Abstract
Down syndrome, caused by the trisomy of chromosome 21, is a complex condition characterized by a number of phenotypic features, including reduced neuron number and synaptic plasticity, early Alzheimer disease-like neurodegeneration, craniofacial dysmorphia, heart development defects, increased incidence of childhood leukemia, and powerful suppression of the incidence of most solid tumors. Mouse models replicate a number of these phenotypes. The Tc1 Down syndrome model was constructed by introducing a single supernumerary human chromosome 21 into a mouse embryonic stem cell, and it reproduces a large number of Down syndrome phenotypes including heart development defects. However, little is still known about the developmental onset of the trisomy 21-induced mechanisms behind these phenotypes or the proteins that are responsible for them. This study determined the proteomic differences that are present in undifferentiated embryonic stem cells and are caused by an additional human chromosome 21. A total of 1661 proteins were identified using two-dimensional liquid chromatography followed by tandem mass spectrometry from whole embryonic stem cell lysates. Using isobaric tags for relative and absolute quantification, we found 52 proteins that differed in expression by greater than two standard deviations from the mean when an extra human chromosome 21 was present. Of these, at least 11 have a possible functional association with a Down syndrome phenotype or a human chromosome 21-encoded gene. This study also showed that quantitative protein expression differences in embryonic stem cells can persist to adult mouse as well as reproduce in human Down syndrome fetal tissue. This indicates that changes that are determined in embryonic stem cells of Down syndrome could potentially identify proteins that are involved in phenotypes of Down syndrome, and it shows that these cell lines can be used for the purpose of studying these pathomechanisms.
Down syndrome (DS)1 is caused by trisomy of human chromosome 21 (HSA21) and has an incidence of 1 in 650 live births (1). Mental retardation, smaller brain size, reduced numbers of neurons, reduced dendritic spine density and plasticity, and early Alzheimer disease-like neurodegeneration are seen in all people with DS, and a plethora of other phenotypes have a variable expression (1, 2). These include phenotypes that are seen only in a subset of DS individuals, such as heart defects, duodenal stenosis, and childhood leukemia. Interestingly there is a lower incidence of most solid tumors in people with DS (3).
Mouse models mimic some of the phenotypes seen in DS. TS65Dn, which contains ∼50% of the genes homologous for HSA21 in three copies, exhibits craniofacial skeletal malformation (4) and reduced cerebellar volume and granular and Purkinje cell densities (5). TS65Dn also displays learning and behavioral deficits (6). Ts1Cje is trisomic for ∼⅔ of the triplicated genes in TS65Dn and displays generally a similar but less severe phenotype than TS65Dn (7, 8). All the orthologous HSA21 genes that are trisomic in TS65Dn and Ts1Cje are present on mouse chromosome 16. HSA21 also has substantial regions of synteny with the mouse chromosomes 10 and 17, and it has therefore been difficult to engineer a mouse DS model that contains all orthologous HSA21 genes in three copies. To get around this problem, a transgenic mouse was derived that contains a freely segregating HSA21 (9); this is the first mouse model with a freely segregating human chromosome. This mouse model for DS was derived from transchromosomic embryonic stem (ES) cells, which contain HSA21 on a wild-type mouse ES cell background (10). The Tc1 mice contain ∼92% of the HSA21 genes and display a number of DS phenotypes (heart defect, learning difficulties, and a reduced cerebellar neuron count) (9).
To date, not a great deal is known about the pathways that are disturbed that lead to the characteristic phenotypes of DS. There have been a limited number of proteomics studies into the aberrant expression of proteins caused by trisomy of genes on HSA21. These were mostly on fully differentiated tissues from mice trisomic for a small number of genes or from people with DS (for instance, see Refs. 11 and 12) with only one study analyzing an ES cell model in neuronal differentiation using two-dimensional (2D) gel electrophoresis (13).
Phenotypic features of DS are retained even in mosaic DS subjects (14) and also in the Tc1 DS mouse model where adult tissues retain <56% trisomic cells (24–55%), having started from a fully trisomic conceptus (9, 15). This implies that many phenotypic features of DS are likely to be determined by events occurring very early in development. Our study utilized the transchromosomic ES cell lines to determine proteomic differences caused by an additional HSA21 in undifferentiated, pluripotent mouse ES cells. The aim was to detect pathways/proteins that are perturbed very early in development and to determine whether changed protein expression can remain during development. The added value of this approach is that transchromosomic ES cells can be used for further manipulation, such as selective switching off of individual supernumerary HSA21 genes using human-specific small interfering RNA reagents. This can help in further genetic dissection of the individual HSA21 gene contributions to the specific phenotypes of DS and open avenues for novel therapeutic concepts.
EXPERIMENTAL PROCEDURES
Cell Lines and Tissues—
The transchromosomic cell line 47-1, described in a previous publication (10), has been produced by tagging the HSA21 with a neomycin resistance marker and introducing the tagged chromosome into a mouse embryonic stem cell line, D3, using microcell-mediated chromosome transfer. ES cells were grown on a layer of mitotically inactivated mouse embryonic fibroblasts (feeder cells) in medium supplemented with leukemia-inhibitory factor (Millipore, Watford, UK). ES medium contained Dulbecco's modified Eagle's medium, 15% FCS, 25,000 units of penicillin/streptomycin, l-glutamine, non-essential amino acids, β-mercaptoethanol, and 5 × 105 units/ml leukemia-inhibitory factor. 47-1 ES cells were also grown in the presence of G418 (500 μg/ml) until one passage before they were lysed. During this last passage the G418 was removed so that the 47-1 and D3 cells had identical culturing conditions. Additionally feeders were removed one passage before lysis. We found that proliferation indexes of 47-1 and D3 grown under these conditions were similar (16). The WA17 mouse-human hybrid cell line, which contained two to three copies of HSA21, was derived from a fusion of mouse A9 cells and human WI-38 fibroblasts (17). They were passaged every 3 days at 3 × 103 cells/cm2 in Dulbecco's modified Eagle's medium and 10% FCS and grown under 5% CO2 at 37 °C. Both the source of HSA21 and the mouse parental cell line are genotypically different from the 47-1/D3 system. Tc1 mice, backcrossed to C57BL/6 (Tc1 two to three times), were maintained at the National Institute for Medical Research in accordance with Home Office regulations (9). Consented terminated human fetal tissue was collected by the Galliera Hospital's Tissue Bank and the MRC-Fetal Tissue Bank. All archived material was consented for use in research, and the project is covered by ethical approval from the North East London Health Authority.
PCR Analysis—
DNA was isolated for analysis using GenElute (Sigma). Analysis of the presence of HSA21 in 47-1 (supplemental Fig. 1A) was carried out as follows. Primer sequences were: USP25-F, AGTAGCGAAACAGTGCATTAC; USP25-R, GATGAGGTCACACCTGAATAG; MIR155b-F, TACTATATGCTGTCACTCCAG; MIR155b-R, AGGTTGAACATCCCAGTGAC; RUNX-F, AAGATGAAACGTGGAGAAATAG; RUNX-R, CTGGACATCACCCACGAGTG; D21S212-F, CATTTTAATGAACACCGCTC; and D21S212-R, GGCCTCCTGGAATAATTCTC. The program used was: 95 °C for 1.5 min; 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s for 35 cycles; and 72 °C for 9.5 min. D21S212 annealing temperature was 53 °C. For analysis of the retention of HSA21 (supplemental Fig. 1B), primer sequences were: 16.6Mb-F, ACCGTGCAAACAGTCTAGAC; and 16.6Mb-R, GCTTACCTCTTTGCAGCCTC. Forward primer contained a 5′ FAM label. The program used was: 95 °C for 7.5 min; 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s for 28 cycles; and 72 °C for 9.5 min. 1 μl of PCR product was mixed with 10 μl of formamide and 0.4 μl of ROX350 size standards (both from Applied Biosystems, Warrington, UK), denatured, and analyzed using a 3130XL automated sequencer according to the manufacturer's instructions (Applied Biosystems). For quantitative RT-PCR, RNA from D3 and 47-1 cells was extracted using an RNeasy Plus Mini kit (Qiagen, Crawley, UK). Quantitative RT-PCR was carried out using an Applied Biosystems 7700 Sequence Detector v1.7 and SYBR green or TaqMan PCR mixture according to the manufacturer's protocol (Applied Biosystems). All transcripts were measured in duplicate against standard curves relative to Gapdh. Primer sequences for quantitative RT-PCR were: Cse1l-F, CATGAGTTCAAGTCAAATGAGC; Cse1l-R, CATTTGCATGGGTACTGCAC; SerpinH1-F, TGCAGTCCATCAACGAGTG; and SerpinH1-R, CTTGTGGTGAAACTTCTCATC.
Labeling—
Lysis of 47-1 and D3 was performed as described previously (18), and 100 μg of total protein (per cell line) was tryptically digested and labeled with isobaric tags for relative and absolute quantification (iTRAQ) reagent 114 (D3 or 47-1) and 117 (47-1 or D3, respectively) according to the manufacturer's recommendation (Applied Biosystems). The labeled peptides were mixed and diluted 10-fold with 0.1% formic acid.
2D LC—
The first dimension of off-line 2D LC was performed using a Dionex UltiMate 3000 LC system (Camberley, Surrey, UK). 200 μg of total digest was injected onto a strong cation exchange (SCX) column (150 × 1 (inner diameter) mm, 5-μm PolySULFOETHYL A, PolyLC, Columbia, MD). The gradient used was 0–500 mm NH4Cl in 2% acetonitrile, and 15 fractions were manually collected. Each fraction was then desalted on a C-18 spin column (Pierce, Perbio, Cramlington, UK) and reconstituted in 25 μl of 0.1% formic acid, and 5-μl aliquots were injected repeatedly onto the reversed phase (RP) nanocolumn. For off-line 2D LC the subsequent RP separation was performed using a Micromass CapLC system (Waters, Elstree, UK) with sample loading onto a trap column (5 × 0.3 mm, 5-μm PepMap C18 guard column, Dionex) at a flow rate of 15 μl/min delivered isocratically with solvent C (0.1% formic acid) by auxiliary pump C. Sample was washed on the trap column for 5 min with solvent C before being switched in line with the RP nanocolumn (150 × 0.075 (inner diameter) mm, 3-μm C18, Dionex), which was equilibrated with 95% solvent A (0.1% formic acid in 5% acetonitrile), 5% solvent B (0.1% formic acid in 95% acetonitrile) at a flow rate of ∼200 nl/min. 5 min after sample loading the proportion of solvent B was increased linearly to 28% over 75 min and then to 80% over 20 min, maintained at 80% solvent B for 15 min (wash phase), and then re-equilibrated at 95% solvent A, 5% solvent B for 10 min. The column effluent was continuously directed into the electrospray ionization source of a Micromass Q-TOF Global mass spectrometer. Four repeat injections were made per SCX fraction, and analysis was performed over successive m/z ranges (19).
Mass Spectrometry—
The Q-TOF Global mass spectrometer was operated in the positive ion electrospray mode using data-dependent analysis (DDA) for the identification and quantification of peptides. DDA uses an initial “survey scan” that identifies the four most abundant multiply charged ions (tryptic peptides usually appear as 2+ and/or 3+ ions), which are then fragmented by MS/MS, which provides amino acid sequence information, before another survey scan is performed, and the cycle is repeated throughout the chromatographic run. DDA analysis was performed using a 0.5-s MS survey scan (m/z range, 420–1000 or e.g. 420–600, 600–700, 700–800, or 800–1000) followed by 1-s MS/MS scans (0.1-s interscan time) on up to four different precursor ions (intensity threshold, 10 counts/s). In the DDA mode, MS/MS spectrum acquisition (in the m/z range 50–1800) was allowed for up to a total of 2.2 s on each precursor ion or stopped when the signal intensity fell below 3 counts/s, and a new MS to MS/MS cycle was started. Precursors were excluded from any further MS/MS fragmentation for 45 s (retention time) to minimize repeated identification of the same peptide; singly charged ions were also excluded as precursors for MS/MS.
Protein Identification—
Each raw data file was first processed by MassLynx 4.1. The parameters for creating pkl files were as follows: spectrum selection criteria: combine sequential scans with same precursor and process all combined scans; mass measure: smooth window, three channels; number of smooths, 2; smooth mode, Savitzky-Golay; centroid: minimum peak width at half-height, 4; centroid mode, centroid top 80%. Peptide and protein identifications were performed using the Mascot search engine (version 2.2, Matrix Science, London, UK) located on a local server. Database searching was restricted to tryptic peptides of mouse or human proteins using the IPI mouse database (July 7, 2007, 56,450 entries) or IPI human database (July 7, 2007, 67,922 entries). The IPI database provides a minimally redundant, yet maximally complete set of proteins for the featured species (one sequence per transcript). Methionine oxidation and N-terminal acetylation of protein were selected as variable modifications, and cysteine blocked with methyl methanethiosulfonate and iTRAQ 114–117 were selected as fixed modifications at the peptide N terminus and side chain of lysine; one missed cleavage was allowed. Precursor and MS/MS tolerances were <0.3 Da (monoisotopic mass).
The level of confidence for peptide identifications was based on the Mascot assignment of “identity” (p < 0.05). Protein identifications were only made when two or more peptides from that protein were identified with Mascot scores above the identity threshold. In cases where the identified protein was a member of a multiprotein family with similar sequences, the protein identified was the one with the highest number of matched peptides and Mascot score. To assess the false positive peptide identification rate, the data were searched as above but against a randomized version of the IPI database.
Protein Quantification—
Quantification was first performed using Mascot 2.2. Using the Mascot quantification method, protein quantification was only performed on proteins identified by two or more peptides with scores above the identity threshold. Protein ratios (47-1/D3) were the “average.” iTRAQ ratios were normalized where a correction factor is applied such that the median for that ratio for all peptide matches in the data set would be unity. The global mean and S.D. of protein ratios (47-1/D3) were calculated for each replicate. The cutoff points for protein differential expression were mean ± 2 S.D. (95% confidence interval). Protein differential expression was also assessed at the peptide level. All peptides (with scores above the identity threshold; approximately 16,000 in experiment 1 and 12,000 in experiment 2) were used to calculate global mean and S.D. of peptide ratios (47-1/D3). The differentially expressed peptides (cutoff points for peptide differential expression were mean ± 2 S.D.) were used to infer differentially expressed proteins.
Western Blotting—
Western blotting was performed according to Laemmli (20) using 12% polyacrylamide gels and 50 μg of total protein/lane. The SERPINH1 (ab13510, 1:500), CSE1L (ab27518, 1:1000), ANXA5 (ab14196, 1:500), and TAGLN (ab14106, 1:500) antibodies were purchased from Abcam (Cambridge, UK); the β-actin antibody was from Sigma (A5316, 1:20,000); the GAPDH antibody (39-8600) was from Invitrogen; and the secondary antibodies (554002 and 554021, 1:5000) were from BD Biosciences. The ECL kit was from Pierce (Perbio).
Analysis of Transcriptome Data—
Data from a previous study (MIAMExpress database, number E-MEXP-654) were imported into Genespring v6.1 (SiliconGenetics), normalized to the 50th percentile of each array, and normalized to the median for each probe set. The data were then filtered for removal of all probe sets that were called present in fewer than four samples and those that changed fewer than two times between the two cell lines. ANOVA was applied to the remaining probe sets (p < 0.05). Additionally a t test was conducted on the data from the four 47-1/D3 pairs. All genes where the differential expression was p < 0.05 are shown (in gray) in Table II.
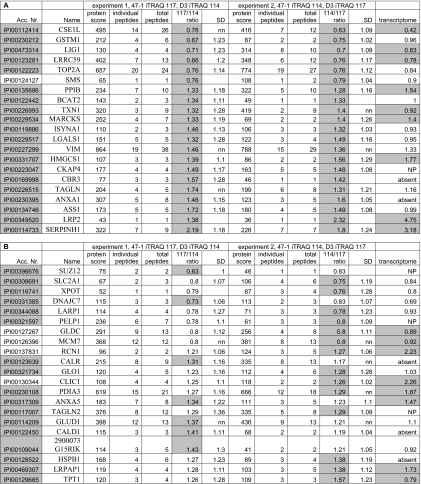
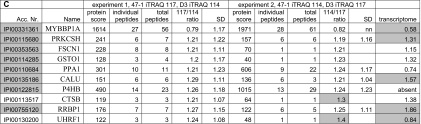
Table II.
The proteins with a significantly altered expression in 47-1 compared with D3 total cell lysates
The columns indicate accession number (Acc. Nr.), name, protein score, individual peptides (number of different peptides per protein), total peptides (number of total peptides), 117/114 ratio (protein ratio 47-1/D3 in experiment 1), 114/117 ratio (protein ratio 47-1/D3 in experiment 2), the geometric S.D. (SD), and transcriptome (comparison with expression results from a parallel gene expression study (24) using Affymetrix MG-U74Av2 arrays). All gray accession numbers have peptides in the 95% confidence intervals. Protein ratios in gray show significance for the protein in the corresponding experiment. Gray transcriptome ratios show significantly changed expression in 47-1 compared with D3 (ANOVA, p < 0.05; t test, p < 0.05; see “Experimental Procedures” for the comparison criteria). nn, not normal distribution. A contains proteins with significantly changed protein ratios in both experiments. B contains proteins with a protein ratio significantly changed in one experiment and nearing significance (changing in the same direction) in the other experiment. C contains proteins with peptide ratios that were significantly changed with protein ratios that neared significance (additionally two proteins are included with a limited number of peptides suggesting a changed expression).
RESULTS
iTRAQ Comparative Proteomics in Mouse ES Cells with Trisomy 21—
Mouse embryonic stem cells containing an intact HSA21 (47-1) and parental cells (D3) were grown as indicated. DNA was isolated, and four loci specific for HSA21 were only PCR-amplified from 47-1 (supplemental Fig. 1A). The number of cells in the 47-1 population that retained HSA21 was between 85 and 90% (supplemental Fig. 1B).
In experiment 1, 100 μg of total protein lysate (from 47-1 and D3) was labeled with either the iTRAQ 117 label (47-1) or the 114 label (D3). These were mixed after which an off-line separation on an SCX column was performed. With an increasing salt concentration 15 fractions were collected. These were subsequently individually injected for RP LC-MS/MS. Every fraction was injected four times to identify peptides in the following mass windows: m/z range 420–600, 600–700, 700–800, and 800–1000. The data generated by the individual injections were merged and analyzed by Mascot. A total of 1445 proteins were detected with at least two peptides above identity threshold and a false discovery rate of 0.9% (supplemental Table 1). A subsequent experiment (experiment 2) was performed in which the labeling of the samples was reversed (47-1, iTRAQ 114; D3, iTRAQ 117). This time 1103 proteins were detected with a false discovery rate of 0.6% (supplemental Table 1). 80.4% of these proteins were also detected in the first experiment. The total number of unique proteins identified in both experiments was 1661.
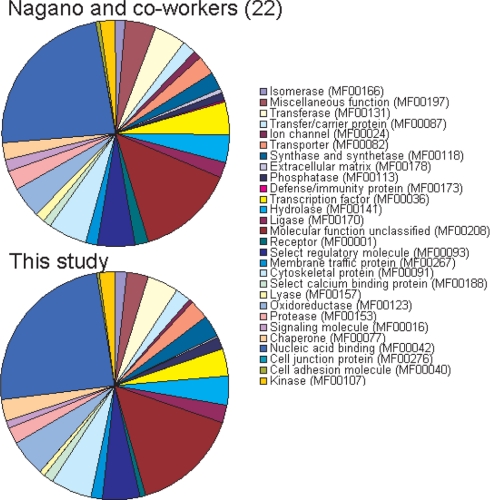
A comparison with a recent study, which identified 5111 proteins in undifferentiated mouse ES cells using stable isotope labeling by amino acids in cell culture and a more sensitive detection system, revealed that 93% of the 1661 proteins detected in our study were also found in this more in-depth study (21). An older study that determined 1790 proteins in E14 ES cells using anion column separation (22) showed a 48% overlap with our study. However, when proteins from both studies are analyzed according to functional category (23) very similar outcomes can be seen (Fig. 1). Both of these studies used ES cell lines different from our starting cell line D3. Taken together, these in silico comparisons with published data provide a solid quality control for the accuracy of detection of the true proteome expression of an undifferentiated mouse ES cell in our raw data.
Fig. 1.
Corroborative external comparison control for the accuracy of detection of the ES cell proteome. Comparison of functional protein classes between our study and Nagano et al. (22) shows that the percentage of proteins per functional class is very similar.
Identification of HSA21-expressed Proteins—
Because of the presence of an entire human chromosome 21 in 47-1, it is predicted that human proteins will be detected. The mass spectrometry data were researched using Mascot against the human IPI database. Because of high sequence homology of proteins derived from mouse and human, 12,000 spectra in experiment 1 and 9,000 spectra in experiment 2 were matched having scores above the identity thresholds. Among these, 99.7% of the spectra matched sequences identical in human and mouse. Four peptides were human-specific and located on HSA21 (Table IA); the spectrum of one of these (cystatin B) can be seen in supplemental Fig. 2. As expected, the 47-1/D3 ratio was increased for each of these four peptides (Table IA). The expression of human HMGN1 is also apparent when comparing the 47-1/D3 ratio between human- and mouse-specific peptides (Table IB; 8.9 versus 1.1, respectively).
Table I.
Identification of HSA21 encoded proteins
A shows the list of four human-specific peptides found in both experiments. Shown is the name of the protein, accession number (Acc. no.), the experiment in which the peptide was detected, the 47-1/D3 ratio (117/114 for experiment 1 and 114/117 for experiment 2), and the peptide sequence. In B proteins possibly expressed from HSA21 are indicated. Average 47-1/D3 ratio was determined for the seven proteins for which no specific HSA21 peptides were found. Shown are the names of the protein, average 47-1/D3 ratio for human specific peptides (Average h), peptides that are identical between mouse and human (Average m/h), and mouse-specific peptides (Average m). A t test was performed to assess whether the contribution of putative HSA21-specific expression is significant (comparison of average m/h with average m). CBR1 is in bold to indicate the significantly changed Average m/h compared to the Average m.
| A
| ||||
|---|---|---|---|---|
| Name | Acc. no. | Experiment | Ratio 47-1/D3 | Peptide sequence |
| HMGN1 | IPI00554761 | 1 | 15.726 | TEESPASDEAGEK |
| CSTB | IPI00021828 | 1 | 2.652 | SQVVAGTNYFIK |
| HMGN1 | IPI00554761 | 1 | 2.033 | QAEVANQETK |
| CSTB | IPI00021828 | 2 | 3.817 | VHVGDEDFVHLR |
| B
| ||||
|---|---|---|---|---|
| Name | Average h | Average m/h | Average m | t test (p value) |
| CSTB | 3.235 | |||
| HMGN1 | 8.88 | 1.078 | ||
| CCT8 | 1.009 | 1.069 | 0.067 | |
| GART | 0.865 | 0.908 | 0.093 | |
| ATP5O | 0.854 | 0.865 | 0.430 | |
| PFKL | 0.958 | 0.936 | 0.230 | |
| CBR1 | 1.214 | 1.040 | 0.044 | |
| GABPA | 1.002 | 0.963 | 0.361 | |
| SON | 0.877 | 0.993 | 0.141 | |
A list containing the IPI accession numbers of all HSA21-coded proteins was generated, and this was compared with the IPI accession numbers that were generated in the Mascot search against the human IPI database. Seven other proteins potentially expressed from HSA21 were detected (GABPA, CCT8, GART, SON, ATP5O, CBR1, and PFKL). No HSA21-specific peptides were detected in these proteins. However, comparison of 47-1/D3 ratios between human/mouse and mouse-specific peptides revealed a significant increase in expression of human/mouse peptides compared with mouse-specific peptides in CBR1 (Table IB). This suggests contribution of expression from HSA21 of CBR1.
Identification of Mouse Proteins with Altered Expression Levels in Trisomy 21—
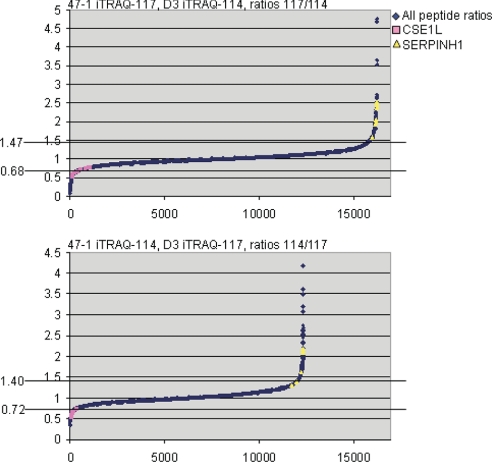
The Mascot data also included the 114/117 ratios for expression from the 47-1 and D3 cell lines. This was used to assess the proteins with an altered expression because of an additional copy of HSA21. The data were assessed in the following two ways. (i) The average protein ratio from the Mascot analysis was used. Proteins whose 114/117 ratios differed by more than two standard deviations from the mean were regarded as differentially expressed. These values were <0.77 or >1.30 for experiment 1 and <0.81 or >1.25 for experiment 2. Proteins that met these criteria in at least one of the two experiments are shown in Table II (114/117 and 117/114 ratios in gray). (ii) All peptides (∼16,000 in experiment 1 and 12,000 in experiment 2) were used to calculate the mean 114/117 ratio, and again ratios that differ by more than two standard deviations from the mean were regarded as different. Fig. 2 shows the ratios of all peptides as well as the confidence intervals (0.68<, >1.47 for experiment 1 and 0.72<, >1.40 for experiment 2). Additionally this figure shows all peptides that identify CSE1L and SERPINH1 (Fig. 2); these two proteins were validated by further experiments (see further results below). Multiple peptides for these two validated proteins fell inside the 95% confidence interval (Fig. 2); this probably means that there is more prospect for extracting further differing proteins from the data by relaxing the 95% confidence interval criterion. As this is likely to lead to an increase of false positives in the detection, the confidence interval was left at 95% for this study. Proteins with peptides outside the 95% confidence interval are shown with gray accession numbers in Table II.
Fig. 2.
The overall distribution of relative expression level ratios for all peptides in trisomy 21 as measured by the forward and reverse labeling in the iTRAQ experiment. The majority of peptides have an expected ratio of close to 1. The 95% confidence intervals are indicated by horizontal lines, and ratios at these intervals are indicated on the left. As examples of proteins with significantly differing expression in trisomy 21, all peptides from the proteins CSE1L (purple) and SERPINH1 (yellow) are indicated, and these show clearly disturbed expression in 47-1 (+HSA21) compared with D3 (euploid control) cells.
Twenty-one proteins were found to have a changed expression where the protein ratio was significantly changed in both experiments (Table IIA). Significant changes are indicated in gray in Table II (gray 117/114 and 114/117 ratios indicate significant protein changes, and the gray accession numbers indicate significant peptide changes). Twenty-one proteins had a protein ratio that was significantly changed in at least one of the two experiments (Table IIB); the other experiment always showed a change in the same direction that neared significance (for example, LRPAP1 protein ratio, 1.28 in experiment 1 and 1.38 in experiment 2 where >1.30 and >1.25, respectively, indicates significance). Eight proteins were only significantly changed when considering peptide ratios (Table IIC), although the respective protein ratios neared significance (supplemental Table 1). For example, MYBBP1A (Table IIC), for which peptide ratios reached significance, showed a protein ratio of 0.79 (experiment 1) and 0.82 (experiment 2) just inside the 95% confidence interval (<0.77 for experiment 1 and <0.81 for experiment 2). Additionally Table IIC contains two proteins with a limited number of peptides with a suggestion that expression was changed in 47-1 compared with D3 (cathepsin B (CTSB) and UHRF1). There were seven instances among the 52 proteins in Table II where at least one of the protein ratios was significantly changed, but the peptide ratios were not. However, on closer examination of individual peptide ratios we found that they were changed for all seven proteins in the same direction as the protein ratios, just narrowly missing the statistical significance thresholds (not shown).
Considering all data in Table II, a total of 15 proteins were down-regulated, and 37 showed higher expression in 47-1 compared with D3. Table II also shows data from a global transcriptome analysis of the same cell lines for the transcripts encoding the detected proteins in which four independent pairs of 47-1/D3 were compared on Affymetrix MG-U74Av2 mouse arrays (24). Ten of the 52 proteins with an altered expression in 47-1 (Table II) were not represented on the mouse arrays (either not present or expressed at insufficient levels to be detected by the arrays). Of the remaining 42 genes, 14 showed a statistically significant expression difference when analyzed by ANOVA at p < 0.05 (see “Experimental Procedures”) (only two were predicted to be found by chance). When a t test (p < 0.05) was performed on the four 47-1/D3 pairs, 22 of 42 proteins had matching transcript changes that reached significance (including the 14 significant changes identified with ANOVA). These are indicated in gray in Table II (transcriptome column). Comparison of these 22 transcript changes with the proteomics data reveals that 19 change in the same direction (six lower and 13 higher expression in 47-1). Intriguingly three proteins showed opposite transcriptome expression ratios (UHRF1, TXN1, and TPT1).
Validation of Altered Expression Levels by Western Blotting—
The different expression profiles were validated by Western blotting for four proteins. Exportin-2/CSE1L is predicted to be a protein that is involved in nuclear export (25) and also in apoptosis (26). SERPINH1 (HSP47/CBP2) is a protein with a classical serpin motif, which functions as a collagen binding factor/chaperonin in the endoplasmic reticulum (27). ANXA5 forms the voltage-dependent Ca2+ channels in phospholipid bilayers and has also been described as an anticoagulant (28, 29). TAGLN is an actin cross-linking protein initially described in fibroblasts and smooth muscle cells (30). CSE1L has a lower expression in 47-1; SERPINH1, ANXA5, and TAGLN show a higher expression in 47-1 compared with D3 (Table II). The altered expression profile was confirmed using three independently grown 47-1/D3 pairs (Fig. 3A) and (for SERPINH1) using a different transchromosomic cell line system (WA17/A9) (Fig. 3B). The WA17 cell line has been engineered to segregate two to three additional copies of HSA21 in a parental mouse fibroblastoid cell line, A9. Both the source of HSA21 and the mouse parental cell line are genotypically different from the 47-1/D3 system.
Fig. 3.
Validation of iTRAQ-detected quantitative differences in protein levels in trisomy 21. A, confirmation of the differential expression of CSE1L, SERPINH1, ANXA5, and TAGLN by Western blotting. Total protein lysates were used from three independently grown 47-1 and D3 cell lines. B, Western blotting of WA17 (mouse fibroblast line with supernumerary HSA21) and A9 (its euploid control) lysates. Antibodies against CSE1L and SERPINH1 were used. C, the expression data (bar chart) contains information from quantitative RT-PCR from n = 7 47-1/D3 pairs (four pairs were the same samples as were applied on the Affymetrix MG-U74Av2 mouse array experiments, and three additional RNA samples were from the same 47-1/D3 pairs as in the Western analysis). Error bars are indicated. ** shows p < 0.01 (highly significant).
To confirm the Affymetrix expression data in Table II, quantitative RT-PCR was performed on the same 47-1/D3 (n = 3) pairs that were used for the Western blotting for CSE1L and SERPINH1 plus 47-1/D3 pairs (n = 4) that were also applied to the microarrays. The overall results (Fig. 3C) show full concordance of the transcriptomics and the proteomics/Western blot analysis (Cse1l down in 47-1 and Serpinh1 up). The change in mRNA expression was highly significant (t test: Cse1l, p = 0.0039; Serpinh1, p = 0.0045).
Quantitative Expression Differences in Mouse DS Model and Human Down Syndrome Fetal Tissues—
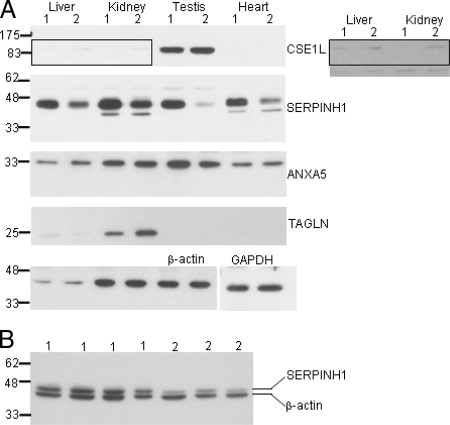
The Tc1 mouse model of DS was used to establish whether the changes in protein expression of CSE1L, SERPINH1, ANXA5, and TAGLN that can be seen in the mouse DS ES cells are also detectable in the transgenic animals. The Tc1 mice were generated from a mouse ES cell, 91-1, which contains a different clone introduced in a different parental mouse ES cell line compared with 47-1. The mouse displays a number of DS phenotypes that it shares with other DS mouse models. It contains ∼90% of the HSA21 genes on a mouse background (9). Total lysates were prepared from adult liver, kidney, testis, and heart from Tc1 mice and non-transgenic litter mates. Western blots were prepared and hybridized with CSE1L, SERPINH1, ANXA5, and TAGLN antibodies. CSE1L was expressed in testis and very slightly in liver and kidney. There was no difference in expression between Tc1 and littermates in testis (Fig. 4A). Although the expression in kidney of CSE1L was low, a difference could be detected (see the panel in different contrast in Fig. 4A). The expression in kidney was higher in non-transgenic littermates; this is in full agreement with the results in ES cells (Table II). SERPINH1 showed increased expression in all tested Tc1 tissues (Fig. 4A). A second experiment where liver, kidney, and testis lysates from a different Tc1/non-transgenic litter mate were tested for SERPINH1 expression gave an identical result (not shown). This is also in full agreement with the results in ES cells (Table II and Fig. 3). ANXA5 showed higher expression in Tc1 transgenics in testis (concordant with the ES cell results) and was unchanged in kidney and heart. Paradoxically a lower expression of ANXA5 was detected in liver in the transgenic animals. This was also the case for TAGLN in kidney. No expression of TAGLN was detected in testis and heart. In liver a very weak but unchanged expression pattern was seen (Fig. 4A).
Fig. 4.
Analysis of the relative levels of selected iTRAQ-differing proteins in tissues from adult DS model mice and human fetal DS tissue. A, Western blot showing total protein lysate from liver, kidney, testis, and heart from transchromosomic Tc1 (1) and normal littermate controls (2). The blot is stained with CSE1L, SERPINH1, ANXA5, TAGLN, and β-actin/GAPDH antibodies. The boxed area in the CSE1L blot is shown to the right in different contrast. B, Western blot showing total protein lysate from human fetal kidneys from DS (four different fetuses) (lanes 1) and from fetal age-matched euploid control (three different fetuses) (lanes 2). The blot is stained with SERPINH1 and β-actin.
Total protein lysates were generated from four DS and three normal, aged-matched, fetal kidneys. Western blots were prepared and hybridized with SERPINH1 antibody. This result also showed an increased expression of SERPINH1 in the four DS samples compared with the three normal samples (Fig. 4B). No difference in expression was detected in CSE1L, ANXA5, and TAGLN in DS fetal kidney compared with aged-matched control (not shown). Taken together, the ability to reproduce some of the significant differences in the WA17/A9 transchromosomic cell line system, the Tc1 mouse model, and human fetal tissues argues against cell line/clone-specific or heterospecific artifacts being the cause of the quantitative proteomic differences observed.
DISCUSSION
We present a quantitative proteomics comparison (using iTRAQ) between a mouse embryonic stem cell line containing an additional HSA21 and the wild-type cell line from which it was derived. Comparison with a published study on systematic identification of proteins in mouse embryonic stem cells (22) showed that 48% of the proteins were shared by our identification procedure. The reason that this is not higher is probably because of a difference in the starting ES cell line (E14 versus D3) as well as differences in fractionation and detection of the peptides (cation versus anion column separation). However, classifying the proteins from both studies according to functional category (23) showed a very similar overall picture (Fig. 1). A proportion of 93% of the list of proteins detected in our study was also detected in a recent, in-depth analysis of mouse ES cell lines (21). The nearly complete overlap with this study (93%) allows us to estimate that the overall sensitivity of detection in our system is of the order of 33% (1661 in our study versus 5111 in the study by Graumann et al. (21)) of all proteins with detectable expression in undifferentiated mouse ES cells.
With the extra HSA21, it is expected that human peptides are present in 47-1, and indeed four human specific peptides were found (from CSTB and HMGN1; Table IA). The increased ratio (higher expression in 47-1) and the fact that a mouse-specific HMGN1 peptide had a ratio that was not increased in 47-1 (ratio, 1.1; Table IB) indicate that the overexpression of these proteins was caused by the expression from HSA21. We did not find any human-specific peptides in seven other proteins that were detected and are present on HSA21 (Table IB). However, comparison of the ratio of peptides identical in human and mouse with mouse-specific peptides revealed that CBR1 had a statistically significant increase in the former. This suggests expression from HSA21. We did not find a difference in the other six proteins despite the fact that specific widespread HSA21 expression in transchromosomic ES cells has been demonstrated at the transcript level (9, 10, 24). This suggests that additional post-transcriptional regulatory mechanisms affect the expression of some proteins from HSA21. It is also highly probable that the expression of a number of HSA21-encoded proteins was below the detection limit in our study.
Fifty-two non-HSA21 proteins were detected with significantly altered levels in the presence of the supernumerary HSA21. Of these, 21 had a protein ratio significantly changed in both experiments (Table IIA), another 21 proteins had a protein ratio significantly changed in one experiment and nearing significance (changing in the same direction) in the other experiment (Table IIB), whereas eight proteins had only peptide ratios reaching the significance threshold (Table IIC). The list of proteins in Table IIA is probably the least likely to include false-positive changes, and this likelihood theoretically increases for Table IIB and further for Table IIC. Interestingly, however, the comparison with the transcriptomics data found 22 of 42 detected transcripts significantly changed (gray in the “transcriptome” column in Table II), and the distribution of these is very similar between Table II, A, B, and C, increasing the probability for validity of the detected protein changes.
A number of proteins with altered expression in trisomy 21 are involved in the same pathways; ASS1 and SMS in arginine metabolism (LINNEA™ pathways, Invitrogen); ASS1 and GLUD1 in arginine and proline metabolism; and PDIA3, CALR, and CTSB in antigen presentation (31). A number of detected differing proteins are involved in remodeling/stabilizing of/binding to the cytoskeleton: CALD1, VIM, MARCKS, FASCIN-1, CKAP4, and TAGLN. Also a relatively large number of proteins that reside in the endoplasmic reticulum are dysregulated: RCN1, PRKCSH, PDIA3, P4HB, CALU, CALR, PPIB, and SERPINH1. Furthermore a large number of proteins are potentially regulated by the RE1-silencing transcription factor REST. In total 20 of the 52 genes from Table II contain a REST binding motif within 50 kb of the transcriptional start site (32); only 14 would have been predicted by chance. REST expression has been shown to be lower in 47-1 compared with D3 (24), which could potentially explain the increased representation of REST-binding site-containing genes in our data.
It is likely that the quantity of a number of proteins remains changed during development and into adulthood as is the case with SERPINH1. For other proteins changed expression could be occurring during particular stages/cell types as could be the case for CSE1L, ANXA5, and TAGLN, expression of which was altered in some tissues but not in others from Tc1 animals (Fig. 4A). It is still possible that cell-autonomous differences in specific protein levels are maintained throughout development but get diluted by the impacts of heterogeneous cell type composition of adult tissue and organs.
Several proteins with a significantly changed expression in this study have associations with pathologies of DS. A number of proteins with higher expression in 47-1 (Table II) might be involved in the premature Alzheimer disease phenotype seen in all people with DS, for example CTSB, LRP2, and LRPAP1. CTSB is the β-secretase responsible for the majority of secreted Aβ42 (33). LRPAP1 is involved in the amount of mature lipoprotein receptor-related protein (LRP) expressed in liver and brain. Indeed the expression of LRP2 was also increased. LRP is the main apoE receptor, which can also bind α2-macroglobulin. This complex is responsible for the clearance of Aβ and therefore prevention of fibril formation (34, 35). Consequences of this dysregulation require further in-depth investigation as it could potentially explain why individuals with DS stay free of Alzheimer disease for many years despite high levels of Aβ42 detected as early as fetal development in DS brains (36).
Mutations in the glucose transporter SLC2A1 (GLUT1) result in GLUT1 deficiency syndrome. GLUT1 is a membrane-bound glycoprotein that is involved in glucose transport across blood-tissue barriers. In brain it exclusively facilitates the entry of d-glucose across the blood-brain barrier (37). This syndrome results in a lower glucose concentration in the cerebrospinal fluid and manifest itself with infantile seizures, microcephaly, and developmental delay (37), phenotypes that are also seen in DS. It would therefore be interesting to measure glucose levels in cerebrospinal fluid in people with DS.
A number of differing proteins have known functional associations with human chromosome 21 gene products. RCN1 has been shown to regulate calcineurin either as inhibitor or activator (38). Calcineurin is part of the same Ca2+-dependent regulatory circuit as calcipressin, the product of the HSA21 gene DSCR1. The actions of the HSA21 genes DSCR1 and DYRK1A cause the dysregulation of the calcineurin/NFAT pathway implicated in the pathogenesis of some of the developmental phenotypes seen in DS (39). The results offer some other suggestions of HSA21 genes that might be responsible for some of the proteome changes. Overexpression of TIAM1, a guanine nucleotide exchange factor that activates RAC, in a colorectal cancer cell line resulted in perturbed expression of 11 proteins in a proteomics experiment (40). Eight proteins in that study had increased expression, and four of these were also increased in our study (FASCIN-1, HSPB1 (HSP27), ANXA5, and GSTO1). It is highly likely that this is not a coincidence but caused by trisomy of TIAM1. A transgenic mouse containing an integrated human yeast artificial chromosome (152F7) also showed an increased proteomic expression of FASCIN-1 (11). This yeast artificial chromosome contains the genes DSCR3, DSCR5, TTC3, and DYRK1A, and one of these genes could therefore also be responsible for the FASCIN-1 overexpression. Table III summarizes, among the 52 significantly differing proteins, all proteins that either are functionally associated with each other in the same pathway or have an association with an HSA21 protein or a DS phenotype.
Table III.
Summary of all proteins that either are functionally associated with each other in the same pathway or have an association with an HSA21 protein or a DS phenotype among the 52 significantly differing proteins shown in Table 1
Aβ, amyloid beta; APP, amyloid precursor protein.
| Protein | Pathway | Association with HSA21 protein | Possible Down syndrome phenotype | Ref. |
|---|---|---|---|---|
| ASS1, SMS, GLUD1 | Proline and arginine metabolism | Unknown | Unknown | 31 |
| PDIA3, CALR, CTSB | Antigen presentation | Unknown | Immune defects | 31 |
| RCN1 | NFAT/calcineurin | DSCR1 | Developmental defects | 38, 39 |
| FASCIN-1, HSPB1 (HSP27), ANXA5, GSTO1 | Cytoskeletal remodeling and regulation | TIAM1 and either DSCR3, DSCR5, TTC3, and/or DYRK1A | Unknown | 11, 40 |
| LRPAP1, LRP2 | Aβ clearance | APP | Alzheimer disease | 34, 35 |
| CTSB | APP cleavage (β-secretase action) | APP | Alzheimer disease | 33 |
Identification of the proteins that are involved in phenotypes of DS is clearly not a trivial undertaking. By studying undifferentiated DS ES cells it is possible to detect proteins with altered expression very early in development. The involvement of the detected proteins in DS phenotypes can be assessed by careful analysis of the affected cell types in DS mouse models. Furthermore our findings can assist in determining the effects of overexpression of (individual) HSA21 genes under physiological conditions on the whole proteome. The added value of this approach is that transchromosomic ES cells can be used for further manipulation, such as selective switching off of individual supernumerary HSA21 genes, using human-specific small interfering RNA reagents (24). This can help in further genetic dissection of the individual HSA21 gene contributions to the specific phenotypes of DS and open avenues for novel therapeutic concepts.
Supplementary Material
Acknowledgments
We thank Chiara Baldo (Galliera Hospital) and the MRC-Fetal Tissue Bank for the human tissue material.
Footnotes
Published, MCP Papers in Press, November 10, 2008, DOI 10.1074/mcp.M800256-MCP200
The abbreviations used are: DS, Down syndrome; HSA21, human chromosome 21; ES, embryonic stem; iTRAQ, isobaric tags for relative and absolute quantification; 2D, two-dimensional; RP, reversed phase; DDA, data-dependent analysis; SCX, strong cation exchange; IPI, International Protein Index; ANOVA, analysis of variance; REST, RE1-silencing transcription factor; CTSB, cathepsin B; LRP, lipoprotein receptor-related protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NFAT, nuclear factor of activated T-cells; ASS1, argininosuccinate synthetase 1; TAGLN, transgelin; GABPA, GA-binding protein transcription factor, alpha subunit 60 kDa; PFKL, phosphofructokinase liver; SMS, spermine synthase; CALR, calreticulin; VIM, vimentin; MARCKS, myristoylated alanine-rich C-kinase substrate; PRKCSH, protein kinase C substrate 80K-H; CALU, calumenin; PPIB, peptidylprolyl isomerase B.
This work was supported by Barts and The London Charitable Foundation Grant RAC405, the AnEUploidy grant from Framework Programme 6 from the European Union Commission, Specialist Programme Grant 06003 from the Leukemia Research Fund-UK, Biotechnology and Biological Sciences Research Council Grants BB/C515771/1 and BB/C511356/1, and grants from the Medical Research Council-UK and The Wellcome Trust.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Epstein, C. ( 2001) The Metabolic and Molecular Bases of Inherited Disease, pp. 1223–1256, McGraw-Hill, New York
- 2.Antonarakis, S. E., Lyle, R., Dermitzakis, E. T., Reymond, A., and Deutsch, S. ( 2004) Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nat. Rev. Genet. 5, 725–738 [DOI] [PubMed] [Google Scholar]
- 3.Yang, Q., Rasmussen, S. A., and Friedman, J. M. ( 2002) Mortality associated with Down's syndrome in the U. S. A. from 1983 to 1997: a population-based study. Lancet 359, 1019–1025 [DOI] [PubMed] [Google Scholar]
- 4.Richtsmeier, J. T., Baxter, L. L., and Reeves, R. H. ( 2000) Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev. Dyn. 217, 137–145 [DOI] [PubMed] [Google Scholar]
- 5.Baxter, L. L., Moran, T. H., Richtsmeier, J. T., Troncoso, J., and Reeves, R. H. ( 2000) Discovery and genetic localization of Down syndrome cerebellar phenotypes using the Ts65Dn mouse. Hum. Mol. Genet. 9, 195–202 [DOI] [PubMed] [Google Scholar]
- 6.Reeves, R. H., Irving, N. G., Moran, T. H., Wohn, A., Kitt, C., Sisodia, S. S., Schmidt, C., Bronson, R. T., and Davisson, M. T. ( 1995) A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat. Genet. 11, 177–184 [DOI] [PubMed] [Google Scholar]
- 7.Sago, H., Carlson, E. J., Smith, D. J., Kilbridge, J., Rubin, E. M., Mobley, W. C., Epstein, C. J., and Huang, T. T. ( 1998) Ts1Cje, a partial trisomy 16 mouse model for Down syndrome, exhibits learning and behavioral abnormalities. Proc. Natl. Acad. Sci. U. S. A. 95, 6256–6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richtsmeier, J. T., Zumwalt, A., Carlson, E. J., Epstein, C. J., and Reeves, R. H. ( 2002) Craniofacial phenotypes in segmentally trisomic mouse models for Down syndrome. Am. J. Med. Genet. 107, 317–324 [DOI] [PubMed] [Google Scholar]
- 9.O’Doherty, A., Ruf, S., Mulligan, C., Hildreth, V., Errington, M. L., Cooke, S., Sesay, A., Modino, S., Vanes, L., Hernandez, D., Linehan, J. M., Sharpe, P. T., Brandner, S., Bliss, T. V., Henderson, D. J., Nizetic, D., Tybulewicz, V. L., and Fisher, E. M. ( 2005) An aneuploid mouse strain carrying human chromosome 21 with Down syndrome phenotypes. Science 309, 2033–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez, D., Mee, P. J., Martin, J. E., Tybulewicz, V. L., and Fisher, E. M. ( 1999) Transchromosomal mouse embryonic stem cell lines and chimeric mice that contain freely segregating segments of human chromosome 21. Hum. Mol. Genet. 8, 923–933 [DOI] [PubMed] [Google Scholar]
- 11.Shin, J. H., Guedj, F., Delabar, J. M., and Lubec, G. ( 2007) Dysregulation of growth factor receptor-bound protein 2 and fascin in hippocampus of mice polytransgenic for chromosome 21 structures. Hippocampus 17, 1180–1192 [DOI] [PubMed] [Google Scholar]
- 12.Bajo, M., Fruehauf, J., Kim, S. H., Fountoulakis, M., and Lubec, G. ( 2002) Proteomic evaluation of intermediary metabolism enzyme proteins in fetal Down's syndrome cerebral cortex. Proteomics 2, 1539–1546 [DOI] [PubMed] [Google Scholar]
- 13.Kadota, M. Nishigaki, R., Wang, C. C., Toda, T., Shirayoshi, Y., Inoue, T., Gojobori, T., Ikeo, K., Rogers, M. S., and Oshimura, M. ( 2004) Proteomic signatures and aberrations of mouse embryonic stem cells containing a single human chromosome 21 in neuronal differentiation: an in vitro model of Down syndrome. Neuroscience 129, 325–335 [DOI] [PubMed] [Google Scholar]
- 14.Hassold, T., and Hunt, P. ( 2001) To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2, 280–291 [DOI] [PubMed] [Google Scholar]
- 15.Shinohara, T., Tomizuka, K., Miyabara, S., Takehara, S., Kazuki, Y., Inoue, J., Katoh, M., Nakane, H., Iino, A., Ohguma, A., Ikegami, S., Inokuchi, K., Ishida, I., Reeves, R. H., and Oshimura, M. ( 2001) Mice containing a human chromosome 21 model behavioral impairment and cardiac anomalies of Down's syndrome. Hum. Mol. Genet. 10, 1163–1175 [DOI] [PubMed] [Google Scholar]
- 16.Mensah, A., Mulligan, C., Linehan, J., Ruf, S., O’Doherty, A., Grygalewicz, B., Shipley, J., Groet, J., Tybulewicz, V., Fisher, E., Brandner, S., and Nizetic, D. ( 2007) An additional human chromosome 21 causes suppression of neural fate of pluripotent mouse embryonic stem cells in a teratoma model. BMC Dev. Biol. 7, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raziuddin, A., Sarkar, F. H., Dutkowski, R., Shulman, L., Ruddle, F. H., and Gupta, S. L. ( 1984) Receptors for human α and β interferon but not for γ interferon are specified by human chromosome 21. Proc. Natl. Acad. Sci. U. S. A. 81, 5504–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unwin, R. D., Pierce, A., Watson, R. B., Sternberg, D. W., and Whetton, A. D. ( 2005) Quantitative proteomic analysis using isobaric protein tags enables rapid comparison of changes in transcript and protein levels in transformed cells. Mol. Cell. Proteomics 4, 924–935 [DOI] [PubMed] [Google Scholar]
- 19.Wang, Y., Muneton, S., Sjovall, J., Jovanovic, J. N., and Griffiths, W. J. ( 2008) The effect of 24S-hydroxycholesterol on cholesterol homeostasis in neurons: quantitative changes to the cortical neuron proteome. J. Proteome Res. 7, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. ( 1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 21.Graumann, J., Hubner, N. C., Kim, J. B., Ko, K., Moser, M., Kumar, C., Cox, J., Schöler, H., and Mann, M. ( 2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 22.Nagano, K., Taoka, M., Yamauchi, Y., Itagaki, C., Shinkawa, T., Nunomura, K., Okamura, N., Takahashi, N., Izumi, T., and Isobe, T. ( 2005) Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics 5, 1346–1361 [DOI] [PubMed] [Google Scholar]
- 23.Thomas, P. D., Campbell, M. J., Kejariwal, A., Mi, H., Karlak, B., Daverman, R., Diemer, K., Muruganujan, A., and Narechania, A ( 2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canzonetta, C., Mulligan, C., Deutsch, S., Ruf, S., O’Doherty, A., Lyle, R., Borel, C., Lin-Marq, N., Delom, F., Groet, J., Schnappauf, F., De Vita, S., Averill, S., Priestley, J. V., Martin, J. E., Shipley, J., Denyer, G., Epstein, C. J., Fillat, C., Estivill, X., Tybulewicz, V. L., Fisher, E. M., Antonarakis, S. E., and Nizetic, D. ( 2008) DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am. J. Hum. Genet. 83, 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutay, U., Bischoff, F. R., Kostka, S., Kraft, R., and Görlich, D. ( 1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061–1071 [DOI] [PubMed] [Google Scholar]
- 26.Brinkmann, U., Brinkmann, E., Gallo, M., and Pastan, I. ( 1995) Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. Proc. Natl. Acad. Sci. U. S. A. 92, 10427–10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikegawa, S., Sudo, K., Okui, K., and Nakamura, Y. ( 1995) Isolation, characterization and chromosomal assignment of human colligin-2 gene (CBP2). Cytogenet. Cell Genet. 71, 182–186 [DOI] [PubMed] [Google Scholar]
- 28.Grundmann, U., Abel, K. J., Bohn, H., Löbermann, H., Lottspeich, F., and Küpper, H. ( 1988) Characterization of cDNA encoding human placental anticoagulant protein (PP4): homology with the lipocortin family. Proc. Natl. Acad. Sci. U. S. A. 85, 3708–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demange, P., Voges, D., Benz, J., Liemann, S., Göttig, P., Berendes, R., Burger, A., and Huber, R. ( 1994) Annexin V: the key to understanding ion selectivity and voltage regulation? Trends Biochem. Sci. 19, 272–276 [DOI] [PubMed] [Google Scholar]
- 30.Shapland, C., Hsuan, J. J., Totty, N. F., and Lawson, D. ( 1993) Purification and properties of transgelin: a transformation and shape change sensitive actin-gelling protein. J. Cell Biol. 121, 1065–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., Katayama, T., Kawashima, S., Okuda, S., Tokimatsu, T., and Yamanishi, Y. ( 2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otto, S. J., McCorkle, S. R., Hover, J., Conaco, C., Han, J. J., Impey, S., Yochum, G. S., Dunn, J. J., Goodman, R. H., and Mandel, G. ( 2007) A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 27, 6729–6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hook, V. Y. ( 2006) Protease pathways in peptide neurotransmission and neurodegenerative diseases. Cell. Mol. Neurobiol. 26, 449–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez, L., Alvarez, V., González, P., González, I., Alvarez, R., and Coto, E. ( 2001) Variation in the LRP-associated protein gene (LRPAP1) is associated with late-onset Alzheimer disease. Am. J. Med. Genet. 105, 76–78 [PubMed] [Google Scholar]
- 35.Nistor, M., Don, M., Parekh, M., Sarsoza, F., Goodus, M., Lopez, G. E., Kawas, C., Leverenz, J., Doran, E., Lott, I. T., Hill, M., and Head, E. ( 2007) α- and β-secretase activity as a function of age and β-amyloid in Down syndrome and normal brain. Neurobiol. Aging 28, 1493–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teller, J. K., Russo, C., DeBusk, L. M., Angelini, G., Zaccheo, D., Dagna-Bricarelli, F., Scartezzini, P., Bertolini, S., Mann, D. M., Tabaton, M., and Gambetti, P. ( 1996) Presence of soluble amyloid β-peptide precedes amyloid plaque formation in Down's syndrome. Nat. Med. 2, 93–95 [DOI] [PubMed] [Google Scholar]
- 37.Klepper, J., and Leiendecker, B. ( 2007) GLUT1 deficiency syndrome—2007 update. Dev. Med. Child Neurol. 49, 707–716 [DOI] [PubMed] [Google Scholar]
- 38.Kishi, T., Ikeda, A., Nagao, R., and Koyama, N. ( 2007) The SCFCdc4 ubiquitin ligase regulates calcineurin signaling through degradation of phosphorylated Rcn1, an inhibitor of calcineurin. Proc. Natl. Acad. Sci. U. S. A. 104, 17418–17423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arron, J. R., Winslow, M. M., Polleri, A., Chang, C. P., Wu, H., Gao, X., Neilson, J. R., Chen, L., Heit, J. J., Kim, S. K., Yamasaki, N., Miyakawa, T., Francke, U., Graef, I. A., and Crabtree, G. R. ( 2006) NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441, 595–600 [DOI] [PubMed] [Google Scholar]
- 40.Liu, L., Zhao, L., Zhang, Y., Zhang, Q., and Ding, Y. ( 2007) Proteomic analysis of Tiam1-mediated metastasis in colorectal cancer. Cell Biol. Int. 31, 805–814 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.