Abstract
Plasma membranes are dynamic compartments with key functions in solute transport, cell shape, and communication between cells and the environment. In mammalian cells and yeast, the plasma membrane has been shown to be compartmented into so-called lipid rafts, which are defined by their resistance to treatment with non-ionic detergents. In plants, the existence of lipid rafts has been postulated, but the precise composition of this membrane compartment is still under debate. Here we were able to experimentally clearly distinguish (i) true sterol-dependent “raft proteins” and (ii) sterol-independent “non-raft” proteins and co-purifying “contaminants” in plant detergent-resistant membranes. We used quantitative proteomics techniques involving 15N metabolic labeling and specific disruption of sterol-rich membrane domains by methyl-β-cyclodextrin. Among the sterol-dependent proteins we found an over-representation of glycosylphosphatidylinositol-anchored proteins. A large fraction of these proteins has functions in cell wall anchoring. We were able to distinguish constant and variable components of plant sterol-rich membrane microdomains based on their responsiveness to the drug methyl-β-cyclodextrin. Predominantly proteins with signaling functions, such as receptor kinases, G-proteins, and calcium signaling proteins, were identified as variable members in plant lipid rafts, whereas cell wall-related proteins and specific proteins with unknown functions make up a core set of sterol-dependent plant plasma membrane proteins. This allows the plant to maintain a balance between static anchoring of cell shape forming elements and variable adjustment to changing external conditions.
Since the first description of microdomains with distinct lipid composition in plasma membranes of mammalian cells (1, 2), the concept of a key role of such “lipid rafts” in cellular processes has been proposed for a range of biological functions. These membrane microdomains are biochemically defined by their resistance to treatment with the non-ionic detergents in low temperature and can be isolated as a distinct fraction in gradient centrifugation.
A role of specific lipid domains composed of sterols and sphingolipids for cellular sorting and trafficking processes was proposed more than 20 years ago (3). It is hypothesized that the distinct lipid composition of lipid rafts creates a special environment that facilitates protein-protein interactions, protein recruitment in cellular trafficking events, and endocytosis and signaling (4–6). Natural sterols and sphingolipids from plants, fungi, or animals have been shown to be capable of inducing the formation of membrane heterogeneities (7). Recent studies on giant blebs produced from cultured mammalian cells allowed visualization of patchy lipid heterogeneity (8). These patches had a temperature-dependent size up to micrometer scales.
In mammalian cells, lipid rafts have been shown to play a role in many different events: endocytosis via caveolae (9), virus budding or pathogen entry (10), regulation of exocytosis (11), actin cytoskeleton organization (12), and apoptosis (13). Cholesterol-dependent segregation of lipid raft proteins from non-raft proteins was visualized in mammalian cells and is consistent with the view that raft domains in the plasma membrane of cells are usually small and highly dispersed, but their size can be modulated by oligomerization of raft components (2).
Although in the mammalian system and in yeast the concept of lipid rafts and their role in cellular processes has been widely studied using a variety of biochemical and cell biological tools, in plants our actual knowledge of the composition and role of such membrane microdomains remained under debate for a long time (14). In recent years, several proteomics studies were carried out on detergent-resistant membrane fractions of Arabidopsis, tobacco, mustard seedlings, or Medicago nodules (15–18). However, most of these studies present long lists of proteins identified in the biochemical preparation of detergent-resistant domains and compare them with whole plasma membrane preparations. Thus, a quantitative proteomics study allowing a distinction of proteins co-purifying in the detergent-resistant membrane (DRM)1 fraction versus true proteins dependent on sterol-rich membrane regions has not yet been carried out in plants.
The aim of this study was to characterize the composition of plasma membrane detergent-resistant domains using comparative quantitative proteomics techniques in combination with selective disruption of the sterol-rich membrane domains by chemical treatment. Thereby a differentiation between co-purifying “non-raft” proteins and sterol-dependent “raft-associated” proteins was possible, giving new insights into the lipid raft model in plants.
Materials and Methods
Arabidopsis Cell Suspension Cultures—
Arabidopsis thaliana Col-0 cell suspension cultures (19) were grown in full medium as described previously (20). For quantitative proteomics experiments, half of the cell cultures were metabolically labeled with 15N by growth under [15N]KNO3 as the only nitrogen source for at least two passages over 2 weeks (21).
Preparation of Plasma Membrane and Detergent-resistant Fraction—
Plasma membranes were purified from the microsomal pellet (100,000 × g) over a two-phase system of dextran and polyethylene glycol (22) using 6.4% dextran, 6.4% polyethylene glycol, 5 mm phosphate buffer, pH 7.8, and 5 mm KCl. Protein concentration was determined by Bradford assay. The plasma membrane pellet was then treated with the non-ionic detergent Triton X-100 for 30 min at 4 °C at a ratio of protein to detergent of 1:15 and shaken at 60 rpm. Treated membranes were then taken up in 2.4 m sucrose to a final concentration of 1.8 m and overlaid by a sucrose step gradient from 1.8 to 0.15 m. After gradient centrifugation at 250,000 × g for 18 h, a ringlike structure was visible at the interface of 1.4 and 0.15 m sucrose concentrations. The fractions of 1 ml above and 1 ml below the interface were collected as the detergent-resistant membrane fraction. The DRM fraction was diluted five times with 10 mm Tris-HCl buffer, pH 8, and membranes were pelleted at 200,000 × g for 1 h.
Isolated plasma membranes (300 μg) were treated with the sterol-disrupting agent methyl-β-cyclodextrin (mβcd) at a final concentration of 5, 15, or 30 mm for 1 h at 37 °C as described previously (23). Membranes were then washed in phosphate buffer and pelleted prior to Triton X-100 treatment for DRM preparation.
Protein Analysis and Identification by LC/MS/MS—
DRMs were pelleted at 100,000 × g. Pellets were resuspended in 6 m urea and 2 m thiourea for denaturation and were subsequently subjected to reduction in DTT, and free SH groups were carbamidomethylated using iodoacetamide (24). This was followed by digestion with Lys-C and with trypsin (24). Digested samples were desalted over C18 Stop And Go Extraction tips (Empore Disk, Varian Inc.) (25).
Tryptic peptide mixtures were analyzed by LC/MS/MS using nanoflow HPLC (Proxeon Biosystems) and an Orbitrap hybrid mass spectrometer (LTQ-Orbitrap, Thermo Electron) as mass analyzer. Peptides were eluted from a 75-μm analytical column (Reprosil C18, Dr. Maisch GmbH) on a linear gradient running from 4 to 64% acetonitrile in 90 min and sprayed directly into the LTQ-Orbitrap mass spectrometer. Proteins were identified by MS/MS by information-dependent acquisition of fragmentation spectra of multiple charged peptides. Up to five data-dependent MS/MS spectra were acquired in the linear ion trap for each FTMS full-scan spectrum acquired at 30,000 full-width half-maximum resolution settings with an overall cycle time of approximately 1 s. Fragment MS/MS spectra from raw files were extracted as DTA files and then merged to peak lists using the default settings of DTASuperCharge version 1.18 (SourceForge, Inc.) with a tolerance for precursor ion detection of 50 ppm.
Fragmentation spectra were searched against a non-redundant Arabidopsis protein database (TAIR8, version April 2008; 31,921 entries) using the Mascot algorithm (version 2.2.0; Matrix Science). The database contained the full Arabidopsis proteome and commonly observed contaminants (human keratin, trypsin, and lysyl endopeptidase); thus no taxonomic restrictions were used during the automated database search. The following search parameters were applied: trypsin as cleaving enzyme; peptide mass tolerance, 10 ppm; MS/MS tolerance, 0.8 Da; and one missed cleavage allowed. Carbamidomethylation of cysteine was set as a fixed modification, and methionine oxidation was chosen as a variable modification. “15N metabolic labeling” was chosen as a quantitative method for Mascot database searching, allowing identification of labeled and unlabeled peptides within the same database search. Only peptides with a length of more than five amino acids were considered.
In general, peptides were accepted without manual interpretation if they displayed a Mascot score greater than 31 (as defined by Mascot p < 0.01 significance threshold). Peptides with a score greater than 24 were manually inspected requiring a series of three consecutive y or b ions to be accepted; in addition, mass accuracy and delta scores were taken into account when single peptides were accepted. Original MS/MS spectra considered for quantitation in this analysis will be deposited in the Promex spectral library, and spectra of accepted “single peptide” hits are included in supplemental Fig. 1. Using the above criteria for protein identification, the rate of false identifications as determined by the “decoy database” function implemented in Mascot version 2.2.0 was 3.4% on a 95% confidence level. The false positive rate in 15N-labeled samples was higher because of increased ambiguity in protein identification resulting from a higher number of isobaric amino acids in labeled samples (26).
Peptide assignment to proteins was done according to the Mascot default settings, i.e. each redundant peptide was primarily assigned to the highest scoring protein. Isoforms of protein only appear in the tables as a separate protein entry if they were assigned at least one unique peptide. A summary of all identified peptides and their respective quantitative values is presented in supplemental Table 2.
Quantitative Protein Analysis—
Ratios between labeled and unlabeled forms of each tryptic peptides were calculated in MSQuant version 1.4.3 (released May 30, 2008; SourceForge, Inc.). Quantitative information was taken from extracted ion chromatograms of the labeled and unlabeled form of each identified peptide. Thereby co-elution of both peptide forms was made a requirement, and it was manually inspected in MSQuant that the pairs of labeled and unlabeled forms actually fit with the expected isotope envelope distributions. Peptides that did not meet these criteria were omitted from the analysis as described previously (21).
Intensity ratios of the labeled 15N form to unlabeled 14N form of each identified peptide were averaged across all peptides belonging to the same protein within one experimental set. Peptides conserved in multiple members of a protein family were identified using the “show subsets” option in Mascot, and the respective peptides present in multiple proteins were excluded from quantitative analysis if the redundant peptides displayed ratios significantly different (p < 0.05; χ2 test) from unique peptides of the same protein (27, 28). Peptides meeting the criteria for sequence identification but for which only 14N forms or only 15N forms were quantified were manually assigned the ratios 0.1 (14N form only) or 10 (15N form only). This affected only peptides for which the pairing labeled or unlabeled peak was at noise level. Because quantitative information was extracted from full-scan spectra with a very low level of noise as obtained in the Orbitrap mass analyzer, no minimum threshold was set for quantitation (29). Protein abundance ratios were converted into log2 values. Only those proteins were considered for further analysis for which intensity ratios were obtained in both paired reciprocal experiments. Ratios of 15N to 14N forms and the respective standard deviation as calculated by MSQuant for each identified peptide and the number of peptides used for quantitation for each protein are presented in supplemental tables. The average relative error of quantitation for all 465 quantified proteins was 10.6%.
Reciprocal Labeling Experimental Setup and Statistical Analysis—
For comparative proteomics analysis, plasma membranes of 15N-labeled cell cultures were treated with mβcd, whereas plasma membranes of control cells were left untreated. In a paired reciprocal experiment using the same starting material, 14N cells were treated with mβcd, whereas 15N cells were left untreated. Labeled and unlabeled plasma membranes were treated with Triton X-100 in one combined sample prior to DRM preparation over a sucrose gradient. The work flow of the experimental setup is shown in Fig. 1. The paired reciprocal experiments (Fig. 1) were independently repeated four times. Protein ratios of each replicate experimental set were z-score-standardized to allow comparison between experiments. In result tables and figures, the averages and S.D. of z-transformed protein ratios from all biological replicate experiments are presented. In general, 60% of all identified proteins were found in at least two independent reciprocal experimental setups, and 25% of all identified proteins were found in all four replicate experiments (supplemental Fig. 2).
Fig. 1.
Work flow of the reciprocal labeling experiments. Two experiments were carried out in parallel. In one case, the 14N cells were subjected to the mβcd treatment, and 15N cells were used as control. In the second case, the 15N cells were used for mβcd treatment, whereas the 14N cells were used as untreated control. In addition, 1:1 mixtures of untreated 14N and 15N cells were used to define inherent differences between the cell cultures and the technical variation. The complete reciprocal experimental design was repeated four times independently. PM, plasma membrane.
The reciprocal labeling setup was chosen over an experimental setup using the same 15N-labeled cultures as repeated internal standard (30) to specifically distinguish which proteins are responding to mβcd (treatment effects) from those proteins that are a priori different between labeled and unlabeled cell cultures (i.e. culture effects) (31).
In summary, the data analysis work flow is based on first determining the variation between cultures based on 15N/14N ratios in independent 1:1 mixtures before mβcd treatment is applied. The ratios in two control experiments showed normal distribution (see Fig. 2B) and were used to define ratio-dependent standard deviations (31). In a second step, the distances to the diagonal in a graphic display of ratios in reciprocal experiments (Fig. 2A) was calculated. Then for each data point the ratio between the distance and the S.D. was calculated, and the p value was determined by a two-tailed t distribution. Subsequently a multiple testing correction was applied to the whole data set using the false discovery rate method introduced by Benjamini and Hochberg (32). Reported proteins correspond to a cutoff false discovery rate of 5%.
Fig. 2.
A, response of proteins to mβcd treatment. Log2 values of 15N to 14N ratios from one experiment were plotted against log2 values of 15N to 14N ratios from the reciprocal experiment. Blue and red symbols indicate those sterol-dependent and sterol-independent proteins that show significant reciprocal response. Yellow symbols indicate those proteins that do not respond to the mβcd treatment (31). B, distribution of ratios from two control experiments (yellow), mβcd-responsive proteins (blue), and other proteins (red). The mβcd-responsive proteins fall into two classes: a core set of very strongly responsive (dark blue) and some less responsive ones (light blue).
Analysis of Sterols by APCI/MS—
Lipids were extracted from equal amounts (150 μg) of mβcd-treated and untreated plasma membrane preparations by a Bligh and Dyer (33) method. A final amount of 10 μg of 3β-hydroxy-5α-cholestane (Sigma-Aldrich) was used as internal standard. Extracted lipids were hydrolyzed with 4 m HCl for 1 h at 80 °C and extracted with 2-hexane. The solvents were evaporated, and samples were diluted in 1:1 chloroform:methanol.
Sterols were subsequently separated by UPLC (Waters) over a C18 column during a 25-min gradient using water and acetonitrile as mobile phases. Eluting peaks were directly ionized by APCI before detection in an LTQ-Orbitrap mass spectrometer (Thermo Fisher). Ions were detected in the m/z range 100–1000, and 500,000 ions were collected in the Orbitrap detector set to a resolution of 30,000 full-width half-maximum. Total ion chromatograms were extracted, and peak volumes were compared relative to the internal standard 3β-hydroxy-5α-cholestane.
Bioinformatics Analysis and Functional Classification—
Proteins were classified to functional groups using the MapMan nomenclature developed for plant genes (34). GPI-anchored proteins were annotated based on published literature and predictions (35–37). Graphs were created using SigmaPlot version 10 (Systat Software), and statistical tests were carried out using Excel Analyze-it package version 2.07.
RESULTS
This study was designed to gain new insights about plant membrane proteins dependent on a sterol-rich membrane environment (so-called “lipid raft-proteins”). The experiments make use of the sterol-disrupting agent mβcd in an untargeted proteomics study. The aim of this work was to disrupt lipid raft-like sterol-rich regions in the plasma membrane and thereby induce a depletion of proteins dependent on a sterol-rich membrane environment. The mβcd-induced depletion from membranes has been described as a characteristic feature for sterol-dependent proteins (38). Thus, peptide intensity ratios between untreated and treated samples will be higher for true sterol-dependent lipid raft proteins (i.e. these proteins will predominantly occur in untreated samples) (23). In contrast, for co-purifying contaminants the intensity ratio from treated and untreated membranes is expected to be close to unity (i.e. these peptides occur in untreated and treated samples in the same amounts).
Effect of mβcd on Plant Plasma Membrane Proteins—
In total, 792 proteins were identified in DRM preparations, and for 465 proteins quantitative information from both reciprocal subsets in at least one independent reciprocal experiment was obtained. About 15% of all the proteins with reciprocal quantitative data were found to be significantly depleted from DRM preparations upon mβcd treatment of plasma membranes (Fig. 2A, blue dots). A full list of proteins that were found to be depleted from DRM preparations upon mβcd treatment in at least one reciprocal experiment is presented in supplemental Table 1. A core set of 37 proteins was identified as mβcd-responsive in DRM preparations of at least two independent reciprocal experimental sets (Table I). These mβcd-responsive proteins are primary candidates for being associated with or located in a sterol-rich membrane microenvironment (lipid rafts). Among all proteins identified as sterol-dependent proteins, we observed a significant (p < 0.03, Fisher exact test) over-representation of single transmembrane domain proteins (supplemental Fig. 3). We found about 2% of the quantified proteins to be enriched in DRM preparations after mβcd treatment in individual experiments (Fig. 2A, red dots).
Table I.
Core list of proteins identified as mβcd-responsive in at least two biological replicate experimental sets
Each protein was functionally classified by MapMan (34). p values indicate the significance of the mβcd response. A cutoff p value of 0.05 was used after correction for multiple testing (32). “DRM” refers to proteins identified as mβcd-responsive based on differential analysis of DRM preparations after mβcd treatment of plasma membranes. “sup” identifies mβcd responsiveness based on release of these proteins to the supernatant after pelleting mβcd-treated plasma membranes. Letters indicate proteins previously published as being GPI-anchored (gpi) (35, 36, 50, 51) or present in DRM in Arabidopsis (drm) (15). Localization is taken from the SUBA database of subcellular localizations (39): PM, plasma membrane; EX, extracellular space; P, plastid; V, vacuole; C, cytosol; N, nucleus; M, mitochondrion. AGI, Arabidopsis gene identifier; DREPP, developmentally regulated plasma membrane polypeptide; v-ATPase, vacuolar type H+-ATPase; LYSM, lysin motif.
| AGI | Description | MapMan functional category | p value | Times found | Experiment | Experimental subcellular localization from SUBA
|
DRM/GPI | |
|---|---|---|---|---|---|---|---|---|
| MS | GFP | |||||||
| At3g07390 | AIR12; extracellular matrix structural constituent | Hormone metabolism | 2.06e−06 | 4 | sup, DRM | ER, EX, P | EX, PM | gpi |
| At4g26690 | MRH5/SHV3; glycerophosphodiester phosphodiesterase/kinase | Lipid metabolism | 2.06e−06 | 4 | sup, DRM | ER, EX, PM, M | EX, PM | gpi |
| At4g31140 | Glycosyl hydrolase family 17 protein | Cell wall | 2.06e−06 | 3 | sup, DRM | ER, EX, PM | EX, PM | gpi |
| At5g58090 | Glycosyl hydrolase family 17 protein | Not assigned, no ontology | 2.06e−06 | 4 | sup, DRM | ER, EX, PM | EX, PM | gpi |
| At5g58480a | Glycosyl hydrolase family 17 protein | Not assigned, no ontology | 2.06e−06 | 2 | DRM | C, ER, EX, PM, M | PM | gpi |
| At3g13560 | Glycosyl hydrolase family 17 protein | Cell wall | 2.54e−06 | 3 | sup, DRM | ER, EX, PM, M | EX, PM | gpi |
| At5g44130 | Fasciclin-like arabinogalactan protein (FLA13) | Cell wall | 7.10e−06 | 6 | sup, DRM | ER, EX, PM, M | EX, PM | gpi |
| At2g25060 | Plastocyanin-like domain-containing protein | Cell wall | 1.29e−05 | 4 | sup, DRM | ER, EX, PM | EX, PM | gpi |
| At4g25240 | SKS1 (SKU5 similar 1); copper ion-binding | Cell wall | 1.56e−05 | 6 | sup, DRM | ER, EX, PM | PM | drm, gpi |
| At5g55480 | Glycerophosphoryldiester phosphodiesterase family protein | Lipid metabolism | 1.56e−05 | 6 | sup, DRM | EX, PM, P, M | EX, PM | drm, gpi |
| At4g31840 | Plastocyanin-like domain-containing protein | Cell wall | 1.72e−05 | 6 | sup, DRM | ER, EX, M, V | PM | gpi |
| At4g12730 | Fasciclin-like arabinogalactan protein (FLA2) | Cell wall | 1.84e−05 | 6 | sup, DRM | ER, EX, PM, M | PM | gpi |
| At3g02740 | Aspartyl protease family protein | Protein degradation | 3.06e−05 | 2 | sup, DRM | ER, EX, V | PM | gpi |
| At1g64760 | Glycosyl hydrolase family 17 protein | Cell wall | 4.62e−05 | 5 | sup, DRM | ER, EX, PM | PM | gpi |
| At5g53870 | Plastocyanin-like domain-containing protein | Cell wall | 5.16e−05 | 4 | sup, DRM | ER, EX, M, N, V | PM | gpi |
| At2g04780 | Fasciclin-like arabinogalactan protein (FLA7) | Cell wall | 9.75e−05 | 6 | sup, DRM | ER, EX, PM | EX, PM | gpi |
| At2g45470 | Fasciclin-like arabinogalactan protein (FLA8) | Cell wall | 2.01e−04 | 5 | sup, DRM | ER, EX | EX, PM | gpi |
| At5g55730 | Fasciclin-like arabinogalactan protein (FLA1) | Cell wall | 2.25e−04 | 5 | sup, DRM | ER, EX, P, M | EX, PM | gpi |
| At3g61260 | DNA-binding family protein/remorin family protein | RNA regulation of transcription | 3.35e−04 | 2 | DRM | N | PM | |
| At3g58100a | Glycosyl hydrolase family 17 protein | Cell wall | 3.99e−04 | 2 | DRM | EX | PM | gpi |
| At2g17120 | LYM2 (LYSM domain GPI-anchored protein 2) | Not assigned, no ontology | 6.53e−04 | 4 | DRM | C, ER, EX, P | EX, PM | gpi |
| At5g51480 | SKS2 (SKU5 similar 2); copper ion binding | Cell wall | 2.20e−03 | 6 | sup, DRM | ER, EX, PM | PM | gpi |
| At2g45820 | DNA-binding protein, putative | RNA regulation of transcription | 3.65e−03 | 4 | sup, DRM | C, N | PM | |
| At5g42100 | Glycosyl hydrolase family 17 protein | Cell wall | 4.64e−03 | 2 | DRM | ER, EX, M | EX, PM | gpi |
| At3g03520a | Phosphoesterase family protein | Not assigned, no ontology | 6.08e−03 | 2 | sup | EX, M, N | V | gpi |
| At4g12420 | SKU5 (skewed 5); copper ion binding | Not assigned, no ontology | 1.28e−02 | 2 | sup | ER, EX, PM | EX, PM | drm, gpi |
| At5g48450 | SKS3 (SKU5 Similar 3); copper ion binding | Cell wall | 1.39e−02 | 2 | DRM | ER; EX, V | PM | |
| At5g25090 | Plastocyanin-like domain-containing protein | Cell wall | 1.88e−02 | 2 | DRM | ER, EX, PM, M | EX, PM | gpi |
| At4g30190 | AHA2 (Arabidopsis H+-ATPase 2); ATPase | Transport, p- and v-ATPases | 1.91e−02 | 2 | sup | C, PM | PM | |
| At2g01210 | Leucine-rich repeat transmembrane protein kinase, putative | Signaling, receptor kinases | 2.05e−02 | 2 | DRM | C, ER, EX, PM | EX, PM, P | |
| At2g05920 | Subtilase family protein | Protein degradation | 2.13e−02 | 2 | DRM | ER, EX, M, P | EX | |
| At4g27520 | Plastocyanin-like domain-containing protein | Cell wall | 2.64e−02 | 3 | sup, DRM | ER, EX, N | EX, PM, P | gpi |
| At1g11820 | Hydrolase, hydrolyzing O-glycosyl compounds | Not assigned, no ontology | 2.99e−02 | 2 | DRM | ER, EX, PM | EX | |
| At2g38940 | ATPT2 (phosphate transporter 2) | Transport phosphate | 3.23e−02 | 3 | sup, DRM | EX, PM, P, M | PM, V | |
| At3g08510 | ATPLC2 (phospholipase C2); phospholipase C | Lipid metabolism | 3.53e−02 | 2 | DRM | C, M, P | PM | |
| At4g36750 | Quinone reductase family protein | Lipid metabolism | 4.54e−02 | 2 | DRM | C, M, N | PM | drm |
| At4g20260 | DREPP plasma membrane polypeptide family protein | Not assigned, no ontology | 4.71e−01 | 4 | sup, DRM | M, N | PM, P | |
Proteins identified based on a single peptide (see supplemental Fig. 1 for annotated spectra).
Specificity of the mβcd Response—
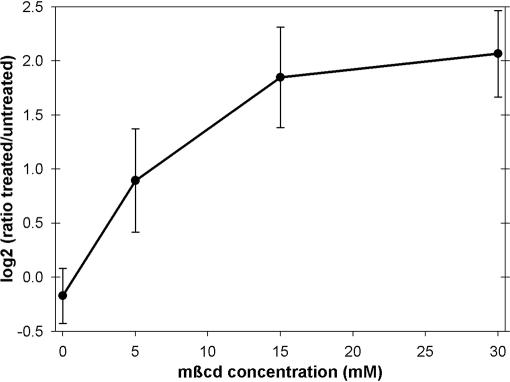
To test whether proteins were removed from the plasma membrane upon mβcd treatment or whether they were rather dissipated within the membrane, mβcd-treated purified plasma membranes were pelleted, and proteins in the supernatant were analyzed. The majority of proteins identified in supernatants after pelleting mβcd-treated plasma membranes were cell wall-related GPI-anchored membrane proteins, remorin, and a phosphate transporter (Table I). An increase in mβcd concentration resulted in an increased release to the supernatant of specifically those proteins that were also classified as sterol-dependent proteins in the reciprocal labeling experiments carried out on DRM preparations. The abundance ratio of these proteins in supernatants of treated versus untreated plasma membranes increased in the mβcd concentration range of 5–15 mm and saturated with an mβcd concentration of 30 mm (Fig. 3). Overall these results support our view that mβcd removes sterol-dependent proteins from the membrane environment in a concentration-dependent manner and that proteins are removed from the membrane by mβcd in a form that does not sediment at 250,000 × g.
Fig. 3.
Concentration dependence of the mβcd-induced depletion of proteins from Arabidopsis plasma membrane. The graph shows the average log2 value (±S.D.) of the 15N to 14N abundance ratios for proteins identified in the supernatant after pelleting of treated versus untreated plasma membranes. Only proteins identified as mβcd-responsive in reciprocal experiments have been considered (n = 37).
Effects of mβcd on Plant Plasma Membrane Sterols—
Sitosterol and campesterol were found to be the most abundant sterols in Arabidopsis plasma membranes as measured by UPLC/APCI/MS (Fig. 4A). Other sterols, such as cholesterol, avenasterols, stigmasterol, and episterol, were present in up to 2 orders of magnitude lower amounts. Upon treatment with mβcd, the identified sterols were removed from DRM preparations in a concentration-dependent manner (Fig. 4B). Treatment of plasma membranes with 30 mm mβcd for 1 h led to a reduction of membrane sterols to about 50% of the levels present in untreated DRM fractions.
Fig. 4.
The effect of mβcd treatment on sterol composition of Arabidopsis plasma membrane. A, abundance of different sterols relative to the standard 3β-hydroxy-5α-cholestane. B, relative changes in sterol content of plasma membrane upon treatment with mβcd at 15 or 30 mm for 1 h. Averages ± S.D. of three biological replicates are shown. d5avenasterol, Δ5-avenasterol; d7avenasterol, Δ7-avenasterol.
Functional Classification of mβcd-responsive Proteins—
Specifically sterol-disrupting treatment of plasma membranes with mβcd caused a clearly significant (p < 0.01; Fisher exact test) depletion of specific classes of proteins from DRM preparations. Fig. 5A displays the distribution of 75 mβcd-responsive and 263 non-responsive proteins to different MapMan functional categories (34). Most (87%) of the proteins showing significant depletion upon mβcd treatments have been localized previously to the plasma membrane by mass spectrometry or by GFP fusions as noted by the SUBA database of subcellular localizations (39). Specifically a third of these responsive plasma membrane proteins has also been annotated as extracellular protein in the SUBA database. In contrast, among the “non-responsive” proteins we found 71% as annotated membrane proteins, and only a total of 3% has been described as extracellular proteins (Fig. 5B).
Fig. 5.
A, distribution of mβcd-responsive proteins (blue) and proteins without an mβcd response (red) to different functional categories. Proteins were classified according to the MapMan classification scheme for plant proteins (34). n = 74 for sterol-dependent proteins and n = 295 for non-responsive proteins. B, subcellular localization from mass spectrometry and/or GFP localization of proteins identified in the mβcd disruption experiments. Subcellular localization was obtained from the SUBA database. RLKs, receptor-like kinases; ABC, ATP-binding cassette; cw, cell wall.
Especially proteins with functions in cell wall structure, lipid modification, vesicle transport, and stress responses and a large number of proteins with unknown functions showed the strongest amplitude in response to mβcd treatment. In contrast, typical co-purifying proteins, such as ribosomal proteins and vacuolar ATPases, were most abundant among proteins that did not show response to mβcd treatment. These and other proteins with no response to mβcd are either sterol-independent DRM proteins or contaminants.
Among the mβcd-responsive (sterol-dependent) proteins a large number of cell wall-related proteins, fasciclin-like arabinogalactan proteins and glycosyl hydrolase family proteins were identified, and most of these proteins have been shown to be GPI-anchored (35). Proteins of primary cell wall synthesis, such as CesA proteins (CesA9 At2g21770 and CesA10 At2g25540) or Cobra (At5g60920) could not be classified because of the lack of a reciprocal quantitative value. Among the non-responsive (non-sterol-dependent) cell wall proteins were mainly proteins with functions in cell wall precursor synthesis and cell wall-degrading enzymes (supplemental Table 1).
Remorins, plant-specific plasma membrane-associated proteins of unknown structure and function, have widely been used as a marker for lipid rafts in plants (16). Two members of this protein family (At2g45820 and At3g61260) have been found as mβcd-responsive in several independent reciprocal experiments. Thus, remorins classify as sterol-dependent proteins in our mβcd treatment experiments.
Surprisingly proteins with functions in signaling such as receptor-like kinases, calcium signaling proteins, and G-protein signaling components showed a strong distribution to non-sterol dependent proteins (not responsive or even enriched by mβcd), and fewer proteins in these functional groups were found to be sterol-dependent (mβcd-responsive) proteins (Fig. 5). In total, four receptor-like kinases (At1g53730, At2g25790, At3g46290, and At5g49760) were identified as members of the sterol-dependent proteins in at least one reciprocal experimental set. In contrast, 10 other receptor-like kinases (At1g25320, At1g29740, At1g73080, At3g24540, At3g28450, At4g08850, At5g46330, At5g46330, At5g65700, and At5g67200) were identified as non-sterol-dependent proteins under the growth conditions for cell cultures in this study. Among non-sterol-dependent receptor kinases, we also found the FLS2 receptor (At5g46330) and two kinases involved in brassinosteroid signaling (At3g13380 and At3g51740). Another 35 receptor-like kinases were identified in DRM fractions but could not be classified into sterol-dependent or non-sterol-dependent proteins as they lacked a reciprocal quantitative value.
Variable and Constant Components of Sterol-rich Membrane Regions—
The identified proteins with signaling functions (receptor-like kinases, calcium signaling proteins, and G-protein signaling proteins) showed a larger experiment-to-experiment deviation from the mean ratio change upon mβcd treatment than did proteins in the functional category of cell wall, lipid-modifying, and vesicle trafficking (Fig. 6). Thus, the effect of mβcd on signaling proteins is significantly more variable for the different proteins within a functional category and independent experimental sets compared with the effect of mβcd on cell wall or lipid-modifying proteins. This is a strong indication for different classes of mβcd-responsive (i.e. sterol-dependent) proteins. We propose that the variability of the mβcd response among signaling proteins is an indication for a more variable and condition-dependent association of signaling proteins with sterol-rich membrane microdomains.
Fig. 6.
A box plot of the deviations from the mean ratio change upon mβcd treatment for each protein in the functional categories “cell wall-related,” “lipid-modifying,” and “vesicle trafficking” as well as for signaling proteins in “receptor kinases,” “calcium signaling,” and “G-protein signaling” across four independent reciprocal experiments is shown. Proteins were classified according to the MapMan classification scheme for plant proteins (34).
DISCUSSION
We used a quantitative proteomics approach to thoroughly characterize sterol-dependent membrane proteins in plant plasma membrane preparations. Our work is the first study on plant DRM that has been carried out up to date allowing such clear differentiation of sterol-dependent and sterol-independent membrane proteins. In addition it reveals novel insights into the concept of the lipid raft model in plants as a dynamic compartment.
Action of mβcd on Plant Plasma Membranes—
The precise effect of mβcd on sterols in the plant has not been studied up to date. Therefore, it is well possible that in our experiments the plant lipid rafts were not completely disrupted by the mβcd treatment as mβcd may not have such a strong effect on plant-specific sterols compared with cholesterol as the dominant sterol in mammalian plasma membranes. It has been shown recently that mβcd acts with different efficiencies on different membrane types, requiring vastly different concentrations and incubation times for different mammalian cell types and tissues (40). It has also been shown on mammalian cells that mβcd removes sterol from the membranes without destroying the bilayer (41).
Our analysis of sterol composition in the DRM preparations after mβcd treatment of plasma membranes indicates that sterols are indeed being removed under the conditions applied in this study. We still observed a strong increase in the removal of sterols when increasing the mβcd concentration from 15 to 30 mm, but this effect was different for individual sterol species. However, we observed a saturation effect on the efficiency with which proteins are being removed from the plasma membrane by mβcd at concentrations between 15 and 30 mm. The mβcd concentration used for the comparative analysis was 25 mm. Thus, it is likely that the mβcd effect on protein was saturated in this study.
Typical contaminants and co-purifying proteins in plasma membrane and DRM preparations such as ribosomal proteins, vacuolar ATPase subunits, or transcription initiation factors (15, 16) did not show any differential response to mβcd treatment. This indicates that the strategy applied here to use depletion of proteins from plasma membranes and DRM by mβcd treatment is a valid means to dissect and characterize the biochemical properties of DRM also in plants. This work is the first detailed global analysis specifically of sterol-rich membrane microdomains in plants, and by its design in reciprocal experiments it allows the precise definition of different categories of proteins in this compartment.
The combination of the analysis of proteins removed from detergent-resistant domain preparations in reciprocal labeling experiments with the subsequent demonstration that these particular proteins are being enriched in the supernatant after pelleting of mβcd-treated membranes is the first evidence in plants that biochemical preparations of detergent-resistant domains and the actual sterol-rich membrane domains (lipid rafts) are not equal. This is an important conclusion drawn from our study, and it is well supported by work from the animal and yeast models where recent findings have strongly suggested that DRMs in standard biochemical preparations are not of the same composition as the in vivo functional lipid rafts in cells (42).
Characterization of Sterol-dependent Proteins—
In our study, cell wall-related proteins as well as lipid- and glucan-modifying proteins were the predominant classes of proteins identified as sterol-dependent in all experiments. In contrast, proteomics studies on mammalian cell cultures using mβcd as a cholesterol-disrupting agent revealed a strong enrichment of signaling proteins, kinases, receptors, and transporters in lipid rafts (23). Although the functional groups of proteins removed by mβcd treatment differ between the animal and plant cells, they share common biochemistry: most proteins depleted after treatment of plant cells are experimentally proven to be GPI-anchored proteins (35, 36). Of the 37 core proteins found as responding to mβcd in plant DRM (Table I), only eight are not modified with a glycosylphosphatidylinositol group, and one of them is predicted to be farnesylated. Furthermore 22 of the GPI-anchored proteins were also found to be released to the supernatant after pelleting of mβcd-treated plasma membrane, and they showed the strongest amplitude of mβcd responses in all experiments. These findings in plants are supported by earlier work on mammalian cells that the GPI anchor may be one of the (important) signals targeting a protein to lipid rafts and that the interaction of these proteins is specific to sterol-rich membrane domains (43–45). Furthermore in agreement with our findings, also quite a large percentage of proteins highly enriched as part of lipid rafts in HeLa cells (23) or human embryonic kidney cells (38) are actually GPI-anchored proteins.
The plant-specific structure of the cell wall requires anchoring and coordinated points of synthesis and modification. In fact, it has clearly been shown in plants that correct membrane targeting of GPI-anchored proteins is essential for cell wall biosynthesis. Mutants with deficient synthesis of GPI anchors display abnormal cell shapes (46). Interestingly in mammalian characterizations of lipid rafts using mβcd, many cytoskeletal proteins were found to be enriched in sterol-rich membrane domains. Thus, possibly in plants the cell wall proteins, which make up the biggest functional group of proteins identified as responding to mβcd, anchor the “plant skeleton” (cell wall) to the plasma membrane in a similar way as mammalian cells anchor their cytoskeleton (47, 48).
The Concept of Constant and Variable Proteins in Sterol-rich Membrane Regions—
Surprisingly a large proportion of proteins published as “lipid raft” proteins as concluded from their proteomics detection in DRM fractions (15, 16), such as many of the receptor-like kinases, did not show drastic depletion from DRM upon mβcd treatment. Moreover we observed an almost equal distribution of signaling proteins to sterol-dependent proteins and not sterol-dependent proteins (Fig. 3). Thus, in our experiments a substantial number of these signaling proteins did not classify as sterol-dependent proteins in our study.
We therefore conclude that sterol-rich membrane regions in plants consist of a “constant” core component mainly containing cell wall-related proteins and lipid-modifying activities and a dynamic component that includes the receptor-like kinases and other signaling proteins. For example, a given receptor-like kinase may be found responsive to mβcd in one experimental set if under these particular conditions it is associated with sterol-rich membrane regions, whereas it may not be classified as mβcd-responsive in the experimental set carried out a few weeks later. Dynamic polarized protein localization has been observed previously in response to pathogen infections (49). When cell cultures are treated with the elicitor peptide flg22, receptor kinase FLS2 displays a strong enrichment in DRM fractions, supporting the concept of stimulation-induced membrane microdomain localization of signaling proteins.2 Thus, diurnal effects of different harvesting times or subtle differences in cell culture conditions may lead to the observed experiment-to-experiment variability in sterol-dependent behavior especially among signaling proteins. The more variable mβcd-responsive proteins such as receptor kinases and transporters may be viewed as condition-dependent raft-associated proteins that may under particular circumstances (e.g. stimulation of any kind) display mβcd-sensitive behavior.
In contrast, cell wall proteins, lipid-modifying proteins, and a set of proteins involved in vesicle transport seem to be a more constant component of plant sterol-rich membrane microdomains independent of external conditions. These findings are in agreement with the model of rafts consisting of a core of mβcd-sensitive proteins, most of which are GPI-anchored, and a peripheral zone with other proteins (38). The majority of proteins we identified in detergent-resistant domains were not responsive to mβcd and thus in our view are not part of the raft model but part of membrane region that is still resistant to Triton X-100 treatment (38).
Conclusion—
In summary, our study is the first comprehensive analysis of plant sterol-rich microdomains making use of the sterol-disrupting drug mβcd, which has already been used in other organisms to characterize and develop models of lipid rafts. We conclusively showed that DRM preparations in plants are not equal to sterol-rich microdomains known as lipid rafts. Rather DRMs include sterol-rich membrane subdomains but also consist of a large fraction of sterol-independent proteins not affected by mβcd treatment. In addition, this work provides the first evidence from plants that the sterol-rich membrane domains are dynamic structures consisting of a core set of structural cell wall-related proteins with GPI anchors and a condition-dependent variable protein component mainly with signaling functions.
Supplementary Material
Acknowledgments
We thank Dirk Hincha and Lothar Willmither for stimulating discussions and critical comments.
Footnotes
Published, MCP Papers in Press, November 25, 2008, DOI 10.1074/mcp.M800346-MCP200
The abbreviations used are: DRM, detergent-resistant membrane; mβcd, methyl-β-cyclodextrin; GPI, glycosylphosphatidylinositol; APCI, atmospheric pressure chemical ionization; UPLC, ultraperformance LC; SUBA database, Arabidopsis Subcellular Database; GFP, green fluorescent protein.
N. Zappel and R. Panstruga, personal communication.
This work was funded by an Emmy-Noether Fellowship of the German Research Foundation (Deutsche Forschungsgemeinschaft) (to W. S.).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Simons, K., and Ikonen, E. ( 1997) Functional rafts in cell membranes. Nature 387, 569–5672 [DOI] [PubMed] [Google Scholar]
- 2.Harder, T., Scheiffele, P., Verkade, P., and Simons, K. ( 1998) Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell. Biol. 141, 929–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karnovsky, M. J., Kleinfeld, A. M., Hoover, R. L., and Klausner, R. D. ( 1982) The concept of lipid domains in membranes. J. Cell Biol. 94, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. A., and London, E. ( 2000) Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275, 17221–17224 [DOI] [PubMed] [Google Scholar]
- 5.Simons, K., and Toomre, D. ( 2000) Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 6.Bickel, P. E. ( 2002) Lipid rafts and insulin signaling. Am. J. Physiol. 282, E1–E10 [DOI] [PubMed] [Google Scholar]
- 7.Xu, X., and London, E. ( 2000) The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry 39, 843–849 [DOI] [PubMed] [Google Scholar]
- 8.Baumgart, T., Hammond, A. T., Sengupta, P., Hess, S. T., Holowka, D. A., Baird, B. A., and Webb, W. W. ( 2007) Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. U. S. A. 104, 3165–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothberg, K. G., Ying, Y. S., Kohlhouse, J. F., Kamen, B. A., and Anderson, R. G. ( 1990) The glycophospholipid-linked folate receptor internalizes folate without entering the clathrin-coated pit endocytotic pathway. J. Cell. Biol. 110, 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fittipaldi, A., Ferrari, A., Zoppe, M., Arcangeli, C., Pellegrini, V., Beltram, F., and Giacca, M. ( 2003) Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J. Biol. Chem. 278, 34141–34149 [DOI] [PubMed] [Google Scholar]
- 11.Salaün, C., James, D. J., and Chamberlain, L. H. ( 2004) Lipid rafts and the regulation of exocytosis. Traffic 5, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falk, J., Thoumine, O., Dequidt, C., Choquet, D., and Faivre-Sarrailh, C. ( 2004) NrCAM coupling to the cytoskeleton depends on multiple protein domains and partitioning into lipid rafts. Mol. Biol. Cell. 15, 4695–4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayllon, V., Fleischer, A., Cayla, X., Garcia, A., and Rebollo, A. ( 2002) Segregation of Bad from lipid rafts is implicated in the induction of apoptosis. J. Immunol. 168, 3387–3393 [DOI] [PubMed] [Google Scholar]
- 14.Bhat, R. A., and Panstruga, R. ( 2005) Lipid rafts in plants. Planta 223, 5–19 [DOI] [PubMed] [Google Scholar]
- 15.Borner, G. H. H., Sherrier, D. J., Weimar, T., Michaelson, L. V., Hawkins, N. D., MacAskill, A., Napier, J. A., Beale, M. H., Lilley, K. S., and Dupree, P. ( 2005) Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mongrand, S., Morel, J., Laroche, J., Caverol, S., Carde, J.-P., Hartmann, M.-A., Bonneu, M., Somon-Plas, F., Lessire, R., and Bessoule, J.-J. ( 2004) Lipid rafts in higher plant cells. Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279, 36277–36286 [DOI] [PubMed] [Google Scholar]
- 17.Morel, J., Claverol, S., Mongrand, S., Furt, F., Fromentin, J., Bessoule, J. J., Blein, J. P., and Simon-Plas, F. ( 2006) Proteomics of plant detergent resistant membranes. Mol. Cell. Proteomics 5, 1396–1411 [DOI] [PubMed] [Google Scholar]
- 18.Lefebvre, B., Furt, F., Hartmann, M. A., Michaelson, L. V., Carde, J. P., Sargueil-Boiron, F., Rossignol, M., Napier, J. A., Cullimore, J., Bessoule, J. J., and Mongrand, S. ( 2007) Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 144, 403–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pauly, M., Eberhard, S., Albersheim, P., Darvill, A., and York, W. S. ( 2001) Effects of the mur1 mutation on xyloglucans produced by suspension-cultured Arabidopsis thaliana cells. Planta 214, 67–74 [DOI] [PubMed] [Google Scholar]
- 20.Jouanneau, J. P., and Peaud-Lenoel, C. ( 1967) Growth and synthesis of proteins in cell suspensions of a kinetin dependent tobacco. Physiol. Plant 20, 834–850 [Google Scholar]
- 21.Engelsberger, W. R., Erban, A., Kopka, J., and Schulze, W. X. ( 2006) Metabolic labeling of plant cell cultures with K15NO3 as a tool for quantitative analysis of proteins and metabolites. Plant Methods 2, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marmagne, A., Salvi, D., Rolland, N., Ephritikhine, G., Joyard, J., and Barbier-Brygoo, H. ( 2006) Purification and fractionation of membranes for proteomic analyses. Methods Mol. Biol. 323, 403–420 [DOI] [PubMed] [Google Scholar]
- 23.Foster, L. J., de Hoog, C., and Mann, M. ( 2003) Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U. S. A. 100, 5813–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen, J. V., Ong, S.-E., and Mann, M. ( 2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3, 608–614 [DOI] [PubMed] [Google Scholar]
- 25.Ishihama, Y., Rappsilber, J., and Mann, M. ( 2006) Modular Stop And Go Extraction Tips with stacked disks for parallel and multidimensional peptide fractionation in proteomics. J. Proteome Res. 5, 988–994 [DOI] [PubMed] [Google Scholar]
- 26.Nelson, C. J., Huttlin, E. L., Hegeman, A. D., Harms, A. C., and Sussman, M. R. ( 2007) Implications of 15N-metabolic labeling for automated peptide identification in Arabidopsis thaliana. Proteomics 8, 1279–1292 [DOI] [PubMed] [Google Scholar]
- 27.Andersen, J. R., Lyon, C. E., Fox, A. H., Leung, A. K. L., Lam, Y. W., Steen, H., Mann, M., and Lamond, A. I. ( 2002) Direct proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 28.Andersen, J. S., Lam, Y. W., Leung, A. K. L., Ong, S.-E., Lyon, C. E., Lamond, A. I., and Mann, M. ( 2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 29.Venable, J. D., Wohlschlegel, J., McClatchy, D. B., Park, S. K., and Yates, J. R., III ( 2007) Relative quantification of stable isotope labeled peptides using a linear ion trap-Orbitrap hybrid mass spectrometer. Anal. Chem. 79, 3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birkemeyer, C., Lüdemann, A., Wagner, C., Erban, A., and Kopka, J. ( 2005) Metabolome analysis: the potential of in vivo labeling with stable isotopes for metabolite profiling. Trends Biotechnol. 23, 28–33 [DOI] [PubMed] [Google Scholar]
- 31.Kierszniowska, S., Walther, D., and Schulze, W. ( 2009) Ratio-dependent significance thresholds in reciprocal 15N-labeling experiments as a robust tool in detection candidate proteins responding to biological treatment. Proteomics, in press [DOI] [PubMed]
- 32.Benjamini, Y., and Hochberg, Y. ( 1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 [Google Scholar]
- 33.Bligh, E. G., and Dyer, W. J. ( 1959) A rapid method for total lipid extraction and purification. Can. J. Biochem. Physiol. 31, 911–917 [DOI] [PubMed] [Google Scholar]
- 34.Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L. A., Rhee, S. Y., and Stitt, M. ( 2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 35.Elortza, F., Nühse, T. S., Foster, L. J., Stensballe, A., Peck, S. C., and Jensen, O. N. ( 2003) Proteomic analysis of glycosylphosphatidylinositol-anchored membrane proteins. Mol. Cell. Proteomics 2, 1261–1270 [DOI] [PubMed] [Google Scholar]
- 36.Elortza, F., Mohammed, S., Bunkenborg, J., Foster, L. J., Nühse, T. S., Brodbeck, U., Peck, S. C., and Jensen, O. N. ( 2006) Modification-specific proteomics of plasma membrane proteins: identification and characterization of glycosylphosphatidylinositol-anchored proteins released upon phospholipase D treatment. J. Proteome Res. 5, 935–943 [DOI] [PubMed] [Google Scholar]
- 37.Marmagne, A., Ferro, M., Meinnel, T., Bruley, C., Kuhn, L., Garin, J., Barbier-Brygoo, H., and Ephritikhine, G. ( 2007) A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 6, 1980–1996 [DOI] [PubMed] [Google Scholar]
- 38.Ilangumaran, S., and Hoessli, D. C. ( 1998) Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 335, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heazlewood, J. L., Verboom, R. E., Tonti-Filippini, J., Small, I., and Millar, A. H. ( 2007) SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res. 35, D213–D218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zidovetzki, R., and Levitan, I. ( 2007) Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim. Biophys. Acta 1768, 1311–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohtani, Y., Irie, T., Uekama, K., Fukunaga, K., and Pitha, J. ( 1989) Differential effects of α-, β- and γ-cyclodextrins on human erythrocytes. Eur. J. Biochem. 186, 17–22 [DOI] [PubMed] [Google Scholar]
- 42.Brown, D. A. ( 2006) Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology 21, 430–439 [DOI] [PubMed] [Google Scholar]
- 43.Cordy, J. M., Hussain, I., Dingwall, C., Hooper, N. M., and Turner, A. J. ( 2003) Exclusively targeting β-secretase to lipid rafts by GPI-anchor addition up-regulates β-site processing of the amyloid precursor protein. Proc. Natl. Acad. Sci. U. S. A. 100, 11735–11740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown, D. A., and Rose, J. K. ( 1992) Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68, 533–544 [DOI] [PubMed] [Google Scholar]
- 45.Legler, D. F., Doucey, M. A., Schneider, P., Chapatte, L., Bender, F. C., and Bron, C. ( 2005) Differential insertion of GPI-anchored GFPs into lipid rafts of live cells. FASEB J. 19, 73–75 [DOI] [PubMed] [Google Scholar]
- 46.Gillmor, C. S., Lukowitz, W., Brininstool, G., Sedbrook, J. C., Hamann, T., Poindexter, P., and Somerville, C. ( 2005) Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17, 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwik, J., Boyle, S., Fooksman, D., Margolis, L., Sheetz, M. P., and Edidin, M. ( 2003) Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. U. S. A. 100, 13964–13969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palazzo, A. F., Eng, C. H., Schlaepfer, D. D., Marcantonio, E. E., and Gundersen, G. G. ( 2004) Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science 303, 836–839 [DOI] [PubMed] [Google Scholar]
- 49.Bhat, R. A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., and Panstruga, R. ( 2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. U. S. A. 102, 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng, Z., Zhang, X., Tang, W., Oses-Prieto, J. A., Suzuki, N., Gendron, J. M., Chen, H., Guan, S., Chalkley, R. J., Peterman, K. T., Burlingame, A. L., and Wang, Z. Y. ( 2007) A proteomic study of brassinosteroid response in Arabidopsis. Mol. Cell. Proteomics 6, 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borner, G. H., Lilley, K. S., Stevens, T. J., and Dupree, P. ( 2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132, 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.