Abstract
Mitogen-activated protein kinases (MAPKs) control neuronal synaptic function; however, little is known about the synaptic substrates regulated by MAPKs. A phosphopeptide library incorporating the MAPK consensus motif (PX(pS/pT)P where pS is phosphoserine and pT is phosphothreonine) was used to raise a phosphospecific antibody that detected MAPK-mediated phosphorylation. The antibody (termed “5557”) recognized a variety of phosphoproteins in the brain, many of which were enriched in postsynaptic density fractions. The immunoblot pattern changed rapidly in response to altered synaptic activity and with the inhibition of specific MAPKs and protein phosphatases. By immunoaffinity purification with 5557 antibody followed by mass spectrometry, we identified 449 putative MAPK substrates of which many appeared dynamically regulated in neuron cultures. Several of the novel candidate MAPK substrates were validated by in vitro phosphorylation assays. Additionally 82 specific phosphorylation sites were identified in 34 proteins, including Ser-447 in δ-catenin, a component of the cadherin adhesion complex. We further raised another phosphospecific antibody to confirm that δ-catenin Ser-447 is modified in neurons by the MAPK JNK in a synaptic activity-dependent manner. Ser-447 phosphorylation by JNK appears to be correlated with δ-catenin degradation, and a δ-catenin mutant defective in Ser-447 phosphorylation showed enhanced ability to promote dendrite branching in cultured neurons. Thus, phosphomotif-based affinity purification is a powerful approach to identify novel substrates of MAPKs in vivo and to reveal functionally significant phosphorylation events.
MAPK1 cascades are well known for their key roles in cell differentiation, proliferation, transcriptional regulation, and development. More recently MAPKs have been implicated in synaptic plasticity in neurons (for a review, see Ref. 1), such as long term potentiation (LTP) and long term depression (LTD), the cellular correlates of learning and memory. In the classical MAPK pathway, which has been characterized primarily in non-neuronal cells, receptor tyrosine kinases or G-protein-coupled receptors signal through small GTPases (e.g. Ras, Rap, or Rac) to multiple tiers of kinases ultimately leading to the activation of the downstream eponymous MAPK. In mammals, three MAPK pathways have been particularly well studied. (i) The prototypical ERKs (ERK1 and ERK2) lie downstream of Ras and Rap1 (2, <3). (ii) The p38 MAPKs (in mammalian brain mainly the isoforms p38α and p38δ) are downstream of Rac1, Cdc42, and Rap1 (2, 4, 5), whereas (iii) the JNKS (mainly JNK1 and JNK3 in brain) are activated by Rac1 and Rap2 (4, 6, 7).
In neurons excitatory synaptic stimulation activates the Ras-ERK pathway (8). ERK activity is necessary (but not sufficient) for LTP in the hippocampus and amygdala and is required for certain memory tasks (2, 9–11). Expression of constitutively active Ras, which activates both ERK1/2 and phosphatidylinositol 3-kinase, is sufficient to induce LTP (2). Recently activation of p38 MAPK (possibly downstream of Rap1) has been implicated in metabotropic glutamate receptor- and NMDA receptor-dependent LTD (2, 5, 11, 12), whereas depotentiation, the depression of recently potentiated synapses, may involve the Rap2-JNK signaling pathway (2, 7). Interestingly hyperphosphorylation of JNK and p38 in neurites surrounding amyloid deposits is a common pathological finding in Alzheimer disease (13) that might contribute to the impairment of LTP by amyloid β peptide (14).
MAPK signaling regulates diverse synaptic functions, such as AMPA receptor trafficking (2, 7, 15) and structural plasticity of dendritic spines (16). Therefore, multiple proteins might be directly regulated by MAPKs at the synapse and in particular within the PSD, the large assembly of signaling and scaffolding molecules that orchestrates the postsynaptic events during synaptic plasticity (17–19). Recently JNK has been reported to phosphorylate AMPA receptor subunits and affect their trafficking (15). In general, however, little is known about the postsynaptic substrates of MAPKs.
Several strategies have been used for identification of kinase substrates (20). Screening for substrates by expression cloning (21) or protein microarrays (22) is prone to false positives. The sequence preference of a kinase determined from phosphorylation of peptide libraries in vitro can be used to scan protein sequences for potential phosphorylation sites but is unreliable by itself (23, 24). MS is a powerful method to discover phosphopeptides in vivo in a large scale and relatively unbiased fashion but cannot identify the kinases involved (25–29). Antibodies raised against a degenerate phosphopeptide mixture representing the consensus phosphorylation site of protein kinase B (Akt) have been used to identify ATP-citrate lyase as an Akt substrate (30, 31). However, phosphomotif antibodies have not been used so far for large scale proteomics identification of kinase substrates.
To discover novel MAPK targets at the synapse, we raised a phosphospecific antibody against a peptide library representing the MAPK consensus phosphorylation motif. Using this antibody we affinity-purified putative MAPK substrates from two different sources: rat brain and cultured hippocampal neurons. Many of the proteins we isolated and identified by sensitive tandem MS are known MAPK substrates or contain excellent consensus MAPK phosphorylation sites. We validated multiple novel candidate MAPK targets with in vitro kinase reactions. More importantly, phosphorylation was confirmed in vivo for a novel phosphorylation site (Ser-447) discovered in δ-catenin, a gene whose deletion is associated with severe cognitive impairment in humans and mice (32). Ser-447 phosphorylation, mediated by JNK MAPK, was bidirectionally regulated by synaptic activity and correlated with δ-catenin stability and function.
EXPERIMENTAL PROCEDURES
Antibodies and DNA Constructs
We purchased antibodies to α-tubulin (mouse, Sigma), β-actin (mouse, Abcam, Cambridge, MA), δ-catenin (mouse, BD Biosciences; and rabbit YV19, Sigma), FLAG (rabbit, Sigma), GFP (rabbit, MBL International, Woburn, MA), Myc (rabbit, Cell Signaling Technology, Danvers, MA), phosphotyrosine (mouse P-Tyr-100, Cell Signaling Technology), and Akt phosphomotif (rabbit 110B7E, Cell Signaling Technology). The PSD-95 antibody (guinea pig) was described previously (33). The Ser(P)-447 δ-catenin-specific antibody was raised against a phosphorylated peptide (CKRTSTAPSpSPGVDSVP where pS is phosphoserine) and affinity-purified on a phosphopeptide column (Sulfolink, Pierce) followed by depletion of non-phosphospecific antibodies on a column coupled with non-phosphorylated cognate peptide.
Plasmids expressing FLAG-Elk-1 (34), GFP-plakophilin 4 (PKP4) (35), and GFP-δ-catenin (36) were kindly provided by Drs. Sharrocks, Hatzfeld, and Kosik, respectively. For expression in neurons, δ-catenin wild type (wt) or S447A were subcloned into a vector with chicken β-actin promoter and N-terminal FLAG tag. The following cDNAs were cloned into GW1 expression vector (British Biotechnology, Oxford, UK) with N-terminal FLAG tag: 4.1N (human, KIAA0338 from Kazusa DNA Research Institute), AbLIM1 (human, KIAA0059), MAGI-2 (human, KIAA0705), Adducin 1 (rat cDNA), and SAPAP4 (rat cDNA).
Phosphomotif Antibody Generation and Purification
Phosphorylated and non-phosphorylated peptide libraries were chemically synthesized with C-terminal cysteine or biotin (see Fig. 1A) and coupled to keyhole limpet hemocyanin for immunization (Covance). The rabbit serum was affinity-purified by negative selection on a streptavidin column (Amersham Biosciences) coupled with non-phosphopeptide library followed by positive selection on a phosphopeptide library column. Bound antibody was eluted with 0.1 m glycine, pH 2.5, 0.5 m NaCl and concentrated by filter centrifugation (Amicon, 10,000 molecular weight cutoff, Millipore, Billerica, MA). For immunoblotting antibody 5557 was used at 0.15 μg/ml in 5% BSA in TBS containing 0.1% Tween 20.
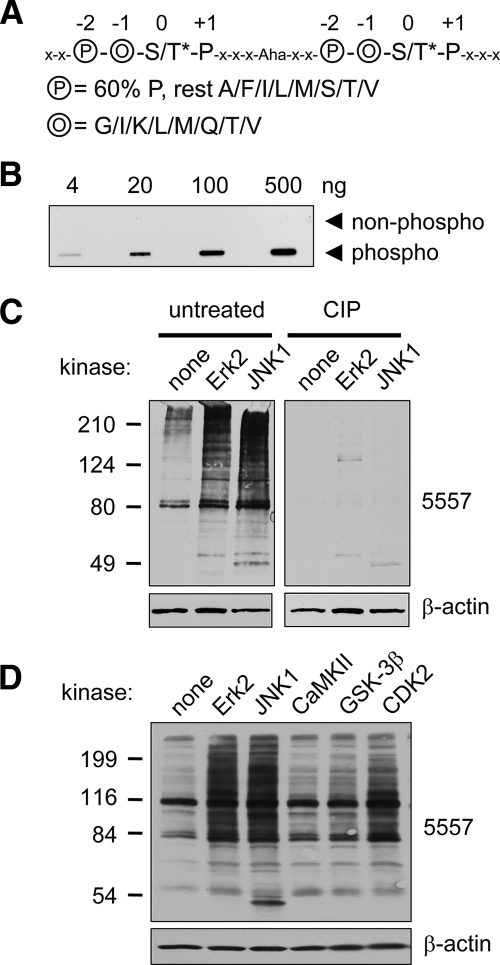
Fig. 1.
Phosphomotif antibody 5557 is phosphospecific and detects MAPK phosphorylation events. A, sequence of phosphopeptide library containing two consecutive consensus MAPK phosphorylation sites used for immunization to produce antibody 5557. The phosphoserine or -threonine (S/T*; 0 position) is followed by proline (1 position) and preceded by two residues, circled O (−1 position; randomly incorporating glycine, isoleucine, lysine, leucine, methionine, glutamine, threonine, or valine) and circled P (−2 position; 60% proline and 40% alanine, phenylalanine, isoleucine, leucine, methionine, serine, threonine, or valine). The linker between the two tandem motifs contains five amino acids (x denotes any amino acid except cysteine) and 6-aminohexoic acid (Aha), which was used to increase the distance between the tandem motifs. B, slot blot showing specificity of affinity-purified antibody 5557 for the phosphopeptide library used for immunization versus an identical library of unphosphorylated peptides. C, PSD1 fraction (prepared in the absence of phosphatase inhibitors) was incubated with recombinant active mitogen-activated protein kinases (30 min at 30 °C) as indicated and immunoblotted with antibody 5557. ERK2 and JNK1 strongly enhanced the 5557 phosphoantibody signal (left panel). Treatment of a duplicate membrane with CIP removed the 5557 signal without affecting β-actin. D, PSD1 fraction was incubated with active ERK2, JNK2, CaMKII (with Ca2+ and calmodulin), GSK-3β, or CDK2 (with cyclin A) for 30 min at 30 °C and immunoblotted with antibody 5557.
Primary Hippocampal and Cortical Neuron Culture, Transfection, and Morphological Analysis
Primary hippocampal cultures were prepared from embryonic day 18 rat brains as described previously (37) and cultured at a density of 80,000/well in 12-well plates. Primary cortical culture was prepared essentially the same way and cultured at a density of 5,500,000 cells/10-cm plate. Hippocampal neurons were transfected with plasmids expressing FLAG-δ-catenin wt or S447A and GFP to outline neuron morphology (37). Neurons were fixed and immunostained with anti-GFP antibody, imaged at 40× magnification (numerical aperture, 1.3) using confocal microscopy (37). Dendritic complexity was analyzed manually as described previously (37).
Preparation of Postsynaptic Density Fractions from Rat Brain
Rat brain was fractionated as described previously (38). Briefly freshly isolated rat forebrain was homogenized in buffer A (5 mm HEPES, pH 7.4, 1 mm MgCl2, 0.5 mm CaCl2, 1 mm NaF, 20 mm β-glycerophosphate, 10 mm pyrophosphate) with a Teflon homogenizer. Postnuclear supernatant (S1) was prepared by pooling the supernatants after a first spin at 1400 × g (10 min) and a second centrifugation (700 × g for 10 min) of the initial pellet resuspended in buffer A. The membrane pellet (P2) after a 13,800 × g spin (10 min) of S1 was resuspended in buffer B (0.32 m sucrose, 6 mm Tris, pH 8.0, 1 mm NaF, 1 mm β-glycerophosphate), loaded on a discontinuous sucrose gradient (0.85 m/1 m/1.15 m sucrose in 8 mm Tris, pH 8), and spun at 82,500 × g (2 h in an SW41 rotor). The synaptosome fraction at the 1 m/1.15 m sucrose interface was harvested by needle puncture and lysed by adding an equal volume of buffer C (6 mm Tris, pH 8.0, 1% Triton X-100). The Triton-insoluble fraction (PSD1) was collected by 33,000 × g centrifugation (1 h) and used for anti-phosphomotif affinity purification (see below). For further fractionation one-third of PSD1 was re-extracted with 0.5% Triton X-100 and spun at 200,000 × g for 1 h to yield PSD2. Another third of PSD1 was extracted with 3% Sarkosyl (in 6 mm Tris, pH = 8.0) and also spun at 200,000 × g to yield PSD3. Protein concentration of all fractions was measured with BCA protein assay (Pierce). All steps were performed at 4 °C in the presence of both protease inhibitor mixture and phosphatase inhibitor mixture I (both 1:200; Sigma). To avoid overloading of the sucrose gradient only one rat forebrain (two for PSD2 and PSD3 preparation) was loaded each run.
Affinity Purification of Phosphoproteins
Synaptosomes from 17 rat forebrains were extracted with 0.5% Triton yielding PSD1 fraction (about 400 μg/brain). PSD1 was boiled for 5 min in 1% SDS (in 150 mm NaCl, 50 mm Tris, 5 mm EDTA, pH 7.2) and then diluted stepwise with 9 volumes of RIPA buffer lacking SDS (1% Triton, 1% deoxycholate, 150 mm NaCl, 10 mm Tris, 5 mm EDTA, 2 mm EGTA, 10 mm β-glycerophosphate, 10 mm pyrophosphate).
A total of 17 10-cm dishes of cortical neuron culture (DIV15–22) treated for 1 h with 0.1 μm okadaic acid (OA; Sigma) or vehicle were Dounce-homogenized in 50 ml of homogenization buffer (0.32 m sucrose, 4 mm HEPES, pH = 7.4, 2 mm EGTA, 10 mm β-glycerophosphate, 10 mm pyrophosphate). Postnuclear supernatant (1000 × g for 10 min) was subjected to 38,500 × g centrifugation (30 min), washed with 30 ml of homogenization buffer, and pelleted again at 38,500 × g to yield crude synaptosomal fraction (P2). Cortical membranes were solubilized in 4 ml of RIPA buffer (as above but also containing 0.1% SDS) yielding about 8 mg of protein.
Insoluble material was removed from rat brain PSD1 and cortical culture P2 lysates by centrifugation (125,000 × g for 25 min). Lysates were precleared with protein A-Sepharose (Sigma) and then immunoprecipitated for 2 h with 50 μg of phosphomotif antibody and 25 μl of protein A-Sepharose slurry. After four consecutive washes with RIPA buffer (1 h total), bound protein was eluted for 1 h with 30 μl of phosphopeptide (10 mg/ml in 20 mm Tris, 300 mm KCl, 1 mm DTT, 0.1% Triton). 2 μl of eluate were analyzed by SDS-PAGE followed by silver staining and comparison with BSA standards by densitometry. All steps were preformed at 4 °C in the presence of both protease and phosphatase inhibitor mixture (1:200; Sigma). The yield of proteins excluding the IgG was ∼4 μg from P2 without okadaic acid treatment, ∼5.5 μg from P2 with the treatment, and ∼2 μg from rat brain PSD1.
Protein Analysis by Mass Spectrometry
Samples were separated for 2 cm by SDS-PAGE to remove the elution peptide and other contaminants. After Coomassie staining, each lane was cut into three pieces to reduce sample complexity (>60 kDa, 50–60 kDa including IgG band, and <50 kDa) followed by in-gel digestion (39). The digested peptides were analyzed by reverse-phase LC-MS/MS (40). The peptide mixtures were dissolved in Buffer A (0.4% acetic acid, 0.005% heptafluorobutyric acid, 5% acetonitrile), loaded onto a 75-μm-inner diameter self-packed column (10 cm of 5-μm C18 resin), and eluted with a 90-min, 10–30% gradient of Buffer B (0.4% acetic acid, 0.005% heptafluorobutyric acid, 95% acetonitrile). The eluted peptides were detected in a high resolution MS survey scan (60,000 resolution) followed by 10 low resolution data-dependent MS/MS scans on an LTQ-Orbitrap hybrid mass spectrometer (Thermo Finnigan). As the Orbitrap had slightly lower sensitivity than the LTQ to detect some weak ions, we reanalyzed the PSD samples using the LTQ to perform MS survey scans.
Acquired MS/MS spectra were searched against the rat reference database of the National Center for Biotechnology Information (Release 21, January 6, 2007) using the SEQUEST algorithm (version 27) (41) as the analyzed samples were prepared from either rat brains or rat neuronal cultures. The common contaminants, such as trypsin and keratins, were added into the database. To evaluate the false discovery rate during the search, all original protein sequences were reversed to generate a decoy database that was concatenated to the original database (42, 43). The total number of protein entries was 48,236. All MS/MS peaks were extracted by the “ExtractMs” program (version 27). Fragment ion m/z ratios were converted to the rounded integer, and therefore m/z tolerance for fragment ions was estimated to be ±0.5 units. Other searching parameters included the mass tolerance of precursor ions (±1.1 units), no enzyme restriction, dynamic mass shifts for oxidized Met (+15.9949 units), phosphorylated Ser/Thr/Tyr (+79.9663 units), four maximal modification sites, and three maximal miscleavages. Only b and y ions were considered during the database match.
To remove false discoveries caused by random matching or spectra of poor quality, we used the number of decoy matches (nd) and the total number of assigned matches (nt) to derive the false discovery rate (FDR = 2 × nd/nt), assuming that the mismatch rate in the original database is the same as in the decoy database (42, 43). Peptide matches were grouped by a combination of trypticity (full, partial, and non-tryptic) and precursor ion charge state (1+, 2+, and 3+). Each group was filtered by mass accuracy (15 ppm for high resolution MS) and dynamically by XCorr (minimal 1.8) and ΔCn (minimal 0.05) values to reduce the false discovery rate to less than 0.5%. As the 0.5% false discoveries were likely found in proteins matched by a single peptide, we validated these peptide matches manually (supplemental Table S5 with links to MS/MS spectra).
In addition, phosphorylation sites in the proteins were identified according to mass shift on Ser/Thr/Tyr residues. As the CID MS/MS spectra of phosphorylated peptides were often less informative because of fewer fragmentation ions, we did not set minimal limitations of XCorr and ΔCn scores. To remove false positives, we filtered phosphopeptides in different groups from non-phosphopeptides according to the target-decoy strategy (42, 43) and lowered the false discovery rate to 0.5%. Furthermore we manually examined all accepted MS/MS spectra as described previously (44) (supplemental Table S2 with links to local MS/MS spectra). For Ser and Thr modification, we removed MS/MS matches without signature phosphate neutral losses (−49 for doubly charged, −32.7 for triply charged, and −24.5 for quadruply charged). If several potential phosphorylation sites were present in the peptides, MS/MS spectra may not enable definitive assignment of modification sites. Therefore, when two top matched phosphopeptides with the same sequence could not be differentiated by ΔCn score (at least 0.08), we considered the phosphorylation site(s) to be ambiguous.
We grouped proteins that shared peptides into clusters as some of the identified peptides were not unique to a single protein (45). This impaired the unequivocal identification of rare proteins detected only by a few peptides. However, clusters sharing multiple peptides mainly consisted of closely related homologs or isoforms. To avoid skewing our further analysis by hand-selecting a single protein within each cluster, we included all possible proteins for the sequence-specific analysis of the complete data set even though we did not identify unique peptides for some of the proteins. This may dilute the true result but might allow detection of rare MAPK substrates.
Label-free Quantification Based on the Integrated Ion Current
Quantitative protein comparison of 5557 immunoprecipitates from untreated and OA-treated cortical cultures was carried out essentially as reported previously (46) in multiple steps using in-house software.
Ion Extraction from MS Scans—
The ion currents for identified peptides were measured in MS survey scans of high resolution based on the isotopic ion selected for MS/MS sequencing. A number of parameters were defined including m/z, charge state, retention time, ion peak range, height, area, and noise level. The noise level for every peak was derived by averaging all peaks in the scan after removing outliers that were at least 2 S.D. away from the mean. All ion currents were then normalized according to the intensity of the noise, assuming that the noise level of MS scans reflected, at least partially, variable ionization efficiency. The peaks used in the analysis had a minimal signal-to-noise ratio of 2.
Peak Alignment between LC Runs—
We first generated two calibration curves for m/z shift and retention time shift along the LC runs based on peptides identified in both runs. Then all detected peaks were matched in comparative runs according to calibrated m/z and retention time. As each pair of gel pieces was analyzed in two consecutive LC-MS/MS runs by an automated system, the variation was generally well controlled (see supplemental Fig. S4). It should be noted that MS survey scans allowed the detection of many peptide ion peaks, but only the most predominant were selected for sequencing by MS/MS analysis. The non-sequenced peaks on MS survey scans were still used for quantification if their matched peptides were sequenced in the comparative runs. If a sequenced peptide could not be matched in the other run, we estimated that the maximum ion current for undetected signal was equal to the noise level and used it to derive the peptide ratio.
Data Integration—
The ratio of every peptide was transformed into logarithmic (log2) values that were averaged over all peptides of a particular protein to determine the enrichment score log2(OA/untreated) and the S.E. (47). If a protein was quantified by both matched peptide ratios and unmatched peptide ratios, only the matched data were averaged.
Statistical Analysis of the Quantitative Data
The statistical analysis was based on null hypothesis as described previously (48). The protein ratios were converted to log ratios (base 2), and the histogram was fitted to Gaussian distribution to evaluate systematic errors (according to the mean) and experimental variations (based on S.D.). The cutoff for significant changes was set to a log2 ratio of 2 (>3.2 S.D.; see Table I). To estimate biological reproducibility, we analyzed one sample from rat brain and two samples from neuronal cell culture. Moreover identified phosphoproteins were evaluated computationally based on the presence of the phosphorylation motif (PX(S/T)P) and MAPK motifs predicted by Scansite at medium stringency level (see Fig. 4C and supplemental Tables S3 and S4). In addition, several of the identified putative substrates were further validated by in vitro kinase assays or by specific phosphoantibodies raised de novo against the identified phosphorylation site.
Table I.
Most promising potential MAPK substrates identified from cortical culture
5557-immunoprecipitated proteins identified from cortical culture were selected based on a high degree of induction upon okadaic acid treatment (log2(OA/untreated) > 2), presence of predicted MAPK phosphorylation site(s) (Scansite, medium stringency), and identification of at least two peptides by MS. For enrichment scores, an S.E. of 0 indicates that the log2 score is derived from a single quantified peptide only. The right column denotes proteins identified previously in the PSD (61). GeneID lists the NCBI gene number for the rat protein, whereas the NCBI gene name is taken from the human homolog. The full list of 410 identified proteins is provided in supplemental Table S3. LIM, Lin-11, Is1-1, Mec-3 zinc-binding domain; FERM = 4.1, Ezrin, Radixin, Moesin protein domain; PTPRF, protein tyrosine phosphatase, receptor type, f polypeptide.
| Accession no. | Human homolog | Description | Peptides | log2(OA/untreated) | Identified in PSD? |
|---|---|---|---|---|---|
| NP_077373.1 | ABI1 | Abl interactor 1 | 3 | 2.01 ± 0.26 | + |
| XP_217645.3 | ABLIM1 | Actin-binding LIM protein 1 long isoform | 9 | 3.34 ± 0.53 | + |
| NP_113968.1 | APBA2 | Amyloid β (A4) precursor protein-binding, family A, member 2 | 2 | 2.11 ± 0.50 | − |
| NP_446192.1 | ARHGEF7 | Rho guanine nucleotide exchange factor 7 | 6 | 2.11 ± 0.43 | + |
| XP_579639.1 | CEP170 | KARP-1 binding protein 1 (long form) | 21 | 2.74 ± 0.25 | − |
| NP_599243.1 | CEP170 | KARP-1 binding protein 1 (short form) | 14 | 3.06 ± 0.32 | − |
| NP_067718.1 | CNKSR2a | Connector enhancer of kinase suppressor of Ras 2; membrane-associated guanylate kinase-interacting protein 2 (MAGUIN2) | 5 | 2.61 ± 0.15 | + |
| XP_227658.3 | FNBP1L | Formin-binding protein 1-like, TOCA1 | 4 | 2.17 ± 0.70 | − |
| XP_240734.3 | FRMD4A | FERM domain-containing protein 4 | 25 | 3.49 ± 0.20 | − |
| XP_232212.3 | FRMD4B | FERM domain-containing 4B/GRP1 binding protein GRSP1 | 2 | 3.80 ± 0.03 | − |
| NP_445869.1 | GAB2 | Growth factor receptor-bound protein 2-associated protein 2 | 2 | 2.73 ± 01p | − |
| NP_067690.1 | HCN4 | Hyperpolarization-activated, cyclic nucleotide-gated K+ channel 4 | 3 | 2.87 ± 01p | − |
| XP_576105.1 | KIAA0284 | FAM68C, homolog of KARP-1; forkhead association domain | 11 | 2.19 ± 0.18 | + |
| XP_576303.1 | KIAA1688 | RhoGAP, MyTH4 and PRP4 domains | 7 | 3.24 ± 0.33 | + |
| XP_343352.2 | KIAA1731 | KIAA1731, no domains, no close homolog | 2 | 4.04 ± 1.01 | − |
| XP_220446.3 | LARP | La ribonucleoprotein domain family, member 1 | 6 | 2.86 ± 0.26 | − |
| NP_851603.1 | MAST1 | Microtubule-associated serine/threonine kinase 1 | 8 | 2.76 ± 0.16 | − |
| XP_343403.2 | NEO1 | Neogenin | 28 | 2.49 ± 0.19 | + |
| XP_215733.3 | PKP4a | Plakophilin 4 | 13 | 2.74 ± 0.26 | + |
| XP_341857.2 | PPFIA3 | PTPRF-interacting protein α3, liprin-α3 | 8 | 2.51 ± 0.41 | + |
| NP_113916.1 | SNAP91 | Synaptosomal-associated protein, 91 kDa | 17 | 2.83 ± 0.22 | + |
| NP_062251.1 | SNIPa | SNAP-25-interacting protein | 44 | 3.88 ± 0.21 | + |
| XP_233820.3 | SOS1 | Son of sevenless 1 | 21 | 3.57 ± 0.20 | + |
| NP_062021.1 | STRN | Striatin, calmodulin-binding protein | 2 | 3.10 ± 0.65 | + |
| NP_446199.2 | UBQLN1 | Ubiquilin 1 | 3 | 2.46 ± 0.80 | − |
| XP_228806.3 | UBQLN2 | Ubiquilin 2 | 11 | 2.71 ± 0.19 | − |
| XP_227405.2 | UBQLN4 | Ubiquilin 4, ataxin-1 ubiquitin-like interacting protein (A1Up) | 3 | 2.46 ± 0.80 | − |
| XP_227587.3 | WDR47 | KIAA0893; WD-40 domain | 8 | 2.15 ± 0.36 | + |
Protein is also listed in Table II.
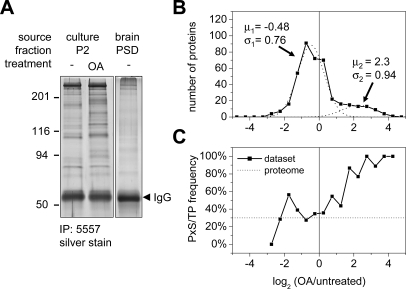
Fig. 4.
Identification of putative MAPK substrates from brain PSD and neuron culture by 5557 immunoprecipitation. A, proteins immunoprecipitated (IP) by antibody 5557 from cortical culture (P2 fraction solubilized in RIPA buffer) or rat brain (PSD1 fraction solubilized by boiling in 1% SDS) were visualized and quantified by silver staining. Okadaic acid treatment of cortical culture (0.1 μm, 1 h) changed the pattern immunopurified by 5557 (lane 2). B, distribution of the abundance ratio log2(OA/untreated) of proteins identified in OA-treated versus untreated cortical culture as an index for OA-induced protein phosphorylation. For all characteristic peptides of a protein the intensity ratios from MS survey scans were transformed into logarithmic (log2) values and averaged. All proteins were binned into 0.5-unit intervals. The distribution was fitted with two summed Gaussian distributions (dashed lines; parameters as indicated: μ1/2, mean, σ1/2, S.D.). C, the percentage of proteins containing potential MAPK motifs (PX(S/T)P) increases with OA-induced enrichment. For comparison the dotted line denotes the basal frequency of 30% within the rat proteome (10,403 of 35,015; NCBI rat reference sequences, May 2007).
In Vitro Kinase Reaction
Epitope-tagged protein was transiently expressed in HEK293 cells (one 10-cm plate), extracted with RIPA buffer (see above), and immunoprecipitated (1–2 h) with epitope tag antibodies and protein A-Sepharose. The beads were washed three times with RIPA buffer followed by MAPK reaction buffer (50 mm Tris, pH = 7.5, 10 mm MgCl2, 1 mm EGTA, 2 mm DTT, 0.01% Brij 35; New England Biolabs) and split into three equal parts. Purified protein on beads was incubated at 30 °C for 30 min in 20 μl of reaction buffer with 5 μCi of [γ-32P]ATP, 100 μm ATP, and active purified ERK2 (100 units; New England Biolabs), JNK1 (200 ng; Millipore), or no kinase. Phosphorylation was detected by autoradiography. Similarly PSD1 fraction (30 μg) was phosphorylated with active ERK2, JNK1, GSK-3β, CaMKII (100 units with Ca2+/calmodulin; New England Biolabs), or cyclin-dependent kinase 2 (CDK2) (100 units with cyclin A; New England Biolabs) in the presence of 100 μm unlabeled ATP and detected by 5557 immunoblotting.
Dephosphorylation with Calf Intestine Phosphatase (CIP)—
For dephosphorylation of phosphoproteins on PVDF membranes, the blots were incubated overnight at 37 °C in CIP buffer with 100 units/ml CIP (Roche Applied Science) after blocking with 5% skim milk and then subjected to the standard Western immunoblotting protocol. A duplicate control membrane was incubated without CIP and then developed in parallel.
RESULTS
Generation and Validation of MAPK Phosphomotif Antibody—
All MAPK family members phosphorylate serine or threonine residues that immediately precede proline within a similar motif of four to nine amino acids. We used a phosphopeptide library representing the core consensus motif of four amino acids to raise a phosphomotif antibody that should recognize proteins phosphorylated by MAPKs (Fig. 1A). Apart from the essential proline (+1 position) following the phosphoserine or phosphothreonine residue (0 position; 50% phosphothreonine and 50% phosphoserine relative abundance in the library), a proline at the −2 position is most favorable for MAPK phosphorylation (23, 49) (Fig. 1A). Therefore, we strongly biased our peptide library such that the −2 position incorporated 60% proline and 40% eight other amino acids (evenly shared between alanine, isoleucine, methionine, leucine, phenylalanine, serine, threonine, and valine). The −1 position consisted of glutamine, glycine, isoleucine, methionine, lysine, threonine, and valine, reflecting the known sequence preference. To increase peptide length and immunogenicity we concatemerized two consecutive MAPK motifs flanked by random residues (labeled “x”) and separated with a single 6-aminohexoic acid spacer (Fig. 1A). Such phosphopeptides should mimic the sites phosphorylated by MAPKs and possibly other proline-directed kinases such as CDK5 that have a partially overlapping consensus motif (49). As a negative control, an equivalent peptide library was also made with non-phosphorylated threonine and serine in the 0 position.
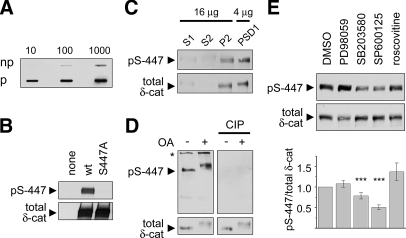
The resulting phosphopeptide library of 20 residues in length (coupled to keyhole limpet hemocyanin) was used to immunize rabbits. The resulting immune serum was first passed through a non-phosphopeptide column and then affinity-purified on a column coupled to the phosphopeptide library. The purified MAPK phosphomotif antibody, termed “5557,” readily detected 4 ng of the phosphopeptide library on “slot blots” but did not react with even 500 ng of the non-phosphorylated library (Fig. 1B). Thus the 5557 antibody is specific for phosphorylated MAPK motif peptides.
To test the antibody on actual MAPK phosphorylation sites, we subjected PSD fractions (PSD1; Fig. 1C) from rat brain to in vitro kinase reaction with recombinant MAPKs ERK2 and JNK1. Immunoblotting with MAPK phosphomotif antibody 5557 revealed significant immunoreactivity in “basal” PSD1 and a dramatic signal increase in PSD fractions incubated with ERK2 and JNK1 (Fig. 1C, left panel). These results demonstrate a plethora of potential MAPK substrates in the PSD (at least under in vitro conditions). Treatment of a duplicate membrane with CIP abolished the kinase-evoked signal as well as the basal immunoreactivity in the PSD (Fig. 1C, right panel). CIP did not affect β-actin immunoreactivity, demonstrating that proteins remain intact on the membrane.
To test the specificity of 5557 antibody for phosphorylation by MAPKs, we incubated the PSD1 fraction with other serine/threonine kinases (Fig. 1D). Incubation with CaMKII (in the presence of Ca2+ and calmodulin) or with GSK-3β barely increased the 5557 signal on immunoblots (CaMKII and GSK-3β phosphorylate RXX(S/T) and (S/T)XXX(pS/pT) motifs at the S/T residues printed in bold, respectively). CDK2/cyclin A, a proline-directed kinase that regulates the cell cycle, moderately increased the 5557 signal when incubated with PSD fractions in vitro. This finding is consistent with the motif preferred by CDK2 ((S/T)PX(R/K)); however, the cell cycle-related cyclin-dependent kinases are not believed to function in postmitotic neurons. Overall these data indicate that phosphoantibody 5557 has specificity for proteins phosphorylated at proline-directed sites and a preference for MAPK phosphomotifs (see also below).
The MAPK Phosphomotif Is Enriched in PSD Fractions—
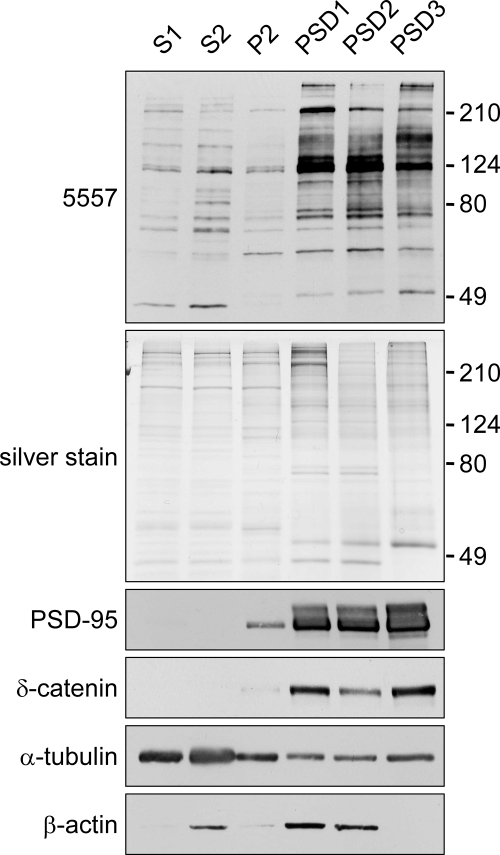
On immunoblots of rat brain fractions, the 5557 phosphomotif antibody detected a weak set of bands in S1 (postnuclear supernatant), S2 (cytosol plus light membranes fraction), and P2 (synaptosome-enriched fraction) (Fig. 2, top panel). Much stronger signals were found in PSD fractions (PSD1–3), suggesting that MAPK-phosphorylated proteins are highly enriched in PSD. Notably consecutive Triton X-100 extraction (PSD2) or Sarkosyl extraction (PSD3) did not deplete 5557 immunoreactivity from PSD fractions (Fig. 2), whereas most of the β-actin was extracted. These data indicate that many MAPK phosphomotif-containing proteins are concentrated in the “core” of the PSD, similar to the fractionation pattern of δ-catenin and the prototypical PSD scaffold PSD-95. Phosphotyrosine-containing proteins, as recognized by a phosphotyrosine antibody (P-Tyr-100), were also enriched in the PSD (supplemental Fig. S1). In contrast, putative Akt-phosphorylated proteins (as recognized by an antibody raised against the Akt motif RXRXX(pS/pT)) were not enriched in the PSD (supplemental Fig. S1).
Fig. 2.
Enrichment of putative MAPK-phosphorylated proteins in postsynaptic density fractions. Shown is an immunoblot of rat brain fractions with MAPK phosphomotif antibody 5557. Brain extracts were fractionated by discontinuous sucrose gradient and detergent extraction into postnuclear supernatant (S1), cytosol plus light membranes (S2), crude synaptosomal fraction (P2), and sucrose gradient-purified synaptosomes extracted once or twice with 0.5% Triton X-100 (PSD1 and PSD2, respectively) or extracted once with 0.5% Triton X-100 followed by 3% Sarkosyl extraction (PSD3). Fractions (5 μg of protein) were immunoblotted with the indicated antibodies or visualized by silver stain. Phosphatase inhibitors were present throughout the PSD fractionation, hence the slightly different pattern compared with Fig. 1C.
Regulation of MAPK Phosphorylation Pattern in Cultured Neurons—
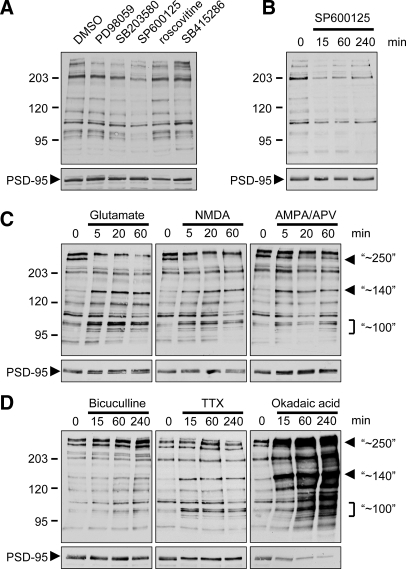
By immunoblotting the 5557 MAPK phosphomotif antibody also detected many bands in lysates of mature hippocampal neuron cultures (DIV21–28) (Fig. 3A). To determine which kinases contribute to this basal phosphorylation pattern we treated cultures with various drug inhibitors of protein kinases. Inhibiting JNK with SP600125 (20 μm; 16 h) broadly reduced 5557 signal (Fig. 3A). Most of this effect occurred within 15 min (Fig. 3B), suggesting that a subset of JNK-mediated phosphorylation sites are rapidly turned over in neurons. Blocking the ERK1/2 pathway with MEK1 inhibitor PD98059 (25 μm), p38 with SB203580 (5 μm), CDK5 with roscovitine (10 μm), or GSK-3β with SB415286 (20 μm) did not decrease 5557 immunoreactivity even after 16 h (Fig. 3A). The 5557 immunoblot signal decreased in a dose-dependent fashion with SP600125 (4-h treatment), ∼70% reduction at 20 μm and ∼50% at 10 μm SP600125 (supplemental Fig. S2). These data are consistent with the reported IC50 value (5–10 μm) of SP600125 for c-Jun phosphorylation (50). Based on these results, JNK MAPKs appear to be responsible for more than half of the phosphosignal recognized by 5557 under basal conditions; this is consistent with the high basal activity of JNK in neurons (51).
Fig. 3.
Effect of synaptic activity and kinase inhibitors on phosphoproteins detected by MAPK phosphomotif 5557 antibody in cultured neurons. Hippocampal neurons (DIV21–25) were treated with various drugs as indicated. Total lysates were immunoblotted with MAPK phosphomotif antibody 5557 and with PSD-95 antibody (below). A, chronic treatment (16 h) with kinase inhibitors as indicated (50 μm PD98059, 5 μm SB203580, 20 μm SP600125, 10 μm roscovitine, and 20 μm SB4152286). B–D, time course with JNK inhibitor SP600125 (B), 100 μm glutamate, 50 μm NMDA, or 100 μm AMPA + 50 μm aminophosphonovaleric acid (APV) (C), and 50 μm bicuculline, 3 μm TTX, or 0.1 μm OA. D, major bands that change with several different treatments are marked with arrowheads or brackets and labeled with the approximate molecular mass (in kDa).
Next we used the 5557 MAPK phosphomotif antibody to explore how the phosphorylation pattern of putative MAPK substrates in neurons is altered by synaptic activity. Stimulation with glutamate (100 μm) or NMDA (50 μm) induced similar rapid changes. Most obviously, several bands around 100 kDa and a single band of ∼140 kDa were strongly induced within 5 min (implying enhanced phosphorylation), whereas a band around 250–300 kDa disappeared with similar kinetics (indicating dephosphorylation) (Fig. 3C). Stimulation of AMPA receptors while blocking NMDA receptors (AMPA + aminophosphonovaleric acid) induced the ∼140-kDa band and some bands in the ∼100-kDa cluster but largely spared the high molecular mass (∼250-kDa) band. Thus activation of NMDA and AMPA receptors has differential effects on MAPK motif phosphorylation.
We also stimulated synaptic activity in cultured neurons by blocking γ-aminobutyric acid type A receptors (bicuculline) or inhibited activity by blocking voltage-gated Na+ channels (tetrodotoxin (TTX)). Bicuculline (50 μm; 15–240 min) had little effect on most bands detected by 5557, but it somewhat induced the two major high molecular mass bands around 250–300 kDa (Fig. 3D); this is in contrast to bath application of NMDA or glutamate (Fig. 3C). TTX (3 μm; 15–240 min) induced phosphorylation of the ∼100- and ∼140-kDa bands similarly to glutamate receptor stimulation but had no effect on the ∼250–300-kDa proteins.
Inhibiting protein phosphatases PP1 and PP2A with OA (100 nm) increased the 5557 signal strongly and progressively over 15–240 min (Fig. 3D). Although most bands were affected, the induction by OA was particularly marked for the bands of ∼100-, ∼140-, and ∼250–300-kDa size that are altered by manipulation of synaptic activity. Adding JNK inhibitor SP600125 10 min before OA treatment halved the increase in 5557 immunoreactivity induced by OA, whereas inhibition of ERK, p38, or CDK5 individually had no significant effect (supplemental Fig. S3). However, blocking JNK, ERK, and p38 in combination prevented the OA-induced phosphorylation almost completely. The rapidity of changes induced by SP600125 and OA implies that there is a high rate of continuous phosphorylation and dephosphorylation of many JNK substrates in steady-state basal conditions.
Affinity Purification and Identification of Putative MAPK Substrates—
To identify potential specific MAPK substrates, we used 5557 phosphoantibody to affinity purify proteins from rat brain or cultured cortical neurons. To increase our yield of synaptic substrates of MAPKs from rat brain, we used the PSD1 fraction, which is highly enriched for 5557-immunoreactive phosphoproteins (see Fig. 2). Prior to immunoprecipitation with 5557 antibody, the PSD was solubilized in 1% SDS to disrupt protein-protein interactions. For the immunoprecipitation from cortical cultures, we used P2 membrane fractions extracted with RIPA buffer (1% Triton X-100, 0.5% deoxycholate) instead of the sucrose gradient-isolated PSD fraction because of limited mass of starting material. Additionally we treated the cultures with protein phosphatase inhibitor OA (100 nm for 1 h) to increase the level of dynamically phosphorylated proteins. After OA treatment, the pattern of 5557-immunoprecipitated proteins changed considerably, and the overall amount increased as visualized by silver staining (Fig. 4A). A protein whose MAPK phosphorylation is increased by OA treatment should be immunoprecipitated to a greater extent by the 5557 phosphomotif antibody after OA treatment. Therefore we estimated the relative abundance of individual proteins immunoprecipitated from untreated versus OA-treated cortical cultures by quantitative MS (see below).
The proteins immunoprecipitated by 5557 antibody from cortical cultures or brain PSD were eluted by competition with excess phosphopeptide (10 mg/ml) and separated by SDS-PAGE (Fig. 4A) followed by in-gel trypsin digestion and LC-MS/MS analysis. After analyzing MS/MS spectra, we accepted a total of 1997 peptide sequences (including 85 phosphorylated peptides representing 82 phosphorylation sites) from up to 449 proteins. The proteins identified from these peptide sequences should be highly enriched for proteins containing the phosphorylated MAPK motif and thus represent putative substrates for MAPKs. Because a few common tryptic peptides are produced by similar or related proteins, we grouped proteins that share peptides into clusters (45). Most clusters (70%) contain only a single protein identified by unique peptide sequences (supplemental Tables S3 and S4).
In 5557 immunoprecipitates from untreated and OA-treated cortical cultures, we identified 224 proteins or clusters of proteins, representing up to 412 proteins in total. 251 proteins were found in both untreated and OA-treated conditions. From the brain PSD fraction, MS analysis of 5557 immunoprecipitates yielded 70 clusters containing up to 115 proteins. There was an overlap of 75 proteins between the cortical culture and brain PSD data sets; i.e. approximately two-thirds of the proteins identified in the smaller PSD data set were also identified from the cortical cultures. Importantly we identified several known MAPK substrates from cortical culture and PSD, e.g. microtubule-associated proteins MAP1B, MAP2, and tau (supplemental Table S1).
Label-free Quantitative Analysis of Proteins Immunopurified by Antibody 5557—
Phosphatase inhibition by OA should increase the levels of a subset of phosphoproteins in cortical cultures, including some putative MAPK substrates. This should be reflected in altered abundance of specific proteins immunopurified by antibody 5557. Indeed several proteins were robustly identified by MS with multiple peptide hits in the affinity-purified fraction from OA-treated but not from untreated neurons (e.g. SNIP, SOS1, and PKP4) (supplemental Table S3).
For a more accurate estimate of the protein abundance, we integrated the ion current over the retention time for individual peptides (52). Although for different precursor peptides the amplitude of the ion currents greatly depends on electrospray ionization efficiency and other factors (e.g. ion suppression), the ion current is highly reproducible when comparative samples have similar protein/peptide contents (supplemental Fig. S4). We therefore determined the -fold enrichment for each peptide before and after OA treatment. The -fold enrichment was further transformed into logarithmic (log2) values that were averaged over all peptides of a particular protein (47).
The histograms of the log ratios could be fitted into two Gaussian curves, representing two populations (Fig. 4B). The main peak, which contains the vast majority of proteins, presumably reflects normal variation around the mean. The fact that this peak is centered around −0.5 (equivalent to a 30% decrease upon OA treatment) rather than 0 probably represents a systematic “error,” such as relative sample loss during the preparation of OA-treated cultures (47). The S.D. of the curve was ∼0.76, and we selected a log2 ratio of 2 (>3.2 S.D.) as the cutoff (Table I). A second smaller broad peak centered at approximately at 2 (4-fold enrichment) represents a subset of proteins whose 5557 immunoprecipitation yield was markedly increased by OA (Fig. 4B).
Interestingly increased 5557 immunopurification of a protein upon OA treatment correlates with the presence of PX(S/T)P motifs in that protein (Fig. 4C). 90% (36 of 40) of the highly induced proteins (log2(OA/untreated) > 2), and 79% (56 of 71) of proteins with log2(OA/untreated) >1 contain sequences conforming to the PX(S/T)P MAPK phosphomotif. In contrast, only 18% (2 of 11) of OA-depleted proteins with log2(OA/untreated) <−2 contain PX(S/T)P sites. These data validate our approach to enrich for MAPK targets by immunopurification with the phosphomotif antibody, and they further argue that the proteins with the highest log2 scores upon OA treatment are most likely to be genuine MAPK substrates.
Some proteins were dramatically increased (e.g. the RasGEF SOS1, 12-fold, log2(OA/untreated) = 3.6) or decreased in abundance (e.g. metabotropic glutamate receptor 5, (GRM5), 4-fold, log2(OA/untreated) = −1.99) upon OA treatment (supplemental Table S3). The most prominent example of proteins increased by OA treatment was the SNAP-25-interacting protein SNIP (also known as p140Cap), which had 44 peptides identified in immunoprecipitates from OA-treated but none from untreated cultures (supplemental Table S3) and a log2 score of 3.9 based on ion current of the precursor peptides (Table I). The function of SNIP is poorly understood: it may connect synaptic vesicles to the cortical actin cytoskeleton and inhibit calcium-dependent vesicle release (53). With an apparent molecular mass of 145 kDa, SNIP is a prime candidate protein for the ∼140-kDa band induced by glutamate, NMDA, and TTX in cultured hippocampal neurons (see Fig. 3). The cadherin-associated protein PKP4 (150 kDa; log2(OA/untreated) = 2.7) was similarly elevated in 5557 immunoprecipitates from OA-treated cultures (Table I). Increased abundance of the molecular scaffolds Striatin (110 kDa; log2(OA/untreated) = 3.1) and connector enhancer of kinase suppressor of Ras 2 (CNKSR2; also known as membrane-associated guanylate kinase-interacting protein (MAGUIN), 118 kDa; log2(OA/untreated) = 2.6) suggests an OA-induced phosphorylation of these proteins at MAPK sites.
Additionally we used in silico prediction of MAPK phosphorylation sites with Scansite at the medium stringency level to identify proteins with suboptimal phosphorylation sites for MAPK that may not conform with the PX(S/T)P motif (23, 24). Table I shows a compilation of the most promising novel MAPK substrates identified from cortical culture by 5557 immunoprecipitation and selected for OA enrichment (log2(OA/untreated) > 2), at least one MAPK site (as predicted by Scansite at medium stringency), and reliable MS identification based on at least two peptides. Table II shows a similar compilation for putative MAPK substrates identified from rat brain PSD.
Table II.
Most promising potential MAPK substrates identified from rat brain PSD
Shown is the list of 5557-immunoprecipitated proteins from rat brain PSD with at least three peptides identified by MS and predicted MAPK phosphorylation site(s) (Scansite, medium stringency). Columns are as in Table I. All 115 identified proteins are listed in supplemental Table S4. SH3, Src homology 3.
| Accession no. | Human homolog | Description | Peptides | Identified in PSD? |
|---|---|---|---|---|
| XP_575594.1 | AAK1 | AP2-associated kinase 1 | 6 | + |
| XP_215761.3 | ACTG1a | Actin, cytoplasmic 2, γ-actin | 13 | − |
| NP_058686.1 | ADD1a | Adducin 1 (α) | 29 | + |
| NP_036623.1 | ADD2a | Adducin 2 (β) | 4 | + |
| NP_062234.1 | ARC | Activity-regulated cytoskeletal associated protein | 3 | + |
| NP_068507.1 | CAMK2Ba | Calcium/calmodulin-dependent protein kinase II β subunit | 12 | + |
| NP_036651.1 | CAMK2Da | Calcium/calmodulin-dependent protein kinase II, δ | 4 | + |
| NP_598289.1 | CAMK2Ga | Calcium/calmodulin-dependent protein kinase II γ | 6 | + |
| NP_067718.1 | CNKSR2a | Connector enhancer of kinase suppressor of Ras 2; MAGUIN2 | 10 | + |
| NP_037064.1 | CRMP1a | Collapsin response mediator protein 1 | 3 | + |
| XP_001064375.1 | CTNND2a | Catenin, δ2 | 8 | + |
| NP_071618.1 | DLG2 | Chapsyn-110; discs, large homolog 2 | 8 | + |
| NP_062567.1 | DLG4 | PSD-95, postsynaptic density protein 95 | 27 | + |
| NP_775168.1 | DLGAP4 | Discs, large homolog-associated protein 4 | 3 | + |
| XP_577204.1 | FLJ22184a | No known homologues, KAR9 domain | 11 | − |
| XP_344572.2 | GPRIN1a | G-protein-regulated inducer of neurite outgrowth 1 | 4 | − |
| NP_476480.1 | HNRPU | Heterogeneous nuclear ribonucleoprotein U | 3 | − |
| NP_113895.1 | HOMER1a | Homer 1 | 7 | − |
| NP_068635.1 | HSPA2a | Heat shock protein 2 | 5 | − |
| XP_224824.3 | HSPA8a | Heat shock protein 8 | 3 | − |
| NP_476483.1 | KIAA1365 | Densin-180 | 3 | − |
| NP_446073.1 | MAGI2a | Membrane-associated guanylate kinase, WW and PDZ domain-containing 2 | 12 | − |
| XP_215469.2 | MAP1Ba | microtubule-associated protein 1B | 13 | + |
| NP_037198.1 | MAP2a | Microtubule-associated protein 2 | 30 | + |
| NP_058900.1 | MAP6a | Microtubule-associated protein 6 | 4 | + |
| NP_058722.1 | MBP | Myelin basic protein | 6 | + |
| XP_574117.1 | MYO7Ba | Myosin-VIIb | 13 | + |
| NP_001009967.1 | PIP5K1Ca | Phosphatidylinositol-4-phosphate 5-kinase, type I, γ | 5 | + |
| XP_215733.3 | PKP4a | Plakophilin 4 | 14 | + |
| NP_445926.1 | PPP1R9Ba | Protein phosphatase 1, regulatory subunit 9B, spinophilin | 7 | + |
| XP_233240.3 | SGIP1a | SH3 domain GRB2-like (endophilin) interacting protein 1 | 12 | − |
| NP_067708.1 | SHANK3 | SH3/ankyrin domain gene 3 | 5 | + |
| NP_062251.1 | SNIPa | SNAP-25-interacting protein | 32 | + |
| NP_062006.1 | SYN1 | Synapsin 1 | 16 | + |
| NP_062032.1 | SYN2a | Synapsin 2 | 4 | + |
| NP_851606.1 | SYNGAP1 | Synaptic Ras GTPase-activating protein 1 | 13 | + |
| NP_067727.1 | SYNPO | Synaptopodin | 7 | + |
| NP_775125.1 | TUBBa | Tubulin, β5 | 13 | − |
| NP_954525.1 | TUBB2Ca | Tubulin, β2c | 14 | − |
| XP_579666.1 | TUBB3a | Tubulin, β3 | 10 | − |
Protein that was also identified from cortical culture (regardless of its enrichment score).
The overlap between these two top scoring lists (Table I versus Table II) consists of SNIP, the scaffold CNKSR2/MAGUIN, and the cadherin-associated PKP4, a relative of δ-catenin, which regulates neuron morphology and synaptic plasticity (32, 54). In total, 59 proteins were identified by at least two peptides from both cortical culture and rat brain PSD by immunopurification with the 5557 antibody.
In addition, from PSD fractions we identified several well known PSD scaffolds such as PSD-95/DLG4, Chapsyn-110/DLG2, SAPAP4/DLGAP4, Shank3, and Homer 1 (Table II). PSD-95/DLG4 has recently been shown to be a functionally important target of JNK1 phosphorylation (55), and we identified the Ser-295 phosphorylation site by 5557 immunoprecipitation and MS (supplemental Table S2), thus validating our approach for discovery of MAPK substrates in the PSD. These PSD scaffold proteins were not found by 5557 immunoprecipitation from cortical culture probably because they are poorly soluble in the RIPA detergent used to extract the cortical cultures. The presence of CaMKII, actin, and tubulin in the PSD data set (Table II) might be due to their high abundance in the PSD and consequent contamination of 5557 immunoprecipitates (38, 48). However, CaMKIIβ indeed contains a proline-directed phosphorylation site, which is not conserved in CaMKIIα/δ/γ (26). The full list of identified proteins annotated with Scansite predictions and OA enrichment score are available on line as supplemental Tables S3 and S4. A more detailed discussion of specific identified proteins is found in the supplemental information (supplemental Fig. S5).
Identification of Phosphorylation Sites—
Although antibody 5557 is highly phosphospecific, the mere identification of immunopurified proteins cannot prove their phosphorylation or reveal the in vivo phosphorylation sites. Thus it is significant that our MS analysis of 5557-immunoprecipitated proteins also identified a large number of phosphopeptide sequences representing 82 phosphorylation sites from 34 proteins (supplemental Table S2). In total, we discovered 58 serine phosphorylation sites, 23 threonine phosphorylation sites, and one tyrosine phosphorylation site, most of which are proline-directed (62 of 83, or 75%). 14 phosphorylation sites exactly match the PX(S/T)P motif predominant in the 5557 library. These data validate the phosphomotif antibody approach for purification of proteins phosphorylated at a MAPK consensus site.
We discovered phosphorylated PX(S/T)P sites in a diversity of proteins, including the cytoskeletal proteins tau (MAPT), MAP2, and Adducin 1 (ADD1), the p21/Cdc42/Rac1-activated kinase 1 (PAK1), doublecortin-like kinase 1 (DCLK1), collapsin response mediator protein 1 (CRMP1/DPYSL1), and δ-catenin (CTNND2). We also uncovered phosphorylation of MAPK motifs on SNIP (e.g. PERSpSPPM) and PKP4 (e.g. RVGpSPLT) (supplemental Table S2). Recalling that the levels of these phosphoproteins change severalfold after OA treatment (see Table I), the identification of these MAPK phosphorylation sites makes SNIP and PKP4 strong candidates for MAPK substrates that are dynamically regulated in neurons.
A fraction of the identified phosphopeptide sequences from 5557 immunoprecipitates do not conform to the MAPK phosphorylation motif; i.e. they are serine/threonine phosphorylation sites lacking a +1 proline residue or tyrosine phosphorylation sites. Most of these phosphopeptide sequences correspond to additional non-MAPK phosphorylation sites on proteins in which we identified bona fide MAPK phosphorylation sites (the latter sites presumably being directly recognized by 5557 antibody). Only for two proteins, CRMP2/DPYSL2 and TUBA6, did we identify exclusively non-proline-directed phosphorylation sites (supplemental Table S2).
Screening of Identified Proteins with in Vitro Kinase Reactions—
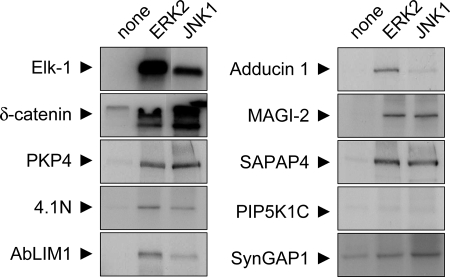
As a further test we asked whether the proteins identified from the 5557 phosphoantibody immunoprecipitation could serve as substrates for MAPKs in vitro. A set of candidate proteins were epitope-tagged, expressed in HEK293 cells, and then immunoprecipitated as substrates for in vitro kinase reactions with recombinant ERK2 or JNK1 and radioactive [γ-32P]ATP (Fig. 5). As a positive control we used the transcription factor Elk-1, which is a known substrate of ERK and JNK (34). ERK2 and JNK1 both readily phosphorylated Elk-1, δ-catenin, PKP4, and to a lesser extent 4.1N, AbLIM1, Adducin 1, MAGI-2, and SAPAP4. Thus our phosphoantibody immunoprecipitation yielded many MAPK substrates at least as defined by in vitro phosphorylation. We found no evidence for phosphorylation of SynGAP1 (synaptic GTPase-activating protein 1) and PIP5K1C (phosphatidylinositol-4-phosphate 5-kinase, type I, γ) by ERK2 or JNK1, although both of them were identified from PSD by 5557 immunoprecipitation. However, it is possible that these proteins are substrates of p38 or other ERK/JNK isoforms not tested here.
Fig. 5.
In vitro phosphorylation of candidate substrates by mitogen-activated protein kinases. Selected candidate proteins (GFP-δ-catenin, GFP-PKP4, FLAG-4.1N, FLAG-AbLIM1, FLAG-MAGI-2, FLAG-SAPAP4, FLAG-PIP5K1C, and FLAG-SynGAP1) were expressed in HEK293 cells, immunoprecipitated with epitope tag antibodies, and incubated with γ-[P32]ATP and different recombinant active MAPKs as indicated. Phosphorylation was visualized by autoradiography. FLAG-tagged transcription factor Elk-1 is used as positive control (see “Results”).
δ-Catenin Is Phosphorylated by Mitogen-activated Protein Kinases in Neurons—
The ultimate validation of our phosphoproteomics screen would be to demonstrate that the identified proteins are really phosphorylated in neurons by a specific MAPK. For PSD-95, we showed indeed that phosphorylation occurs in vivo on residue Ser-295 as identified by MS (see supplemental Table S2) and that it is mediated by the MAPK JNK1 and regulated by activity (55).
δ-Catenin was chosen as a second protein for validation. We raised and affinity-purified a phosphospecific antibody (termed pS-447) against Ser-447, a novel phosphorylation site in δ-catenin (sequence STAPSpSPGVD; see supplemental Table S2). Purified pS-447 antibody displayed high specificity for the phosphopeptide over the non-phosphopeptide in slot blots (Fig. 6A) and detected wild type δ-catenin expressed in HEK293 cells but not a mutant δ-catenin in which Ser-447 is substituted with alanine (S447A-δ-catenin) (Fig. 6B). The phosphorylated δ-catenin was also enriched in the P2 and PSD1 fractions of rat brain (Fig. 6C). In immunoblots of cultured cortical neurons, pS-447 antibody detected a ∼140-kDa band co-migrating with δ-catenin. This band smeared and shifted upward upon treatment of cultures with OA (100 nm; 1 h) and was associated with a slight increase in integrated pS-447 signal (Fig. 6D). Immunoblotting with δ-catenin antibody also revealed a smearing of the band and slower migration for total δ-catenin. The pS-447 bands were quantitatively removed by treating the membrane with CIP, whereas total δ-catenin signal was unchanged, thus proving the specificity of the pS-447 antibody for phosphorylated δ-catenin (Fig. 6D). These data establish that endogenous δ-catenin is phosphorylated on Ser-447 in cultured neurons and rat brain.
Fig. 6.
δ-Catenin serine 447 is phosphorylated by JNK in neurons. A, affinity-purified antibody raised against δ-catenin Ser(P)-447 (pS-447) was tested against the phosphopeptide used for immunization (p) and an identical non-phosphorylated peptide (np) in a slot blot (10, 100, or 1000 ng of peptide). B, GFP-tagged wt δ-catenin or a mutant with serine 447 substituted by alanine (S447A) was expressed in HEK293 cells and immunoblotted with pS-447 phosphoantibody or GFP antibody. C, immunoblot of rat brain fractions with pS-447 and total δ-catenin antibodies. Brain extracts were fractionated into postnuclear supernatant (S1), cytosol plus light membranes (S2), crude synaptosomal fraction (P2), and synaptosomes extracted with 0.5% Triton X-100 (PSD1). D, extracts from untreated or OA-treated (100 nm, 1 h) cortical cultures (DIV25) were probed with antibodies as indicated. CIP treatment removed pS-447 phosphoantibody signal but spared total δ-catenin (δ-cat). The asterisk denotes a cross-reactive band detected by the phosphoantibody. E, hippocampal cultures (DIV23–27) treated with DMSO or kinase inhibitors were immunoblotted with antibodies to Ser(P)-447 δ-catenin and total δ-catenin. Histogram shows immunoblot intensity of Ser(P)-447/total δ-catenin normalized to DMSO control (mean ± S.E. of seven independent experiments). Statistical analysis was performed by two-sided t test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Which kinase is responsible for phosphorylation of Ser-447 in δ-catenin? Treatment with SP600125 (JNK inhibitor; 20 μm, 16 h) and to a lesser extent SB203580 (p38 inhibitor; 5 μm) reduced serine 447 phosphorylation of δ-catenin but not total δ-catenin levels (Fig. 6E). In the same experiments, PD98059 and roscovitine had no effect (Fig. 6E). These data indicate that JNK is the main mediator of basal Ser-447 phosphorylation in neurons.
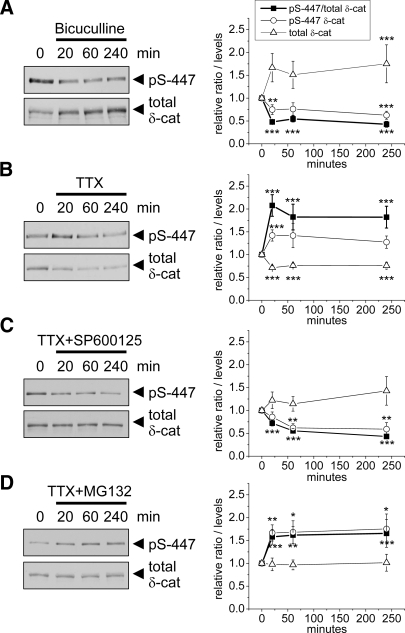
The pS-447 phosphoantibody also allowed us to measure the effect of synaptic activity on phosphorylation of δ-catenin. Stimulating activity in cultured neurons with bicuculline (50 μm) led to rapid dephosphorylation of Ser-447 occurring within 20 min and associated with accumulation of total δ-catenin (Fig. 7A). In contrast, silencing activity with TTX treatment (3 μm) increased Ser-447 phosphorylation and reduced total δ-catenin levels within 20 min (Fig. 7B). The regulation of δ-catenin Ser-447 phosphorylation by TTX and bicuculline is bidirectional and qualitatively similar to that of PSD-95 Ser-295 (55) consistent with both of these sites being targets of JNK MAPK.
Fig. 7.
Bidirectional regulation of δ-catenin Ser-447 phosphorylation by neuronal activity. Hippocampal neurons (DIV23–27) were treated with bicuculline (50 μm) (A) or TTX (3 μm) (B) for the indicated times and immunoblotted with pS-447 δ-catenin phosphoantibody and for total δ-catenin (δ-cat). In C and D, SP600125 (20 μm) or MG132 (10 μm) was added 10 min prior to TTX (3 μm). Graphs at right show immunoblot intensity of Ser(P)-447 δ-catenin, total δ-catenin, and their ratio normalized to DMSO control (mean ± S.E., n = 9). *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student's t test).
To test whether TTX-induced phosphorylation of Ser-447 is dependent on JNK, we pretreated neurons for 10 min with JNK inhibitor SP600125 (20 μm) before adding TTX (3 μm). SP600125 prevented the effect of TTX on δ-catenin; the combination actually led to decreased Ser-447 phosphorylation (Fig. 7C).
Because TTX-induced phosphorylation at Ser-447 was accompanied by reduced δ-catenin levels, we asked whether δ-catenin is degraded by the proteasome upon phosphorylation. Blocking proteasome function with MG132 (10 μm) prior to adding TTX (3 μm) did not block Ser-447 phosphorylation but prevented the reduction of δ-catenin levels (Fig. 7D). These results are consistent with the idea that JNK-dependent phosphorylation of δ-catenin at Ser-447 targets δ-catenin for proteasomal degradation.
Functional Significance of δ-Catenin Ser-447 Phosphorylation—
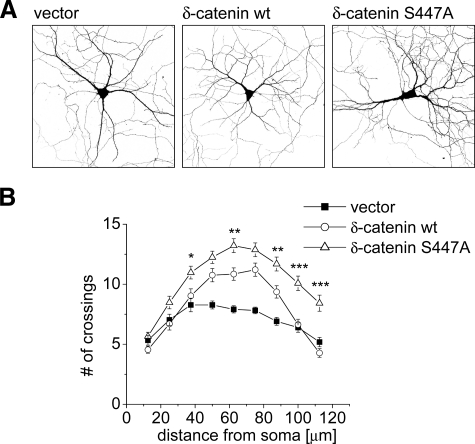
Because δ-catenin promotes dendrite branching (56, 57), we asked whether phosphorylation at Ser-447 affects this aspect of δ-catenin function. We transfected hippocampal neurons after 7 days in culture with either wild type or mutant S447A-δ-catenin, which cannot be phosphorylated at Ser-447 (Fig. 8A). 5 days after transfection we quantified dendritic complexity by Sholl analysis, counting the number of dendritic branches crossing equidistant concentric circles around the cell body. In normal neurons the highest number of dendrite crossings is found at 40–80-μm distance from the cell soma. Compared with the vector control, δ-catenin wild type-transfected neurons had significantly more branches at 50–87.5 μm (p < 0.01). Importantly neurons transfected with δ-catenin S447A mutant showed a significantly greater increase in dendritic branching than those transfected with wild type δ-catenin, and their dendrites also extended further to the periphery (Fig. 8B). These data indicate that mutation of Ser-447 enhances the dendrite branching function of δ-catenin perhaps by preventing JNK-dependent proteasome-mediated degradation of δ-catenin in neurons.
Fig. 8.
δ-Catenin S447A stimulates dendrite branching more than wild type. A, hippocampal neurons (DIV7) were co-transfected with GFP and either empty vector, wild type δ-catenin, or mutant δ-catenin-S447A and imaged by confocal microscopy after 5 days. B, Sholl analysis. The number of dendrite branches crossing concentric circles around the cell body was manually counted (mean ± S.E., n = 33 to 38). Asterisks indicate significance of two-way analysis of variance comparing δ-catenin wt and S447A: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
DISCUSSION
Identification of Putative MAPK Substrates—
LC-MS/MS is increasingly powerful at identifying proteins and phosphopeptides in complex mixtures. Although a few peptides are generally sufficient to identify a protein, current methods are far from covering all tryptic peptides, let alone all posttranslational modifications within a complex biological sample. Therefore we used affinity purification with a phosphomotif antibody directed against phosphorylated MAPK consensus sequences to enrich for in vivo MAPK substrates. Naturally the immunopurification approach is biased toward the consensus sequence of the phosphopeptide library used to generate the 5557 antibody.
The above caveats notwithstanding, the yield was rich and diverse. Using immunopurification with phosphomotif antibody, we identified 82 phosphorylation sites and up to 449 candidate substrates for MAPKs, including a large number of known targets of MAPKs (supplemental Table S1). More importantly, we identified numerous proteins with predicted MAPK sites or directly sequenced phosphorylation sites that had not been associated previously with MAPK signaling. Specific examples of these candidate substrates and their significance are discussed in the supplemental information available on line.
The rapid increase of 5557 immunoblot signal induced by OA suggests dynamic phosphorylation and dephosphorylation of many neuronal proteins. Within 30 min the 5557 signal increased ∼3-fold in hippocampal extracts, suggesting that during that time many of the 5557 epitopes normally undergo phosphorylation and dephosphorylation (supplemental Fig. S3). Assuming that our antibody 5557 is not biased toward phosphorylation sites produced by any individual MAPK, it appears that mainly JNK is responsible for basal phosphorylation events in cultured neurons (Fig. 3, A and B). The JNK inhibitor SP600125 removed up to 70% of the basal 5557 signal and ∼50% of the 5557 signal induced by OA. The preponderance of JNK-mediated phosphorylation detected by 5557 phosphoantibody is consistent with the known high “constitutive” activity of JNK MAPKs in neurons (51, 55).
Direct sequencing of a peptide containing a phosphorylated serine/threonine next to a proline provides additional evidence that the corresponding protein is a MAPK substrate. Of the 82 phosphorylation sites determined in our analysis, most (62 of 82, or 76%) of them were proline-directed. Two unbiased phosphoproteomics studies of the PSD fraction (which identified 625 phosphorylation sites) (26) and of synaptosomes (974 sites) (27) yielded a smaller fraction of phosphorylated proline-directed (S/T)P sites (both ∼50%). Notably there is a rather small overlap of 16 and 12 phosphorylation sites, respectively, between these unbiased phosphoproteomics analyses and our study (26, 27). The result emphasizes that motif-specific affinity enrichment allows in-depth analysis of a specific subset of the phosphorylated proteome, complementing the unbiased IMAC or TiO2 methods for phosphoprotein isolation. An additional advantage of phosphomotif antibodies is that they allow rapid investigation of the broad regulation of phosphorylation under many experimental conditions (see Figs. 1 and 3 for examples).
Validation of Putative MAPK Substrates—
We used in vitro kinase assays to prescreen several interesting candidate proteins and found that seven of the proteins tested were indeed phosphorylated by one or more of the MAPKs ERK and JNK in vitro. Because in vitro phosphorylation is prone to false-positive artifacts, it is imperative to confirm phosphorylation in vivo. We raised phosphoantibodies against specific phosphorylation sites in δ-catenin and PSD-95. Indeed these proteins were endogenously phosphorylated at the identified sites in neurons: serine 295 of PSD-95 is a target of JNK1 (55); serine 447 of δ-catenin is mainly phosphorylated by JNK and perhaps p38 (this study). PSD-95 is a PSD scaffold that controls synaptic strength by binding to glutamate receptors. JNK phosphorylation of Ser-295 promotes PSD-95 accumulation at synapses, and dephosphorylation of Ser-295 is required for LTD (55).
δ-Catenin belongs to the p120 catenin family of proteins that stabilize cadherin cell adhesion molecules at the cell surface and regulate actin dynamics via Rho GTPases. The p120 catenin family affects dendrite branching, AMPA receptor trafficking, and synaptic plasticity (32, 56, 58, 59). The significance of serine/threonine phosphorylation of p120 catenin family members in neurons is not understood. Here we identified a novel phosphorylation site in δ-catenin at Ser-447 that is principally mediated by JNK and that is associated with depletion of δ-catenin levels probably via proteasome degradation. A δ-catenin mutant mutated at the Ser-447 phosphorylation site showed enhanced ability to promote dendrite branching in cultured hippocampal neurons, affirming a functional significance of this phosphorylation. Most likely, phosphorylation at Ser-447 reduces δ-catenin stability, but the possibility that it alters the activity of δ-catenin cannot be excluded.
From the same protein family we also identified PKP4 as a putative MAPK substrate with multiple proline-directed phosphorylation sites. MAPK regulation of δ-catenin and PKP4 has not been reported previously. Because cadherin-mediated adhesion and/or signaling is important for morphological and functional plasticity of synapses (60), it will be interesting to determine the physiological significance of MAPK phosphorylation of these cadherin-associated proteins, especially in the brain in vivo. Further study of the other proteins discovered in our novel phosphoproteomics screen should shed additional light on the mechanisms of MAPKs in synaptic plasticity and disease.
Supplementary Material
Acknowledgments
We thank Drs. Hatzfeld, Kosik, and Sharrocks and the Kazusa DNA Research Institute for providing plasmids. We thank Baris Bingol, Yelin Chen, Honor Hsin, Natasha Hussain, and Christopher Nelson for critical reading of the manuscript.
Footnotes
Published, MCP Papers in Press, December 3, 2008, DOI 10.1074/mcp.M800233-MCP200
The abbreviations used are: MAPK, mitogen-activated protein kinase; PSD, postsynaptic density; LTP, long term potentiation; LTD, long term depression; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; CDK, cyclin-dependent kinase; NMDA, N-methyl-d-aspartic acid; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; TTX, tetrodotoxin; OA, okadaic acid; CIP, calf intestine phosphatase; DIV, days in vitro; GFP, green fluorescent protein; PSD-95, postsynaptic density protein 95; PKP4, plakophilin 4; AbLIM1, actin-binding LIM protein 1; MAGI-2, membrane-associated guanylate kinase, WW and PDZ domain-containing 2; SAPAP4, SAP90/PSD-95-associated protein 4, disks large-associated protein 4; SNIP, SNAP-25-interacting protein; wt, wild type; CaMKII, calcium/calmodulin-dependent protein kinase II; CNKSR2, connector enhancer of kinase suppressor of Ras 2; MAGUIN, membrane-associated guanylate kinase-interacting protein; NCBI, National Center for Biotechnology Information.
This work was supported, in whole or in part, by National Institutes of Health Grants DA019937 and AG025688 (to J. P.).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Thomas, G. M., and Huganir, R. L. ( 2004) MAPK cascade signalling and synaptic plasticity. Nat. Rev. Neurosci. 5, 173–183 [DOI] [PubMed] [Google Scholar]
- 2.Zhu, J. J., Qin, Y., Zhao, M., Van Aelst, L., and Malinow, R. ( 2002) Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell 110, 443–455 [DOI] [PubMed] [Google Scholar]
- 3.Morozov, A., Muzzio, I. A., Bourtchouladze, R., Van-Strien, N., Lapidus, K., Yin, D., Winder, D. G., Adams, J. P., Sweatt, J. D., and Kandel, E. R. ( 2003) Rap1 couples cAMP signaling to a distinct pool of p42/44MAPK regulating excitability, synaptic plasticity, learning, and memory. Neuron 39, 309–325 [DOI] [PubMed] [Google Scholar]
- 4.Minden, A., Lin, A., Claret, F. X., Abo, A., and Karin, M. ( 1995) Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell 81, 1147–1157 [DOI] [PubMed] [Google Scholar]
- 5.Huang, C. C., You, J. L., Wu, M. Y., and Hsu, K. S. ( 2004) Rap1-induced p38 mitogen-activated protein kinase activation facilitates AMPA receptor trafficking via the GDI. Rab5 complex. Potential role in (S)-3,5-dihydroxyphenylglycene-induced long term depression. J. Biol. Chem. 279, 12286–12292 [DOI] [PubMed] [Google Scholar]
- 6.Machida, N., Umikawa, M., Takei, K., Sakima, N., Myagmar, B. E., Taira, K., Uezato, H., Ogawa, Y., and Kariya, K. ( 2004) Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J. Biol. Chem. 279, 15711–15714 [DOI] [PubMed] [Google Scholar]
- 7.Zhu, Y., Pak, D., Qin, Y., McCormack, S. G., Kim, M. J., Baumgart, J. P., Velamoor, V., Auberson, Y. P., Osten, P., van Aelst, L., Sheng, M., and Zhu, J. J. ( 2005) Rap2-JNK removes synaptic AMPA receptors during depotentiation. Neuron 46, 905–916 [DOI] [PubMed] [Google Scholar]
- 8.Fiore, R. S., Murphy, T. H., Sanghera, J. S., Pelech, S. L., and Baraban, J. M. ( 1993) Activation of p42 mitogen-activated protein kinase by glutamate receptor stimulation in rat primary cortical cultures. J. Neurochem. 61, 1626–1633 [DOI] [PubMed] [Google Scholar]
- 9.English, J. D., and Sweatt, J. D. ( 1997) A requirement for the mitogen-activated protein kinase cascade in hippocampal long term potentiation. J. Biol. Chem. 272, 19103–19106 [DOI] [PubMed] [Google Scholar]
- 10.Blum, S., Moore, A. N., Adams, F., and Dash, P. K. ( 1999) A mitogen-activated protein kinase cascade in the CA1/CA2 subfield of the dorsal hippocampus is essential for long-term spatial memory. J. Neurosci. 19, 3535–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolshakov, V. Y., Carboni, L., Cobb, M. H., Siegelbaum, S. A., and Belardetti, F. ( 2000) Dual MAP kinase pathways mediate opposing forms of long-term plasticity at CA3-CA1 synapses. Nat. Neurosci. 3, 1107–1112 [DOI] [PubMed] [Google Scholar]
- 12.Brown, T. C., Tran, I. C., Backos, D. S., and Esteban, J. A. ( 2005) NMDA receptor-dependent activation of the small GTPase Rab5 drives the removal of synaptic AMPA receptors during hippocampal LTD. Neuron 45, 81–94 [DOI] [PubMed] [Google Scholar]
- 13.Pei, J. J., Braak, E., Braak, H., Grundke-Iqbal, I., Iqbal, K., Winblad, B., and Cowburn, R. F. ( 2001) Localization of active forms of C-jun kinase (JNK) and p38 kinase in Alzheimer's disease brains at different stages of neurofibrillary degeneration. J. Alzheimer's Dis. 3, 41–48 [DOI] [PubMed] [Google Scholar]
- 14.Wang, Q., Walsh, D. M., Rowan, M. J., Selkoe, D. J., and Anwyl, R. ( 2004) Block of long-term potentiation by naturally secreted and synthetic amyloid beta-peptide in hippocampal slices is mediated via activation of the kinases c-Jun N-terminal kinase, cyclin-dependent kinase 5, and p38 mitogen-activated protein kinase as well as metabotropic glutamate receptor type 5. J. Neurosci. 24, 3370–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas, G. M., Lin, D. T., Nuriya, M., and Huganir, R. L. ( 2008) Rapid and bi-directional regulation of AMPA receptor phosphorylation and trafficking by JNK. EMBO J. 27, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu, G. Y., Deisseroth, K., and Tsien, R. W. ( 2001) Spaced stimuli stabilize MAPK pathway activation and its effects on dendritic morphology. Nat. Neurosci. 4, 151–158 [DOI] [PubMed] [Google Scholar]
- 17.Kennedy, M. B., Beale, H. C., Carlisle, H. J., and Washburn, L. R. ( 2005) Integration of biochemical signalling in spines. Nat. Rev. Neurosci. 6, 423–434 [DOI] [PubMed] [Google Scholar]
- 18.Sheng, M., and Kim, M. J. ( 2002) Postsynaptic signaling and plasticity mechanisms. Science 298, 776–780 [DOI] [PubMed] [Google Scholar]
- 19.Sheng, M., and Hoogenraad, C. C. ( 2007) The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu. Rev. Biochem. 76, 823–847 [DOI] [PubMed] [Google Scholar]
- 20.Johnson, S. A., and Hunter, T. ( 2005) Kinomics: methods for deciphering the kinome. Nat. Methods 2, 17–25 [DOI] [PubMed] [Google Scholar]
- 21.Fukunaga, R., and Hunter, T. ( 1997) MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu, H., Klemic, J. F., Chang, S., Bertone, P., Casamayor, A., Klemic, K. G., Smith, D., Gerstein, M., Reed, M. A., and Snyder, M. ( 2000) Analysis of yeast protein kinases using protein chips. Nat. Genet. 26, 283–289 [DOI] [PubMed] [Google Scholar]
- 23.Yaffe, M. B., Leparc, G. G., Lai, J., Obata, T., Volinia, S., and Cantley, L. C. ( 2001) A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 19, 348–353 [DOI] [PubMed] [Google Scholar]
- 24.Obenauer, J. C., Cantley, L. C., and Yaffe, M. B. ( 2003) Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins, M. O., Yu, L., Coba, M. P., Husi, H., Campuzano, I., Blackstock, W. P., Choudhary, J. S., and Grant, S. G. ( 2005) Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 280, 5972–5982 [DOI] [PubMed] [Google Scholar]
- 26.Trinidad, J. C., Specht, C. G., Thalhammer, A., Schoepfer, R., and Burlingame, A. L. ( 2006) Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics 5, 914–922 [DOI] [PubMed] [Google Scholar]
- 27.Munton, R. P., Tweedie-Cullen, R., Livingstone-Zatchej, M., Weinandy, F., Waidelich, M., Longo, D., Gehrig, P., Potthast, F., Rutishauser, D., Gerrits, B., Panse, C., Schlapbach, R., and Mansuy, I. M. ( 2007) Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol. Cell. Proteomics 6, 283–293 [DOI] [PubMed] [Google Scholar]
- 28.Collins, M. O., Yu, L., Campuzano, I., Grant, S. G., and Choudhary, J. S. ( 2008) Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol. Cell. Proteomics 7, 1331–1348 [DOI] [PubMed] [Google Scholar]
- 29.Trinidad, J. C., Thalhammer, A., Specht, C. G., Lynn, A. J., Baker, P. R., Schoepfer, R., and Burlingame, A. L. ( 2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 30.Berwick, D. C., Hers, I., Heesom, K. J., Moule, S. K., and Tavare, J. M. ( 2002) The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J. Biol. Chem. 277, 33895–33900 [DOI] [PubMed] [Google Scholar]
- 31.Kane, S., Sano, H., Liu, S. C., Asara, J. M., Lane, W. S., Garner, C. C., and Lienhard, G. E. ( 2002) A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J. Biol. Chem. 277, 22115–22118 [DOI] [PubMed] [Google Scholar]
- 32.Israely, I., Costa, R. M., Xie, C. W., Silva, A. J., Kosik, K. S., and Liu, X. ( 2004) Deletion of the neuron-specific protein δ-catenin leads to severe cognitive and synaptic dysfunction. Curr. Biol. 14, 1657–1663 [DOI] [PubMed] [Google Scholar]
- 33.Kim, E., and Sheng, M. ( 1996) Differential K+ channel clustering activity of PSD-95 and SAP97, two related membrane-associated putative guanylate kinases. Neuropharmacology 35, 993–1000 [DOI] [PubMed] [Google Scholar]
- 34.Yang, S. H., Whitmarsh, A. J., Davis, R. J., and Sharrocks, A. D. ( 1998) Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 17, 1740–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatzfeld, M., and Nachtsheim, C. ( 1996) Cloning and characterization of a new armadillo family member, p0071, associated with the junctional plaque: evidence for a subfamily of closely related proteins. J. Cell Sci. 109, 2767–2778 [DOI] [PubMed] [Google Scholar]
- 36.Lu, Q., Paredes, M., Medina, M., Zhou, J., Cavallo, R., Peifer, M., Orecchio, L., and Kosik, K. S. ( 1999) δ-Catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 144, 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoogenraad, C. C., Feliu-Mojer, M. I., Spangler, S. A., Milstein, A. D., Dunah, A. W., Hung, A. Y., and Sheng, M. ( 2007) Liprinα1 degradation by calcium/calmodulin-dependent protein kinase II regulates LAR receptor tyrosine phosphatase distribution and dendrite development. Dev. Cell 12, 587–602 [DOI] [PubMed] [Google Scholar]
- 38.Peng, J., Kim, M. J., Cheng, D., Duong, D. M., Gygi, S. P., and Sheng, M. ( 2004) Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 279, 21003–21011 [DOI] [PubMed] [Google Scholar]
- 39.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. ( 1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 40.Peng, J., and Gygi, S. P. ( 2001) Proteomics: the move to mixtures. J. Mass Spectrom. 36, 1083–1091 [DOI] [PubMed] [Google Scholar]
- 41.Eng, J., McCormack, A. L., and Yates, J. R., III ( 1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 42.Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J., and Gygi, S. P. ( 2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 43.Elias, J. E., and Gygi, S. P. ( 2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 44.Xia, Q., Cheng, D., Duong, D. M., Gearing, M., Lah, J. J., Levey, A. I., and Peng, J. ( 2008) Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J. Proteome Res. 7, 2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldwin, M. A. ( 2004) Protein identification by mass spectrometry: issues to be considered. Mol. Cell. Proteomics 3, 1–9 [DOI] [PubMed] [Google Scholar]
- 46.Liao, L., Cheng, D., Wang, J., Duong, D. M., Losik, T. G., Gearing, M., Rees, H. D., Lah, J. J., Levey, A. I., and Peng, J. ( 2004) Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J. Biol. Chem. 279, 37061–37068 [DOI] [PubMed] [Google Scholar]
- 47.Listgarten, J., and Emili, A. ( 2005) Statistical and computational methods for comparative proteomic profiling using liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 4, 419–434 [DOI] [PubMed] [Google Scholar]
- 48.Cheng, D., Hoogenraad, C. C., Rush, J., Ramm, E., Schlager, M. A., Duong, D. M., Xu, P., Wijayawardana, S. R., Hanfelt, J., Nakagawa, T., Sheng, M., and Peng, J. ( 2006) Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics 5, 1158–1170 [DOI] [PubMed] [Google Scholar]
- 49.Songyang, Z., Lu, K. P., Kwon, Y. T., Tsai, L. H., Filhol, O., Cochet, C., Brickey, D. A., Soderling, T. R., Bartleson, C., Graves, D. J., DeMaggio, A. J., Hoekstra, M. F., Blenis, J., Hunter, T., and Cantley, L. C. ( 1996) A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 16, 6486–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bennett, B. L., Sasaki, D. T., Murray, B. W., O'Leary, E. C., Sakata, S. T., Xu, W., Leisten, J. C., Motiwala, A., Pierce, S., Satoh, Y., Bhagwat, S. S., Manning, A. M., and Anderson, D. W. ( 2001) SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. U. S. A. 98, 13681–13686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu, X., Raber, J., Yang, D., Su, B., and Mucke, L. ( 1997) Dynamic regulation of c-Jun N-terminal kinase activity in mouse brain by environmental stimuli. Proc. Natl. Acad. Sci. U. S. A. 94, 12655–12660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Old, W. M., Meyer-Arendt, K., Aveline-Wolf, L., Pierce, K. G., Mendoza, A., Sevinsky, J. R., Resing, K. A., and Ahn, N. G. ( 2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 53.Chin, L. S., Nugent, R. D., Raynor, M. C., Vavalle, J. P., and Li, L. ( 2000) SNIP, a novel SNAP-25-interacting protein implicated in regulated exocytosis. J. Biol. Chem. 275, 1191–1200 [DOI] [PubMed] [Google Scholar]
- 54.Elia, L. P., Yamamoto, M., Zang, K., and Reichardt, L. F. ( 2006) p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron 51, 43–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim, M. J., Futai, K., Jo, J., Hayashi, Y., Cho, K., and Sheng, M. ( 2007) Synaptic accumulation of PSD-95 and synaptic function regulated by phosphorylation of serine-295 of PSD-95. Neuron 56, 488–502 [DOI] [PubMed] [Google Scholar]
- 56.Martinez, M. C., Ochiishi, T., Majewski, M., and Kosik, K. S. ( 2003) Dual regulation of neuronal morphogenesis by a δ-catenin-cortactin complex and Rho. J. Cell Biol. 162, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arikkath, J., Israely, I., Tao, Y., Mei, L., Liu, X., and Reichardt, L. F. ( 2008) Erbin controls dendritic morphogenesis by regulating localization of δ-catenin. J. Neurosci. 28, 7047–7056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silverman, J. B., Restituito, S., Lu, W., Lee-Edwards, L., Khatri, L., and Ziff, E. B. ( 2007) Synaptic anchorage of AMPA receptors by cadherins through neural plakophilin-related arm protein AMPA receptor-binding protein complexes. J. Neurosci. 27, 8505–8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perez-Moreno, M., and Fuchs, E. ( 2006) Catenins: keeping cells from getting their signals crossed. Dev. Cell 11, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kosik, K. S., Donahue, C. P., Israely, I., Liu, X., and Ochiishi, T. ( 2005) δ-Catenin at the synaptic-adherens junction. Trends Cell. Biol. 15, 172–178 [DOI] [PubMed] [Google Scholar]
- 61.Collins, M. O., Husi, H., Yu, L., Brandon, J. M., Anderson, C. N., Blackstock, W. P., Choudhary, J. S., and Grant, S. G. ( 2005) Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, Suppl. 1, 16–23 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.