Abstract
The ciliated cells of the node of the mouse embryo contribute to the establishment of left-right patterning via generation of leftward laminar fluid flow and initiation of a left-sided morphogen gradient. Here, we identify FOXJ1CreER2T mice in which expression of Cre recombinase is directed to ciliated node cells. The data demonstrate that foxj1 is expressed specifically in the node throughout the developmental window critical for left-right patterning. In transgenic embryos, Cre expression is detected by immunohistochemistry in ciliated cells of the node. Rosa26R reporter mice, in which expression of lacZ is activated only after Cre-mediated recombination, demonstrate strong and uniform labeling at the node when crossed with FOXJ1CreER2T mice. Cell labeling occurred as early as 0–2 somite stages specifically within cells of the node and recombination was highly efficient in response to tamoxifen. FOXJ1CreER2T transgenic mice represent a new genetic tool for the analysis of node-specific gene expression and will also be valuable in the study of node cell lineage and temporal cell fate mapping.
Generation of mice in which tissue-specific loss of gene function occurs is a powerful tool for dissecting gene function. The use of the site specific recombination using the Cre/loxP recombinase system to excise genes flanked by loxP sites has allowed cell type- and tissue-specific gene knockout (Lewandoski, 2001; Nagy, 2000). However, this technology is limited by the availability of transgenic mouse lines expressing Cre recombinase in the cells or tissues of interest.
The node of the mouse embryo is a transient embryonic structure that is important for the generation of left-right patterning. The cells of the node contain monocilia that function in the establishment of left-right asymmetry by generating nodal flow (McGrath et al., 2003; Nonaka et al., 2002; reviewed in Hirokawa et al., 2006). There is direct evidence in zebrafish that impairment of cilia function in the organizer, using morpholinos targeted to dorsal forerunner cells/Kupffer’s vesicle, disrupts left-right patterning (Essner et al., 2005). In mouse, strains have been created to allow inactivation of gene activity specifically within large, non-ciliated crown cells surrounding the node, thus disrupting important asymmetric regulatory pathways (Brennan et al., 2002; Saijoh et al., 2003). However, delineation of the specific role(s) of the ciliated cells of the node has been difficult to study in mouse due to lack of reagents for cell type specific gene inactivation. Here we examine the ability of FOXJ1CreER2T mice to generate node cell-specific recombination.
Foxj1 is a member of the forkhead box family of transcription factors which is expressed in ciliated cells of the respiratory, reproductive and central nervous systems (Hackett et al., 1995; Lim et al., 1997). This transcription factor has an important role in ciliogenesis and is necessary for basal bodies to anchor to the apical cytoskeleton (Gomperts et al., 2004). Foxj1 has previously been shown to be expressed in the node of the presomite embryo (Zhang et al., 2004), and targeted deletion of foxj1 results in abnormal cardiac looping and randomization of nodal and pitx2 gene expression in the lateral plate mesoderm, indicating the requirement of foxj1 for the development of proper left-right patterning (Brody et al., 2000; Chen et al., 1998).
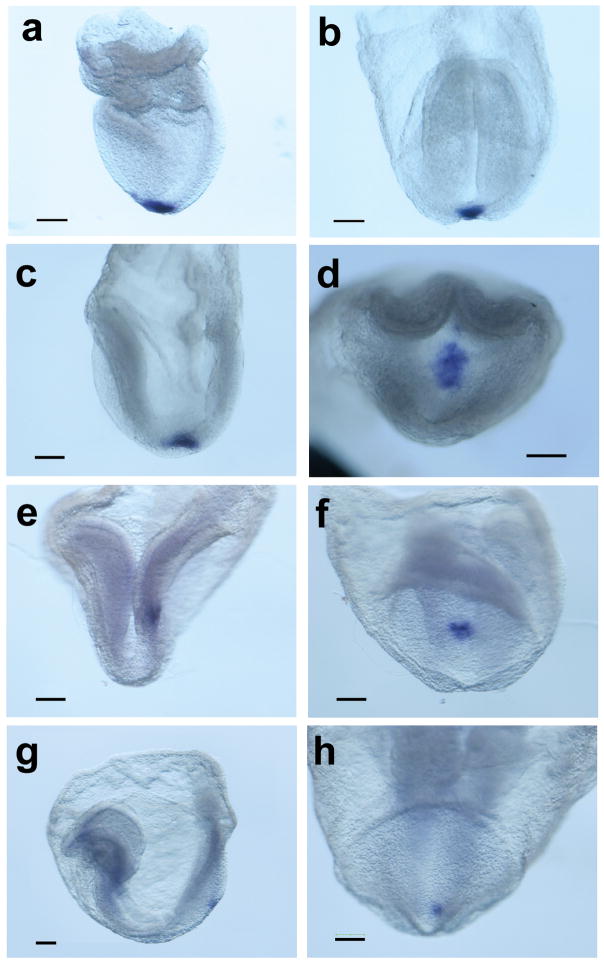
To further evaluate gene expression during critical stages of left-right patterning, a foxj1 riboprobe was generated and sense and antisense probes were tested in late headfold (LHF) and E9.5 embryos. The antisense probe showed specific staining in the region of the node (n = 2/2) and in the choroid plexus (n = 6/6) at the respective stages (data not shown), in accordance with published data (Zhang et al., 2004). Subsequently, gene expression was evaluated from late streak through 14 somite stage by whole mount in situ hybridization (WISH)(Fig. 1 and Table 1). The data indicate that foxj1 is expressed at least as early as late streak stages of gastrulation and continues to be highly expressed in early head fold (EHF), LHF and 0–4 somite stage embryos. Expression subsequently decreases, but continues to be present in a majority of embryos until 8 somite stage. These results indicate that foxj1 is expressed during the initiation of left-right signaling at the node and persists during critical stages of signal transduction in the lateral plate mesoderm. Expression at these stages was specifically restricted to the node region, suggesting that the foxj1 promoter might be a useful reagent for directing node-specific gene expression.
FIG. 1.
Node-specific expression of foxj1. Whole mount in situ hybridization using a foxj1 riboprobe demonstrates expression exclusively in cells of the ventral node in (a) late streak stage (b) late bud stage (c) early headfold (d) late headfold/0 somite (e, f) 0–2 somite (g, h) 4–6 somite and (i, j) 8–10 somite embryos. Panels f, h, and j show distal or posterior views of the node of the embryos pictured in a left lateral view in e, g, and i. Scale bar, 100 μm.
Table 1.
Stage specific expression of foxj1 in the node
| Stage | # analyzed | # staining | Percent |
|---|---|---|---|
| EHF-LHF | 9 | 9 | 100 |
| 0–4 somite | 12 | 12 | 100 |
| 4–6 somite | 6 | 5 | 83 |
| 6–8 somite | 10 | 8 | 80 |
| 9–10 somite | 4 | 1 | 25 |
| 11–14 somite | 3 | 0 | 0 |
EHF, early headfold; LHF, late headfold
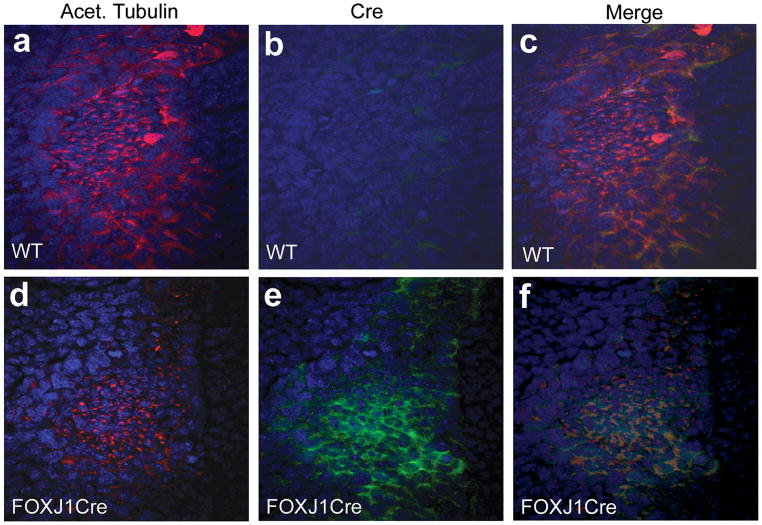
Previous studies have demonstrated the ability of a foxj1 promoter to direct expression to ciliated structures of the lung epithelium (Rawlins et al., 2007; Zhang et al., 2007). To further investigate node-specific expression, we utilized transgenic mice in which a 1 Kb FOXJ1 human promoter drives expression of a tamoxifen-inducible Cre recombinase. The node of a mouse embryo is morphological distinguishable by its teardrop shape and small cell size. In addition, the node stains strongly with anti-acetylated tubulin, which labels microtubules as well as cilia at the node. Surrounding the node are large crown cells which lack monocilia. In these cells, molecular evidence of asymmetry can be identified at E8. Immunohistochemistry was performed at 0–2 somite stages using an anti-Cre antibody and anti-acetylated tubulin to label the node (Fig. 2). Cre expression was detected in cells of the node at this stage (n = 7), but not in non-transgenic littermate control embryos (n = 5).
FIG. 2.
Cre immunohistochemistry. The node of E7.75–E8 embryos is highlighted by staining with anti-acetylated tubulin (a, d) in wild type and FOXJ1CreER2T embryos. Cre staining is identified within the node of FOXJ1CreER2T embryos (e), but is not found in wt embryos (b). WT, wild type.
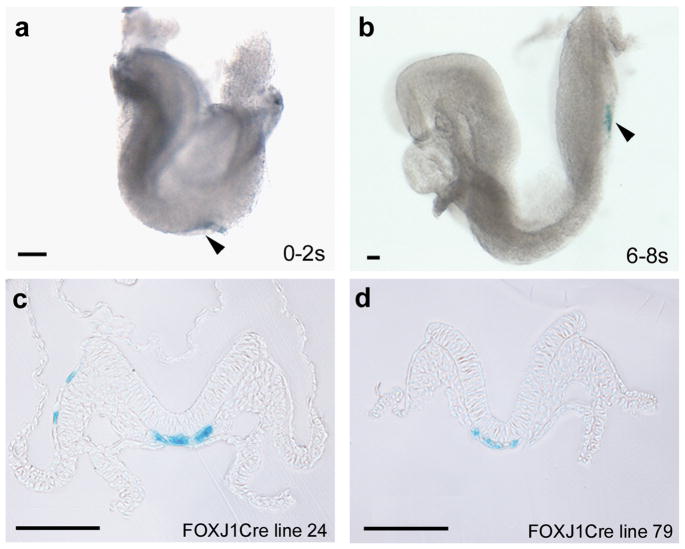
To assess whether Cre expression at the node mediates recombination, FOXJ1CreER2T mice were mated to Rosa26R reporter mice. Embryos were harvested at E7.75 through E8.5 and processed for X-gal staining. The results indicate that recombination occurs in node cells as early as 0 somite stage, with robust recombination occurring by 4 somite stage (Fig. 3). Sections through the node indicate greater than 80% labeling, suggesting high levels of recombination in response to the tamoxifen dosing regimen. Two transgenic lines were evaluated and both showed similar levels of recombination. Line 79 had no extra-nodal lacZ expression in embryos analyzed from 0–18 somites (n = 10), whereas line 24 exhibited extra-nodal staining in approximately 15% of embryos at these stages (n = 20)(Fig. 3c). These transgenic mice should therefore be useful for node-specific gene deletion. It should be noted that transgenic mice which utilize a FOXJ1 promoter to drive Cre without tamoxifen induction have also been shown to direct expression to ciliated epithelium of the lung (Zhang et al., 2007), and it would be of interest to test node-specific expression in these mice as well.
FIG. 3.
X-gal staining is present in the node in FOXJ1CreER2T::R26R embryos. X-gal staining is visible in the node of whole embryos (a, b at arrowhead) and in sections (c, d). In a small subset of embryos of line 24, extra-nodal X-gal staining was present in a few cells (c). Scale bar, 100 μm.
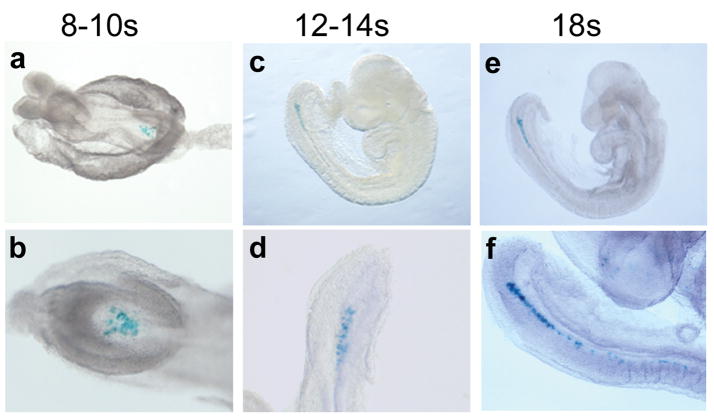
In addition to the functional importance of FOXJ1CreER2T mice for gene inactivation, this tool can be exploited for lineage labeling due to the irreversible nature of recombination. As proof of principle, FOXJ1CreER2T embryos were harvested at 12–18 somite stages and used for whole mount in situ hybridization (WISH) with a Cre riboprobe. As expected from foxj1 WISH data, in which expression is lost after the 11 somite stage, no node or notochord Cre transcript expression was identified (data not shown). In contrast, X-gal staining is present at these stages in FOXJ1CreER2T::Rosa26R mice (Fig. 4), indicating that this strategy will be useful in cell fate mapping experiments. The inducible nature of Cre recombinase in these mice in response to tamoxifen will allow independent investigation of temporal versus lineage specific gene expression. The ability to lineage label using FOXJ1CreER2T mice therefore provides a tool to address technically difficult questions related to ciliated node cell progeny.
FIG. 4.
Fate mapping of ciliated cells from the node. Females were dosed with tamoxifen at E6.5 and embryos harvested at somite stages as shown. Labeled cells are found in the node at 8–10 somite stage (a, whole embryo; b, high power view of the node) and in the node, notochord, and tailbud at later somite stages (c–f).
In summary, the FOXJ1CreER2T transgenic mice direct Cre expression to ciliated cells of the ventral node in a specific fashion. Onset of Cre expression and recombination occurs during developmental windows in which left-right patterning signal transduction pathways function, indicating that these mice will be a useful tool to further test gene function in ciliated node cells.
MATERIALS AND METHODS
Transgenic mice and tamoxifen induced Cre recombination
FOXJ1CreER2T mice have been described previously (Rawlins et al., 2007). Two independent lines of FOXJ1CreER2T mice were used for all experiments. Both lines showed similar levels of recombination. The Rosa26R (Gt(ROSA)26Sortm1Sor) reporter line (Soriano, 1999) was obtained from Jackson Laboratories. Tamoxifen free base (Sigma) was diluted at a concentration of 10mg/ml in corn oil. Pregnant females were injected with two doses of tamoxifen intraperitoneally. The first dose of 75 mg/kg body weight was injected at E5.5 and the second dose of 40 mg/kg body weight was injected at E6.5.
Whole mount in situ hybridization and X-gal staining
A riboprobe was created using the following primers for PCR amplification of a 236 bp fragment of foxj1 exon 1 (NM_008240) upstream of the conserved forkhead domain: 5′ GTCAGGATCCGCTGCAGGAATTCTCCATTC 3′; 5′TAATCTCGAGTGGTGGCATAGTCCACGTC 3′. The PCR fragment was cloned into BamHI and Xba1 sites of pBluescript (restriction sites in primers underlined). Sense and antisense riboprobes were prepared using a digoxigenin RNA labeling kit (Roche) as per manufacturer’s instructions. WISH was performed as described previously (Purandare et al., 2002). X-gal staining was performed according to standard procedures after a 20 minute fixation in 4% paraformaldehyde (PFA) in PBS. For sections, X-gal stained embryos were embedded in plastic and sectioned at 8um with a glass knife as described previously (Ware et al., 2006).
Whole-mount Immunohistochemistry
WT and FOXJ1CreER2T embryos were dissected at E7.75–E8 in cold PBS and fixed with 4% PFA in PBS overnight. After fixation, embryos were dehydrated with a graded methanol series and embryos with 0–2 somites were selected for further analysis. After stepwise rehydration, embryos were permeabilized with 0.2% Triton X-100 in PBS (PBT), blocked in 3% bovine serum albumin in PBT for 1 hour, and subsequently incubated with primary antibodies overnight at 4°C with gentle rocking. After thorough washing with PBT, embryos were stained with secondary antibodies. After washing, embryos were mounted in VECTASHIELD HardSet mounting medium (Vector Laboratories Inc.) and imaged on a confocal laser-scanning microscope (Nikon PCM 2000) at 60X magnification. Primary antibodies include mouse monoclonal anti-acetylated tubulin clone 6-11B-1 (Sigma) used at 1:200 dilution and rabbit polyclonal anti-Cre antibody (Novagen, San Diego, CA) used at 1:1000 for whole mount embryos. Secondary antibodies include goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 568 (Molecular Probes) used at 1:150 dilution.
Acknowledgments
We thank Brigid Hogan for FOXJ1CreER2T mice and advice on tamoxifen dosing and Brian Hackett for advice on riboprobe design. The Cre plasmid was the gift of Katherine Yutzey.
References
- Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiro SD. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am J Respir Cell Mol Biol. 2000;23:45–51. doi: 10.1165/ajrcmb.23.1.4070. [DOI] [PubMed] [Google Scholar]
- Chen J, Knowles HJ, Hebert JL, Hackett BP. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J Clin Invest. 1998;102:1077–1082. doi: 10.1172/JCI4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- Gomperts BN, Gong-Cooper X, Hackett BP. Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. J Cell Sci. 2004;117:1329–1337. doi: 10.1242/jcs.00978. [DOI] [PubMed] [Google Scholar]
- Hackett BP, Brody SL, Liang M, Zeitz ID, Bruns LA, Gitlin JD. Primary structure of hepatocyte nuclear factor/forkhead homologue 4 and characterization of gene expression in the developing respiratory and reproductive epithelium. Proc Natl Acad Sci U S A. 1995;92:4249–4253. doi: 10.1073/pnas.92.10.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left-right asymmetry. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Lim L, Zhou H, Costa RH. The winged helix transcription factor HFH-4 is expressed during choroid plexus epithelial development in the mouse embryo. Proc Natl Acad Sci U S A. 1997;94:3094–3099. doi: 10.1073/pnas.94.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418:96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
- Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, Deng JM, Vogel H, Behringer RR, Belmont JW, Casey B. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129:2293–2302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci U S A. 2007;104:410–417. doi: 10.1073/pnas.0610770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijoh Y, Oki S, Ohishi S, Hamada H. Left-right patterning of the mouse lateral plate requires nodal produced in the node. Dev Biol. 2003;256:160–172. doi: 10.1016/s0012-1606(02)00121-5. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Ware SM, Harutyunyan KG, Belmont JW. Zic3 is critical for early embryonic patterning during gastrulation. Dev Dyn. 2006;235:776–785. doi: 10.1002/dvdy.20668. [DOI] [PubMed] [Google Scholar]
- Zhang M, Bolfing MF, Knowles HJ, Karnes H, Hackett BP. Foxj1 regulates asymmetric gene expression during left-right axis patterning in mice. Biochem Biophys Res Commun. 2004;324:1413–1420. doi: 10.1016/j.bbrc.2004.09.207. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huang G, Shornick LP, Roswit WT, Shipley JM, Brody SL, Holtzman MJ. A transgenic FOXJ1-Cre system for gene inactivation in ciliated epithelial cells. Am J Respir Cell Mol Biol. 2007;36:515–519. doi: 10.1165/rcmb.2006-0475RC. [DOI] [PMC free article] [PubMed] [Google Scholar]