Abstract
Although prognostic gene expression signatures for survival in early stage lung cancer have been proposed, for clinical application it is critical to establish their performance across different subject populations and in different laboratories. Here we report a large, training-testing, multi-site blinded validation study to characterize the performance of several prognostic models based on gene expression for 442 lung adenocarcinomas. The hypotheses proposed examined whether microarray measurements of gene expression either alone or combined with basic clinical covariates (stage, age, sex) can be used to predict overall survival in lung cancer subjects. Several models examined produced risk scores that substantially correlated with actual subject outcome. Most methods performed better with clinical data, supporting the combined use of clinical and molecular information when building prognostic models for early stage lung cancer. This study also provides the largest available set of microarray data with extensive pathological and clinical annotation for lung adenocarcinomas.
Introduction
In the United States and in many western countries lung cancer represents the leading cause of cancer-related death1. The five-year, overall survival rate is 15% and has not improved over many decades. This is mainly because approximately two-thirds of lung cancers are discovered at advanced stages, for which cure by surgical resection is no longer an option. Furthermore, even among early stage subjects who are treated primarily by surgery with curative intent, 30–55% will develop and die of metastatic recurrence. Recent multi-national clinical trials conducted in several continents have demonstrated that adjuvant chemotherapy significantly improved the survival of early stage (IB-II) subjects (IALT, JBR10, ANITA, UFT, LACE)2. Nevertheless, it is clear that a proportion of stage I subjects have poorer prognosis and may benefit significantly from adjuvant chemotherapy, while some relatively good prognosis stage II subjects may not benefit significantly from adjuvant chemotherapies. It remains possible that the latter subjects could potentially derive additional benefit from adjuvant targeted therapies2–4. Therefore, there is an urgent need to establish new diagnostic paradigms for improving the selection of stage I–II subjects who are most likely to benefit from receiving adjuvant chemotherapy, and for identifying such subjects as candidates for clinical trials.
Global gene expression profiling using microarray technologies has helped to improve our understanding of the histological heterogeneity of non-small cell lung cancer and has identified novel potential biomarkers and gene signatures for classifying subjects with significantly different survival outcomes5–11. However, the performance and general applicability of published classifiers has not been easy to establish due to small numbers of subjects examined and inclusion of heterogeneous tumor types. Furthermore, there have not been uniform criteria for sample inclusion, annotation, sample processing, and data analyses. To address these concerns and to generate a large microarray database of NSCLC samples that have been collected and studied using a common protocol12, we conducted a large retrospective, multi-site, blinded study. The study included a blinded validation step to characterize the performance of several newly-developed prognostic models using a total of 442 lung adenocarcinomas, the specific type of lung cancer that is increasing in incidence13.
To ensure scientific validity of the results, subject samples along with all relevant clinical, pathological and outcome data were collected by investigators at four institutions using data from six lung cancer treatment sites with a priori defined inclusion criteria. Gene expression data on subsets of lung adenocarcinomas were generated by each of four different laboratories using a common platform and following a protocol previously demonstrated to be robust and reproducible12. We considered four separate hypotheses: 1. gene expression alone can predict outcomes for all samples; 2. gene expression and basic clinical covariates (stage, age, sex) can predict outcomes for all samples; 3. gene expression alone can predict outcomes for stage 1 samples; and 4. gene expression and basic clinical covariates can predict outcomes for stage 1 samples. Note that prediction on stage 1 samples is more difficult than on the full study set as these samples are relatively homogeneous. The consideration of clinical covariates is highly relevant as the basic variables considered here will always be available in practice, and gene expression-based prediction is relevant in practice only if it provides additional information to these measures. We followed a strict protocol for the data collection, data analysis, and performance evaluation phases of our study. Data generated at two sites was used as a training set and the results were validated using the independent datasets from the other two participating sites following a blinded protocol. The results from this study provide not only valid assessment of outcome prediction in the multi-institutional setting but also a rich dataset for future analysis and provide an example of how large datasets can be generated and tested by cooperation and pooling of resources among many investigators.
Results
Consortium and classifier development
A total of 442 lung adenocarcinomas have been collected with high quality gene expression data, pathological data, and clinical information describing the severity of the disease at surgery and the clinical course of the disease after sampling. These samples, collected from 6 contributing treatment institutions, were grouped into four sets of data based on the laboratory where samples were processed for microarray analysis. The distribution of several clinical variables for these 4 data sets is shown in Table 1. The first two data sets, UM and HLM, were released to members of the consortium for the development of classifiers appropriate for our four hypotheses. Details of our protocol for developing and evaluating classifiers are provided in Supplementary Materials section 1.
Table 1.
Summary statistics of data
| UM | HLM | CAN/DF | MSK | |
|---|---|---|---|---|
| Sample size | 177 | 79 | 82 | 104 |
| Age (mean, sd) | 64 (10) | 67 (10) | 61 (10) | 65 (10) |
| Gender (% male) | 56% | 51% | 56% | 36% |
| Stage I | 66% | 54% | 68% | 61% |
| Stage II | 16% | 26% | 32% | 19% |
| Stage III | 18% | 19% | 0% | 20% |
| Median follow-up (months) | 54 | 39 | 40 | 43 |
| Number of deaths | 75 | 50 | 28 | 34 |
Eight classifiers producing either categorical or continuous risk scores were developed by investigators using the training data, and were tested for effectiveness on the two remaining data sets (MSK and CAN/DF). Most of these classifiers incorporate techniques that have repeatedly been applied in gene expression-based prognosis and found to work well in at least some instances. As an overview, data reduction was carried out using gene clustering (method A), univariate testing (methods B, C, D, E, F, G), or on a mechanistic basis (method H). Final scoring/classification was done based on Cox regression modeling using ridging penalties on gene cluster summaries (method A), on individual genes (method B), or on principal components (methods F,G); on cluster membership (methods C,D), or on voting (method H). A number of other factors such as subselection of the training samples, gene filtering, and data transformation were handled in various ways as described in detail in Supplementary Materials section 2. We note that all classifiers started with the same set of DChip-processed expression summaries, so handling of the data at the CEL file level was uniform across the methods.
Classifier performance without and with clinical covariates
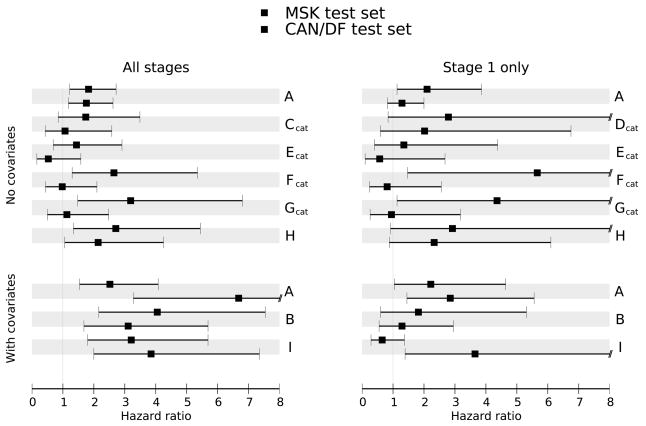
The estimated hazard ratios for the risk scores produced by the eight prognosis methods, with 95% confidence intervals, are shown for the two validation sets in Figure 1. Hazard ratios substantially greater than 1.0 indicate that subjects in the validation set with high predicted risk have poor outcomes. Confidence intervals in Figure 1 and the corresponding p-values given in Supplementary Materials section 3a indicate which of the methods perform significantly better than expected by chance. As another performance measure, we calculated the concordance probability estimate (CPE), which measures how well the subject outcomes agree with the predicted risk scores. CPE values close to 0.5 indicate no concordance (poor predictivity) while CPE values approaching 1.0 indicate strong concordance (good predictivity). Based on these measures, most of the classifiers performed well in at least some situations. Finally, for 3-year survival we constructed ROC curves for continuous predictors and tables of sensitivity/specificity estimates for categorical predictors. These are shown in Supplementary Materials section 6.
Figure 1.
Classifier Performance: Hazard Ratios of methods A–I on validation datasets for four hypotheses, along with 95% confidence intervals.
There are some notable observations about the classifiers as a group. Most methods performed much better on sample sets containing all stages compared to just stage 1 subjects. This reflects an ability to stratify by stage even when stage is not explicitly included in the model. Including clinical covariates improves the performance of most of the models. In fact, without clinical covariates, no model achieved a hazard ratio significantly greater than 1 in both validation sets for the stage 1 samples. An important criterion was that a model should perform well in both validation sets as an indication of robust performance in routine clinical testing. For prediction on all stages using gene expression data, only methods A and H performed with consistent statistical significance. For prediction on all stages using both gene expression and with clinical covariates, methods A and B produced hazard ratios exceeding two for both validation sets. For prediction on subjects with stage 1 disease using gene expression data only, three of the methods (A, D and H) gave hazard ratios exceeding one for both validation sets. Of these, only method A had a hazard ratio significantly greater than one for one of the datasets. For prediction on subjects with stage 1 disease using gene expression data and clinical covariates method A gives hazard ratios that exceed two and are statistically significant for both datasets. For many of the classifiers, good performance in one setting was offset by poor performance in a different setting. Thus method A seemed to have the best overall performance across the four hypotheses.
Kaplan-Meier analyses indicate several subgroups based on subject survival
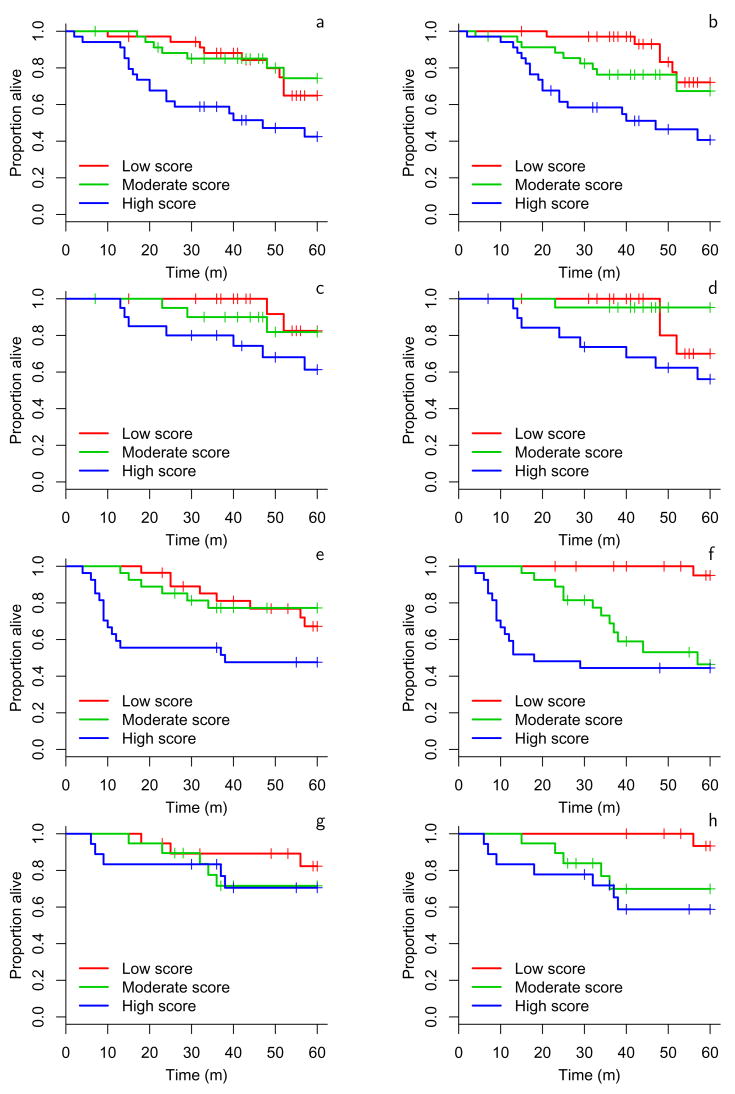
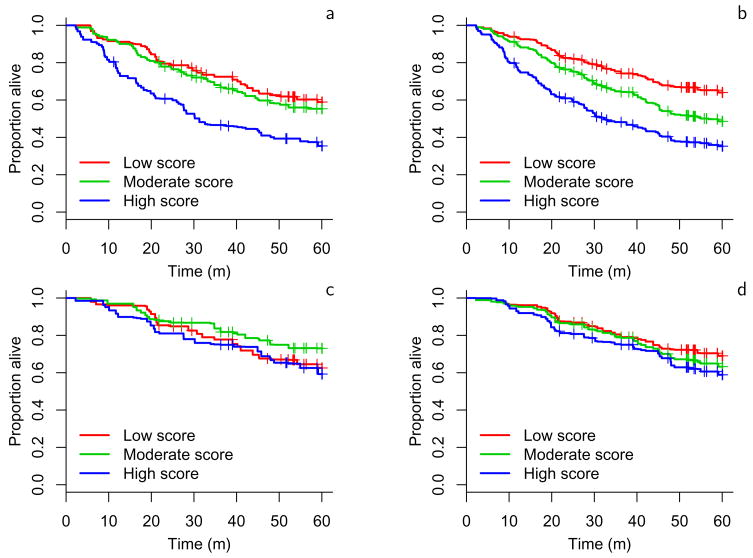
Using method A to stratify subjects into 3 groups, we generated Kaplan-Meier plots to illustrate the survival differences among the groups determined by this classification scheme for both the validation (Figure 2) and the training datasets (Figure 3). This illustrates that lung adenocarcinomas can be divided into groups with different survival rates. Kaplan-Meier plots showing the performance of the other classifiers on the validation datasets are available Supplementary Figure S1. The plots developed from method A again illustrate that risk predictors evaluated on all subjects performed better than those evaluated on subjects with stage 1 disease. Furthermore, using clinical covariates together with the gene expression data improved outcome prediction compared to using gene expression data alone. Method A included the null value 1 in its 95% hazard ratio confidence interval in only 1 of 8 situations considered (Figure 2). The one hypothesis where method A did not give significant prediction was stage 1 subjects scored using only gene expression measures. As noted above, no method gave significant results for both validation sets in this setting. This suggests that stage 1 tumors may be classified more efficiently using clinical parameters along with gene expression data.
Figure 2.
Kaplan-Meier estimates of the survivor function for method A on each validation dataset for the 4 hypotheses. a: MSK test set all stages, b: MSK test set with covariates all stages, c: MSK test set stage 1 only, d: MSK test set stage 1 only with covariates, e: CAN/DF test set all stages, f: CAN/DF test set with covariates all stages, g: CAN/DF test set stage 1 only, h: CAN/DF test set stage 1 only with covariates.
Figure 3.
Kaplan-Meier estimates of the survivor function for method A (cross-validated) on training sets UM and MSK. a: all stage, b: all stages with covariates, c: stage 1 only, d: stage 1 only with covariates.
Analyses of additional classifiers
The additional classifiers shown in Supplementary Materials section 3 (J, K, L, M, and N) were derived from the probesets listed in the Potti et al9, and the Chen et al10, articles. While it was not possible to reconstruct the classifiers reported in the original papers, we utilized the reported probesets to construct classifiers, and we tested them on our validation data. The performances of these classifiers were generally comparable to, although slightly poorer than those for methods A–H developed for this paper. As shown in Supplementary Materials section 3, the hazard ratios are in most cases larger than one, but they did not give statistically significant hazard ratios consistently for both validation datasets. For these classifiers the addition of clinical covariates improved the predictive ability.
We considered two other ways to compare the classifiers developed for this study. Supplementary Materials section 4 shows how each tumor sample was classified by each of the methods. The graphs show that a number of subjects could be correctly classified by many different methods. These may represent extreme cases that can be easily recognized. There were a number of tumors where the classifiers disagree, which could reflect classifier quality or tumors that are more ambiguous in terms of the available data. This highlights the greatest problem facing expression-based classification of tumors --are misclassifications due to inaccurate clinical information, tissue sampling problems, bad classifiers, or do they simply reflect the continuum of tumor types that can arise? The overlap in predictivity is not explained by a high overlap in the probesets used for classification (Supplementary Materials section 5). There was overlap between the genes used in method H and those in one of the clusters observed to be important in method A. Many of these genes were associated with proliferation which is consistent with more aggressive lung adenocarcinomas demonstrating increased proliferative potential. For all the other newly developed classifiers the overlap is reflective of similarities in the methods used to select genes. The variety of probesets showing some predictive capacity suggests that information about lung adenocarcinoma outcomes may not be concentrated in just a few exceptional genes.
Discussion
Several studies of primary lung adenocarcinoma or NSCLC have reported the ability to generate expression signatures capable of grouping subjects according to their survival outcomes. However, most studies are small (approximately 100 subjects or fewer) and typically drew data from a single treatment institution. Gene expression profiles with real clinical applicability must be recognizable despite variability that might occur in the processing of samples at different institutions. So far, little has been published on the ability of prognostic methods for lung cancer to perform in larger data sets or with independent validation samples. Often the published signatures show little overlap in the genes identified as significant predictors of outcome. Thus there is a strong possibility that sample collection methods, processing protocols, single-institution subject cohorts, small sample sizes, and peculiarities of the different microarray platforms are contributing significantly to the results. To address these issues, a multi-institutional collaborative study was conducted to generate gene expression profiles from a large number of samples with a priori determined clinical features that could be used to fully evaluate proposed prognostic models for potential clinical implementation.
The design and execution of the present study was performed recognizing the specific issues discussed above. Significant emphasis was placed on reducing technical variability by using similar protocols, reagents and platforms12, so that the major uncontrolled variables represent the biology of the lung cancers and associated clinical data. The sample sizes used for training and validation were determined to be of sufficient size, and two blinded external validation sets were used to provide a realistic assessment of the performance of each prognostic method. This is in contrast to the more common approach of obtaining all the data from a single source and randomly assigning samples to training and validation sets for the development and assessment of classifiers. Furthermore, great care was taken to standardize the pathological assessment of each tumor sample and the collection of clinical information across all institutions involved in this study. The lessons learned from this coordinated effort will likely influence the research practice for future profiling efforts in lung cancer.
Several classifiers were developed from the training data and tested on the independent data sets. These classifiers represent many of the established techniques for classifier development, with novel approaches also represented. The classifiers had various levels of success in stratifying subjects according to risk. Two of the methods (C and E) showed little predictive capacity. The poor performance of method E was expected as one individual gene parameter is too sensitive to noise to perform well in gene expression data collected from multiple institutions. More complex classifiers showed better success, with a few classifiers demonstrating the ability to classify across different institutional data sets, and within the stage 1 tumors. The most successful classifiers at stratifying stage 1 samples were trained on samples from all stages. This suggests that heterogeneity of aggressiveness exist in stage 1 tumors, and the pattern of gene expression in higher stage tumors is informative for predicting the risk of stage 1 tumors. We note that the power for comparing classifiers tends to be lower than the power for identifying differentially expressed genes. This study was not adequately powered to draw sharp lines between the performances of different classifiers.
Method A, which worked with all tumor samples or with Stage I samples alone, both with and without clinical covariates, showed the best overall predictive ability. Method H also had good performance without clinical covariates. The genes in these classifiers may provide insight into the biology of aggressive tumors. Method A relied on the correlated expression of multiple gene clusters to predict subject outcome. Relatively higher expression of genes in cluster 6 of method A (545 genes) was associated with poor subject outcome. This cluster included cell proliferation-related genes including cyclin A (CCNA2) and other cyclins, BUB1B, topoisomerases, check point genes (CHEK1), chromosomal and spindle protein genes. Method H also relied heavily on these genes for classification. This is consistent with elevated cell proliferation and loss of cell cycle control being associated with poor outcomes7. Greater expression of genes in cluster 4 of method A (262 genes), cluster 5 (82 genes), and cluster 12 (427 genes) were associated with better survival. Cluster 4 includes several differentiation related genes such as thyroid transcription factor 1 (TITF1), pulmonary-associated surfactant protein B (SFTPB), as well as G protein-coupled receptor 116 (GPR116) and MAP3K12 binding inhibitory protein 1 (MBIP) while cluster 12 contains many immunological-related genes. This is consistent with tumors showing some aspects of recognition by the subject’s immune system having better outcomes14. The variety of genes found useful for classification suggests that multiple mechanisms contribute to the clinical progression of lung adenocarcinomas and that multiple classifiers may be equally effective.
This study provides a realistic assessment of the challenges in developing prognostic models for early stage lung cancer. A significant degree of outcome prediction accuracy was observed using gene expression data alone, yet the hazard ratios for most of our models increase with the inclusion of clinical data (Figure 1). Conversely, gene expression data improves the predictive performance of clinical parameters alone (method I), compared to method A which uses gene expression and clinical variables. We note that even this uniquely large study was not adequately powered to make comparisons between classification methods with high statistical confidence. Nevertheless, some interesting trends emerge. For the all-stage analysis, method I (clinical variables only) was competitive with most of the procedures using gene expression data without clinical variables, consistent with gene expression largely recapitulating stage. However it is notable that method A with covariates performs substantially better on the CAN/DF samples than either method A without covariates, or method I. In the stage 1 analysis, the clinical variables reduce to age and sex. In the MSK test set, these variables are uninformative about disease risk, so the fact that gene expression appears to risk stratify subjects in method A is important. The predictive performance of method I in the stage 1 CAN/DF test set is driven by a strong association with age. However it is unclear how far this relationship will generalize. Therefore, an integrated approach using gene expression together with associated clinical, pathological, and other information may be more promising for future work, as has previously been pointed out in studies examining prostate and breast cancer15,16. While it is not possible to attribute the slightly better results across the hypotheses and test sets with method A compared to the other methods to specific classifier properties, we do note that method A did utilize substantially more genes than the other approaches and incorporated an initial gene clustering procedure. These properties may have contributed to its more consistent performance. We have provided a detailed discussion of the challenges in using gene expression profiling for lung cancer prognosis in practice in Supplementary Materials section 2. Our findings suggest that clinical covariates should be collected with the same care as utilized for obtaining gene expression signatures.
The present study was designed to address three key issues in the field of gene expression based outcome prediction. First, this study provides the largest gene expression data set with pathological and clinical annotation for lung adenocarcinomas to date. Because of the large sample size, additional analyses of prognostic genes associated with specific histological subtypes, such as bronchioalvelolar carcinomas, can now be undertaken. Extensive pathological and mutational annotation of each specimen is ongoing and this careful assessment will provide an extremely valuable resource for hypothesis generation. Secondly, this data was used to test in a rigorous manner the current methodologies used to predict tumor biology and, by inference, subject prognosis from gene expression signatures. Finally, this study was used to identify issues relevant to the use of gene expression profiles that should be taken into consideration in designing future studies. We had observed previously12 that the biological variation between tumors exceeds the technical variation introduced by microarray analysis. We have observed in this study that clinical covariates improve upon gene expression alone as a mechanism for stratifying tumor samples. We have also learned that coordinating the collection of clinical and pathological data across several institutions is an important task for prospective studies designed to further refine prognostic signatures. There are also limitations in using subject survival as an end-point that may be overcome by using time to tumor recurrence as the primary endpoint in place of overall survival. Although there still remain significant challenges to the use of gene expression-based classifiers in the clinical setting, the potential that these tools can improve subject care and increase survival provides a strong impetus to continue to refine these approaches for eventual clinical utilization.
Methods
Investigator consortium: Four institutions (University of Michigan Cancer Center (UM), Moffitt Cancer Center (HLM), Memorial Sloan-Kettering Cancer Center (MSK) and the Dana-Farber Cancer Institute (DFCI)) formed a consortium with support and collaboration of NCI investigators to develop and validate gene expression signatures of lung adenocarcinomas. Details of the specimens, criteria for inclusion, clinical covariates collected, mRNA processing and hybridization are described in Supplementary Materials section 1. Consent was obtained for all patients and the protocols approved by the respective Institutional Review Board of each institution. The cel files for the study are available at the following URL:
https://caarraydb.nci.nih.gov/caarray/publicExperimentDetailAction.do?ex
pId=1015945236141280
Links to the pathology and clinical data are also available at this site.
Training and validation sets: Initial evaluation of the gene expression data suggested that the data from the UM, HLM and MSK was broadly similar although distinguishable, but the data from CAN/DF showed some systematic differences from the other three sites mainly due to reduced signal intensity. The CAN/DF set was also distinguished in that it lacked stage 3 samples. To give a realistic evaluation of how a prognostic method might be used in practice, it was decided that the combined data from UM and HLM would be used as the training set, with MSK held out as a similar but external validation set. The CAN/DF data was held out as a second and more challenging external validation set.
Analysis protocol
A strict protocol for analysis was followed, with data for the two validation sets held by a third party “honest broker” during analysis of the training data. Risk scoring procedures were developed on the training data for four distinct hypotheses described above. The available clinical variables were AJCC stage, age, and gender. Prognostic models were developed on the training data by each of the four groups, with each group submitting one or more candidate models for some or all of the four hypotheses defined above. After the models were defined and documented, the honest broker released the validation set gene expression and clinical data (but not the outcome data) for the two validation data sets to the four groups and each candidate prognostic model was used to predict outcomes for each subject. It was permitted for methods to standardize gene expression levels within each test set or refer to percentile points of summary features in a test set, but otherwise predictions were made for each test sample in isolation. Some models produced a continuous risk score for each subject while others grouped the subjects into a finite number of ordered risk categories. These predictions were then passed back to the honest broker, allowing evaluation of the performance of the prognostic models. Results for all methods we considered are presented in this paper.
For performance evaluation, we used each predicted risk score as the covariate in a univariate Cox proportional hazards model, with overall survival (censored at 60 months) as the outcome variable. The continuous risk scores were standardized to have unit interquartile range to make the hazard ratios from the proportional hazards model comparable to each other, and approximately comparable to those from binary predictors. The estimated hazard ratio and its 95% confidence interval and p-value (shown in Supplementary Materials section 3) provided a means to directly compare the performances of different procedures on a unidimensional scale. For graphical representation, continuous risk scores were binned into tertiles, and Kaplan-Meier estimates of the survivor function were plotted for each subgroup. This allows for assessment of any “dose-response” relationship and also facilitates graphical comparison between different predictors.
An alternative measure of performance is provided by the concordance probability estimate (CPE)17. The CPE estimates the concordance probability, which is the probability that for a given pair of subjects selected at random from the study population, the subject with better prognosis has a better outcome. CPE values close to 0.5 indicate poor predictive accuracy and values approaching 1.0 indicate increasingly good predictive accuracy.
Finally, we constructed ROC curves for the continuous predictors and tables of sensitivity and specificity values for the categorical predictors. Sensitivity and specificity were calculated using Bayes’ theorem and Kaplan-Meier estimates of the survivor function to appropriately handle censoring. Details are provided in Supplementary Materials section 6 and Supplementary Figure S2.
Risk scoring procedures
A variety of strategies were employed to construct prognostic models. All of the methods used an initial step to reduce the amount of data for final modeling of the outcomes. Detailed descriptions of each method are provided in Supplementary Materials section 2.
Supplementary Material
Acknowledgments
We thank M Orringer, A Pickens, F Taylor, N Liu, D Lau, M Whitehead, L Chen, L Vargas, Y Xiao, M Maddaus, C Hoang. This work was supported by NCI Grants: CA84953, CA84999, CA84995, CA85052, CA46592 and Contracts: 263-MQ-319735, 263-MQ-319740, 263-MQ-319746, 263-MQ-510430 from the Canadian Cancer Society.
References
- 1.Jemal A, Seigel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer Statistics 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Booth CM, Shepherd FA. Adjuvant chemotherapy for resected non-small cell lung cancer. J Thorac Oncol. 2006 Feb;1(2):180–187. [PubMed] [Google Scholar]
- 3.Gandara DR, Wakelee H, Calhoun R, Jablons D. Adjuvant chemotherapy of stage I non-small cell lung cancer in North America. J Thorac Oncol. 2007 Jul;2(7 Suppl 3):S125–127. doi: 10.1097/JTO.0b013e318074fe31. [DOI] [PubMed] [Google Scholar]
- 4.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabárbara P, Seymour L National Cancer Institute of Canada Clinical Trials Group. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005 Jul 14;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98(24):13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S. Gene-expression profiles predict survival of subjects with lung adenocarcinoma. Nature Med. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 8.Wigle DA, Jurisica I, Radulovich N, Pintilie M, Rossant J, Liu N, Lu C, Woodgett J, Seiden I, Johnston M, Keshavjee S, Darling G, Winton T, Breitkreutz BJ, Jorgenson P, Tyers M, Shepherd FA, Tsao MS. Molecular profiling of non–small cell lung cancer and correlation with disease-free survival. Cancer Res. 2002;62:3005–3008. [PubMed] [Google Scholar]
- 9.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, Kratzke R, Watson MA, Kelley M, Ginsburg GS, West M, Harpole DH, Nevins JR. A genomic strategy to refine prognosis in early-stage non–small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 10.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, Chan WK, Li HN, Liu CC, Singh S, Chen WJ, Chen JJ, Yang PC. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007 Jan 4;356(1):11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y, Lemon W, Liu PY, Yi Y, Morrison C, Yang P, Sun Z, Szoke J, Gerald WL, Watson M, Govindan R, You M. A gene expression signature predicts survival of subjects with stage I non-small cell lung cancer. PLoS Med. 2006 Dec;3(12):e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbin KK, Beer DG, Meyerson M, Yeatman TJ, Gerald WL, Jacobson JW, Conley B, Buetow KH, Heiskanen M, Simon RM, Minna JD, Girard L, Misek DE, Taylor JM, Hanash S, Naoki K, Hayes DN, Ladd-Acosta C, Enkemann SA, Viale A, Giordano TJ. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin Cancer Res. 2005 Jan 15;11(2 Pt 1):565–572. [PubMed] [Google Scholar]
- 13.Fry WA, Phillips JL, Menck HR. Ten-year survey of lung cancer treatments and survival in hospitials in the United States. Cancer. 1999;86:1867–1876. doi: 10.1002/(sici)1097-0142(19991101)86:9<1867::aid-cncr31>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Moran CJ, Arenberg DA, Huang CC, Giordano TJ, Misek DE, Iannettonni MD, Orringer MB, Hanash S, Beer DG. Rantes expression by lung adenocarcinomas is a predictor of survival in stage I subjects. Clin Cancer Res. 2002;8:3803–3812. [PubMed] [Google Scholar]
- 15.Stephenson AJ, Smith A, Kattan MW, Satagopan J, Reuter VE, Scardino PT, Gerald WL. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer. 2005 Jul 15;104(2):290–298. doi: 10.1002/cncr.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sotirou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to subject care? Nature Rev Cancer. 2007;7:545–553. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 17.Gonen M, Heller G. Concordance probability and discriminatory power in proportional hazards regression. Biometrika. 2005;92:965–970. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.