoxyphenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3h)
Yield: 65%; 1H NMR (300 MHz, DMSO-d6) δ 3.62 (s, 3 H, OMe), 3.67 (s, 3 H, OMe), 6.93 (s, 1 H), 7.06 (dd, J = 8.4 and 1.8 Hz, 1 H), 7.11 (d, J = 8.4 Hz, 1 H), 8.69 (s, 1 H); HRMS calcd for C13H12N5O5S2 (M+H) 382.0274, found 382.0282.
5-(5-nitrothiazol-2-ylthio)-4-(4-(trifluoromethoxy)phenyl)-4H-1,2,4-triazol-3-ol (3i)
Yield: 67%; 1H NMR (300 MHz, DMSO-d6) δ 7.39–7.57 (m, 4 H), 8.63 (s,1 H), MS m/z 427 (M+Na)+, 405 (M+H)+, 364, 338, 306, 284, 192, 147, 106, 102, 97; HRMS calcd for C12H7F3N5O4S2 (M+H) 405.9886, found 405.9891.
4-(4-nitrophenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3j)
Yield: 60%; 1H NMR (300 MHz, DMSO-d6) δ 7.74 (d, J = 8.7 Hz, 2 H), 8.33 (d, J = 8.7 Hz, 2 H), 8.62 (s, 1 H); MS m/z 367 (M+H)+, 360, 355, 85; HRMS calcd for C11H6N6O5S2 366.9914 (M+H), found 366.9900.
4-(3,4-difluorophenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3k)
Yield: 58%; 1H NMR (300 MHz, DMSO-d6) δ 7.28–7.59 (m, 3 H), 8.64 (s, 1 H); MS m/z 357 (M+H)+, 356, 355, 288, 226, 174, 130, 125, 102, 84, 56; HRMS calcd for C11H6F2N5O3S2 (M+H) 357.9875, found 357.9866.
4-(3-fluorophenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3l)
Yield: 68%; 1H NMR (300 MHz, DMSO-d6) δ 7.25–7.45 (m, 3 H), 7.49–7.56 (m, 1 H), 8.66 (s, 1 H); MS m/z 361 (M+Na)+, 339 (M+H)+, 324, 229, 173, 158, 125, 116, 84, 74; HRMS calcd for C11H7FN5O3S2 (M+H) 339.9969, found 339.9970.
4-(3-methoxyphenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3m)
Yield: 61%; 1H NMR (300 MHz, DMSO-d6) δ 3.72 (s, 3 H, OMe), 6.95 (d, J = 8.1 Hz, 1 H), 7.01 (s, 1 H), 7.02 (d, J = 8.1 Hz, 1 H), 7.37 (t, J = 7.8 Hz, 1 H), 8.68 (s, 1 H); MS m/z 725 (2M+Na)+, 373 (M+Na)+, 352 (M+H)+, 338, 306, 284, 209, 201, 147, 106, 102, 97; HRMS calcd for C12H10N5O4S2 (M+H) 352.0169, found 352.0177.
4-(2-methoxyphenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3n)
Yield: 69%; 1H NMR (300 MHz, DMSO-d6) δ 3.69 (s, 3 H, OMe), 7.03 (t, J = 7.5 Hz, 1 H), 7.18 (d, J = 8.4 Hz, 1 H), 7.30 (d, J = 7.8 Hz, 1 H), 7.47 (t, J = 7.8 Hz, 1 H), 8.69 (s, 1 H); HRMS calcd for C12H10N5O4S2 (M+H) 352.0169, found 352.0172.
5-(5-nitrothiazol-2-ylthio)-4-(3-(trifluoromethyl)phenyl)-4H-1,2,4-triazol-3-ol (3o)
Yield: 95%; 1H NMR (300 MHz, DMSO-d6) δ 7.73–7.75 (m, 2 H), 7.83 (d, J = 3.0 Hz, 1 H), 7.86 (s, 1 H), 8.66 (s, 1 H); MS m/z 412 (M+Na)+, 390 (M+H)+, 159, 141, 129, 102, 98, 90, 74; HRMS calcd for C12H6F3N5O3S2 389.9937 (M+H), found 389.9943.
5-(5-nitrothiazol-2-ylthio)-4-(4-(trifluoromethyl)phenyl)-4H-1,2,4-triazol-3-ol (3p)
Yield: 94%; 1H NMR (300 MHz, DMSO-d6) δ 7.55 (d, J = 9.0 Hz, 2 H), 8.02 (d, J = 9 Hz, 2 H) 8.65 (s, 1 H); MS m/z 412 (M+Na)+, 390 (M+H)+, 366, 221, 138, 74; HRMS calcd for C12H6F3N5O3S3 389.9936 (M+H), found 389.9936.
4-cyclohexyl-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3q)
Yield: 72%; 1H NMR (300 MHz, DMSO-d6) δ 1.01–1.26 (m, 3 H), 1.56–1.79 (m, 5 H), 2.01–2.10 (m, 2 H), 3.82–3.87 (m, 1 H), 8.75 (s, 1 H); MS m/z 677 (2M+Na)+, 350 (M+Na)+, 328 (M+H)+, 310, 306, 284, 281, 245, 204, 179, 138, 134, 129, 106, 100, 88, 60; HRMS calcd for C11H14N5O3S2 (M+H) 328.0533, found 328.0548.
4-(4-methoxyphenyl)-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-ol (3r)
Yield: 71%; 1H NMR (300 MHz, DMSO-d6) δ 3.76 (s, 3 H, OMe), 7.01 (d, J = 8.7 Hz, 2 H), 7.30 (d, J = 8.7 Hz, 2 H), 8.68 (s, 1 H); HRMS calcd for C12H10N5O4S2 (M+H) 352.0169, found 352.0174.
Similarly, compounds (4a–d) were prepared using different aryl and alkyl bromides.
5-(butylthio)-4-phenyl-4H-1,2,4-triazol-3-ol (4a)
Yield: 68%; 1H NMR (300 MHz, CDCl3) δ 0.90 (t, J = 7.2 Hz, 3 H), 1.39 (sextet, J = 7.8 Hz, 2 H), 1.66 (quintet, J = 7.5 Hz, 2 H), 3.01 (t, J = 7.5 Hz, 2 H), 7.35–7.57 (m, 5 H), 10.42 (br.s, 1 H, OH); MS m/z 272 (M+Na)+, 250 (M+H)+, 233, 190, 149, 102, 85; HRMS calcd for C12H16N3OS (M+H) 250.1009, found 250.1007.
5-(ethylthio)-4-phenyl-4H-1,2,4-triazol-3-ol (4b)
Yield: 72%; 1H NMR (300 MHz, DMSO-d6) δ 1.35 (t, J = 7.2 Hz, 3 H), 3.02 (t, q, J = 7.2 Hz, 2 H), 7.35–7.56 (m, 5 H), 10.51 (br. s, 1 H, OH); MS m/z 244 (M+Na)+, 222 (M+H)+, 200, 149, 121, 85; HRMS calcd for C10H12N3OS (M+H) 222.0696, found 222.0697.
5-(benzylthio)-4-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-4H-1,2,4-triazol-3-ol (4c)
Yield: 75%; 1H NMR (300 MHz, DMSO-d6) δ 4.16 (s, 2 H), 4.26 (s, 4 H), 6.75 (d, J = 8.4 Hz, 1 H(, 6.82 (s, 1 H), 6.94 (d, J = 8.7 Hz, 1 H), 7.22–7.31 (m, 5 H); MS m/z 705 ((2M+Na)+, 683 (2M+H)+, 364 (M+Na)+, 342 (M+H)+, 326, 233, 158, 107, 87; HRMS calcd for C17H16N3O3S (M+H) 342.0907, found 342.0912.
5-(5-nitrothiophen-2-ylthio)-4-phenyl-4H-1,2,4-triazol-3-ol (4d)
Yield: 71%; 1H NMR (300 MHz, DMSO-d6) δ 7.06 (d, J = 4.2 Hz, 1 H), 7.31–7.38 (m, 2 H), 7.41–7.49 (m, 3 H), 7.94 (d, J = 4.5 Hz, 1 H); MS m/z 321 (M+H)+, 277, 233, 200, 149, 97, 85; HRMS calcd for C12H9N4O3S2 (M+H) 321.0111, found 321.0116.
2-benzoyl-N-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)hydrazinecarbothioamide (5a)
To a suspension of sodium acetate (226 mg, 2.76 mmol) and benzoylhydrazine (532 mg, 2.76 mmol) in acetonitrile (5 mL) was added 2,3-dihydro-1,4-benzodioxin-6-yl-isothiocyanate (532 mg, 2.76 mmol) and the reaction mixture was stirred for 24 h at room temperature. The solvent was removed by rotary evaporation and the resulting crude was dissolved in 2 M NaOH solution. The filtrate was acidified with 1 N HCl. The precipitate was collected and washed with water (3 × 20 mL), and hexanes (2 × 20 mL) to afford 5a as a white solid (873 mg, 96%). 1H NMR (300 MHz, DMSO-d6) δ 4.23 (s, 4 H), 6.81 (m, 2 H), 6.99 (s, 1 H, NH), 7.49 (d, J = 6.9 Hz, 1 H), 7.52 (d, J = 7.8 Hz, 1 H), 7.58 (d, J = 7.2 Hz, 1 H), 7.96 (d, J = 6.9 Hz, 2 H), 9.62 (s, 1 H), 9.68 (s, 1 H, NH), 10.50 (s, 1H, NH); MS m/z 352 (M+Na)+, 330 (M+H)+, 306, 296, 284, 179, 138, 100, 83; HRMS calcd for C16H15N3O3S 330.0907 (M+H), found 330.0902.
2-benzoyl-N-(3-methoxyphenyl)hydrazinecarbothioamide (5c)
Yield: 78%; 1H NMR (300 MHz, DMSO-d6) δ 3.73 (s, 3 H, OMe), 6.72 (d, J = 7.8 Hz, 1 H), 7.04 (d, J = 7.8 Hz, 1 H), 7.22 (t, J = 8.4 Hz, 1 H), 7.45–7.59 (m, 3 H), 7.94 (d, J = 7.5 Hz, 2 H), 9.72 (s, 1 H, NH), 9.73 (s, 1 H, NH), 10.53 (s, 1 H, NH).
4-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-5-phenyl-4H-1,2,4-triazole-3-thiol (6a)
Compound 5a (1.15 g, 3.50 mmol) in 2 M NaOH aqueous solution (35 mL) was stirred at reflux for 6 h. The reaction mixture was allowed to cool to room temperature and then acidified with 1 N HCl. The resulting precipitate was collected by filtration and washed with water (3 × 20 mL), hexanes (2 × 20 mL), and 1:1 hexanes: diethyl ether (20 mL) respectively and dried in vacuo to afford 6a as a white solid (655 mg, 60%). 1H NMR (300 MHz, DMSO-d6) δ 4.29 (s, 4 H), 6.75 (dd, J = 2.4, 8.1 Hz, 1 H), 7.93 (d, J = 8.7 Hz, 1 H), 7.94 (s, 1 H), 7.34–7.45 (m, 5 H); MS m/z 334 (M+Na)+, 312 (M+H)+, 280, 209, 179, 136, 100, 83; HRMS calcd for C16H13N3O2S 312.0801 (M+H), found 312.0803.
4-(3-methoxyphenyl)-5-phenyl-4H-1,2,4-triazole-3-thiol (6c)
Yield: 71%; 1H NMR (300 MHz, DMSO-d6) δ 3.72 (s, 3 H, OMe), 6.87 (d, J = 7.8 Hz, 1 H), 7.02–7.08 (m, 2 H), 7.32–7.42 (m, 6 H); MS m/z 306 (M+Na)+, 284 (M+H)+, 252, 149, 129, 106, 97, 85; HRMS calcd for C15H14N3OS (M+H) 284.0852, found 284.0854. Compounds (6b, 6d–g) were commercially available.
2-(4-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-5-phenyl-4H-1,2,4-triazol-3-ylthio)-5-nitrothiazole (7a)
To a solution of 6a (173 mg, 0.556 mmol) in MeOH (1.4 mL) was added MeONa (1.3 mL, 0.5 M solution in MeOH) and stirred. After 5 minutes, 2-bromo-5-nitrothiazole (127 mg, 0.608 mmol) was added to the reaction mixture and stirred until deemed complete by TLC (18 h). The reaction mixture was acidified with 1 N HCl and the resulting precipitate was collected by filtration and washed with water (2 × 30 mL), haxanes (2 × 30 mL), and 10% ethyl acetate in hexanes (2 × 30 mL) to give a solid. The residue was chromatographed over silica gel (50% ethyl acetate in hexane) to afford the 7a (167 mg, 68%). 1H NMR (300 MHz, DMSO-d6) δ 4.29 (s, 4 H), 6.91 (dd, J = 2.4, 7.8 Hz, 1 H), 6.89 (d, J = 2.4 Hz, 1 H), 7.44–7.52 (m, 5 H), 8.73 (s, 1 H); MS m/z 462 (M+Na)+, 440 (M+H)+, 160, 150, 142, 138, 125, 102, 98, 60; HRMS calcd for C19H13N5O4S2 440.0482 (M+H), found 440.0478.
Compounds (7b–g) were synthesized the following procedure as mentioned for 7a.
2-(4-(2,4-dichlorophenyl)-5-(1-methyl-1H-pyrrol-2-yl)-4H-1,2,4-triazol-3-ylthio)-5-nitrothiazole (7b)
Yield: 91%; 1H NMR (300 MHz, DMSO-d6) δ 3.96 (s, 3 H), 5.59 (dd, J = 1.5, 3.9 Hz, 1 H), 6.00 (dd, J = 1.5, 3.6 Hz, 1 H) 7.06 (t, J = 1.8 Hz, 1 H); MS m/z 475 (M+Na)+, 452 (M+H)+, 397, 369, 338, 159, 110, 98, 79; HRMS calcd for C1cH10Cl2N6O2S2 452.9756 (M+H), found 452.9759.
2-(4-(3-methoxyphenyl)-5-phenyl-4H-1,2,4-triazol-3-ylthio)-5-nitrothiazole (7c)
Yield: 72%; 1H NMR (300 MHz, DMSO-d6) δ 3.71 (s, 3 H, OMe), 6.97 (d, J = 7.8 Hz, 1 H), 7.10–7.16 (m, 2 H), 7.33–7.50 (m, 6 H), 8.70 (s, 1 H); MS m/z 434 (M+Na)+, 412 (M+H)+, 364, 354, 298, 252, 141, 138, 129, 102, 60; HRMS calcd for C18H14N5O3S2 (M+H) 412.0533, found 412.0541.
2-(4-(furan-2-ylmethyl)-5-(naphthalen-1-yl)-4H-1,2,4-triazol-3-ylthio)-5-nitrothiazole (7d)
Yield: 13%; 1H NMR (300 MHz, DMSO-d6) δ 5.16 (s, 2 H), 6.00 (d, J = 3.3 Hz, 1 H), 6.14 (d, J = 2.4 Hz, 1 H), 7.38 (s, 1 H), 7.62–7.78 (m, 5 H), 8.10 (d, J = 6.9 Hz, 1 H), 8.23 (d, J = 6.9 Hz, 1 H), 8.73 (s, 1 H); MS m/z 871 (2M+H)+, 458 (M+Na)+, 436 (M+H)+, 326, 179, 102, 88, 84; HRMS calcd for C20H13N5O3S2 436.0533 (M+H), found 436.0543.
5-nitro-2-(4-phenyl-5-(thiophen-2-yl)-4H-1,2,4-triazol-3-ylthio)thiazole (7e)
Yield: 72%; 1H NMR (300 MHz, DMSO-d6) 6.86–7.06 (m, 2 H), 7.53–7.74 (m, 6 H), 8.68 (s, 1 H); MS m/z 409 (M+Na)+, 387 (M+H)+, 354, 338, 326, 310, 284, 179, 147, 138, 121, 106, 97, 60; HRMS calcd C15H10N5O2S3 (M+H) 387.991, found 387.9996.
5-nitro-2-(4-phenyl-5-(pyridin-4-yl)-4H-1,2,4-triazol-3-ylthio)thiazole (7f)
Yield: 62%; 1H NMR (300 MHz, DMSO-d6) δ 7.36 (d, J = 6 Hz, 2 H), 7.50–7.61 (m, 5 H), 8.62 (d, J = 5.4 Hz, 2 H), 8.69 (s, 1 H); MS m/z 405 (M+Na)+, 383 (M+H)+, 359, 310, 226, 158, 141, 138, 129, 121, 109, 97, 90, 60; HRMS calcd for C16H11N6O2S2 (M+H) 383.0379, found 383.0381.
5-((4-methyl-5-(5-nitrothiazol-2-ylthio)-4H-1,2,4-triazol-3-yl)methyl)-3-(pyrazin-2-yl)-1,2,4-oxadiazole (7g)
Yield: 79%; 1H NMR (300 MHz, DMSO-d6) δ 1.24 (s, 2 H), 3.50 (s, 3 H), 8.78 (s, 1 H), 8.80 (s, 1 H), 8.87 (s, 2 H), 9.31 (s, 1 H); MS m/z 871 (2M+H)+, 458 (M+Na)+, 436 (M+H)+, 326, 179, 102, 88, 84; HRMS calcd for C20H13N5O3S2 436.0533 (M+H), found 436.0543.
2,5-bis(benzylthio)-1,3,4-thiadiazole (8a)
To a solution of 1,3,4-thiadiazole-2,5-dithiol (93 mg, 0.62 mmol) in MeOH (3 mL) was added MeONa (2.8 mL, 0.5 M solution in MeOH) and stirred. After 5 minutes, benzyl bromide (233 mg, 1.364 mmol) was added to the reaction mixture and stirred until deemed complete by TLC (6 h). The reaction mixture was acidified with 1 N HCl and the resulting precipitate was collected by filtration and washed with water (2 × 30 mL), haxanes (2 × 30 mL), and 10% ethyl acetate in hexanes (2 × 30 mL) to give a white solid. The residue was chromatographed over silica gel (30% ethyl acetate in hexane) to afford the 8a (204 mg, 80%). 1H NMR (300 MHz, DMSO-d6) δ 4.49 (s, 4 H), 7.25–7.45 (m, 10 H), MS m/z 353 (M+Na)+, 331 (M+H)+, 192, 124, 102, 85; HRMS calcd for C16H15N2S2 (M+H) 331.0392, found 331.0398.
2,5-bis(4-fluorobenzylthio)-1,3,4-thiadiazole (8b)
Yield: 75%; 1H NMR (300 MHz, DMSO-d6) δ 4.49 (s, 4 H), 7.11–7.19 (m, 4 H), 7.42–7.46 (m, 4 H), MS m/z 389 (M+Na)+, 367 (M+H)+, 338, 284, 192, 147, 106, 102, 97, 85; HRMS calcd for C16H13F2N2S3 (M+H) 367.0203, found 367.0208.
2,5-bis(4-methoxybenzylthio)-1,3,4-thiadiazole (8c)
Yield: 76%; 1H NMR (300 MHz, DMSO-d6) δ 3.72 (s, 6 H), 4.43 (s, 4 H), 6.87 (d, J = 8.4 Hz, 4 H), 7.32 (d, J = 8.4 Hz, 4 H); MS m/z 413 (M+Na)+, 391 (M+H)+, 305, 261, 226, 158, 149, 138, 106, 102, 97, 84; HRMS calcd for C18H19N2O2S3 (M+H) 391.0603, found 391.0606.
2,5-bis(3-nitrobenzylthio)-1,3,4-thiadiazole (8d)
Yield: 73%; 1H NMR (300 MHz, DMSO-d6) δ 4.64 (s, 4 H), 7.61(t, J = 8.1 Hz, 2 H), 7.86 (d, J = 7.8 Hz, 2 H), 8.12 (d, J = 8.4 Hz, 2 H), 8.29 (s, 2H); HRMS calcd for C16H13N4O4S3 (M+H) 421.0093, found 421.0095.
2,5-bis(4-nitrobenzylthio)-1,3,4-thiadiazole (8e)
Yield: 72%; 1H NMR (300 MHz, DMSO-d6) δ 4.63 (s, 4 H), 7.68 (d, J = 8.7 Hz, 4 H), 8.17 (d, J = 8.7 Hz, 4 H); MS m/z 442 (M+Na)+, 421 (M+H)+, 397, 364, 338, 306, 277, 201, 192, 147, 129, 118, 106, 102, 97; HRMS calcd for C16H13N4O4S3 (M+H) 421.0093, found 421.0097.
2,5-bis(5-nitrothiazol-2-ylthio)-1,3,4-thiadiazole (8f)
Yield: 90%; 1H NMR (300 MHz, DMSO-d6) δ 8.86 (s, 2H); MS m/z 429 (M+Na)+, 406 (M+H)+, 227, 159, 102, 56; HRMS calcd for C8H2N6O4S5 406.8814 (M+H), found 406.8814.
2,2′-(1,3,4-thiadiazole-2,5-diyl)bis(sulfanediyl)dithiazole-5-carboxylic acid (8g)
Yield: 78%; 1H NMR (300 MHz, DMSO-d6) δ 8.36 (s, 2 H); MS m/z 404 (M+H)+, 306, 253, 233, 169, 146, 102, 95, 87, 85; HRMS calcd for C10H5N4O4S5 (M+H) 404.8909, found 404.8908.
5-(5-nitrothiazol-2-ylthio)-1,3,4-thiadiazol-2-amine (9)
To a solution of 5-amino-1,3,4-thiadiazole-2-thiol (211 mg, 1.586 mmol) in MeOH (3 mL) was added MeONa (3.8 mL, 0.5 M solution in MeOH) and stirred. After 5 minutes, 2-bromo-5-nitrothiazole (331 mg, 1.586 mmol) was added to the reaction mixture and stirred until deemed complete by TLC (12 h). The reaction mixture was acidified with 1 N HCl and the resulting precipitate was collected by filtration and washed with water (2 × 30 mL), haxanes (2 × 30 mL), and 10% ethyl acetate in hexanes (2 × 30 mL) to give a white solid. The residue was chromatographed over silica gel (70% ethyl acetate in hexane) to afford the 9 (281 mg, 68%). 1H NMR (300 MHz, DMSO-d6) δ 7.94 (br s, 2 H, NH2), 8.74 (s, 1 H); MS m/z 283 (M+Na)+, 261 (M+H)+, 229, 193, 158, 126, 97, 84, 47; HRMS calcd C5H4N5O2S3 (M+H) 261.9522, found 261.9515.
4-fluoro-N-(5-(5-nitrothiazol-2-ylthio)-1,3,4-thiadiazol-2-yl)benzamide (10)
A solution of 9 (75 mg, 0.288 mmol), 4-fluorobenzoic acid (40 mg, 0.288 mmol), EDC (66 mg, 0.34 mmol), HOBt (47 mg, 0.34 mmol), DIEA (0.15 mL, 0.86 mmol) in DMF (2 mL) was stirred at 50 °C for 16 h. The reaction mixture was extracted with ethyl acetate (50 mL), washed with 10% NaHCO3 solution (30 mL), brine (2 × 30 mL), and water (2 × 30 mL), dried (MgSO4), and concentrated in vacuo. The residue was chromatographed over silica gel (30% ethyl acetate in hexane) to afford the 10 (82 mg, 75%). 1H NMR (300 MHz, DMSO-d6) δ 7.42–7.46 (m, 2 H), 7.67 (s, 1 H, NH), 8.20–8.22 (m, 2 H), 8.87 (s, 1 H); MS m/z 381 (M-H)+, 306, 248, 204, 180, 174, 154, 138, 96; HRMS calcd for C12H5FN5O3S3 (M-H) 381.9544, found 381.9535.
5-(benzylthio)-1,3,4-thiadiazol-2-amine (11)
Yield: 69%; 1H NMR (300 MHz, DMSO-d6) δ 4.28 (s, 2 H), 7.11–7.19 (m, 2 H), 7.34–7.42 (m, 5 H, Ar-H, NH2); MS m/z 264 (M+Na)+, 242 (M+H)+, 192, 147, 102, 97, 85; HRMS calcd C9H9H9FN3S2 (M+H) 242.0216, found 242.0215.
DELFIA Assay (dissociation enhanced lanthanide fluoro-immuno assay
To each well of 96-well streptavidin-coated plates (Perkin-Elmer) 100 μL of a 100 ng/ml solution of biotin-labeled pep-JIP11 (Biotin-lc-KRPKRPTTLNLF, where lc indicates a hydrocarbon chain of 6 methylene groups) was added. After 1 hr incubation and elimination of unbound biotin-pep-JIP11 by 3 washing steps, 87 μL of Eu-labeled anti-GST antibody solution (300 ng/ml; 1.9 nM), 2.5 μL DMSO solution containing test compound, and 10 μL solution of GST-JNK for a final protein concentration of 10 nM was added. After 1 hr incubation at 0 °C, each well was washed 5 times to eliminate unbound protein and the Eu-antibody if displaced by a test compound. Subsequently, 200 μL of enhancement solution (Perkin-Elmer) was added to each well and fluorescence measured after 10 min incubation (excitation wavelength, 340 nm; emission wavelength, 615 nm). Controls include unlabeled peptide and blanks receiving no compounds. Protein and peptide solutions were prepared in DELFIA buffer (Perkin-Elmer).
In Vitro Kinase Assay
The LanthaScreen™ assay platform from Invitrogen was utilized. The time-resolved fluorescence resonance energy transfer assay (TR-FRET) was performed in 384 well plates. Each well received JNK (35 ng/mL), ATF2 (400 nM), and ATP (200 μM) in 50mM HEPES, 10mM MgCl 2, 1mM EGTA and 0.01% Brij-35, pH 7.5 and test compounds. The kinase reaction was performed at room temperature for 1 hr. After which, the terbium labeled antibody and EDTA were added into each well. After an additional hour incubation, the signal was measured at 520/495 nm emission ratio on a fluorescence plate reader (Victor 2, Perkin-Elmer).
Cell based assays for c-Jun
The cell based kinase assays for c-Jun and ATF2 phosphorylation carried out using the LanthaScreen c-Jun (1–79) Hela (Invitrogen, Carlsbad, CA) which stably express GFP-c-Jun 1–79. Phosphorylation was determined by measuring the time resolved FRET (TR-FRET) between a terbium labeled phospho-specific antibody and the GFP-fusion protein12. The cells were plated in white tissue culture treated 384 well plates at a density of 10000 cell per well in 32 μl assay medium (Opti-MEM®, supplemented with 1% charcoal/dextran-treated FBS, 100 U/mL penicillin and 100 μg/mL streptomycin, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 25 mM HEPES pH 7.3, and lacking phenol red). After overnight incubation, cells were pretreated for 60 min with compound (indicated concentration) followed by 30 min of stimulation with 2 ng/ml of TNF-α–which stimulates both JNK and p38. The medium was then removed by aspiration and the cells were lysed by adding 20 μl of lysis buffer (20 mM TRIS-HCl pH 7.6, 5 mM EDTA, 1% NP-40 substitute, 5 mM NaF, 150 mM NaCl, 1:100 protease and phosphatase inhibitor mix, SIGMA P8340 and P2850 respectively). The lysis buffer included 2 nM of the terbium labeled anti-pc-Jun (pSer73) detection antibodies (Invitrogen). After allowing the assay to equilibrate for 1 hour at room temperature, TR-FRET emission ratios were determined on a BMG Pherastar fluorescence plate reader (excitation at 340 nm, emission 520 nm and 490 nm; 100 μs lag time, 200 μs integration time, emission ratio = Em520/Em 490).
Molecular Modeling
Molecular modeling studies were conducted on a Linux workstation and a 64 3.2-GHz CPUs Linux cluster. Docking studies were performed using the X-ray coordinates of JNK1 (PDB code 2H96).25 The complexed JIP peptide was extracted from the protein structure and was used to define the binding site for docking of small molecules. The genetic algorithm (GA) procedure in the GOLD docking software performed flexible docking of small molecules whereas the protein structure was static.26–30 For each compound, 20 solutions were generated and subsequently ranked according to Chemscore.26 The protein surface was prepared with the program MOLCAD as implemented in Sybyl and was used to analyze the binding poses for studied small molecules. 26–30
In Vivo experiments
Insulin Tolerance Test
Eleven week old male BKS.Cg-+Leprdb/+Leprdb/OlaHsd db/db mice from Harlan (Harlan Sprague Dawley, Inc.; Indianapolis, IN) were randomized based on blood glucose levels acclimated three day prior to drug dosing. Blood glucose was read using a hand-held glucose meter (OneTouch Ultra, LifeScan, a Johnson & Johnson company, UK) after tail snipping. Mice were fasted 6 hours before intraperitoneal (i.p.) administration of 25 mg/kg of compounds 9, 7b, and 8f. Thirty minutes after test article administration, Bovine Insulin (Sigma, I-0516 at 0.75 mg/kg) was administered via i.p. injection. Blood samples were taken at designated time points and blood glucose levels were measured as previously described. Food was returned three hours after test article administration.
Supplementary Material
Acknowledgments
In vivo experiments related to the insulin tolerance were performed by Explora Biolabs. Elemental analysis was performed by NuMega lab, San Diego. Financial support was obtained thanks to NIH grants DK073274, DK080263, and by a CSRA by Syndexa pharmaceuticals (to M.P).
Abbreviations
- JNK
c-Jun N-terminal protein kianse
- MAPK
mitogen-activated protein kinase
- JIP1
JNK-interacting protein 1
- EDC
N-ethyl-N′-(3-dimethylaminopropyl)-corbodiimide
- DMF
N,N-dimethyl formamide
Footnotes
Supporting Information Available: Proton NMR spectra, HPLC traces of key compounds (purity), elemental analysis of key compounds, and supporting figures. This material is available free of charge via the internet at http://pubs.acs.org
References
- 1.Manning G. The protein kinase complement of human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Manning AM, Davis RJ. Targeting JNK for therapeutic benefit: from junk to gold? . Nat Rev Drug discovery. 2003;2:554–565. doi: 10.1038/nrd1132. [DOI] [PubMed] [Google Scholar]
- 3.Bogoyevitch MA. Therapeutic promise of JNK ATP –noncompetive inhibitors. Trends in Molecular Medicine. 2005;11:232–239. doi: 10.1016/j.molmed.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Gupta S, Barret T, Whitmarsh AJ, Cavanagh J, Sluss HK, De’rijard B, Davis RJ. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 5.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 6.Pearson G, Robinson F, Gibson TB, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein kinase pathways: regulation and physiochemical functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 7.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 8.Yang S-H, Whitmarsh AJ, Davis RJ, Sharrocks AD. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barr RK, Kendrick TS, Bogoyevitch MA. Identification of the critical features of a small peptide inhibitor of JNK activity. J Biol Chem. 2002;277:10987–10997. doi: 10.1074/jbc.M107565200. [DOI] [PubMed] [Google Scholar]
- 10.Bonny C, Oberson A, Negri S, Sauser C, Schorderet DF. Cell-permeable peptide inhibitors of JNK: novel blockers of beta-cell death. Diabetes. 2001;50:77–82. doi: 10.2337/diabetes.50.1.77. [DOI] [PubMed] [Google Scholar]
- 11.Dickens M, Roger JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ. A Cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 12.Heo YS, Kim S-KY-K, Sung B-J, Lee HS, Lee JI, Park S-Y, Kim JH, Hwang KY, Hyun Y-L, Jeon YH, Ro S, Cho JM, Lee TG, Yang C-H. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 2004;23:2185–2195. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneto H, Nakatani Y, Miyatsuka T, Kawamori D, Matsuoka T, Matsuhisa M, Kajimoto Y, Ichijo H, Yamasaki Y, Hori M. Possible novel therapy for diabetes with cell-permeable JNK-inhibitory peptide. Nature Medicine. 2004;10:1128–1132. doi: 10.1038/nm1111. [DOI] [PubMed] [Google Scholar]
- 14.Swahn B-M, Xue Y, Arzel E, Kallin E, Magnus A, Plobeck N, Viklund J. Design synthesis of 2′-anilino-4 4-bipyridines as selective inhibitors of JNK-3. Bioorg Med Chem Lett. 2006;16:1397–1401. doi: 10.1016/j.bmcl.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 15.Graczyk PP, Khan A, Bhatia GS, Palmer V, Medland D, Numata H, Oinuma H, Catchick J, Dunne A, Ellis M, Smales C, Whitfield J, Neame SJ, Shah B, Wilton D, Morgan L, Patel T, Chung R, Desmond H, Staddon JM, Sato N, Inoue A. The neuroprotective action of JNK3 inhibitors based on 6,7-dihydro-5-H-pyrrolo[1,2-a]imidazole scaffold. Bioorg Med Chem Lett. 2005;15:4666–4670. doi: 10.1016/j.bmcl.2005.07.076. [DOI] [PubMed] [Google Scholar]
- 16.Gaillard P, Jeanclaude-Etter I, Ardissone V, Arkinstall S, Cambet Y, Camps M, Chabert C, Church D, Cirillo R, Gretener D, Halazy S, Nichols A, Szyndralewiez C, Vitte P-A, Gotteland J-P. Design and synthesis of the first generation of novel potent, selective, and in vivo active benzothiazol-2-yl-acetonitrile inhibitors of JNK. J Med Chem. 2005;48:4596–4607. doi: 10.1021/jm0310986. [DOI] [PubMed] [Google Scholar]
- 17.Szczepankiewicz BG, Kosogof C, Nelson TJ, Liu G, Liu B, Zhao H, Serby MD, Serby MD, Xin Z, Liu M, Gum RJ, Haasch DL, Wang S, Clampit JE, Johnson EF, Lubben TH, Stashko MA, Olejniczak ET, Sun C, Dorwin SA, Haskins K, Abad-Zapatero C, Fry EH, Hutchins CW, Sham HL, Rondinone CM, Tervillyan JM. Aminopyridine-based JNK inhibitors with cellular activity and minimal cross-kinase activity. J Med Chem. 2006;49:3563–3580. doi: 10.1021/jm060199b. [DOI] [PubMed] [Google Scholar]
- 18.Swahn B-M, Huerta F, Kallin E, Malmstrom J, Weigelt T, Viklund J, Womack P, Xue Y, Ohberg L. Design and synthesis of 6-anilinoindazoles as selective inhibitors of JNK. Bioorg Med Chem Lett. 2005;15:5095–5099. doi: 10.1016/j.bmcl.2005.06.083. [DOI] [PubMed] [Google Scholar]
- 19.Ruckle T, Biamonte M, Grippi-Vallotton T, Arkinstall S, Cambet Y, Camps M, Chabert C, Church DJ, Halazy S, Jiang X, Martinou I, Nichols A, Sauer W, Gotteland J-P. Design and synthesis and biological activity of novel, potent, and selective, (benzoylaminomethyl)thiophene sulfonamide inhibitors of JNK. J Med Chem. 2004;47:6921–6934. doi: 10.1021/jm031112e. [DOI] [PubMed] [Google Scholar]
- 20.Bennett BL, Sasaki DT, Murray BW, O’Leary EC, Sakata ST, Xu W, Leisten JC, Motiwala A, Pierce S, Satoh Y, Bhagwat SS, Manning AM, Anderson DW. SP600125, an anthrapyrazolone inhibitor of JNK. Proc Natl Acad Sci USA. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stebbins JL, De SK, Machleidt T, Becattini B, Vazquez J, Kuntzen C, Chen L-H, Cellitti JF, Riel-Mehan M, Emdadi A, Solinas G, Karin M, Pellecchia M. Identification of a novel JNK inhibitor targeting JNK-JIP interaction. Proc Natl Acad Sci, USA. 2008;105:16809–16813. doi: 10.1073/pnas.0805677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vazquez J, De SK, Chen L-H, Riel-Mehan M, Emdadi A, Cellitti J, Stebbins JL, Rega MF, Pellecchia M. Development of paramagnetic probes for molecular recognition in protein kinases. J Med Chem. 2008;51:3460–3465. doi: 10.1021/jm800068w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suni MM, Nair VA, Joshua CP. Heterocyclization of 1-aryl/alkyl-2-thiobiureas to 4-aryl/alkyl-3-substituted- 2-1,2,4-triazolin-5-ones. Tetrahedron. 2001;57:2003–2009. [Google Scholar]
- 24.Rostamizadeh S, Mollahoseini K, Moghadasi S. A one pot synthesis of 4,5-disubstituted 1,2,4-triazole -3-thions on solid support under microwave irradiation. Phosphorus, Sulfur, and Silicon and the Related Elements. 2006;181:1839–1845. [Google Scholar]
- 25.Zhao H, Serby MD, Xin Z, Szczepankiewicz BG, Liu M, Kosogof C, Liu B, Nelson LTJ, Johnson EF, Wang S, Pederson T, Gum RJ, Clampit JE, Haasch DL, Abad-Zapatero C, Fry EH, Rondinone C, Trevillyan JM, Sham HL, Liu G. Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c-Jun NH2-terminal kinase inhibitors. J Med Chem. 2006;49:4455–4458. doi: 10.1021/jm060465l. [DOI] [PubMed] [Google Scholar]
- 26.GOLD, Version, 2.1; The Cambridge Crystallographic Data Center: 12, Union Road, Cambridge, CB2 1EZ, UK
- 27.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 28.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput-Aided Mol Des. 1997;11:425–455. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 29.Teschner M, Henn C, Vollhardt H, Reiling S, Brickmann J. Texture mapping: A new tool for molecular graphics. J Mol Graphics. 1994;12:98–105. doi: 10.1016/0263-7855(94)80074-x. [DOI] [PubMed] [Google Scholar]
- 30.Pearlman, R. S. “concord,” distributed by Tripos Internation, St. Louis, MO; 1998;
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.