Abstract
Exposure to environmental pollutants such as polychlorinated biphenyls (PCBs) is now taken into account to partly explain the worldwide decline of amphibians. PCBs induce deleterious effects on developing amphibians including deformities and delays in metamorphosis. However, the molecular mechanisms by which they express their toxicity during the development of tadpoles are still largely unknown. A proteomics analysis was performed on developing Xenopus laevis tadpoles exposed from 2 to 5 days postfertilization to either 0.1 or 1 ppm Aroclor 1254, a PCB mixture. Two-dimensional DIGE with a minimal labeling method coupled to nanoflow liquid chromatography-tandem mass spectrometry was used to detect and identify proteins differentially expressed under PCBs conditions. Results showed that 59 spots from the 0.1 ppm Aroclor 1254 condition and 57 spots from the 1 ppm Aroclor 1254 condition displayed a significant increase or decrease of abundance compared with the control. In total, 28 proteins were identified. The results suggest that PCBs induce mechanisms against oxidative stress (peroxiredoxins 1 and 2), adaptative changes in the energetic metabolism (enolase 1, glycerol-3-phosphate dehydrogenase, and creatine kinase muscle and brain types), and the implication of the unfolded protein response system (glucose-regulated protein, 58 kDa). They also affect, at least at the highest concentration tested, the synthesis of proteins involved in normal cytogenesis (α-tropomyosin, myosin heavy chain, and α-actin). For the first time, proteins such as aldehyde dehydrogenase 7A1, CArG binding factor-A, prolyl 4-hydroxylase β, and nuclear matrix protein 200 were also shown to be up-regulated by PCBs in developing amphibians. These data argue that protein expression reorganization should be taken into account while estimating the toxicological hazard of wild amphibian populations exposed to PCBs.
Over the last few decades, many populations of amphibians have declined in a number of geographical locations worldwide (1–3). Causes of this decline are assumed to result from man-made alterations of the environment, and exposure to environmental pollutants such as polychlorinated biphenyls (PCBs)1 is now taken into account (4). PCBs were manufactured in the 1950s for use in electrical insulators, plasticizers, and carbonless copy paper (5). Twenty years after their production ban in most industrialized countries, PCBs are still persistent and widely distributed in the environment (6).
It has already been reported that PCBs induce deleterious effects on wild organisms. In developing amphibians, they cause mortality (7), developmental deformities (8–11), delays in metamorphosis (12), immunological effects (13), and disruption of gonad development (14–16).
It is admitted that PCBs exert part of their toxicity by binding to the cytosolic aryl hydrocarbon receptor (AhR). In the nucleus, the activated AhR forms a heterodimer with the aryl hydrocarbon nuclear translocator, and the complex binds to the xenobiotic-responsive elements or aryl hydrocarbon response element I (AHREI), which regulates the expression of numerous genes involved in physiological and developmental processes (17, 18). The AhR-aryl hydrocarbon nuclear translocator heterodimer also acts as a coactivator of the transcription of responsive genes via the interaction with another response element, AHREII (19). However, in developing tadpoles of numerous frog species, an age-dependent insensitivity to chlorinated compounds linked to a low affinity for the AhR has been reported. The AhR machinery is present but requires high levels of inducer to provoke physiological changes (20–22). So far, the molecular mechanisms by which PCBs induce their toxicity during the development of tadpoles are still largely unknown. This hampers the risk assessment for developing tadpoles when they are environmentally exposed to these pollutants.
Proteomics is one of the possible strategies to gain better insight into the molecular responses to PCBs. Proteomics has been initially used successfully in drug discovery, biomarker identification, and protein-protein interaction studies in human disease processes (23, 24). This approach has been recently applied in ecotoxicology. It has been reported that environmental stresses such as variations of salinity and temperature and exposure to environmental contaminants like heavy metals, xenoestrogen, and chlorinated compounds have an impact on protein expression in different tissues of relevant aquatic organisms (25–31). Nevertheless such studies are scarce, and most of them focused on non-model organisms with the consequence of low output of protein identification (31).
The alteration of the genes expression in Xenopus laevis tadpoles exposed to PCBs has been explored in different studies (10, 32, 33). For example, 18-day postfertilization (pf) tadpoles exposed for 3 days to 50 ppb Aroclor 1254 showed significant up-regulation of several genes such as nerve growth factor, glyceraldehyde-3-phosphate dehydrogenase, interleukin-1β-converting enzyme, proopiomelanocortin, and p53 (32). However, no correlation between the mRNA and protein levels has been reported so far as the impact of PCBs on protein expression profiles in developing organisms has not been documented. Because the understanding of the molecular mechanisms by which PCBs interact with normal amphibian development is of special interest, the potential effects of a mixture of these environmental pollutants on the protein expression of developing X. laevis tadpoles were evaluated. To achieve this goal, the 2D DIGE minimal labeling approach coupled to nanoflow LC-MS/MS was applied to detect and identify proteins differentially expressed in PCB conditions. Identification of these proteins provides insight into the potential molecular mechanisms by which PCBs are interfering with amphibian development and will eventually lead to the proposal of candidate biomarkers for environmental pollution assessment.
MATERIALS AND METHODS
Animals, Breeding, and Housing—
Adult African clawed frogs (X. laevis) were obtained in 2004 from the National Breeding Laboratory of Xenopus, University of Rennes, France. Animals were maintained in dechlorinated water at 22 ± 1 °C with a 12:12 hour photoperiod schedule. Fresh water was changed every other day. Animals were fed three times a week with commercial trout food (Trouw) or chironomid larvae. Breeding was induced by subcutaneous injection of adults with 750 IU of human chorionic gonadotropin (Sigma). Cleaving embryos of stage 8–13 (34) were placed in FETAX medium (625 mg of NaCl, 96 mg of NaHCO3, 30 mg of KCl, 15 mg of CaCl2, 60 mg of CaSO4·2H2O, and 75 mg of SO4·7H2O/liter of distilled water) until they hatched (48 h pf).
Chemical Exposure—
Normally developing tadpoles (stage 35/36) were placed in glass bowls filled with 200 ml of FETAX medium. Each experimental condition included three replicates of 20–25 tadpoles. The PCB mixture Aroclor 1254 (Alltech Associates Inc.) was added to the medium using DMSO (Sigma) (final concentration of 0.05%) as a solvent, resulting in nominal concentrations of 0.1 and 1 ppm. A medium control group and a DMSO solvent control group (0.05%) were included in each experiment. During the assay, the temperature was maintained at 22 ± 1 °C, the solutions were changed every day, and dead tadpoles were removed daily. When tadpoles reached stage 45 (4 days pf), they were fed a mixture of Spirulina algae. After 72 h of exposure, the survival rate was recorded, and tadpoles were pooled and weighed, snap frozen, and stored at −80 °C. For each treatment, one replicate was assigned for proteomics analysis, and another was assigned for chemical analysis. Each experiment was repeated four times with tadpoles obtained from independent spawnings. Animals and tadpoles used in the present work were treated in accordance with an animal use protocol (code FUNDP 07/089) approved by the Ethic Commission of the Facultés Universitaires Notre-Dame de la Paix.
Protein Extraction and CyDye Labeling—
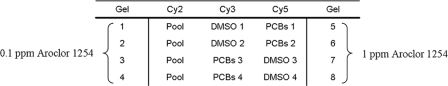
Proteins were extracted in 1:3 (w/v) lysis buffer (7 m urea, 2 m thiourea, 4% CHAPS, 30 mm Tris; GE Healthcare) and solubilized by sonication on ice. Samples were then centrifuged for 15 min at 12,000 × g. The pH of the soluble protein extract was adjusted to 8.5 by addition of 50 mm NaOH, and protein concentration was measured using the Bio-Rad protein assay. For DIGE minimal labeling, 25 μg of protein sample were labeled with 200 pmol of CyDye (GE Healthcare). Protein samples from DMSO control and PCB conditions (0.1 and 1 ppm Aroclor 1254) were labeled with Cy3 and Cy5. The reverse labeling of the test samples with Cy3 and of the control DMSO with Cy5 was done as well. A mixed sample composed of equal amounts of proteins from both Aroclor 1254-contaminated groups and DMSO control was minimally labeled with Cy2 and used as an internal standard (Fig. 1). Four independent replicates (tadpoles obtained from four independent spawnings) were used for each experimental condition. Labeling was performed on ice for 30 min in the dark and quenched with 1 mm lysine for 10 min on ice. The labeled mixtures were combined, and the total proteins (75 μg) were mixed v/v with the reduction solution (7 m urea, 2 m thiourea, 2% DTT, 2% CHAPS, 2% IPG 4–7 buffer; GE Healthcare) for 15 min at room temperature.
Fig. 1.
Schematic overview of the experimental conditions. Control DMSO and test PCB samples were labeled with either Cy3 or Cy5, reversing the labeling for half of the samples. The internal standard corresponding to a mixture of equal amounts of control and test samples was labeled with Cy2. Four replicates were used per experimental condition.
Separation of Proteins by 2D DIGE—
Prior to electrofocusing, IPG strips (24 cm, pH 4–7; GE Healthcare) were passively rehydrated overnight with 450 μl of a standard rehydration solution (7 m urea, 2 m thiourea, 2% CHAPS, 0.5% IPG 4–7 buffer, 2% DTT). The eight sample sets containing the labeled mixtures were then cup-loaded onto the IPG strips, and isoelectric focusing was performed with an Ettan™ IPGphor II isoelectric focusing unit (GE Healthcare). The electrophoresis conditions were as follows: 20 °C for 18 h; step 1, 300 V for 3 h; step 2, 1000 V for 6 h; step 3, 8000 V for 6 h; step 4, 8000 V for 6 h for a total of 68,000 V-h. Focused IPG strips were reduced (1% DTT) and alkalized (2.5% iodoacetamide) in equilibration buffer (50 mm Tris, 6 m urea, 30% glycerol, 2% SDS, pH 8.8) just before loading onto a 12.5% 24-cm, 1-mm-thick acrylamide gel. The strips were overlaid with 1% agarose in SDS running buffer (25 mm Tris, 192 mm glycine, 0.1% SDS) and run in an Ettan DALTsix electrophoresis unit (GE Healthcare) at a constant 3 watts/gel at 15 °C until the blue dye front had run off the bottom of the gels.
Image Analysis and Statistics—
Labeled CyDye gels were scanned with the Typhoon 9400 scanner (GE Healthcare) at wavelengths specific to the CyDyes. Resolution was 100 μm. Image analysis was carried out with DeCyder software (GE Healthcare). The differential in-gel analysis module co-detected and differentially quantified the protein spots in each image using the internal standard sample as a reference to normalize the data. At a second step, biological variation analysis was used to calculate ratios between samples and internal standard abundances by performing a gel-to-gel matching of the internal standard spot maps from each gel. Protein spots that showed a statistically significant (p < 0.01) Student's t test for an increased or decreased intensity were accepted as being differentially expressed between Aroclor 1254-contaminated and DMSO control groups.
Mass Spectrometry and Protein Identification—
For peptide sequencing and protein identification, preparative gels including 350 μg of proteins of mixed samples were performed following the protocol described above except that they were poststained with ruthenium(II) tris(bathophenanthroline disulfonate) overnight (7 μl of ruthenium/1 liter of 20% ethanol) after 6 h of fixation in 30% ethanol, 10% acetic acid and 3 × 30 min in 20% ethanol at 20 °C (35).
Peptides were analyzed by using a nanoflow LC-ESI-MS/MS (Waters) instrument on a CapLC Q-TOF2 mass spectrometer (Waters). Spots were excised from preparative gels by using the Ettan Spot Picker (GE Healthcare), and proteins were cleaved with trypsin by in-gel digestion. The gel pieces were twice washed with distilled water and then shrunk with 100% acetonitrile. The proteolytic digestion was performed by the addition of 3 μl of modified trypsin (Promega) suspended in 100 mm NH4HCO3 cold buffer. Proteolysis was performed overnight at 37 °C. The supernatant was collected and combined with the eluate of a subsequent elution step with 5% formic acid. The eluates were kept at −20 °C prior to analysis.
The digests were separated by reverse phase liquid chromatography using a 75-μm × 150-mm reverse phase NanoEase column (Waters) in a CapLC (Waters) liquid chromatography system. Mobile phase A was 95% 0.1% formic acid in water and 5% acetonitrile. Mobile phase B was 0.1% formic acid in acetonitrile. The digest (1 μl) was injected, and the organic content of the mobile phase was increased linearly from 5% B to 40% in 40 min and from 40% B to 100% B in 5 min. The column effluent was connected to a PicoTip emitter (New Objective) inside the Q-TOF source. Peptides were analyzed in data-dependent acquisition mode on a Q-TOF2 (Waters) instrument. In a survey scan, MS spectra were acquired for 1 s in the m/z range between 450 and 1500. When the intensity of 2+ or 3+ ions increased above 20 counts/s there was an automatic switch to the MS/MS mode. The CID energy was automatically set according to m/z and charge state of the precursor ion. Acquisition in MS/MS mode was stopped when the intensity fell below 5 counts/s or after 15 s. Q-TOF2 and CapLC systems were piloted by MassLynx 4.0 (Jasco). For the electrospray survey, background was subtracted with a threshold of 35%, polynomial 5. For smoothing, the Savitzky-Golay method with two iterations and a window of three channels was used. Finally we assigned the mass of peaks with a threshold of 3%, a minimum peak with four channels, and a centroid top method at 80%. For MS/MS raw data, a rigorous deisotoping method with a threshold of 3% was performed. Peak lists were created using ProteinLynx Global Server 2.2.5 (Waters) and saved as a PKL file for use with Mascot 2.1 (Matrix Science). Enzyme specificity was set to trypsin, and the maximum number of missed cleavages per peptide was set at 1. Carbamidomethylation was allowed as a fixed modification, and oxidation of methionine was allowed as a variable modification. Mass tolerance for the monoisotopic precursor peptide window and MS/MS tolerance window were set to ±0.3 Da. We also specified ESI-Q-TOF as instrument. The peak lists were searched against the Xenopus subset of the National Center for Biotechnology Information non-redundant (NCBInr) database (15,569 entries in September 2007). Control searches of all the files against the whole NCBInr database (5,454,477 entries in September 2007) was used to confirm the identification.
For all protein identifications, a minimal individual peptide score of 20 (below this score no identity or homology was found for the analyzed peptides) and expect value below 1 were used for the initial identification criteria (all peptide sequences linked to protein identification are reported in Table I). For single peptide-based protein identifications, the sequence identified and the precursor m/z and charge observed as well as the score for this peptide are given in the supplemental data. In the case of redundant protein identifications, the protein identification with the highest score was selected. However, if all the individual peptides completely matched to more than one UniProt database accession number, we aligned the sequences using BLAST (basic local alignment search tool). If the alignment showed 100% sequence identity, the UniProt accession number with the best description was chosen. When peptides matched to different isoforms or to different members of the same protein family, the following criteria were applied for selecting which isoform to report; if one peptide with a high score matched exclusively to a specific isoform or protein member, the identification could be made unambiguously. Moreover the correlation between theoretical pI and molecular mass of the protein with the position of the corresponding spot in the 2D gel was also taken into account.
Table I.
List of the responsive proteins showing different abundance in tadpoles exposed to 0.1 and 1 ppm Aroclor 1254 versus control DMSO
No., spot number as given by the DeCyder software on the 2D gels; Accession no., accession number in UniProt/TrEMBL; Peptide fragments, unique peptides analyzed by Mascot ion search software, charge state always equal to 1, M represents oxidized methionine; Score, Mascot probability based on Mowse score calculated for MS/MS results, significance is reached for score ≥20; Expect, the expect value reflects the probability that the sequence match is a random event, significance is reached for expect value <1; SC, sequence coverage (in %); Mr, molecular weight; Fc, -fold change where a positive value indicates an increase in protein spot intensity and a negative value indicates a decrease in intensity in tadpoles exposed to PCBs versus control DMSO.
| No. | Accession no. | Identification | Peptide fragments | Score | Expect | SC | pI | Mr | Fc |
|---|---|---|---|---|---|---|---|---|---|
| 0.1 ppm Aroclor 1254 | |||||||||
| Cytoskeleton | |||||||||
| 957 | P16878 | Keratin, type 2 cytoskeletal (X. laevis) | GKLEGELR | 23 | 0.23 | 19 | 5.5 | 55,287 | 1.93 |
| FLEQQNR | 35 | 0.013 | |||||||
| TEISELNR | 40 | 0.0044 | |||||||
| ALYEAELR | 62 | 2.4e−005 | |||||||
| SVSYGVSSGR | 30 | 0.028 | |||||||
| LQAEIESVK | 50 | 0.00043 | |||||||
| WELLQNQK | 23 | 0.18 | |||||||
| YEDEINKR | 21 | 0.25 | |||||||
| LAELEAALQK | 71 | 2.3e−006 | |||||||
| SAVPNAGFSQMR | 35 | 0.0088 | |||||||
| ALDMDSIIAEVK | 81 | 2.3e−007 | |||||||
| 988 | P16878 | Keratin, type 2 cytoskeletal (X. laevis) | FLEQQNR | 34 | 0.018 | 32 | 5.5 | 55,287 | 1.85 |
| LLEGEENR | 46 | 0.001 | |||||||
| TEISELNR | 41 | 0.0035 | |||||||
| ALYEAELR | 28 | 0.056 | |||||||
| SVSYGVSSGR | 30 | 0.032 | |||||||
| LQAEIESVK | 37 | 0.0082 | |||||||
| AQYEDIANK | 49 | 0.00042 | |||||||
| WELLQNQK | 29 | 0.044 | |||||||
| ANAESAYQSK | 57 | 5.7e−005 | |||||||
| LAELEAALQK | 69 | 3.5e−006 | |||||||
| KLLEGEENR | 34 | 0.017 | |||||||
| FQELQAAAGR | 46 | 0.0011 | |||||||
| EYQELMNVK | 45 | 0.001 | |||||||
| SYSVTTTSSSR | 52 | 0.00021 | |||||||
| TGAENEFVVLK | 34 | 0.014 | |||||||
| NMQDLVEDFK | 42 | 0.002 | |||||||
| STKTEISELNR | 44 | 0.0013 | |||||||
| SAVPNAGFSQMR | 48 | 0.00044 | |||||||
| ALDMDSIIAEVK | 79 | 3.7e−007 | |||||||
| TGAENEFVVLKK | 97 | 5.4e−009 | |||||||
| 1563 | A1DPL0 | Capping protein β subunit (X. laevis) | LVEDMENK | 20 | 0.43 | 8 | 5.7 | 30,864 | 1.72 |
| RLPPQQIEK | 39 | 0.0043 | |||||||
| TGSGTMNLGGSLTR | 102 | 2e−009 | |||||||
| Protein synthesis and degradation | |||||||||
| 851 | Q7ZWU3 | Glucose-regulated protein, 58 kDa (X. laevis) | QAGPASVDLR | 43 | 0.0016 | 9 | 5.7 | 56,486 | 1.57 |
| LADDPNIVIAK | 25 | 0.089 | |||||||
| LAPEYEIAATK | 25 | 0.1 | |||||||
| VDCTANSNICNK | 76 | 6.3e−007 | |||||||
| 893 | Q7ZWU3 | Glucose-regulated protein, 58 kDa (X. laevis) | LNFAVANR | 33 | 0.023 | 19 | 5.7 | 56,486 | 1.56 |
| SADGIVSTMK | 54 | 0.00013 | |||||||
| QAGPASVDLR | 75 | 1.1e−006 | |||||||
| SADGIVSTMKK | 70 | 3e−006 | |||||||
| LADDPNIVIAK | 66 | 7.8e−006 | |||||||
| FVMQEEFSR | 56 | 8.1e−005 | |||||||
| LAPEYEIAATK | 58 | 4.8e−005 | |||||||
| DGEDSGSYDGPR | 58 | 5.3e−005 | |||||||
| KLAPEYEIAATK | 40 | 0.003 | |||||||
| VDCTANSNICNK | 95 | 7.4e−009 | |||||||
| EATNPPVVKEDEKPK | 22 | 0.14 | |||||||
| 946 | Q5XGB9 | Leucine aminopeptidase 3 (X. tropicalis) | TLIEFATR | 24 | 0.14 | 8 | 8.4 | 53,788 | 1.54 |
| FAEIFEQK | 50 | 0.00033 | |||||||
| SGGACTAAAFLK | 89 | 4.1e−008 | |||||||
| GVLYAEGQNLAR | 47 | 0.00058 | |||||||
| TIQVDNTDAEGR | 81 | 2.2e−007 | |||||||
| 1659 | Q68A89 | Proteasome subunit α type (X. laevis) | GVNTFSPEGR | 41 | 0.0028 | 4 | 4.8 | 26,613 | 1.48 |
| Glucose metabolism, neoglucogenesis | |||||||||
| 1089 | Q7SZ25 | Enolase (X. laevis) | IEEELGSK | 54 | 0.00019 | 9 | 6.2 | 47,817 | 2.01 |
| ACNCLLLK | 48 | 0.00066 | |||||||
| AREIFDSR | 35 | 0.012 | |||||||
| NLNVVEQEK | 38 | 0.0055 | |||||||
| IGAEVYHNLK | 62 | 2.5e−005 | |||||||
| 1106 | Q7SZ25 | Enolase (X. laevis) | IEEELGSK | 58 | 6.2e−005 | 10 | 6.2 | 47,817 | 1.56 |
| ACNCLLLK | 45 | 0.0013 | |||||||
| AREIFDSR | 22 | 0.25 | |||||||
| NLNVVEQEK | 28 | 0.049 | |||||||
| GAEVYHNLK | 27 | 0.079 | |||||||
| 1109 | Q7SZ25 | Enolase (X. laevis) | IEEELGSK | 49 | 0.00051 | 10 | 5.9 | 47,930 | 1.83 |
| ACNCLLLK | 48 | 0.00067 | |||||||
| DGKYDLDFK | 26 | 0.072 | |||||||
| IGAEVYHNLK | 68 | 5.7e−006 | |||||||
| LMIEMDGTENK | 52 | 0.00021 | |||||||
| 1432 | Q66KM4 | Glyoxylate reductase/ hydroxypyruvate reductase (X. tropicalis) | RLPPEGQK | 41 | 0.0028 | 7 | 5.9 | 35,326 | 1.85 |
| TAVFINTSR | 38 | 0.0053 | |||||||
| VPEAMEEVR | 25 | 0.12 | |||||||
| RVPEAMEEVR | 33 | 0.018 | |||||||
| 1440 | Q7ZYM3 | GPD1 protein (X. laevis) | EAFGMSLIK | 20 | 0.36 | 8 | 6.3 | 38,342 | 1.56 |
| GVDEGPEGLR | 41 | 0.003 | |||||||
| LISDIIQER | 35 | 0.012 | |||||||
| Oxidative stress | |||||||||
| 1190 | Q7ZX44 | Txndc5 (X. laevis) | EFSGMSDVK | 20 | 0.34 | 9 | 5.8 | 45,889 | 2.45 |
| NGEKVDQYK | 23 | 0.16 | |||||||
| LFKPGQEAVK | 37 | 0.0074 | |||||||
| IAKVDCTAER | 45 | 0.00098 | |||||||
| 1787 | Q5XH88 | Peroxiredoxin 1 (X. tropicalis) | SKEYFNK | 21 | 0.27 | 13 | 5.9 | 22,640 | 1.92 |
| AVMPDGQFK | 23 | 0.17 | |||||||
| IGQPAPDFTAK | 47 | 0.00072 | |||||||
| 1824 | Q6P8F2 | Peroxiredoxin 2 (X. tropicalis) | DSKEFFSK | 52 | 0.06 | 9 | 5.9 | 22,640 | 1.74 |
| QITINDLPVGR | 34 | 0.32 | |||||||
| Metabolism | |||||||||
| 1196 | Q7ZYQ9 | Ckm (X. laevis) | FEEILTR | 37 | 0.0073 | 4 | 6.2 | 42,905 | 2.28 |
| GQTIDDMMPAQK | 58 | 4.7e−005 | |||||||
| 1244 | Q8AVH2 | Ckb (X. laevis) | TDINSANLK | 29 | 0.048 | 5 | 6.1 | 42,442 | 2.42 |
| GGNMKEVFNR | 56 | 6.7e−005 | |||||||
| 1245 | Q8AVH2 | Ckb (X. laevis) | VLTLDMYK | 41 | 0.0025 | 7 | 6.1 | 42,442 | 2.03 |
| GGNMKEVFNR | 55 | 9.3e−005 | |||||||
| LSTEEEYPDLSK | 59 | 3.4e−005 | |||||||
| 1263 | Q8AVH2 | Ckb (X. laevis) | GGNMKEVFNR | 34 | 6.7e−005 | 5 | 6.1 | 42,442 | 1.97 |
| TDINSANLK | 29 | 0.048 | |||||||
| Other function | |||||||||
| 992 | Q28GS6 | Aldehyde dehydrogenase 7 family member A1 (X. tropicalis) | QGLSSSIFTK | 64 | 1.2e−005 | 8 | 6.2 | 55,139 | 1.94 |
| STCTINYSK | 30 | 0.039 | |||||||
| CEGGTVVCGGK | 40 | 0.0036 | |||||||
| GAPTTSLTSVAVTK | 40 | 0.0034 | |||||||
| 1344 | Q7ZYE9 | Hnrpab (X. laevis) | DLKDYFAK | 26 | 0.083 | 5 | 5.7 | 35,785 | 1.9 |
| FGEVSDCTIK | 46 | 0.00078 | |||||||
| 1372 | Q98UD3 | CArG-binding factor A (X. laevis) | FGEVSDCTIK | 53 | 0.00014 | 21 | 5.7 | 35,785 | 1.5 |
| GAGGGQNDAEGDQINASK | 64 | 1.1e−005 | |||||||
| 1 ppm Aroclor 1254 | |||||||||
| Cytoskeleton | |||||||||
| 799 | P16878 | Keratin, type 2 cytoskeletal (X. laevis) | GKLEGELR | 23 | 0.23 | 19 | 5.5 | 55,287 | 1.75 |
| FLEQQNR | 35 | 0.013 | |||||||
| TEISELNR | 40 | 0.0044 | |||||||
| ALYEAELR | 62 | 2.4e−005 | |||||||
| SVSYGVSSGR | 30 | 0.028 | |||||||
| LQAEIESVK | 50 | 0.00043 | |||||||
| WELLQNQK | 23 | 0.18 | |||||||
| YEDEINKR | 21 | 0.25 | |||||||
| LAELEAALQK | 71 | 2.3e−006 | |||||||
| SAVPNAGFSQMR | 35 | 0.0088 | |||||||
| ALDMDSIIAEVK | 81 | 2.3e−007 | |||||||
| 820 | P16878 | Keratin, type 2 cytoskeletal (X. laevis) | FGSGGSSGVK | 45 | 0.001 | 26 | 5.5 | 55,287 | 1.56 |
| FLEQQNR | 38 | 0.0074 | |||||||
| LLEGEENR | 31 | 0.035 | |||||||
| TEISELNR | 36 | 0.011 | |||||||
| ALYEAELR | 27 | 0.074 | |||||||
| SVSYGVSSGR | 22 | 0.2 | |||||||
| LQAEIESVK | 45 | 0.0013 | |||||||
| AQYEDIANK | 34 | 0.012 | |||||||
| ANAESAYQSK | 53 | 0.00015 | |||||||
| LAELEAALQK | 70 | 2.8e−006 | |||||||
| FQELQAAAGR | 24 | 0.17 | |||||||
| EYQELMNVK | 35 | 0.0094 | |||||||
| SYSVTTTSSSR | 48 | 0.00061 | |||||||
| STKTEISELNR | 77 | 6.3e−007 | |||||||
| SAVPNAGFSQMR | 59 | 3.6e−005 | |||||||
| 968 | Q7SY65 | Keratin, type 1 cytoskeletal 18-B (X. laevis) | ESELVQVR | 26 | 0.12 | 10 | 5.2 | 47,974 | 1.41 |
| NSVTELRR | 30 | 0.04 | |||||||
| AQYDGLAQK | 24 | 0.15 | |||||||
| LIDDTNISR | 44 | 0.0013 | |||||||
| VVAESNDTEVLKA | 38 | 0.0055 | |||||||
| 989 | Q05AX6 | Keratin 19 (X. laevis) | LAADDFR | 32 | 0.029 | 6 | 4.9 | 45,326 | 1.52 |
| IVLQIDNAR | 26 | 0.076 | |||||||
| TLETANSGLELK | 46 | 0.00083 | |||||||
| 1000 | Q28IM9 | Keratin 12 (X. tropicalis) | LAADDFR | 27 | 0.088 | 18 | 4.9 | 41,846 | 1.67 |
| LATYLEK | 29 | 0.042 | |||||||
| SEITELRR | 43 | 0.0021 | |||||||
| FENELTLR | 32 | 0.021 | |||||||
| ADYEVLAEK | 30 | 0.035 | |||||||
| VLDELNLAR | 38 | 0.0047 | |||||||
| TIVEEVVDGK | 55 | 0.00012 | |||||||
| ALEAANAELEVK | 66 | 8.5e−006 | |||||||
| Protein synthesis and degradation | |||||||||
| 744 | Q7ZWU3 | Glucose-regulated protein, 58 kDa (X. laevis) | QAGPASVDLR | 65 | 1e−005 | 4 | 5.7 | 56,486 | 1.46 |
| LADDPNIVIAK | 32 | 0.018 | |||||||
| 700 | Q7ZWU3 | Glucose-regulated protein, 58 kDa (X. laevis) | QAGPASVDLR | 43 | 0.0016 | 9 | 5.7 | 56,486 | 1.3 |
| LADDPNIVIAK | 25 | 0.089 | |||||||
| LAPEYEIAATK | 25 | 0.1 | |||||||
| VDCTANSNICNK | 76 | 6.3e−007 | |||||||
| 787 | Q6DIK2 | Chaperonin containing TCP1 subunit 2 (β) (X. tropicalis) | LAVEAVLR | 36 | 0.0078 | 12 | 5.8 | 57,727 | 1.45 |
| CDLLNISR | 38 | 0.0063 | |||||||
| ESVAMESFAK | 22 | 0.19 | |||||||
| AGADEEKAETAR | 62 | 2e−005 | |||||||
| VAEIELAEKEK | 41 | 0.0028 | |||||||
| GATQQILDEAER | 51 | 0.00023 | |||||||
| TPGKESVAMESFAK | 47 | 0.0006 | |||||||
| Myofibrillogenesis and muscle contraction | |||||||||
| 1152 | Q6DIV8 | Actin α1 skeletal muscle (X. tropicalis) | IIAPPERK | 38 | 0.0057 | 18 | 5.2 | 41,988 | −2.24 |
| AGFAGDDAPR | 66 | 8.9e−006 | |||||||
| DLTDYLMK | 29 | 0.053 | |||||||
| GYSFVTTAER | 31 | 0.025 | |||||||
| EITALAPSTMK | 32 | 0.023 | |||||||
| DSYVGDEAQSK | 59 | 3.9e−005 | |||||||
| HQGVMVGMGQK | 39 | 0.0047 | |||||||
| DSYVGDEAQSKR | 56 | 8.3e−005 | |||||||
| 1173 | Q5Y819 | Myosin heavy chain α isoform (X. laevis) | ADIAESQVNK | 25 | 0.12 | 8 | 5.6 | 39,382 | −1.94 |
| LDEAEQIAMK | 60 | 3.5e−005 | |||||||
| EQDTSAHLER | 41 | 0.0025 | |||||||
| 1330 | Q01173 | Tropomyosin-1 α chain (X. laevis) | SLEAQAEK | 37 | 0.0084 | 11 | 4.4 | 32,630 | −1.97 |
| ATDAEGDVASLNR | 72 | 2e−006 | |||||||
| LEEAEKAADESER | 49 | 0.00041 | |||||||
| 420 | Q5M901 | Myosin-binding protein h (X. tropicalis) | DCAFIKK | 20 | 0.33 | 9 | 5.4 | 56,235 | 1.43 |
| FTQALANR | 38 | 0.0058 | |||||||
| ALENFVQIR | 26 | 0.09 | |||||||
| AINSLGEASVDCR | 68 | 3.9e−006 | |||||||
| IQNLNTGDKVTVR | 59 | 3.5e−005 | |||||||
| Metabolism | |||||||||
| 1026 | Q7ZYQ9 | Ckm (X. laevis) | FEEILTR | 37 | 0.0073 | 4 | 6.2 | 42,905 | 1.42 |
| GQTIDDMMPAQK | 58 | 4.7e−005 | |||||||
| 1056 | Q8AVH2 | Ckb (X. laevis) | FCTGLTK | 22 | 0.19 | 5 | 6.1 | 42,442 | 1.55 |
| FGEILKR | 55 | 0.00015 | |||||||
| LLVEMEK | 21 | 0.31 | |||||||
| LLVEMEKR | 20 | 0.34 | |||||||
| 1071 | Q8AVH2 | Ckb (X. laevis) | GGNMKEVFNR | 34 | 0.011 | 4 | 6.1 | 42,442 | 1.55 |
| LLVEMEKR | 20 | 0.34 | |||||||
| FCTGLTK | 22 | 0.19 | |||||||
| Other function | |||||||||
| 478 | Q802B7 | NADH-ubiquinone oxidoreductase 75-kDa subunit (X. laevis) | VAGVLQGVQGK | 32 | 0.019 | 2 | 6.1 | 79,575 | 1.62 |
| SATYVNTEGR | 26 | 0.07 | |||||||
| 483 | Q802B7 | NADH-ubiquinone oxidoreductase 75-kDa subunit (X. laevis) | SNYLLNSR | 34 | 0.012 | 7 | 6.1 | 79,575 | 1.29 |
| VAGVLQGVQGK | 49 | 0.00034 | |||||||
| SATYVNTEGR | 33 | 0.013 | |||||||
| LQEVSPNLVR | 27 | 0.066 | |||||||
| GNEMQVGTYVEK | 61 | 2.1e−005 | |||||||
| 553 | Q8JHX7 | Dihydrolipoamide acetyltransferase precursor (X. laevis) | ILVAEGTR | 30 | 0.043 | 1 | 7.2 | 66,849 | 1.41 |
| 745 | Q7ZXW4 | Nmp200 (X. laevis) | FLASTGMDR | 31 | 0.023 | 1 | 5.9 | 54,772 | 1.52 |
| 780 | Q7ZTJ5 | P4hb protein (X. laevis) | VVDYNGER | 37 | 0.0082 | 5 | 4.6 | 57,980 | 1.55 |
| LITLEEEMTK | 43 | 0.0014 | |||||||
| MDSTANEIEAVK | 82 | 1.9e−007 | |||||||
| 786 | Q7ZTJ5 | P4hb protein (X. laevis) | ALAPEYEK | 26 | 0.09 | 5 | 4.6 | 57,980 | 1.57 |
| VADYNGER | 36 | 0.008 | |||||||
| LITLEEEMTK | 39 | 0.0035 | |||||||
| 797 | Q7ZTJ5 | P4hb protein (X. laevis) | ALAPEYEK | 21 | 0.3 | 7 | 4.6 | 57,980 | 1.18 |
| VVDYNGER | 26 | 0.094 | |||||||
| LITLEEEMTK | 46 | 0.00069 | |||||||
| MDSTANEIEAVK | 80 | 2.8e−007 | |||||||
| 833 | Q28GS6 | Aldehyde dehydrogenase 7 family member A1 (X. tropicalis) | QGLSSSIFTK | 64 | 1.2e−005 | 8 | 6.2 | 55,139 | 1.54 |
| STCTINYSK | 30 | 0.039 | |||||||
| CEGGTVVCGGK | 40 | 0.0036 | |||||||
| GAPTTSLTSVAVTK | 40 | 0.0034 |
Polychlorinated Biphenyl Analysis—
PCBs were extracted according to a slight modification of Environmental Protection Agency method 608 as described previously with modifications for analysis with tadpoles (36, 37). Twenty-four PCB congeners (from di- to nonachlorinated) (IUPAC numbers 28, 44, 52, 66, 70, 87, 95, 101, 105, 110, 118, 128, 138, 149, 153, 156, 170, 180, 183, 187, 194, 195, 206, and 209) were identified and quantified. PCB concentrations were transformed in Aroclor 1254 equivalent and expressed in μg/g of lipids and in μg/g of body weight. All the tadpoles assigned for chemical analysis were pooled to obtain about 100 mg of lipid after extraction. Extraction of lipids was performed with hexane using an ASE 200 Accelerated Solvent Extractor (Dionex). All the extracts were used for lipid content determination: solvent was evaporated using a Turbovap LV (Zymarck) until a constant weight was obtained. Samples were then diluted in 3 ml of n-hexane, and a surrogate (PCB 112 with a final concentration of 50 pg/μl) was added to quantify possible loss of PCBs during the procedure. The extracts were subjected to cleanup with sulfuric acid to remove organic matter (lipids, lipoproteins, and glucides), and 2 ml of a mixture of concentrated (95%) and fuming (30%) sulfuric acid (3:1; v/v) were added to the extract. The mixture was shaken and centrifuged at 1750 × g. The supernatant was removed, and 3 ml of n-hexane were added to the decanted acid. Shaking, centrifugation, and removal were performed a second time before evaporation of the solvent. A cleanup column (Superclean™ ENVI Florisil solid phase extraction tubes (6 ml), Supelco) was also used to remove polar molecules. Columns were eluted with 5 ml of acetone, 5 ml of acetone-hexane, and 12 ml of hexane before the extracts were eluted with 6 ml of hexane. After the addition of 125 μl of a surrogate (PCB 30; 100 pg/μl diluted in hexane) and 125 μl of an internal standard (Mirex; 100 pg/μl diluted in hexane), the extracts were analyzed using a high resolution gas chromatograph (Thermoquest) equipped with a 63Ni electron capture detector. PCBs were separated by progressive temperature increase. Congeners were identified and quantified according to their retention time using the software Chrom-Card for Windows 4.0. The quantification limit of PCBs in tissue was 1 ng/g (w/w) and 200 ng/g of lipids.
Statistical Analysis—
Results for the survival and growth parameters were expressed as the mean (n = 4) ± S.D. Normality analysis of data was assessed by Kolmogorov-Smirnov test. Homogeneity of variances was tested by Bartlett test. Differences between groups were compared using one-way analysis of variance followed by post hoc least significant difference multiple comparison test at a 5% significance level. All statistical analyses were performed using Statistica™ software for Windows (StatSoft).
RESULTS
General Impact on Animals and Level of PCBs in Tissues
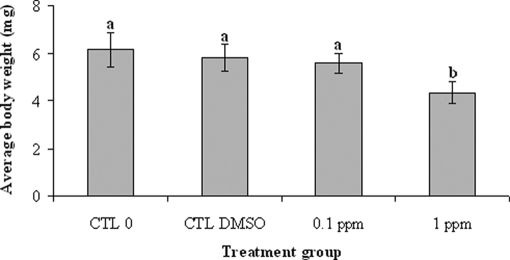
The percentage of surviving tadpoles was 92.5 ± 4.5% for the medium control group and 90.9 ± 3.6% for the DMSO control group. Exposure to 0.1 and 1 ppm Aroclor 1254 had no effect (p = 0.84) on the survival of tadpoles (93.1 ± 5.2 and 89.7 ± 5.1%, respectively). Regarding their final average body weight (Fig. 2), tadpoles exposed to 1 ppm Aroclor 1254 weighed 4.3 ± 0.5 mg, which was significantly (p < 0.05) lower compared with the average body weight of tadpoles from the control 0, the DMSO control, and the 0.1 ppm-treated groups (6.2 ± 0.7, 5.8 ± 0.5, and 5.5 ± 0.4 mg, respectively). Exposure to PCBs did not impact the developmental stages as all tadpoles from the different experimental conditions reached stages 44/45 by the end of the experiment.
Fig. 2.
Final average body weight (mg) of X. laevis tadpoles (3 days posthatching) following a 3-day exposure to Aroclor 1254. Data are presented in all figures as mean (n = 4) ± standard deviation (error bars) of the mean. Columns sharing at least one common superscript letter (a or b) are not significantly different, whereas the other differ at p < 0.05. CTL, control.
Levels of PCBs in tadpoles exposed to 0.1 and 1 ppm Aroclor 1254 increased in a dose-dependent manner and were as high as 13,693 μg/g of lipids in the 0.1 ppm group and 39,653 μg/g of lipids in the 1 ppm group. Low quantities of PCBs were also found in tissues of untreated control and DMSO control groups (2669 and 1737 μg/g of lipids, respectively).
Proteome Analysis
Protein Expression—
To understand how PCBs could affect amphibian development, the effects of these pollutants on the protein expression pattern in developing X. laevis tadpoles was investigated. 2D DIGE technique was used to compare tadpoles from the DMSO control group with tadpoles exposed for 72 h to 0.1 and 1 ppm Aroclor 1254. The number of spots detected in the four gels of the 0.1 ppm group was 1659 ± 170, whereas 1622 ± 159 spots were detected in the 1 ppm group. Only the 1083 spots from the 0.1 ppm group and the 937 spots from the 1 ppm group that were commonly matched between the four gels were selected for further statistical analysis. Protein spots that showed significant differences (p < 0.01) in intensity between tadpoles exposed to PCBs and DMSO were selected for MS/MS identification.
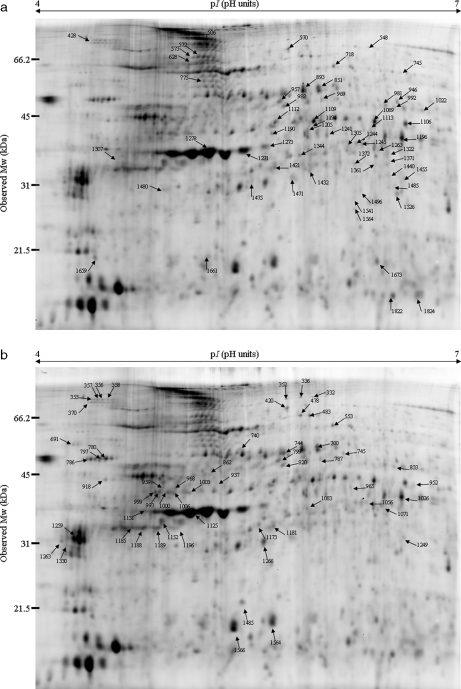
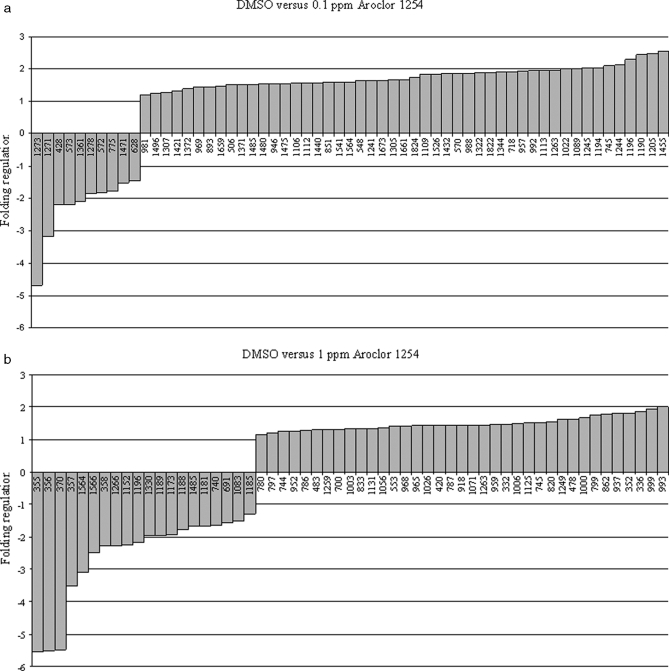
Changes in the protein expression pattern in tadpoles exposed to Aroclor 1254 are presented in Fig. 3. In the 0.1 ppm condition, 59 spots (Fig. 3a) showed significant differences (p < 0.01) in intensity in all gels corresponding to a change in protein abundance. An increase in abundance was observed for 83% of the protein spots (Fig 4a). The increase was between 1.2 and 2 for 39 spots and between 2 and 2.5 for 10 spots. 17% of protein spots showed a decrease in abundance with stronger variations because the -fold decrease reached 2–4 for four spots and even more for one spot.
Fig. 3.
Representative 2D gels showing the protein expression profiles obtained from X. laevis tadpoles exposed for 72 h to the PCB mixture Aroclor 1254. Proteins of the samples obtained for the different experimental conditions were differentially labeled with Cy3 and Cy5. An internal standard composed of equal amounts of each sample and labeled with Cy2 was added. Labeled samples (25 μg of each of the Cy3 and Cy5 labeled samples and of the Cy2 labeled internal standard) were loaded on 24-cm pH 4–7 non-linear IPG strips and subjected to IEF. Proteins were further separated by SDS-PAGE (12.5%) in the second dimension. Arrows and numbers allocated by the DeCyder software indicate spots with significant changes in intensity (p < 0.01, Student's t test in four independent gels). a, 2D gel image with proteins differentially expressed in the 0.1 ppm condition. b, 2D gel image with proteins differentially expressed in the 1 ppm condition.
Fig. 4.
Sets of protein spots showing differences in intensity between the PBC experimental groups and the DMSO control group. The y axis represents the -fold change intensity of the protein spots where a positive value indicates an increase in abundance and a negative value indicates a decrease in abundance. Data are organized on the x axis with the down-regulated proteins on the left side and the up-regulated proteins on the right side. a, tadpoles exposed to 0.1 ppm Aroclor 1254 versus control DMSO. b, tadpoles exposed to 1 ppm Aroclor 1254 versus DMSO control group.
The second experimental group of tadpoles exposed to 1 ppm Aroclor 1254 showed a similar profile with 57 protein spots (Fig. 3b) displaying significant differences (p < 0.01) in intensity. Among these spots, 20 corresponded to a decreased abundance of proteins (35%), and 37 (65%) corresponded to an increased abundance after PCB exposure compared with the control tadpoles (Fig. 4b). The -fold increase ranged between 1.15 and 2. For the 35% of spots with a decreased abundance, the -fold change corresponded again to stronger variations with the -fold decrease ranging between 1.3 and 2 for 10 spots, between 2 and 4 for seven spots, and over 4 for three spots. The comparative analysis of protein data sets enabled selecting six protein spots commonly up-regulated between both experimental conditions.
Protein Identification—
For the mass spectrometry analysis, preparative gels were run and stained with ruthenium (ruthenium(II) tris(bathophenanthroline disulfonate)). In these gels, 15–18% of the protein spots could not be matched with certainty in comparison with the 2D DIGE pattern. Only 48 and 47 protein spots clearly identified in the 0.1 and 1 ppm condition, respectively, were selected and excised for mass spectrometry analysis.
In total, the analysis of 45 protein spots allowed the identification of 28 different proteins (Table I). Single peptide-based protein identifications are illustrated in the supplemental data. The identified proteins can be divided into different groups, the first one corresponding to the six protein spots up-regulated within both PCB-exposed groups. Spots 957/799 and 988/820 (first and second spot numbers correspond to 0.1 and 1 ppm conditions, respectively) were commonly identified in X. laevis as cytokeratin type 2 that forms intermediate filaments of the cytoskeleton. Spot 992/833 was identified by homology to a Xenopus tropicalis protein as aldehyde dehydrogenase 7A1 (ALDH7A1). Spots 1196/1026 and 1263/1071 corresponded to X. laevis creatine kinase muscle type (Ckm) and brain type (Ckb), respectively. The creatine kinase isoenzymes catalyze the synthesis of phosphocreatine and its subsequent use in the regeneration of ATP. Finally spot 851/700 was identified in X. laevis as glucose-regulated protein, 58 kDa (GRP58); GRP58 has a protein-disulfide isomerase (PDI)-like activity and plays an important role in oxidative protein folding in the endoplasmic reticulum.
The second group included 16 protein spots up-regulated after the exposure of tadpoles to 0.1 ppm Aroclor 1254. Spot 1563 was identified in X. laevis as capping protein β subunit; capping protein binds to the barbed ends of actin filaments and is involved in the cytoskeleton regulation. Spots 893 and 1659 were identified in X. laevis as GRP58 and proteasome subunit α type, respectively. Proteasomal α subunits are major components of the 20 S proteasome, which is involved in the cytosolic proteolytic machinery. Spot 946 was identified by homology to X. tropicalis as leucine aminopeptidase 3 (LAP3) which is a metallopeptidase that cleaves N-terminal residues from proteins and peptides. Spots 1089, 1106, and 1109 were identified in X. laevis as enolase (ENO). ENO catalyzes the dehydration of 2-phospho-d-glycerate to phosphoenolpyruvate in the glycolytic pathway and the reverse reaction in gluconeogenesis. Spot 1432 corresponded to glyoxylate reductase/hydroxypyruvate reductase (Grhpr), which is similar to the corresponding protein of X. tropicalis. Grhpr functions both as glyoxylate reductase and as hydroxypyruvate reductase and plays a key role in directing the carbon flux to gluconeogenesis by its ability to convert hydroxypyruvate to d-glycerate. Spot 1440 was identified as X. laevis glycerol-3-phosphate dehydrogenase 1 (GPD1) involved in the branch point of the glycolytic pathway by converting dihydroxyacetone phosphate into glycerol 3-phosphate. Spot 1190 corresponded to X. laevis thioredoxin domain-containing 5 (Txndc5) whose biological function is not well described. Spots 1787 and 1824 were identified by homology to X. tropicalis as peroxiredoxin 1 (Prx Ι) and peroxiredoxin 2 (Prx ΙΙ), respectively. Peroxiredoxins are members of the thiol-specific antioxidant proteins that catalyze the reduction of hydrogen peroxide with the use of electrons provided by thioredoxin. Spots 1244 and 1245 were both identified as X. laevis Ckb. Finally spots 1344 and 1372 were identified, respectively, as X. laevis heterogeneous nuclear ribonucleoprotein A/B (Hnrpab) proteins and CArG binding factor-A (CBF-A). The Hnrpab proteins comprise numerous proteins with a general packing role in RNA processing and transport. More precisely, CBF-A belongs to the subfamily of Hnrpab proteins and functions in both transcriptional and post-transcriptional processes of gene regulation.
The third group was made up of 17 proteins up- or down-regulated after the exposure of tadpoles to 1 ppm Aroclor 1254. Spots 1152, 1173, and 1330 were the only down-regulated identified proteins. Spot 1152 was similar to X. tropicalis actin α1 skeletal muscle, whereas spots 1173 and 1330 were identified as X. laevis myosin heavy chain α isoform and α-tropomyosin, respectively. Actin α, myosin heavy chain, and tropomyosin α are muscle-specific proteins that play essential roles in myofibril assembly and muscle contraction. Spot 420 was similar to X. tropicalis myosin-binding protein h, which appears to function in the assembly of thick filaments during myofibrillogenesis. Spots 989 and 968 corresponded to X. laevis keratin 19 and keratin type 1 cytoskeletal 18-B, respectively, whereas spot 1000 was identified by homology to X. tropicalis as keratin 12. Keratins are major components of the cytoskeleton intermediate filaments. Spot 744 was identified in X. laevis as GRP58. Spot 787 was assigned to X. tropicalis chaperonin containing TCP1, subunit 2 that is involved in protein folding. Spots 478 and 483 were both identified in X. laevis as NADH-ubiquinone oxidoreductase 75-kDa subunit, which is a major component of the mitochondrial respiratory chain complex 1 and catalyzes electron transfer from NADH to ubiquinone. Spot 1056 was identified as X. laevis Ckb. Spot 553 was assigned to X. laevis dihydrolipoamide acetyltransferase E2 precursor that is a member of the pyruvate dehydrogenase complex controlling the conversion of pyruvate to acetyl-CoA and NADH. Spot 745 corresponded to nuclear matrix protein 200 (Nmp200) of X. laevis known in mammals to be involved in pre-mRNA splicing, ubiquitylation, and DNA double strand break repair. Finally spots 780, 786, and 797 were all identified in X. laevis as prolyl 4-hydroxylase β (P4hb). P4h is a α2β2 tetramer in which the β subunits are multifunctional polypeptides identical to the enzyme PDI.
DISCUSSION
The molecular mechanisms by which PCBs induce their toxicity during the development of tadpoles remain largely unknown. The present study is the first to investigate the potential effects of relevant environmental concentrations of these pollutants on the protein expression profiles of developing X. laevis tadpoles.
PCBs are known to affect the survival of numerous species, including amphibians. In the present study, the exposure of 5-day pf X. laevis tadpoles to 0.1 and 1 ppm Aroclor 1254 for 72 h did not impair their survival. This observation is in agreement with the data of Fisher et al. (11) that established that the survival of 9-day pf X. laevis tadpoles was not affected by the exposure to 1 ppm Aroclor 1254. The same observation was made on 7-day pf X. laevis tadpoles (10). However, another study conducted on X. laevis highlighted that 18-day pf tadpole exposed to 0.7 ppm Aroclor 1254 for 48 h showed a survival rate around 55% (32). Moreover the incidence of mortality was 10 times higher in the study performed by Zhou et al. (38) in which all tadpoles of 9 days pf died after 4 days of exposure to 1 ppm Aroclor 1254. This heterogeneity of the reported survival rates could be explained by an age-dependent insensitivity to chlorinated compounds in developing tadpoles. Indeed it has been reported that the embryos and tadpoles of green frogs (Rana clamitans), leopard frogs (Rana pipiens), and American toads (Bufo americanus) are 100–1000-fold less sensitive to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced lethality than most fish species (20). The crux of this insensitivity is that TCDD binds with very low affinity to the frog AhR especially during early development (22). Despite the presence of the AhR machinery, high levels of inducer are required to provoke cytochrome P4540 1A1 (CYP1A1) induction (21). The same reason could explain a low affinity of PCBs for the AhR during the early development of tadpoles.
The present study has brought to light that the exposure of X. laevis tadpoles, a relevant aquatic organism used as a model in ecotoxicological and developmental studies, to 0.1 or 1 ppm Aroclor 1254 led to significant changes in the abundance of 59 and 57 protein spots, respectively. The use of mass spectrometry downstream of 2D DIGE allowed the identification of different sets of PCB-responsive proteins. The function of these proteins can provide new clues on the molecular mechanisms by which PCBs induce toxicity during the development of amphibians. The set of proteins commonly identified between both Aroclor 1254 concentrations was limited to six. This uncoordinated response to different concentrations of the same compounds could be explained by the principle that the severity, or the probability of the effect, must be related to the dose or exposure level (40). This is true for the toxicogenomics and toxicoproteomics fields in which investigations of toxicant exposure indicated that dose-dependent changes are currently highlighted. For example, Poynton et al. (41) showed that in Daphnia magna exposed to different concentration of heavy metals each concentration produced a distinct gene expression profile. At the protein level, it has been highlighted that in Mytilus edulis exposed to graded copper concentrations only 11 protein spots were jointly regulated between experimental conditions (27).
Oxidative stress has been postulated to play a role in the toxic manifestations of PCBs and could thus induce an antioxidant response in exposed cells and tissues (42–44). In a previous study, modification of the activity of antioxidant enzymes such as superoxide dismutases, catalase, and glutathione redox cycle enzymes in 5-day pf X. laevis tadpoles exposed to 0.1 and 1 ppm Aroclor 1254 could not be readily observed, but the data do not exclude the induction of other antioxidant systems in response to PCBs (45). This is indeed the case because it was shown in this study that Prx Ι and Prx ΙΙ were up-regulated in tadpoles exposed to 0.1 ppm Aroclor 1254. Prx Ι and Prx ΙΙ are known as stress-inducible antioxidant enzymes as various stress agents are able to up-regulate the genes encoding Prx I and Prx II both in vitro and in vivo. Moreover the stress-inducible Prx I gene is activated through the antioxidant/electrophile response element (ARE/EpRE) present in the promoter region (46). The ARE/EpRE is a cis-acting regulatory element found in the 5′-flanking regions of numerous genes such as detoxifying phase 2 enzymes (several GSTs, heme oxygenase I, and cyclooxygenase 2) and is activated by redox-cycling phenols via the production of reactive oxygen species (47, 48). To our knowledge, very few studies have linked PCBs with the up-regulation of genes activated through the ARE/EpRE. However, Aroclor 1254 significantly induced the expression of the glutathione S-transferase Mu1 (GSTM1) gene, which is regulated through the ARE/EpRE (49), in the gills and digestive tract of the abalone Haliotis discus (50). Moreover some studies on rat hepatoma cells have reported that TCDD, whose chemical structure is similar to co-planar PCBs, induced the expression of different proteins in an ARE-dependent manner (51, 52). Thus, the up-regulation of antioxidant enzymes such as Prx I with ARE-containing gene promoters is compatible with the hypothesis that PCBs could induce oxidative stress.
Endoplasmic reticulum (ER) stress is induced when high levels of misfolded proteins accumulate in the ER, generating the unfolded protein response (UPR). The UPR results in the up-regulation of chaperones such as GRP58, GRP74, GRP98, and PDI to prevent protein aggregation and cell death (53). In the ecotoxicological field, very few studies have described a possible induction of the UPR by toxicants (31, 54–56). In the present report, GRP58, also known as PDIA3, was one of the proteins up-regulated in both Aroclor 1254 conditions, suggesting a possible induction of the UPR by PCBs. PDIA3 is known to be overexpressed in TCDD-sensitive Long-Evans rat (52) and in the thymus of male marmosets (Callithrix jacchus) exposed to dioxin (57). It has also been highlighted that the gene encoding for PDIA3 contains AHREI, AHREII, and ARE motifs (52). Moreover the up-regulation of the proteasomal subunit α type, whose gene contains AHREI-AHREII motifs (19), and LAP3 in the 0.1 ppm condition could be linked to an overproduction of oxidized proteins within the cells. During mild oxidative stress, the 20 S proteasome degrades modified proteins (58). Oxidized proteins are continuously produced in cells as a result of the aerobic metabolism, but the protein oxidation can be increased by xenobiotic exposure (59). However, the link between PCBs and an overproduction of oxidized proteins has not been described in detail. Livingstone (60) reported an increase in oxidative damages such as oxidized proteins in both fish and invertebrates exposed to single or mixed contaminants including PCBs. Our findings on GRP58/PDIA3, the proteasomal subunit α type, and LAP3 are compatible with the hypothesis that PCBs could favor protein oxidation within the ER and cytosol, launching the UPR and increased proteolysis to protect the cell against aggregated and damaged proteins that can provoke cell death.
After exposure to 0.1 ppm Aroclor 1254, tadpoles displayed up-regulation of ENO, Grhpr, and GPD1, all enzymes involved in glycolytic and/or gluconeogenesis pathways. The impact of PCBs on glycolysis and gluconeogenesis has been poorly studied. Glycerol-3-phosphate dehydrogenase is known to be up-regulated in the thymus of male marmosets exposed to TCDD (57). In mice, exposure to PCBs induced an up-regulation of lactate dehydrogenase (61). However, Weber et al. (62) showed a reduced activity of phosphoenolpyruvate carboxykinase in mice exposed to TCDD. Also Kraemer and Schulte (63) highlighted that the exposure of the common mummichog Fundulus heteroclitus to PCBs induced a down-regulation of the equilibrium enzymes of glycolysis and gluconeogenesis. In the reported cases, the down-regulation of the enzymes was hypothesized to play a role in the toxic effects of PCBs, particularly those related to the wasting syndromes. Presently the overexpression of such enzymes could be linked to the increased requirements of both energy and protein synthesis/degradation pathways (64). This hypothesis is strengthened by the fact that Ckm and Ckb are up-regulated in both conditions. Creatine kinases are crucial enzymes for high energy-consuming tissues like the brain, heart, and muscle, and their abundance is commonly correlated with muscle injury. These enzymes work as a buffering system for cellular ATP levels playing a central role in energy metabolism (65). Except from the serum of KANEMI YOSHU patients where high levels of creatine kinase have been correlated with high concentration of PCBs (66), few studies investigated the impact of PCBs on rapid energy metabolism. Finally the effects of PCB exposure should also be evaluated not only on the abundance of these enzymes but also on their enzymatic activities.
In the present study, the average body weight of tadpoles was significantly reduced by the exposure to 1 ppm Aroclor 1254 in agreement with other studies highlighting the interferences of PCBs with the growth of tadpoles (8, 10, 11). In a previous study, the reduction of weight in contaminated tadpoles was linked to an impairment of the energetic pathways in response to an increased energy demand associated with stress (45). Another explanation of the observed body weight reduction might be that PCBs could affect muscle development by interfering with normal myogenesis as actin α, myosin heavy chain α, and α-tropomyosin were down-regulated in tadpoles exposed to 1 ppm Aroclor 1254. Coletti et al. (67) showed that in vitro exposure of a rat myogenic cell line to Aroclor 1254 resulted in a decreased differentiation and fusion of myoblasts into myotubes. This response suggested that developing muscles could also be targets of PCBs. This hypothesis was also expressed by Fisher et al. (11) to explain the obscured or absent myotomal boundaries in the tail muscle of tadpoles exposed to Aroclor 1254. Two general mechanisms can be involved to explain the synthesis of myofibrillar proteins in the immature skeletal muscle: regulation may occur at the level of transcription or at the level of translation (68). The global effect of PCBs on the muscle development regulation has been poorly studied at the molecular level. The gene encoding for cardiac α-actin was reported to be slightly suppressed in the cardiovascular system of zebrafish (Danio rerio) embryos exposed to TCDD (69). However, Borlak and Thum (70) reported an up-regulation of the genes encoding the skeletal α-actin and α-myosin heavy chain in primary cardiomyocytes exposed in vitro to PCBs. Our data suggest a possible down-regulation of some of these genes. However, further analyses monitoring the expression of cytoskeletal genes at both the mRNA and protein levels are required to get a better insight into the response of tadpoles to PCBs.
Other biological functions evenly seem to be affected in tadpoles exposed to PCBs. ALDH7A1, also known as antiquitin in humans, was up-regulated in both Aroclor 1254 conditions. Antiquitin is assumed to play a role in osmoregulation and/or detoxification, but these functions have not been experimentally substantiated in animals (71). However, in plants, antiquitin is known to play a role in detoxification as the enzyme catalyzes the oxidation of endogenous and exogenous aldehydes to their corresponding carboxylic acids (72). Some authors have already reported that PCBs and TCDD are able to induce the expression of genes encoding for aldehydes (19, 52, 73, 74). Moreover those genes are known to contain AHREI, AHREII, and ARE elements and are responsive to the aryl hydrocarbon receptor (52, 75). The present up-regulation of ALDH7A1 might be linked to a detoxification function even if the latter has not yet been confirmed in animals. The data also suggest that even if developing tadpoles are less sensitive to chlorinated compounds because of a low affinity for the AhR putative enzymes involved in detoxification processes such as ALDHs are overexpressed in developing amphibians exposed to PCBs.
CBF-A was originally described as a ubiquitously expressed protein that binds to CArG box motifs (76). CBF-A is also able to bind the Ha-ras element sequence with high affinity in rat carcinoma cells (77). The Ha-ras proto-oncogene is constitutively expressed in all cell types and can be induced in response to a wide number of mitogenic stimuli (78). PCBs are assumed to promote and perhaps to initiate malignant tumor formation. Among other things, PCBs that are AhR agonists show tumor-promoting activity in rodent liver (79). It has already been demonstrated that PCBs increase the expression of genes encoding proto-oncogenes such as Ha-ras (80–82). The present results highlight that PCBs are able to increase the abundance of CBF-A in contaminated tadpoles. However, it would be worthwhile to investigate whether the CBF-A up-regulation could be linked with an up-regulation of Ha-ras mRNA expression promoting carcinogenesis.
In tadpoles exposed to 1 ppm Aroclor 1254, an increase in the abundance of the P4hb subunit was also observed. This subunit is required for the proper tetramer formation of P4h. An increase of the β subunit could thus promote the formation of the active enzyme, essentially favoring collagen biosynthesis (83). However, this hypothesis is in contradiction with previous studies highlighting a reduction of collagen synthesis in the presence of PCBs (84, 85). Because a protein-disulfide isomerase activity has also been assumed for prolyl 4-hydroxylase β (86, 87), the overexpression of the P4hb subunit is in agreement with the up-regulation of GPR58 and reinforces the hypothesis that tadpole cells and tissues exposed to PCBs initiate defensive responses for protection against modified and misfolded proteins.
Lastly the exposure of tadpoles to 1 ppm Aroclor 1254 also induced the overexpression of Nmp200. Nmp200 is up-regulated after exposure to genotoxic agents and seems to be involved in the repair of DNA double strand breaks (88, 89). PCBs are known to be genotoxic as they induce intrachromosomal recombinations in vitro and in vivo. This genotoxicity might be explained by an oxidative stress as oxidation activities linked to the presence of PCBs might induce DNA strand breaks (39). The up-regulation of Nmp200 that we observed supports the genotoxic role of PCBs.
In conclusion, the present study is the first to highlight impacts of environmentally relevant concentrations of a PCB mixture on the proteome of developing tadpoles. It has been shown that PCB toxicity could be related to interactions with well known mechanisms such as oxidative stress, energy metabolism, myogenesis, and UPR. It has also been found that proteins such as ALDH7A1, CBF-A, P4hb, and Nmp200 could be associated with detoxification and toxicity processes in developing amphibians. The comparative analysis of protein data sets enabled selecting only six protein spots commonly up-regulated between both experimental conditions. Those proteins are linked to the UPR, energy metabolism, and detoxification processes, suggesting that those responses are of preferential concern when tadpoles are facing PCB exposure. These data demonstrate that environmentally relevant exposure to PCBs can deeply modify the amphibian proteome and suggest that these changes have to be taken into account while estimating the toxicological hazard of wild amphibian populations exposed to those chemicals.
Supplementary Material
Acknowledgments
We thank Marie-Claire Forget from Unité de Recherche en Biologie des Organismes, University of Namur (Namur, Belgium), Catherine Demazy from Unité de Recherche en Biologie Cellulaire (URBC), University of Namur (Namur, Belgium), and Murielle Louvet from the Laboratoire d'Ecologie animale et d'Ecotoxicologie, University of Liège (Liège, Belgium) for valuable help during biochemical, proteomics, and chemical analysis. We are also grateful to Rachel Madison from the University of California Davis for proofreading the manuscript. The proteomic platform of the URBC is supported by the Fonds National de la Recherche Scientifique/Fonds de la Recherche Fondamentale et Collective.
Footnotes
Published, MCP Papers in Press, November 16, 2008, DOI 10.1074/mcp.M800323-MCP200
The abbreviations used are: PCB, polychlorinated biphenyl; AhR, aryl hydrocarbon receptor; AHRE, aryl hydrocarbon response element; ARE, antioxidant response element; EpRE, electrophile response element; ALDH, aldehyde dehydrogenase; Ckb, creatine kinase brain type; CBF-A, CArG binding factor-A; FETAX, frog embryo teratogenesis assay for Xenopus; GRP, glucose-regulated protein; LAP3, leucine aminopeptidase 3; Ckm, creatine kinase muscle type; Nmp200, nuclear matrix protein 200; PDI, protein-disulfide isomerase; Prx, peroxiredoxin; P4hb, prolyl 4-hydroxylase β; TCDD, 2,3,7,8-tetrachlorodibenzo-p-dioxin; UPR, unfolded protein response; pf, postfertilization; 2D, two-dimensional; ENO, enolase; Grhpr, glyoxylate reductase/hydroxypyruvate reductase; GPD1, glycerol-3-phosphate dehydrogenase 1; Hnrpab, heterogeneous nuclear ribonucleoprotein A/B; ER, endoplasmic reticulum.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Blaustein, A. R., and Dobson, A. ( 2006) Extinctions: a message from the frogs. Nature 439, 143–144 [DOI] [PubMed] [Google Scholar]
- 2.Pasmans, F., Mutschmann, F., Halliday, T., and Zwart, P. ( 2006) Amphibian decline: the urgent need for amphibian research in Europe. Vet. J. 171, 18–19 [DOI] [PubMed] [Google Scholar]
- 3.Stuart, S. N., Chanson, J. S., Cox, N. A., Young, B. E., Rodrigues, A. S., Fischman, D. L., and Waller, R. W. ( 2004) Status and trends of amphibian declines and extinctions worldwide. Science 306, 1783–1786 [DOI] [PubMed] [Google Scholar]
- 4.Glennemeier, K. A., and Denver, R. J. ( 2001) Sublethal effects of chronic exposure to an organochlorine compound on northern leopard frog (Rana pipiens) tadpoles. Environ. Toxicol. 16, 287–297 [PubMed] [Google Scholar]
- 5.Safe, S. H. ( 1994) Polychlorinated biphenyls (PCBs): environmental impact, biochemical and toxic responses, and implications for risk assessment. Crit. Rev. Toxicol. 24, 87–149 [DOI] [PubMed] [Google Scholar]
- 6.Ulbrich, B., and Stahlmann, R. ( 2004) Developmental toxicity of polychlorinated biphenyls (PCBs): a systematic review of experimental data. Arch. Toxicol. 78, 252–268 [DOI] [PubMed] [Google Scholar]
- 7.Savage, W. K., Quimby, F. W., and DeCaprio, A. P. ( 2002) Lethal and sublethal effects of polychlorinated biphenyls on Rana sylvatica tadpoles. Environ. Toxicol. Chem. 21, 168–174 [PubMed] [Google Scholar]
- 8.Gutleb, A. C., Appelman, J., Bronkhorst, M. C., van den Berg, J. H. J., Spenkelink, A., Brouwer, A., and Murk, A. J. ( 1999) Delayed effects of pre- and early-life time exposure to polychlorinated biphenyls on tadpoles of two amphibian species (Xenopus laevis and Rana temporaria). Environ. Toxicol. Pharmacol. 8, 1–14 [DOI] [PubMed] [Google Scholar]
- 9.Gutleb, A. C., Appelman, J., Bronkhorst, M. C., van den Berg, J. H. J., and Murk, A. J. ( 2000) Effects of oral exposure to polychlorinated biphenyls (PCBs) on the development and metamorphosis of two amphibian species (Xenopus laevis and Rana temporaria). Sci. Total Environ. 262, 147–157 [DOI] [PubMed] [Google Scholar]
- 10.Jelaso, A. M., Lehigh-Shirey, E., Predenkiewicz, A., Means, J., and Ide, C. F. ( 2002) Aroclor 1254 alters morphology, survival, and gene expression in Xenopus laevis tadpoles. Environ. Mol. Mutagen. 40, 24–35 [DOI] [PubMed] [Google Scholar]
- 11.Fisher, M. A., Jelaso, A. M., Predenkiewicz, A., Schuster, L., Means, J., and Ide, C. F. ( 2003) Exposure to the polychlorinated biphenyl mixture Aroclor® 1254 alters melanocyte and tail muscle morphology in developing Xenopus laevis tadpoles. Environ. Toxicol. Chem. 22, 321–328 [PubMed] [Google Scholar]
- 12.Lehigh-Shirey, E. A., Jelaso-Langerveld, A., Mihalko, D., and Ide, C. F. ( 2006) Polychlorinated biphenyl exposure delays metamorphosis and alters thyroid hormone system gene expression in developing Xenopus laevis. Environ. Res. 102, 205–214 [DOI] [PubMed] [Google Scholar]
- 13.Linzey, D. W., Burroughs, J., Hudson, L., Marini, M., Robertson, J., Bacon, J., Nagarkatti, M., and Nagarkatti., P. S. ( 2003) Role of environmental pollutants on immune functions, parasitic infections and limb malformations in marine toads and whistling frogs from Bermuda. Int. J. Environ. Health. Res. 13, 125–148 [DOI] [PubMed] [Google Scholar]
- 14.Qin, Z. F., Zhou, J. M., Chu, S. G., and Xu, X. B. ( 2003) Effects of chinese domestic polychlorinated biphenyls (PCBs) on gonadal differentiation in. Xenopus laevis. Environ. Health. Perspect. 111, 553–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin, Z. F., Zhou, J. M., Cong, L., and Xu, X. B. ( 2005) Potential ecotoxic effects of polychlorinated biphenyls on Xenopus laevis. Environ. Toxicol. Chem. 24, 2573–2578 [DOI] [PubMed] [Google Scholar]
- 16.Reeder, A. L., Ruiz, M. O., Pessier, A., Brown, L. E., Levengood, J. M., Phillips, C. A., Wheeler, M. B., Warner, R. E., and Beasley, V. R. ( 2005) Intersexuality and the cricket frog decline: historic and geographic trends. Environ. Health. Perspect. 113, 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denison, M. S., and Nagy, S. R. ( 2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334 [DOI] [PubMed] [Google Scholar]
- 18.Dalton, T. P., Puga, A., and Shertzer, H. G. ( 2002) Induction of cellular oxidative stress by aryl hydrocarbon receptor activation. Chem.-Biol. Interact. 141, 77–95 [DOI] [PubMed] [Google Scholar]
- 19.Boutros, P. C., Moffat, I. D., Franc, M. A., Tijet, N., Tuomisto, J., Pohjanvirta, R., and Okey, A. B. ( 2004) Dioxin-responsive AHRE-ΙΙ gene battery: identification by phylogenetic footprinting. Biochem. Biophys. Res. Commun. 321, 707–715 [DOI] [PubMed] [Google Scholar]
- 20.Jung, R. E., and Walker, M. K. ( 1997) Effects of 2,3,7,8-tetrachlorodibenzo-p dioxin (TCDD) on development of anuran amphibians. Environ. Toxicol. Chem. 16, 230–240 [Google Scholar]
- 21.Bello, S. M., Franks, D. G., Stegeman, J. J., and Hahn, M. E. ( 2001) Acquired resistance to Ah receptor agonists in a population of Atlantic killifish (Fundulus heteroclitus) inhabiting a marine superfund site: in vivo and in vitro studies on the inducibility of xenobiotic metabolizing enzymes. Toxicol. Sci. 60, 77–91 [DOI] [PubMed] [Google Scholar]
- 22.Lavine, J. A., Rowatt, A. J., Klimova, T., Whitington, A. J., Dengler, E., Beck, C., and Powell, W. H. ( 2005) Aryl hydrocarbon receptors in the frog Xenopus laevis: two AhR1 paralogs exhibit low affinity for 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Toxicol. Sci. 88, 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanash, S. ( 2003) Disease proteomics. Nature 422, 226–232 [DOI] [PubMed] [Google Scholar]
- 24.Walgren, J. L., and Thompson, D. C. ( 2004) Application of proteomic technologies in the drug development process. Toxicol. Lett. 149, 377–385 [DOI] [PubMed] [Google Scholar]
- 25.Kimmel, D. G., and Bradley, B. P. ( 2001) Specific proteins response in the calanoid copepod Eurytemora affinis (Poppe, 1880) to salinity and temperature variation. J. Exp. Mar. Biol. Ecol. 266, 135–149 [Google Scholar]
- 26.Shepard, J. L., Olsson, B., Tedengren, B. P., and Bradley, B. P. ( 2000) Protein expression signatures identified in Mytilus edulis exposed to PCBs, copper and salinity stress. Mar. Environ. Res. 50, 337–340 [DOI] [PubMed] [Google Scholar]
- 27.Shepard, J. L., and Bradley, B. P. ( 2000) Protein expression signatures and lysosomal stability in Mytilus edulis exposed to graded copper concentrations. Mar. Environ. Res. 50, 457–463 [DOI] [PubMed] [Google Scholar]
- 28.Hogstrand, C., Balesaria, S., and Glover, C. N. ( 2002) Application of genomics and proteomics for study of the integrated response to zinc exposure in a non-model fish species, the rainbow trout. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 133, 523–535 [DOI] [PubMed] [Google Scholar]
- 29.Shrader, E. A., Henry, T. R., Greely, M. S., and Bradley, B. P. ( 2003) Proteomics in Zebrafish exposed to endocrine disrupting chemicals. Ecotoxicology 12, 485–488 [DOI] [PubMed] [Google Scholar]
- 30.Apraiz, I., Mi, J., and Cristobal, S. ( 2006) Identification of proteomic signatures of exposure to marine pollutants in mussels (Mytilus edulis). Mol. Cell. Proteomics 5, 1274–1285 [DOI] [PubMed] [Google Scholar]
- 31.Silvestre, F., Dierick, J.-F., Dumont, V., Dieu, M., Raes, M., and Devos, P. ( 2006) Differential protein expression profiles in anterior gills of Eriocheir sinensis during acclimation to cadmium. Aquat. Toxicol. 76, 45–58 [DOI] [PubMed] [Google Scholar]
- 32.Jelaso, A. M., Lehigh-Shirey, E., Means, J., and Ide, C. F. ( 2003) Gene expression patterns predict exposure to PCBs in developing Xenopus laevis tadpoles. Environ. Mol. Mutagen. 42, 1–10 [DOI] [PubMed] [Google Scholar]
- 33.Jelaso, A. M., Delong, C., Means, J., and Ide, C. F. ( 2005) Dietary exposure to Aroclor 1254 alters gene expression in Xenopus laevis frogs. Environ. Res. 98, 64–72 [DOI] [PubMed] [Google Scholar]
- 34.Nieuwkoop, P. D., and Faber, J. ( 1994) Normal Table of Xenopus laevis (Daudin), Garland Publishing, Inc., New York
- 35.Rabilloud, T., Strub, J. M., Luche, S., van Dorsselaer, A., and Lunardi, J. ( 2001) A comparison between SYPRO Ruby and ruthenium II tris (bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics 1, 699–704 [DOI] [PubMed] [Google Scholar]
- 36.Hugula, J. L., Philippart, J. C., Kremers, P., Goffinet, G., and Thomé, J. P. ( 1995) PCB contamination of the common barbel, Barbus barbus (Pisces, Cyprinidae), in the river Meuse in relation to hepatic monooxygenase activity and ultrastructural liver changes. Aquat. Ecol. 29, 125–145 [Google Scholar]
- 37.Debier, C., Pomeroy, P. P., Dupont, C., Joiris, C., Comblin, V., Le Boulengé, E., Larondelle, Y., and Thomé, J.-P. ( 2003) Quantitative dynamics of PCB transfer from mother to pup during lactation in UK grey seals Halicheorus grypus. Mar. Ecol. Prog. Ser. 247, 237–248 [Google Scholar]
- 38.Zhou, J. M., Qin, Z. F., Cong, L., and Xu, X. B. ( 2004) Toxicity of PCBs (Aroclor-1221, 1254) to embryos and larvae of Xenopus laevis. Bull. Environ. Contam. Toxicol. 73, 379–384 [DOI] [PubMed] [Google Scholar]
- 39.Schiestl, R. J., Aubrecht, J., Yap, W. Y., Kandikonda, S., and Sidhom, S. ( 1997) Polychlorinated biphenyls and 2,3,7,8-tetrachlorodibenzo-p-dioxin induce intrachromosomal recombination in vitro and in vivo. Cancer. Res. 57, 4378–4383 [PubMed] [Google Scholar]
- 40.Bernard, A. ( 2008) Biomarkers of metal toxicity in population studies: research potential and interpretation issue. J. Toxicol. Environ. Health Part A 71, 1259–1265 [DOI] [PubMed] [Google Scholar]
- 41.Poynton, H. C., Loguinov, A. V., Varshavsky, J. R., Chan, S., Perkins, E. J., and Vulpe, C. D. ( 2008) Gene expression profiling in Daphnia magna part I: concentration-dependent profiles provide support for the no observed transcriptional effect level. Environ. Sci. Technol. 42, 6250–6256 [DOI] [PubMed] [Google Scholar]
- 42.Jin, X., Kennedy, S. W., Di Muccio, T., and Moon, T. W. ( 2001) Role of oxidative stress and antioxidant defense in 3,3′,4,4′,5-pentachlorobiphenyl-induced toxicity and species-differential sensitivity in chicken and duck embryos. Toxicol. Appl. Pharmacol. 172, 241–248 [DOI] [PubMed] [Google Scholar]
- 43.Katynski, A. L., Vijayan, M. M., Kennedy, S. W., and Moon, T. W. ( 2004) 3,3′,4,4′,5-Pentachlorobiphenyl (PCB 126) impacts hepatic lipid peroxidation, membrane fluidity and β-adrenoceptor kinetics in chick embryos. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 137, 81–93 [DOI] [PubMed] [Google Scholar]
- 44.Muthuvel, R., Venkataraman, P., Krishnamoorthy, G., Gunadharini, D. N., Kanagaraj, P., Jone Stanley, A., Srinivasan, N., Balasubramanian, K., Aruldhas, M. M., and Arunakaran, J. ( 2006) Antioxidant effect of ascorbic acid on PCB (Aroclor 1254) induced oxidative stress in hypothalamus of albino rats. Clin. Chim. Acta 365, 297–303 [DOI] [PubMed] [Google Scholar]
- 45.Gillardin, V., Silvestre, F., Divoy, C., Thomé, J.-P., and Kestemont, P. ( 2009) Effects of Aroclor 1254 on oxidative stress in developing Xenopus laevis tadpoles. Ecotoxicol. Environ Saf. 72, 546–551 [DOI] [PubMed] [Google Scholar]
- 46.Ishii, T., and Yanagawa, T. ( 2007) Stress-induced peroxiredoxins. Subcell. Biochem. 44, 375–384 [DOI] [PubMed] [Google Scholar]
- 47.Moelenkamp, J. D., and Johnson, J. A. ( 1999) Activation of antioxidant/electrophile-responsive elements in IMR-32 human neuroblastoma cells. Arch. Biochem. Biophys. 363, 98–106 [DOI] [PubMed] [Google Scholar]
- 48.Chen, C., and Kong, A. N. ( 2004) Dietary chemopreventive compounds and ARE/EpRE signalling. Free. Radic. Biol. Med. 36, 1505–1516 [DOI] [PubMed] [Google Scholar]
- 49.Ohtsuji, M., Katsuoka, F., Kobayashi, A., Aburatani, H., Hayes, J. D., and Yamamoto, M. ( 2008) NRF1 and NRF2 play distinct roles in activation of antioxidant response element-dependent genes. J. Biol. Chem. 283, 33554–33562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan, Q., Whang, I., and Lee, J. ( 2008) Molecular characterization of mu class glutathione-s-transferase from disk abalone (Haliotis discus discus), a potential biomarker of endocrine-disrupting chemicals. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 150, 187–199 [DOI] [PubMed] [Google Scholar]
- 51.Sarioglu, H., Brandner, S., Haberger, M., Jacobsen, C., Lichtmannegger, J., Warmke, M., and Andrae, U. ( 2008) Analysis of 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced proteome changes in 5L rat hepatoma cells reveals novel targets of dioxin action including the mitochondrial apoptosis regulator VDAC2. Mol. Cell. Proteomics 7, 394–410 [DOI] [PubMed] [Google Scholar]
- 52.Pastorelli, R., Carpi, D., Campagna, R., Airoldi, L., Pohjanvirta, R., Viluksela, M., Hakansson, H., Boutros, P. C., Moffat, I. D., Okey, A. B., and Fanelli, R. ( 2006) Differential expression profiling of the hepatic proteome in a rat model of dioxin resistance: correlation with genomic and transcriptomic analyses. Mol. Cell. Proteomics 5, 882–894 [DOI] [PubMed] [Google Scholar]
- 53.Yoshimura, F. K., and Luo, X. ( 2007) Induction of endoplasmic reticulum stress in thymic lymphocytes by the envelope precursor polyprotein of a murine leukemia virus during the preleukemic period. J. Virol. 81, 4374–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hiramatsu, N., Kasai, A., Du, S., Takeda, M., Hayakawa, K., Okamura, M., Yao, J., and Kitamura, M. ( 2007) Rapid, transient induction of ER stress in the liver and kidney after acute exposure to heavy metal: evidence from transgenic sensor mice. FEBS Lett. 581, 2055–2059 [DOI] [PubMed] [Google Scholar]
- 55.Lui, F., Inageda, K., Nishitai, G., and Matsuoka, M. ( 2006) Cadmium induces the expression of Grp78, an endoplasmic reticulum molecular chaperone, in LLC-PK1 renal epithelial cells. Environ. Health. Perspect. 114, 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skandrani, D., Gaubin, Y., Beau, B., Murat, J.-C., Vincent, C., and Croute, F. ( 2006) Effect of selected insecticides on growth rate and stress protein expression in cultured human A549 and SH-SY5Y cells. Toxicol. In Vitro 20, 1378–1386 [DOI] [PubMed] [Google Scholar]
- 57.Oberemm, A., Meckert, C., Brandenburger, L., Herzig, A., Lindner, Y., Kalenberg, K., Krause, A., Ittrich, C., Kopp-Schneider, A., Stahlmann, R., Richter-Reichhelm, H. B., and Gundert-Remy, U. ( 2005) Differential signatures of protein expression in marmoset liver and thymus induced by single-dose TCDD treatment. Toxicology 206, 33–48 [DOI] [PubMed] [Google Scholar]
- 58.Davies, K. J. A. ( 2001) Degradation of oxidized proteins by the 20S proteasome. Biochimie (Paris) 83, 301–310 [DOI] [PubMed] [Google Scholar]
- 59.Gibson, J. D., Pumford, N. R., Samokyszyn, V. M., and Hinson, J. A. ( 1996) Mechanism of acetaminophen-induced hepatotoxicity: covalent binding versus oxidative stress. Chem. Res. Toxicol. 9, 580–585 [DOI] [PubMed] [Google Scholar]
- 60.Livingstone, D. R. ( 2001) Contaminant-stimulated reactive oxygen species production and oxidative damage in aquatic organisms. Mar. Pollut. Bull. 42, 656–666 [DOI] [PubMed] [Google Scholar]
- 61.Buu-Hoi, N. P., Changh, P. H., Sesque, G., Azum-Gelade, M. C., and Saint-Ruf, G. ( 1972) Enzymatic functions as targets of the toxicity of “dioxin” (2,3,7,8-tetrachlorodibenzo-p-dioxin). Naturwissenschaften 59, 173–174 [DOI] [PubMed] [Google Scholar]
- 62.Weber, L. W., Lebofsky, M., Stahl, B. U., Gorski, J. R., Muzi, J. R., and Rozman, K. ( 1991) Reduced activities of key enzymes of gluconeogenesis as possible cause of acute toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in rats. Toxicology 66, 133–144 [DOI] [PubMed] [Google Scholar]
- 63.Kraemer, L. D., and Schulte, P. M. ( 2004) Prior PCB exposure suppresses hypoxia-induced up-regulation of glycolytic enzymes in Fundulus heteroclitus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 139, 23–29 [DOI] [PubMed] [Google Scholar]
- 64.Zhang, D. H., Tai, L. K., Wong, L. L., Chiu, L. L., Sethi, S. K., and Koay, E. S. C. ( 2005) Proteomic study reveals that proteins involved in metabolic and detoxification pathways are highly expressed in HER-2/neu-positive breast cancer. Moll. Cell. Proteomics 4, 1686–1696 [DOI] [PubMed] [Google Scholar]
- 65.Bessman, S. P., and Carpenter, C. L. ( 1985) The creatine-creatine phosphate energy shuttle. Annu. Rev. Biochem. 56, 831–862 [DOI] [PubMed] [Google Scholar]
- 66.Yoshimura, T., Okita, M., Nakano, J., Shiraishi, H., Iwanaga, H., Tomori, K., and Okamoto, M. ( 2003) Elevation of serum creatine kinase and low serum aldolase in the patients with KANEMI YUSHOU. Fukuoka Igaku Zasshi 94, 97–102 [PubMed] [Google Scholar]
- 67.Coletti, D., Palleschi, S., Silvestroni, L., Cannavo, A., Vivarelli, E., Tomei, F., Molinaro, M., and Adamo, S. ( 2001) Polychlorobiphenyls inhibit skeletal muscle differentiation in culture. Toxicol. Appl. Pharmacol. 175, 226–233 [DOI] [PubMed] [Google Scholar]
- 68.Fiorotto, M. L., Davis, T. A., and Reeds, P. J. ( 2000) Regulation of myofibrillar protein turnover during maturation in normal and undernourished rat pups. Am. J. Physiol. 278, R845–R854 [DOI] [PubMed] [Google Scholar]
- 69.Handley-Goldstone, H. M., Grow, M. W., and Stegeman, J. J. ( 2005) Cardiovascular gene expression profiles of dioxin exposure in zebrafish embryos. Toxicol. Sci. 85, 683–693 [DOI] [PubMed] [Google Scholar]
- 70.Borlak, J., and Thum, T. ( 2002) PCBs alter gene expression of nuclear transcription factors and other heart-specific genes in cultures of primary cardiomyocytes: possible implications for cardiotoxicity. Xenobiotica 32, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 71.Fong, W. P., Cheng, C. H. K., and Tang, W. K. ( 2006) Antiquitin, a relatively unexplored member in the superfamily of aldehyde dehydrogenase with diversified physiological functions. CMLS Cell. Mol. Life Sci. 63, 2881–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vasiliou, V., Pappa, A., and Petersen, D. R. ( 2000) Role of aldehyde dehydrogenase in endogenous and xenobiotic metabolism. Chem.-Biol. Interact. 129, 1–19 [DOI] [PubMed] [Google Scholar]
- 73.Dragnev, K. H., Nims, R. W., Fox, S. D., Lindahl, R., and Lubet, R. A. ( 1995) Relative potencies of induction of hepatic drug-metabolizing enzyme genes by individual PCB congeners. Toxicol. Appl. Pharmacol. 132, 334–342 [DOI] [PubMed] [Google Scholar]
- 74.Frueh, F. W., Hayashibara, K. C., Brown, P. O., and Whitlock, J. P., Jr. ( 2001) Use of cDNA microarrays to analyse dioxin-induced changes in human liver gene expression. Toxicol. Lett. 122, 189–203 [DOI] [PubMed] [Google Scholar]
- 75.Takimoto, K., Lindahl, R., and Pitot, H. C. ( 1992) Regulation of 2,3,7,8-tetrachlorodibenzo-p-dioxin-inducible expression of aldehyde dehydrogenase in hepatoma cells. Arch. Biochem. Biophys. 298, 493–497 [DOI] [PubMed] [Google Scholar]
- 76.Kamada, S., and Miwa, T. ( 1992) A protein binding to CArG box motifs and to single-stranded DNA functions as a transcriptional repressor. Gene (Amst.) 119, 229–236 [DOI] [PubMed] [Google Scholar]
- 77.Mikheev, A. M., Inoue, A., Jing, L., Mikheeva, S. A., Li, V., Leanderson, T., and Zarbl, H. ( 2004) Frequent activation of CArG binding factor-A expression and binding in N-methyl-N-nitrosourea-induced rat mammary carcinomas. Breast Cancer Res. Treat. 88, 95–102 [DOI] [PubMed] [Google Scholar]
- 78.McCormick, F. ( 1995) Ras-related proteins in signal transduction and growth control. Mol. Reprod. Dev. 42, 500–506 [DOI] [PubMed] [Google Scholar]
- 79.Buchmann, A., Kunz, W., Wolf, C. R., Oesch, F., and Robertson, L. W. ( 1986) Polychlorinated biphenyls, classified as either phenobarbital- or 3-methylcholanthrene-type inducers of cytochrome P-450, are both hepatic tumor promoters in diethylnitrosamine-initiated rats. Cancer Lett. 32, 243–253 [DOI] [PubMed] [Google Scholar]
- 80.Jenke, H. S., Michel, G., Hornhardt, S., and Berndt, J. ( 1991) Protooncogene expression in rat liver by polychlorinated biphenyls (PCB). Xenobiotica 21, 945–960 [DOI] [PubMed] [Google Scholar]
- 81.Borlakoglu, J. T., Scott, A., Henderson, C. J., Jenke, H. J., and Wolf, C. R. ( 1993) Transplacental transfer of polychlorinated biphenyls induces simultaneously the expression of P450 isoenzymes and the protooncogenes c-Ha-ras and c-raf. Biochem. Pharmacol. 45, 1373–1386 [DOI] [PubMed] [Google Scholar]
- 82.Borlak, J. T., Scott, A., Henderson, C. J., Jenke, H. J., and Wolf, C. R. ( 1996) Transfer of PCBs via lactation simultaneously induces the expression of P450 isoenzymes and the protooncogenes c-Ha-ras and c-raf in neonates. Biochem. Pharmacol. 51, 517–529 [DOI] [PubMed] [Google Scholar]
- 83.Zoeller, J. J., and Iozzo, R. V. ( 2008) Proteomic profiling of endorepellin angiostatic activity on human endothelial cells. Proteome Sci. 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramajayam, G., Sridhar, M., Karthikeyan, S., Lavanya, R., Veni, S., Vignesh, R. C., Ilangovan, R., Sitta Djody, S., Gopalakrishnan, V., Arunakaran, J., and Srinivasan, N. ( 2007) Effects of Aroclor 1254 on femoral bone metabolism in adult male Wistar rats. Toxicology 241, 99–105 [DOI] [PubMed] [Google Scholar]
- 85.Lind, P. M., Larsson, S., Oxlund, H., Håkansson, H., Nyberg, K., Eklund, T., and Örberg, J. ( 2000) Change of bone tissue composition and impaired bone strength in rats exposed to 3,3′,4,4′,5-pentachlorobiphenyl (PCB126). Toxicology 150, 41–51 [DOI] [PubMed] [Google Scholar]
- 86.Lovat, P. E., Corazzari, M., Armstrong, J. L., Martin, S., Pagliarini, V., Hill, D., Brown, A. M., Piacentini, M., Birch-Machin, M. A., and Redfern, C. P. ( 2008) Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res. 68, 5363–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Noiva, R. ( 1999) Protein disulfide isomerase: the multifunctional redox chaperone of the endoplasmic reticulum. Semin. Cell Dev. Biol. 10, 481–493 [DOI] [PubMed] [Google Scholar]
- 88.Mahajan, K. N., and Mitchell, B. S. ( 2003) Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc. Natl. Acad. Sci. U. S. A. 100, 10746–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang, N. R., Kaur, R., Lu, X., Shen, X., Li, L., and Legerski, R. J. ( 2005) The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J. Biol. Chem. 280, 40559–40567 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.