Abstract
Huntington disease (HD) is fatal in humans within 15–20 years of symptomatic disease. Although late stage HD has been studied extensively, protein expression changes that occur at the early stages of disease and during disease progression have not been reported. In this study, we used a large two-dimensional gel/mass spectrometry-based proteomics approach to investigate HD-induced protein expression alterations and their kinetics at very early stages and during the course of disease. The murine HD model R6/2 was investigated at 2, 4, 6, 8, and 12 weeks of age, corresponding to absence of disease and early, intermediate, and late stage HD. Unexpectedly the most HD stage-specific protein changes (71–100%) as well as a drastic alteration (almost 6% of the proteome) in protein expression occurred already as early as 2 weeks of age. Early changes included mainly the up-regulation of proteins involved in glycolysis/gluconeogenesis and the down-regulation of the actin cytoskeleton. This suggests a period of highly variable protein expression that precedes the onset of HD phenotypes. Although an up-regulation of glycolysis/gluconeogenesis-related protein alterations remained dominant during HD progression, late stage alterations at 12 weeks showed an up-regulation of proteins involved in proteasomal function. The early changes in HD coincide with a peak in protein alteration during normal mouse development at 2 weeks of age that may be responsible for these massive changes. Protein and mRNA data sets showed a large overlap on the level of affected pathways but not single proteins/mRNAs. Our observations suggest that HD is characterized by a highly dynamic disease pathology not represented by linear protein concentration alterations over the course of disease.
Huntington disease (HD)1 is an autosomal dominantly inherited disorder that usually presents in midlife and causes death 15–20 years after the first symptoms occur (1). The disease-triggering mutation consists of an unstable, elongated cytosine-adenine-guanine (CAG) trinucleotide repeat at the 5′-end of the HD gene in the first exon. The CAG repeat codes for a variable number of glutamines (2). The length of the polyglutamine repeat in the gene product of HD, Huntingtin (Htt), is one of the most important factors that determines the age of onset of the disorder (2). Individuals with 6–35 CAG repeats are not affected. Those with 36–39 CAG repeats have an increased risk of acquiring the disease, and repeats of 40 and above always cause disease (3–5). Onset of the motor disorder usually occurs between 35 and 45 years, but the age of onset can range anywhere from 2 to 80 years (2). In general, the larger the size of the polyglutamine repeat, the earlier the age of onset (6).
HD has been studied extensively at the level of single (interacting) proteins (7–9) and well characterized pathways, such as cell death (10–12). However, these experimental approaches have been rather limited in their identification of proteins that contribute to disease pathogenesis. Protein-protein interaction network studies have increased the number of proteins available (13, 14). In addition, unbiased, large scale approaches have identified a large number of genes potentially involved in HD because microarrays probe a very large number of expression alterations simultaneously at the mRNA level (15, 16). Two-dimensional gel electrophoresis (2-DE)- (17, 18) or liquid chromatography-based proteomics approaches in combination with mass spectrometry are able to investigate many protein alterations at the same time (19, 20) increasing the number of proteins known to be involved in HD and complementing the protein interaction data.
HD has already been studied extensively at the transcriptomic level using brain tissue from a large number of mouse models (21–25) and humans (26) as well as human blood cells (27) using mostly late stages with visible disease phenotype. Proteome data on expression changes for HD are more scarce and so far only available for the R6/2 model (28–31). A common denominator of these studies is that only stages with disease phenotype were investigated (28–31). To understand the disease pathology in more detail, early changes need to be investigated. Here we present protein expression changes that occur at 2, 4, 6, 8, and 12 weeks of age in the well established R6/2 mouse model of HD representing the transition from the absence of disease-related phenotypes to a pronounced symptomatic disease state. Our results show that a large number of protein alterations are present prior to the developments of disease phenotypes. In addition most protein alterations were found to be disease stage-specific, and there were no proteins that were found to be altered at every stage investigated.
EXPERIMENTAL PROCEDURES
Experimental Animals—
Hemizygous R6/2 transgenic mice (32) were bred and reared in our colony by backcrossing R6/2 males to CBA × C57BL/6 F1 females (B6CBAF1/OlaHsd, Harlan Olac, Bicester, UK). R6/2 mice were always housed with wild-type mice and were subject to a 12-h light:12-h dark cycle. All animals had unlimited access to water and breeding chow, and housing conditions and environmental enrichment were as described previously (33). R6/2 transgenic mice and wild-type littermate controls were sacrificed by cervical dislocation, and brains were rapidly removed, flash frozen in liquid nitrogen, and stored at −80 °C until use. We analyzed total brain from 2-, 4-, 6-, 8-, and 12-week-old R6/2 mice (n = 5–8). Genotyping and CAG repeat sizing were performed as described previously (34). Early changes in development at embryonic days 16 and 18; birth; and 1, 2, and 4 weeks were studied using brains of C57BL/6 mice (n = 3; Charles River WITA GmbH, Sulzfeld, Germany).
Protein Extraction Procedure—
Total protein extracts were prepared from entire brains. The extraction procedure has been published previously and validated (30, 35). Frozen tissue, 1.6 parts (v/w) buffer P (50 mm Trizma® (Tris base) (Sigma-Aldrich), 50 mm KCl, and 20% (w/v) glycerol at pH 7.5) supplemented with a final CHAPS concentration of 4% (w/v) in the sample, 0.08 parts protease inhibitor solution I (one Complete™ tablet (Roche Applied Science) dissolved in 2 ml of buffer 1), and 0.02 parts protease inhibitor solution II (1.4 μm pepstatin A and 1 mm phenylmethylsulfonyl fluoride in ethanol) were ground to a fine powder in a mortar precooled in liquid nitrogen. The tissue powder was transferred into a 2-ml tube (Eppendorf, Hamburg, Germany), quickly thawed, and supplied with glass beads (0.034 units of glass beads/combined weight of tissue, buffers, and inhibitors in mg; glass beads, 2.5 ± 0.05-mm diameter; Worf Glaskugeln GmbH, Mainz, Germany). Each sample was sonicated six times in an ice-cold water bath for 10 s each with cooling intervals of 1 min 50 s in between. The homogenate was stirred for 30 min in buffer P without CHAPS at 4 °C in the presence of 0.025 parts (v/w) Benzonase (Merck) and a final concentration of 5 mm magnesium chloride in the sample. Subsequently 6.5 m urea and 2 m thiourea were added, and stirring was continued for 30 min at room temperature until urea and thiourea were completely dissolved. The protein extract was supplied with 70 mm dithiothreitol (Bio-Rad), 2% (v/w) ampholyte mixture (Servalyte pH 2–4, Serva, Heidelberg, Germany), corrected by the amount of urea added (correction factor = sample weight prior to addition of urea/sample weight after addition of urea), and stored at −80 °C. Protein concentrations were determined in sample aliquots without urea using Bio-Rad DC Protein Assay according to the protocol supplied by the manufacturer.
2-DE—
After genotyping, mice were allocated to the HD or control group. Sample pairs were randomly selected choosing one mouse brain from each group. 2-D gels were run in batches of two. Sample pairs consisting of an HD and a control were run in parallel in both dimensions of 2-DE, IEF, and SDS-PAGE. Different sample pairs were processed at different days to avoid confounding of the experiment due to the same processing date. Protein samples were separated by the large gel 2-DE technique developed in our laboratory as described previously (17, 18). The gel format was 40 cm (isoelectric focusing) × 30 cm (SDS-PAGE) × 0.75 mm (gel width). For IEF using the carrier ampholyte technique, we applied 6 μl (20 μg/μl) of protein extract of each sample to the anodic end of an IEF gel (40 cm) and used a carrier ampholyte mixture to establish a pH gradient from 3 to 10. Proteins were visualized in SDS-PAGE polyacrylamide gels by high sensitivity silver staining (18, 36). For SDS-PAGE the IEF gels were cut in half and run as acidic and basic sides. 2-D gels were dried (described extensively in Ref. 36) and scanned at 300 dpi and 16-bit gray scale using a scanner (Microtek Scan Maker 9800XL, Evestar GmbH, Willich, Germany). The 2-D gel images were subsequently saved in Tiff format to avoid loss of quality due to compression.
Quantitative Analysis of Protein Expression—
After uploading the 2-D gel images, protein spot patterns were evaluated by Delta2D imaging software version 3.5 (DECODON, Greifswald, Germany) as was already described recently in detail elsewhere (37). Delta2D is our standard 2-D gel evaluation software and was validated already in many of our studies (30, 35, 38–41). Briefly 2-D spot patterns of HD and control mouse brains were matched using the Delta2D “exact” mode matching protocol. First sample pairs (HD and control) were matched individually. Subsequently all HD gels were matched to create a “match link” between all 2-D spot patterns using match vectors (37). Using “union” mode a fusion image was generated, including the visible spots for each 2-D gel from each time point (2, 4, 6, 8, and 12 weeks) creating 10 fusion gels (five time points, one fusion gel for the acidic side and one for the basic side of 2-D gels). Only the fusion image was used for spot detection using the following settings for Delta2D: local background region, 100; average spot size, 1; and sensitivity, 100%. Spots were not edited manually after spot detection. About 2000 spots on the fusion image were transferred to all other 2-D gel images for each time point. This ensures that for each stage investigated the identity for each spot on a gel is identical. Relative spot volume intensities (fractions of 100%) were used for quantitative protein expression analysis. After background subtraction, normalized spot intensity values were copied into Excel spreadsheets for statistical analysis. Data sets were analyzed applying a Student's t test. In the case of HD versus control comparisons a paired t test was used to compare sample pairs run side by side during both electrophoresis runs. In longitudinal studies where different age stages of HD or control were compared with one another, we used an unpaired t test because no natural pairing exists. Pairs were randomly selected from each group (time points) compared. The 2-D gel evaluation procedure by Delta2D remained the same as in HD versus control comparisons.
To determine changes in total protein concentration we determined the protein amount (gray value determined by Delta2D 2-D gel evaluation software) changed for all up- or down-regulated proteins for each stage. Now the sum of the protein amounts for e.g. all significantly up-regulated proteins from controls was subtracted from the sum of all HD proteins. Therefore we obtained the actual amount of changed protein. To determine the total protein amount changed we added the amount for up- and down-regulation. To determine whether total amount of protein changed was significantly different between stages we used an unpaired t test. The total amounts changed for each 2-D gel pair (five HD versus control repeats) were calculated separately. We subsequently compared the total amounts of each stage with its adjacent stages by t test. All protein amounts are relative as they are altered as compared with control, and although changes in gray scale value (spot volume) are proportional to protein concentration changes the absolute concentration values are not available. Sample size comprised at least five biological sample pairs. The Student's t test was used because the data investigated were normally distributed. The Kolmogorov-Smirnov Z test provided by the statistical analysis software SPSS 16.0 (SPSS Inc., Chicago, IL) was used to determine the “normal” distribution of our data. SPSS calculates a “two-tailed significance level,” testing the probability that the observed distribution is significantly deviant from the expected normal distribution. That is, a finding of non-significance means that the sample distribution is normal. We used the data set of statistically significant protein isoforms (p < 0.05, paired Student's t test) at 8 weeks to check for normal distribution. This data (sub)set was chosen because (i) testing all data points (4816 spots) was very difficult using the program available (SPSS) and (ii) the significant changes were the relevant ones for this study. We investigated the up- and down-regulated spots separately. The two-tailed significance level for up- and down-regulated protein isoforms was on average 0.81 ± 0.21 and 0.86 ± 0.17, respectively. That is, the null hypothesis (normal distribution) is true (p > 0.05), and the data are therefore normally distributed. In addition, all protein isoform changes tested were distributed normally.
The rate of false positives was estimated according to the following equation for unpaired t tests.
 |
Eq. 1 |
We assumed that the null hypothesis that there is no difference between samples tested is valid. In addition, x1 and x2 are the sample means of the distributions, and s1 and s2 are the corresponding standard deviations. n1 and n2 indicate the number of sample pairs used in our study. Because we assumed an equal standard normal distribution of both data sets to be tested for false positives we assume that the standard deviation s1 = s2. Z denotes the desired confidence and was determined to be 2.015 for Q(0.95) (0.95 quantiles) and n = n1 = n2 = 5 according to the “quantiles of Student's t distribution.” The false positive rate was therefore determined to be smaller than 15% of the significant changes for an unpaired t test. This value is even smaller for paired t tests.
Evaluation of False Positive Protein Changes—
To determine the reliability of the results obtained by our HD time course investigation it was important to establish a base line of protein alterations that occur even if no disease is present. Therefore we selected a representative time point, 8 weeks, and compared eight control gels with each other. We randomly allocated the eight 2-D gels into two groups of four each. Those groups were analyzed in the same way as HD and control gels were. We analyzed a total of 4031 protein spots by an unpaired Student's t test (p < 0.05) and found that 17 spots were up-regulated and 22 were down-regulated in expression. This makes a total of 39 spots changed at 8 weeks without disease present. When comparing this result with the changes obtained during our time course study where 205, 42, 40, 157, and 240 spots were altered between HD and control at 2, 4, 6, 8, and 12 weeks, respectively (Table I), we found a percentage of false positives of 19, 93, 98, 25, and 16%, respectively. In addition, a direct comparison of changes at 8 weeks (157 spots) shows that 25% of the proteins may be false positives, or at least 118 changes were identified correctly.
Table I.
Disease progression in the HD mouse model R6/2 is only reflected late in disease by altered protein isoforms
Shown is the number of differentially expressed protein spots. Only spots altered by >10% of spot volume and p < 0.05 are indicated. A paired Student's t test was used to determine statistical significance. All spots differentially expressed on the 2-D gels at each respective stage are listed.
| Age | Number of differentially expressed spots
|
||
|---|---|---|---|
| Up | Down | Total | |
| weeks | |||
| 2 | 117 | 88 | 205 |
| 4 | 19 | 23 | 42 |
| 6 | 17 | 23 | 40 |
| 8 | 93 | 64 | 157 |
| 12 | 120 | 120 | 240 |
Protein Identification—
For protein identification by mass spectrometry, 40 μl of extract was separated by 2-DE and stained using an MS-compatible silver staining protocol (42). Protein spots of interest were excised from 2-D gels and subjected to in-gel tryptic digestion. Tryptic fragments were analyzed by nanoflow HPLC (Dionex/LC Packings, Amsterdam, Netherlands)/ESI-MS and -MS/MS on an LCQ Deca XP ion trap instrument (Thermo Finnigan, Waltham, MA). Nanoflow HPLC was directly coupled to ESI-MS analysis. Protein spot eluates of 15 μl were loaded onto a PepMap100 C18 precolumn (5 μm, 100 Å, 300-μm-inner diameter × 5 mm; Dionex/LC Packings) using 0.1% (v/v) trifluoroacetic acid at a flow rate of 20 μl/min. Peptides were separated onto a PepMap100 C18 100 column (3 μm, 100 Å, 75-μm-inner diameter × 15 cm; Dionex/LC Packings). The elution gradient was created by mixing 0.1% (v/v) formic acid in water (solvent A) and 0.1% (v/v) formic acid in acetonitrile (solvent B) and run at a flow rate of 200 nl/min. The gradient was started at 5% (v/v) solvent B and increased linearly up to 50% (v/v) solvent B after 40 min. ESI-MS data acquisition was performed throughout the LC run. Three scan events, (i) full scan, (ii) zoom scan of most intense ion in full scan, and (iii) MS/MS scan of the most intense ion in full scan, were applied sequentially. No MS/MS scan on single charged ions was performed. Raw data were extracted by the TurboSEQUEST algorithm, and trypsin autolytic fragments and known keratin peptides were subsequently filtered. All DTA (peak list files for mass spectrometry results generated by the SEQUEST search algorithm) files generated by BioWorks version 3.2 (Thermo Scientific, Waltham, MA) were merged and converted to MASCOT generic format files (MGF). Mass spectra were analyzed using our in-house MASCOT software package license version 2.1 automatically searching the NCBInr database for Mus musculus (house mouse) (NCBInr_20061206, 107,853 sequences). The M. musculus subset of the NCBInr database was used because only mouse samples were investigated. In rare cases, hits were researched using the Mammalia subset of the NCBInr database. All non-M. musculus proteins are indicated in supplemental Table 1 by addition of either “Homo sapiens” or “Rattus norvegicus” after their protein name. To reduce the length of the protein names for the large majority of M. musculus identifications because of space constraints the species label was omitted in many cases. MS/MS ion search was performed with the following set of parameters: (i) taxonomy, M. musculus (house mouse); (ii) proteolytic enzyme, trypsin; (iii) maximum of accepted missed cleavages, 1; (iv) mass value, monoisotopic; (v) peptide mass tolerance, 0.8 Da; (vi) fragment mass tolerance, 0.8 Da; and (vii) variable modifications, oxidation of methionine and acrylamide adducts (propionamide) on cysteine. No fixed modifications were considered. Only proteins with scores corresponding to p < 0.05 with at least two independent peptides identified were considered. The cutoff score for individual peptides using ESI identification was equivalent to p < 0.05 for each peptide and usually in a MOWSE (molecular weight search) score range from 32 to 37. This number was calculated by the MASCOT software. Furthermore theoretical and practical molecular weight and pI for each protein identified by database search were compared to remove proteins with deviating masses and pI values.
Pathway Enrichment Analysis in the Protein Data Set—
Official gene symbols and gene names (Mouse Genome Informatics) were used to investigate similarities in protein expression alterations between stages. The gene names were retrieved using the GI numbers supplied by MASCOT after a database search. To investigate an enrichment of specific pathways in the altered protein expression data set, we used the “Web-based gene set analysis toolkit” (WEBGESTALT) tool supplied by Vanderbilt University. We used the “Gene set analysis tool” and selected the gene set analysis option “Function” and the category “KEGG table and maps.” KEGG is the abbreviated form of Kyoto Encyclopedia of Genes and Genomes and is a bioinformatics database containing information on genes, proteins, reactions, and pathways. The following parameters were used to create the KEGG tables: reference set, “WEBGESTALT_MOUSE”; significance level, p < 0.01; and minimum number of genes, 2. Statistical methods available were “hypergeometric test” and “Fisher's exact test.” For our data the results were the same with either test.
Analysis of mRNA Data Sets for Co-regulation with Protein Expression Data Sets—
We used an mRNA data set for the R6/2 HD mouse model for 6, 9, and 12 weeks that had been published previously (22, 43). Only the striatal data set was utilized. The data sets were already analyzed for statistical significance in their respective studies (22, 43). Briefly Affymetrix microarrays were normalized using robust multiarray averaging. Analysis was performed using R version 2.3 and the Bioconductor packages Affy and Limma. Differential gene expression in each array set was assessed relative to unaffected controls using paired t tests. Random matching generated six HD-control sample pairs (43). The mRNA data set consists of 22,626 probe set identities. From the entire mRNA data set only genes with differential regulation at the protein level were selected for analysis. On the single mRNA/protein level we found that 88% of 371 altered proteins could be correlated to corresponding mRNA data. A common gene symbol (mouse variant) of protein and mRNA data was used as the selection criterion. We now had mRNA expression data for 328 of our proteins at 6, 9, and 12 weeks for HD and control at our disposal. We now determined whether any significant changes occurred in any of the 328 mRNAs at each of the three stages. The significance level for altered mRNA expression selected for our study was p < 0.05. If probe sets were not altered significantly in more than 66% of cases (note that more than one probe set per gene name is present on the mRNA chip) they were discarded. In the three data sets at 6, 9, and 12 weeks of age, one (Rbmx), three (Eef1a2, Mapre2, and Nono), and zero genes showed opposite regulatory behavior, respectively. Opposite regulatory behavior was present when individual probe sets coding for the same gene name on the mRNA array showed opposite expression behavior. The number of statistically significant mRNA alterations of the total of 328 was determined for each stage. The gene names for altered proteins were compared with the gene names of mRNAs to determine the overlap in expression changes between protein and mRNA on the single mRNA/protein level.
To investigate the overlap in cellular pathways between the mRNA and the protein data sets we used WEBGESTALT using the same parameters as already described earlier under “Pathway Enrichment Analysis in the Protein Data Set.” To determine enriched pathways we used all significantly changed probe sets from the mRNA data sets at the stage investigated and not just those altered also in the protein data set.
RESULTS
We investigated the HD mouse model R6/2 for changes in the expression levels of proteins during phenotype onset and progression using a 2-DE gel-based proteomics approach. We observed two stages at which an extraordinarily large number of protein alterations had occurred. These peak alterations were present prior to onset and after the development of pronounced HD-related phenotypes. In addition, most of the changes were found to be stage-specific.
Altered Protein Expression during HD Progression in R6/2 Mice—
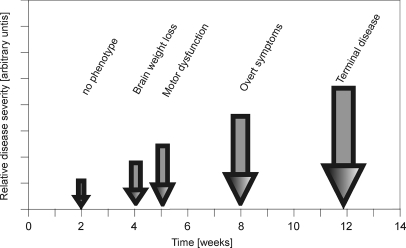
To cover onset and all stages of disease progression in the well characterized R6/2 HD mouse model, we selected ages starting where no phenotype is present (2 weeks) and finishing where mice demonstrate pronounced symptoms (12 weeks). In our colony, disease end point is defined by 20% loss of body weight that occurs at 14–15 weeks of age. Further time points were selected to correspond to stages where hallmarks of disease progression appear. Loss of brain weight occurs from 4 weeks of age (44), an impairment of motor function as measured by RotaRod analysis is present from ∼6 weeks (45), and a visible phenotype is present from ∼8 weeks (45) (Fig. 1). After comparing R6/2 with control mice for 2, 4, 6, 8, and 12 weeks separately the number of differentially expressed proteins for each time point investigated was determined (Table I). For the 2-, 4-, 6-, 8-, and 12-week time points, a total of 3821, 4006, 4030, 4816, and 4283 protein isoforms were analyzed, respectively, to determine significant protein expression differences. As expected very few protein isoforms were altered in R6/2 mice at 4 (42 isoforms, 1.0% of total isoforms investigated at this age) and 6 weeks (40 isoforms, 1.0%) of age because the phenotype at these stages is very mild. The number of altered protein isoforms increased more than 3-fold at 8 weeks (157 isoforms, 3.3%) and a further 1.5-fold at 12 weeks (240 isoforms, 5.6%).
Fig. 1.
Scheme of disease progression in the HD mouse model R6/2. Pathological hallmarks are marked on the time scale by arrows, and the observed phenotype is indicated on top of each arrow.
Unexpectedly we found a large number of protein isoforms altered at 2 weeks of age (205 isoforms, 5.4%). This number was almost as high as those detected at 12 weeks (Table I). We suspected that the number of protein isoforms changed at 2 weeks might not necessarily reflect a drastic change in the total amount of altered protein. Therefore, we determined the relative amount of protein changed (Fig. 2) and found that 3.1% of the total spot volume (protein concentration) was altered at 2 weeks comparable to 3.2% at 12 weeks of age. Again the values were almost identical. The y axis of Fig. 2 indicates the amount of protein changed relative to a total protein concentration of 100% for a 2-D gel.
Fig. 2.
Relative protein concentration changes during HD progression. Total brain extracts of R6/2 mice were studied at 2, 4, 6, 8, and 12 weeks of age. Protein concentration alterations were calculated for all significantly altered protein isoforms (p < 0.05, <0.9- or >1.1-fold change). The total spot volume (spot intensity × spot area) on each 2-D gel was set to 100%. The y axis indicates the amount of protein changed relative to a total protein concentration of 100%. Asterisks indicate statistical significance (p < 0.05). Black bars indicate up-regulated proteins, and gray bars indicate down-regulated proteins. The numbers above each bar indicate percentage of change. The line above the bars indicates the sum of up- and down-regulated protein concentration changes that is also indicated by numbers.
We subsequently identified the altered protein isoforms by mass spectrometry. Table II shows the number of identified proteins for each stage separately. The identification rates were in general very high, ranging from 83 to 94% except for at 6 weeks (53%). However, at 6 weeks, only a very small number of proteins were altered (40), and therefore, the failure to identify specific proteins makes a huge difference to the percentage identified. Because of the generally high identification ratio, the proteins changed represent the entire data set of altered protein spots (Table I). A low identification rate may leave an important subset of proteins beyond scrutiny and may bias the study for easily identifiable proteins. It is well established that a protein may be represented by more than one protein isoform (spot) on a 2-D gel. Therefore we determined the number of non-redundant proteins that were changed for each stage. We used the gene name as a selection criterion: proteins sharing the same gene name were considered as one non-redundant protein. Therefore, proteins with a different protein name but the same gene name and a protein altered in more than one isoform (seen on the 2-D gel as a protein spot (18)) on the 2-D gel were considered to be the same protein and counted only once per stage. Table IIIA shows the number of non-redundant proteins changed at each disease stage. As might have been expected, at each stage proteins were represented by more than one protein spot on the 2-D gel. The number of non-redundant proteins was 73, 89, 86, 77, and 70% of the total number of proteins identified at 2, 4, 6, 8, and 12 weeks, respectively. In total 371 individual, non-redundant proteins were identified. Interestingly almost all proteins represented by more than one spot showed the same regulatory pattern in all isoforms; that is, they were either up- or down-regulated (Table IIIB).
Table II.
Number of identified proteins changed in R6/2 disease progression
| Age | Identified differentially expressed spots
|
||
|---|---|---|---|
| Up | Down | Totala | |
| weeks | |||
| 2 | 91 | 80 | 171 (83%) |
| 4 | 16 | 21 | 37 (88%) |
| 6 | 10 | 11 | 21 (53%) |
| 8 | 93 | 45 | 138 (88%) |
| 12 | 114 | 112 | 226 (94%) |
The percentage of identified proteins as compared with the numbers in Table I is given in parentheses. The number of differentially expressed proteins is listed. Only proteins altered by >10% of spot volume and p < 0.05 are indicated. A paired Student's t test was used to determine statistical significance.
Table III.
Number of non-redundant proteins changed at each stage during R6/2 disease progression
| Age | A. Number of proteins
|
||
|---|---|---|---|
| Up | Down | Totala | |
| weeks | |||
| 2 | 65 | 59 | 124 (73%) |
| 4 | 15 | 18 | 33 (89%) |
| 6 | 10 | 8 | 18 (86%) |
| 8 | 74 | 31 | 105 (77%) |
| 12 | 84 | 74 | 158 (70%) |
| Age | B. Number of proteins with more than one spot
|
||
|---|---|---|---|
| Sameb | Differentb | Total | |
| weeks | |||
| 2 | 47 | 0 | 47 |
| 4 | 4 | 0 | 4 |
| 6 | 3 | 0 | 3 |
| 8 | 33 | 2 | 35 |
| 12 | 68 | 3 | 71 |
The percentage of non-redundant proteins as compared with the number of proteins identified in Table II is indicated in parentheses.
Regulation of redundant spots (up/down) is in the same or opposite direction. Only proteins altered by >10% of spot volume and p < 0.05 are indicated. A paired Student's t test was used to determine statistical significance.
Drastic Protein Expression Changes Early in Disease Precede HD-related Phenotypes—
The most unexpected observation was that an early peak in protein alterations in terms of numbers (Tables I–III) and amount (Fig. 1) was observed prior to phenotype onset, and we sought to identify the mechanism underlying this early peak. It is already known that protein changes related to development that are present at 2 weeks of age in the mouse are drastically reduced in adulthood (38). We investigated the longitudinal changes in R6/2 and wild-type mice to elucidate the magnitude of protein changes during development in the presence and absence of disease (Table IV). This means we compared each stage within a group (HD or control) with its adjacent stages. The number of protein isoform changes between 2 and 4 weeks, 318 in HD and 298 in wild type, were more numerous than all subsequent changes during longitudinal development and its interaction with HD disease progression. Interestingly the number of changes in wild-type and R6/2 mice reached another peak between 6 and 8 weeks (Table IV). These changes were reproduced when alterations in protein amount (concentration) were considered: again alterations were most dramatic between 2 and 4 weeks in HD and wild-type mice (16.3 and 13.3%, respectively) (Table V). All mice with nominal age 2 weeks were sacrificed exactly 14 days after birth, but to rule out a difference in “developmental age” between R6/2 and control mice at 2 weeks of age we compared the body weight of both groups. R6/2 mice had an average weight of 7.3 ± 0.75 g, and controls had an average weight of 7.54 ± 1.15 g. Brain weights were normally distributed and showed no statistically significant difference (p = 0.814, paired Student's t test).
Table IV.
Longitudinal changes during wild-type and R6/2 development: number of proteins
Only proteins altered by >10% of spot volume and p < 0.05 are indicated. An unpaired Student's t test was used to determine statistical significance. Changes within the group of HD or control were considered. Adjacent age stages were compared with each other.
| Age | Number of protein isoforms
|
||
|---|---|---|---|
| Up | Down | Total | |
| weeks | |||
| A. R6/2 | |||
| 2/4 | 137 | 181 | 318 |
| 4/6 | 39 | 36 | 75 |
| 6/8 | 26 | 105 | 131 |
| 8/12 | 66 | 23 | 89 |
| B. Wild type | |||
| 2/4 | 162 | 136 | 298 |
| 4/6 | 30 | 80 | 110 |
| 6/8 | 78 | 230 | 308 |
| 8/12 | 110 | 32 | 142 |
Table V.
Longitudinal changes during wild-type and R6/2 development: amount of protein
Only proteins altered by >10% of spot volume and p < 0.05 are indicated. An unpaired Student's t test was used to determine statistical significance. Changes within the group of HD or control were considered. Adjacent age stages were compared with each other.
| Age | Amount of protein isoforms altered
|
||
|---|---|---|---|
| Up | Down | Total | |
| weeks | % | ||
| A. R6/2 | |||
| 2/4 | 8.5 | 7.8 | 16.3 |
| 4/6 | 0.9 | 1.2 | 2.1 |
| 6/8 | 2.7 | 3.2 | 5.9 |
| 8/12 | 2.2 | 2.0 | 4.2 |
| B. Wild type | |||
| 2/4 | 6.8 | 6.5 | 13.3 |
| 4/6 | 0.8 | 2.2 | 3.0 |
| 6/8 | 8.0 | 6.1 | 14.1 |
| 8/12 | 3.4 | 2.8 | 6.2 |
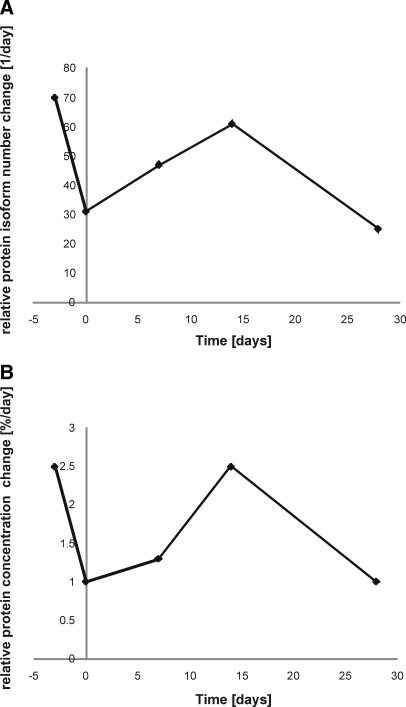
Therefore, the number and extent of protein expression changes occurring between 2 and 4 weeks were large regardless of the presence of disease. We now used C57BL/6 mice to study expression changes during early development before 2 weeks of age (Table VI). When comparing the results for wild-type CBA × C57BL/6 and inbred C57BL/6 mice, we found 298 versus 352 protein isoforms altered and 13.3 versus 14.2% changes in protein amount; this represents a very high degree of similarity (Tables IV, V, and VI). When looking at earlier changes it becomes clear that the amount of change is considerably higher between time points in earlier mouse developmental stages (35). We analyzed protein changes at embryonic days 16 and 18 and postnatal day (P) 0, P7, P14, and P28 (n = 3 per time point). We then calculated the number of isoforms and protein amount changed between consecutive time points and converted this to the average number of isoforms changed per day for comparison purposes (Table VI). A peak in the change in protein amount and isoform number was observed between P7 and P14 (1 and 2 weeks after birth; Table VI and Fig. 3). These data suggest that during normal development the changes in protein expression that occur at early stages (before 2 weeks) are already very pronounced. Generally the changes per day are very high except shortly before birth (Table VIB). A perturbation such as the expression of the transgenic fragment of the HD gene in R6/2 mice (32) may easily disturb a delicate equilibrium of expression changes during normal development. This may explain the large number of protein expression changes observed early in disease at 2 weeks (Tables I–III and Fig. 2).
Table VI.
Longitudinal changes during C57BL/6 mouse development
Only proteins that are altered >10% of spot volume and p < 0.05 are indicated. An unpaired Student's T-test was used to determine statistical significance. E, embryonic day; P, postnatal day.
| Age | A. Number of protein isoforms
|
|||
|---|---|---|---|---|
| Up | Down | Total | Total/day | |
| weeks | ||||
| E16/E18 (−3 days) | 52 | 87 | 139 | 70 |
| E18/P0 (0 days) | 37 | 57 | 94 | 31 |
| P0/P7 (7 days) | 157 | 170 | 327 | 47 |
| P7/P14 (14 days) | 245 | 181 | 426 | 61 |
| P14/P28 (14 days) | 155 | 197 | 352 | 25 |
| Age | B. Amount of protein
|
|||
|---|---|---|---|---|
| Up | Down | Total | Total/day | |
| weeks | % | |||
| E16/E18 (−3 days) | 2.6 | 2.3 | 4.9 | 2.5 |
| E18/P0 (0 days) | 1.1 | 1.8 | 2.9 | 1.0 |
| P0/P7 (7 days) | 4.8 | 4.3 | 9.1 | 1.3 |
| P7/P14 (14 days) | 7.7 | 9.6 | 17.3 | 2.5 |
| P14/P28 (14 days) | 8.8 | 5.4 | 14.2 | 1.0 |
Fig. 3.
Longitudinal changes of protein expression during development. Changes in protein expression per day in terms of number of proteins (A) and protein concentration (B) were determined. Total brain extracts of embryonic day 16 and 18 and neonate P0, P7, P14, and P28 wild-type C57BL/6 mice were compared. Altered protein numbers and concentration were calculated for all significantly altered protein isoforms (p < 0.05, <0.9- or >1.1-fold change). For a complete list of all alterations see Table VI.
Overlap of Protein Expression Changes between Different Stages of Disease Progression—
After studying the possible causes of the protein expression overlap, the identity and properties of the proteins generating this early peak in differential expression were investigated. First the degree of similarity between the proteins that change in disease was investigated. We compared the gene names corresponding to the non-redundant proteins identified for each stage and found a protein expression overlap of 6–38% depending on the stages compared (Table VIIA). The protein expression overlap is defined as the percentage of gene names shared between two stages compared. Interestingly the overlap between 2 and 12 weeks of age was very high (38%). We then considered whether the direction of change in expression (i.e. up or down) was concordant or discordant between stages (Table VIIB). It became clear that even if the same proteins were differentially regulated at both 2 and 12 weeks the direction of expression was not necessarily the same. Correction for the direction of expression, that is the same genes in the stages compared with opposite regulation (i.e. up versus down) were removed, reduced the overlap to 27%. This means that 73% of the protein changes were specific for 2 and 12 weeks of age. The overlap in differentially expressed proteins was corrected to account for the direction of change for all comparisons and found to range from 0 to 29% with the lowest being between 6 and 8 weeks of age (Table VIIA). Although an expression overlap of up to 29% is still large, overall between 71 and 100% of the proteins that were differentially expressed were stage-specific (Table VII).
Table VII.
Protein expression overlap between R6/2 stages investigated
A, non-redundant proteins (Table III) were compared between the time points indicated. Note that the time point with the lower number of differentially expressed proteins was set to 100% to ensure that changes can reach 100%. B, the number of overlapping protein changes (A, overlapping) was investigated for changes sharing the same or opposite direction of expression (up/down). Only proteins altered by >10% of spot volume and p < 0.05 are indicated. An unpaired Student's t test was used to determine statistical significance.
| Age | A. Number of non-redundant protein isoforms
|
||
|---|---|---|---|
| Overlapping | Total | Relative overlapa | |
| weeks | % | ||
| 2/4 | 10 | 33 | 30 (18) |
| 4/6 | 3 | 18 | 16 (11) |
| 6/8 | 1 | 18 | 6 (0) |
| 8/12 | 35 | 106 | 33 (29) |
| 2/12 | 46 | 124 | 38 (27) |
| Age | B. Number of overlapping protein changes
|
||
|---|---|---|---|
| Same | Opposite | Relative similarity | |
| weeks | % | ||
| 2/4 | 6 | 4 | 60 |
| 4/6 | 2 | 1 | 67 |
| 6/8 | 0 | 1 | 0 |
| 8/12 | 31 | 4 | 89 |
| 2/12 | 32 | 14 | 70 |
The expression overlap corrected by the relative similarity of expression orientation between mRNA and protein datasets (number in parentheses).
Early Changes in Energy Metabolism during HD Pathology—
To obtain a deeper understanding of the processes involved in disease progression it is important to determine whether specific pathways are enriched within the altered protein data set for which we carried out a KEGG analysis (46). We analyzed all time points separately and only included pathways where at least three proteins were enriched. This cutoff was chosen to ensure that a reasonable number of proteins were altered for a given pathway thereby providing strong evidence for altered regulation. Pathways enriched in up-regulated and down-regulated proteins were identified (Table VIII). Interestingly glycolysis/gluconeogenesis was found to be at the top of the up-regulated categories at all stages except at 12 weeks of age when most up-regulated proteins were found in pathways involving proteasome function. In contrast some proteins involved in glycolysis/gluconeogenesis were down-regulated at 12 weeks. Of other down-regulated pathways, the “regulation of actin cytoskeleton” was at the top of the list at 2 and 12 weeks of age, and proteasome function was down-regulated at 2 weeks of age. In summary, “glycolysis/gluconeogenesis” was mostly up-regulated, whereas “regulation of cytoskeleton” was down-regulated. The regulation of proteasome function seems to be stage-specific.
Table VIII.
Pathways enriched at each stage and during disease progression
A KEGG pathway analysis was carried out, and only pathways with three or more proteins and p < 0.01 were included. Pathways marked bold were altered in at least four of five time points studied.
| Age | Pathways | Number of proteins |
|---|---|---|
| weeks | ||
| A. Pathways up-regulated | ||
| 2 | Glycolysis/gluconeogenesis | 9 |
| Pentose phosphate pathway | 4 | |
| Citrate cycle (tricarboxylic acid cycle) | 4 | |
| Glutamate metabolism | 4 | |
| 4 | Glycolysis/gluconeogenesis | 3 |
| Oxidative phosphorylation | 3 | |
| 6 | No pathways | |
| 8 | Glycolysis/gluconeogenesis | 5 |
| Citrate cycle (tricarboxylic acid cycle) | 4 | |
| Proteasome | 3 | |
| Tryptophan metabolism | 3 | |
| 12 | Proteasome | 10 |
| Glycolysis/gluconeogenesis | 5 | |
| Citrate cycle (tricarboxylic acid cycle) | 3 | |
| Cell communication | 3 | |
| B. Pathways down-regulated | ||
| 2 | Regulation of actin cytoskeleton | 3 |
| Proteasome | 3 | |
| 4 | Gap junction | 2 |
| 6 | No pathways | |
| 8 | Oxidative phosphorylation | 3 |
| 12 | Regulation of actin cytoskeleton | 6 |
| Glycolysis/gluconeogenesis | 5 | |
| Axon guidance | 3 | |
| Pyruvate metabolism | 3 |
When the overlap between proteins of altered expression was considered, those overlapping between 2 and 12 weeks of age (Table VII) belong to glycolysis/gluconeogenesis, the “pentose phosphate pathway,” and “proteasome”. In contrast, when the protein expression overlap from 8 and 12 weeks was compared, glycolysis/gluconeogenesis was still the top ranking enriched pathway followed by “Parkinson disease” (Ube1x and Uchl1), and “metabolism of xenobiotics by cytochrome P450” as well as “MAPK signaling pathway” were also included. At the level of individual proteins, only nine gene names, Cops4, Efhd2, Mtpn, Phpt1, Sept7, Slc25a12, Stmn1, Tppp, and Uchl1, were identified in the expression overlap data sets of 2 versus 12 weeks and 8 versus 12 weeks. Therefore, because a total of 49 genes overlapped between 2 and 12 weeks and 35 overlapped between 8 and 12 weeks, there is an overlap of 23% between both data sets at the individual protein level.
We now investigated the possibility that because some proteins within an altered pathway may be up-regulated and others may be down-regulated those pathways may be lost in an analysis that separates up- and down-regulated proteins. Table X shows that all pathways found with separate analysis (Table VIII) were also found when up- and down-regulated proteins were subjected to KEGG analysis simultaneously. Because some numbers of proteins of the pathways involved are higher in Table X, proteins of the same pathway show opposite regulation. Still most proteins were either up- or down-regulated. Still more altered pathways per stage were found with simultaneous analysis, and importantly in addition to glycolysis/gluconeogenesis, “oxidative phosphorylation” was found to be altered at all stages except 6 weeks (Table X).
Table X.
Pathways enriched at each stage and during disease progression: all proteins
A KEGG pathway analysis was carried out, and only pathways with three or more proteins and p < 0.01 were included. Pathways marked bold were altered in at least four of five time points studied.
| Age | Pathways | Number of proteins |
|---|---|---|
| weeks | ||
| 2 | Glycolysis/gluconeogenesis | 9 |
| Citrate cycle (tricarboxylic acid cycle) | 6 | |
| Regulation of actin cytoskeleton | 5 | |
| Proteasome | 5 | |
| Oxidative phosphorylation | 5 | |
| Pentose phosphate pathway | 4 | |
| Glutamate metabolism | 4 | |
| Propanoate metabolism | 4 | |
| Pyruvate metabolism | 4 | |
| Fructose and mannose metabolism | 4 | |
| 4 | Glycolysis/gluconeogenesis | 4 |
| Oxidative phosphorylation | 4 | |
| 6 | No pathways | |
| 8 | Glycolysis/gluconeogenesis | 6 |
| Oxidative phosphorylation | 6 | |
| Citrate cycle (tricarboxylic acid cycle) | 5 | |
| Glutathione metabolism | 4 | |
| Fatty acid metabolism | 3 | |
| MAPK signaling pathway | 3 | |
| Proteasome | 3 | |
| Valine, leucine, and isoleucine degradation | 3 | |
| 12 | Glycolysis/gluconeogenesis | 10 |
| Proteasome | 10 | |
| Regulation of actin cytoskeleton | 7 | |
| Pyruvate metabolism | 6 | |
| Fructose and mannose metabolism | 5 | |
| Axon guidance | 4 | |
| Cell communication | 4 | |
| Oxidative phosphorylation | 3 | |
| Focal adhesion | 3 | |
| Neurodegenerative disorder | 3 | |
| Parkinson disease | 3 |
This common enrichment in altered pathways is clearly contrasted by the fact that only two proteins, represented by the gene names glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and pleckstrin homology (PH) and SEC7 domain-containing protein 3 (Psd3), were altered at four of the stages and that none were altered in all five stages. Gapdh catalyzes d-glyceraldehyde 3-phosphate, phosphate, and NAD+ to 3-phospho-d-glyceroyl phosphate and NADH and is involved in glycolysis, but a nuclear function has also been described. Psd3 acts as a guanine nucleotide exchange factor for ARF6 and is located at cell junctions, the presynapse, the postsynaptic cell membrane, and the postsynaptic density.
In summary, when analyzing our proteomics data set, we found two peaks in protein alteration, one early (5.4% of all protein isoforms changed, 2 weeks) and one late in disease (5.6%, 12 weeks). In addition, most changes at each time point investigated were stage-specific (71–100%; see Table VII).
Correlation of mRNA Expression Kinetics during HD Progression in R6/2 Mice—
Recently mRNA expression data for R6/2 mice at 6, 9, and 12 weeks of age have been published (22, 47), and we used these data sets to compare mRNA and protein expression data to determine the degree by which altered mRNA regulates protein expression. The data sets were generated from the striatum of 6-, 9-, and 12-week-old R6/2 mice. Of the 371 non-redundant proteins that we identified, only 39 were not represented in the mRNA data. Therefore we were able to correlate the expression profiles of 328 mRNAs (88% of the proteins) with their protein expression. Only two of the stages studied at the mRNA level (6 and 12 weeks) coincided directly with the stages studied on the proteome level (2, 4, 6, 8, and 12 weeks). We compared the mRNA expression data with adjacent protein expression data on the single mRNA/protein level. 6-week mRNA expression data were compared with 4-, 6-, and 8-week protein data, the 9-week mRNA data were compared with 8- and 12-week protein data, and the 12-week mRNA data were compared with 8- and 12-week protein data.
The comparison of altered mRNAs and proteins at 6 weeks revealed an overlap of 3 of 13 (23%), and at 12 weeks the overlap was 42% when the direction of alteration was not taken into account. At 6 weeks all three mRNAs that showed a statistically significant alteration were regulated in the opposite direction to the alteration in protein expression (up/down). At 12 weeks this was the case for 16 of the 58 altered mRNAs. Interestingly this opposite regulation was lower, 3 of 31 (10%), when the 9-week mRNA data set was considered (Table IX). Because the pathways that are differentially regulated at 2 and 12 weeks of age are quite similar, mRNA alterations at 12 weeks were compared with the proteins that were altered at 2 weeks. 42% of the mRNAs altered at 12 weeks were also altered at the protein level at 2 weeks. If those changing in the opposite direction are excluded the overlap is still 23% (Table IX). Therefore although there is co-regulation between protein and mRNA expression it is generally low on the level of individual proteins/mRNA.
Table IX.
Comparison of significantly altered protein with mRNA expression in R6/2 mice
| Age | Number of mRNAs
|
|||
|---|---|---|---|---|
| Total mRNAsa | 6 weeksb | 9 weeksb | 12 weeksb | |
| weeks | ||||
| 2 | 112 (12) | NDc | ND | 47 (21) |
| 4 | 33 (0) | 5 (2) | ND | ND |
| 6 | 13 (5) | 3 (3) | 6 (4) | ND |
| 8 | 99 (7) | 15 (5) | 25 (10) | 35 (6) |
| 12 | 137 (21) | ND | 31 (3) | 58 (16) |
Total mRNAs were determined by using the number of non-redundant proteins (see also Table IIIA (Total)) minus the number that are not represented in the mRNA data set (parentheses).
Number of mRNAs regulated in the opposite direction (up/down) to the proteins is indicated.
ND, not done.
We now investigated the overlap between pathways of the mRNA and protein data sets. We determined the number of differentially expressed proteins considering mRNAs with p < 0.05. Subsequently we carried out a KEGG analysis using 2775, 4338, and 2940 significantly altered, non-redundant genes for 6, 9, and 12 weeks, respectively. We compared up- and down-regulated mRNAs and proteins for each stage together (Tables X and XI). Stage “8 weeks” of the protein data set was compared with “9 weeks” of the mRNA data set, and stage “12 weeks” of the protein data set was compared with 12 weeks of the mRNA data set. We found an overlap of 6 of 8 pathways at 8/9 weeks and 6 of 10 pathways at 12 weeks. No pathways were altered in the protein data set at 12 weeks. The top five pathways in Table XI are those listed at the top of a KEGG pathway analysis when using the mRNA datasets for each of the three age stages investigated. The additional pathways listed are altered in at least one of the stages from the protein data set. Interestingly the top scoring pathways of the mRNA data set such as oxidative phosphorylation and regulation of actin cytoskeleton were also altered in the protein data set (Tables X and XI).
Table XI.
Pathways enriched at each stage and during disease progression: mRNA
The first five pathways selected were the top pathways in the KEGG analysis of the mRNA data set. The following pathways are those that overlap with any pathway in the protein data sets. The cutoff for inclusion into the table for the mRNA data sets was 10 mRNAs. Pathways marked bold overlap with protein expression data at 8 weeks (mRNA at 9 weeks) and 12 weeks. Only pathways with three or more proteins and p < 0.01 were included. Numbers in parentheses were obtained from a second dataset for 12-week-old R6/2 mice.
| Age | Pathways | Number of proteins |
|---|---|---|
| weeks | ||
| 6 | MAPK signaling pathway | 70 |
| Regulation of actin cytoskeleton | 43 | |
| Insulin signaling pathway | 40 | |
| Focal adhesion | 40 | |
| Neuroactive ligand-receptor interaction | 38 | |
| Axon guidance | 27 | |
| Oxidative phosphorylation | 26 | |
| Cell communication | 15 | |
| Glycolysis/gluconeogenesis | 10 | |
| Fatty acid metabolism | 10 | |
| Neurodegenerative disorders | 10 | |
| Proteasome | 10 | |
| 9 | MAPK signaling pathway | 102 |
| Focal adhesion | 68 | |
| Regulation of actin cytoskeleton | 64 | |
| Insulin signaling pathway | 56 | |
| Calcium signaling pathway | 52 | |
| Oxidative phosphorylation | 49 | |
| Axon guidance | 44 | |
| Cell communication | 20 | |
| Glycolysis/gluconeogenesis | 16 | |
| Citrate cycle (tricarboxylic acid cycle) | 15 | |
| Neurodegenerative disorders | 14 | |
| Glutamate metabolism | 13 | |
| Fatty acid metabolism | 10 | |
| 12 | MAPK signaling pathway | 64 (80) |
| Focal adhesion | 51 (50) | |
| Regulation of actin cytoskeleton | 46 (55) | |
| Purine metabolism | 41 (47) | |
| Cell cycle | 39 (44) | |
| Axon guidance | 32 (43) | |
| Oxidative phosphorylation | 19 (23) | |
| Cell communication | 17 (23) | |
| Valine, leucine, and isoleucine degradation | 13 (17) | |
| Proteasome | 14 (16) |
DISCUSSION
In this study we investigated early changes in protein expression and followed these during disease progression in the R6/2 HD mouse model. We observed two peaks of altered protein expression, one at 2 weeks of age prior to the onset of phenotypes and one at 12 weeks when symptoms are pronounced. These changes corresponded to about 6% of the entire proteome (protein isoforms) studied. Although most alterations were stage-specific, in some cases e.g. proteins involved in glycolysis/gluconeogenesis were dysregulated at every stage. In addition there was a pronounced similarity between early and late changes at the protein and mRNA level. When comparing the mRNA and protein changes they showed a small overlap (<30%); that is over 70% of the changes were specific to mRNAs or proteins.
The identification of early disease-related alterations will play a key role in our understanding of HD. However, most studies focus on late stages of disease in mouse models once an overt phenotype has occurred or study patients with manifest disease or postmortem brains of people who have died at late stage disease. In this study, we investigated the disease kinetics of altered protein expression levels, starting prior to phenotype onset and continuing through known stages of the disease.
Peak in Early Changes at 2 Weeks of Age—
The most startling result of this study was the extent to which protein alterations had already occurred at 2 weeks of age. The number of protein alterations was almost equivalent to those found at a late stage of disease and showed a significant overlap in terms of gene names. The pathway identified using the KEGG database that is most altered during disease progression is glycolysis/gluconeogenesis. Interestingly at 2 weeks, almost all glycolytic enzymes (phosphofructokinase (Pfkm), aldolase (Aldoa and Aldoc), Gapdh, phosphoglycerate kinase 1 (Pgk1), and pyruvate kinase (Pkm2)) were up-regulated in their expression level (supplemental Table 1). In addition, an up-regulation of energy metabolism was further supported by up-regulation of key enzymes of the citrate cycle: aconitase 2 (Aco2), fumarate hydratase 1 (Fh1), and malate dehydrogenase 2 (Mdh2). This is in accordance with a weight loss that occurs early in HD despite a high caloric intake (48). Weight loss has clearly been established in pre- and early symptomatic patients, and a significant reduction in the concentration of branched chain amino acids could be detected in their plasma by nuclear magnetic resonance spectroscopy suggesting that a perturbed mitochondrial energy metabolism is relevant to early pathogenesis (49). These changes are also consistent with an early disturbance of the transcription factor PGC-1α. The suppression of PGC-1α leads to mitochondrial dysfunction and therefore a dysfunction of the energy metabolism and neurodegeneration (50, 51).
At 2 weeks of age, an increase in “glutamate metabolism” was observed (Table VIIIA). Glutamate-ammonia ligase (Glul), glutamate dehydrogenase 1 (Glud1), glutamate oxalacetate transaminase 2 (Got2), and 4-aminobutyrate aminotransferase (Abat) were all up-regulated in expression. Glul converts glutamate to glutamine in astrocytes after glutamate-mediated neuronal signaling. Glutamate is removed by astrocytes after signaling from the synaptic cleft (52, 53), and glutamate conversion to glutamine is directly coupled to glutamate signaling (52, 53). Consequently this increased Glul activity may indicate an increase in glutamate signaling at a very early stage in HD progression at least in the R6/2 mouse model studied. Increased glutamate signaling at the N-methyl-d-aspartate subclass of ionotropic glutamate receptors has been linked to excitotoxicity, especially in striatal neurons, which has been proposed as a pathogenic disease mechanism for HD (54).
Furthermore at 2 weeks of age proteins involved in exo- and endocytosis were altered in expression. Cplx1, Cplx2 (both down-regulated), Syn1, and Syn2 (both up-regulated) are involved in exocytosis. Pacsin1 (up-regulated; three isoforms) and Dnm1 (down-regulated) are involved in endocytosis (55). In addition, Arpc1a and Arpc5, members of the Arp2/3 complex linking Pacsin1 and N-Wasp to actin to ensure functional endocytosis (56), were up- and down-regulated, respectively. Homer1 (two isoforms) located in the postsynaptic density (57) was up-regulated in expression. This dysregulation of proteins involved in exo- and endocytosis suggests that synapses are perturbed early in the R6/2 HD model. These changes were not detected by KEGG pathway analysis because up- and down-regulated proteins were investigated separately.
So far up- and down-regulated pathways were studied separately. Because proteins may be up- or down-regulated in the same pathways in disease we also considered pathway changes when up- and down-regulated proteins were subjected to KEGG analysis simultaneously. We found all pathways altered when we studied up- and down-regulated proteins separately. In addition we found that not only glycolysis/gluconeogenesis but also “oxidative phosphorylation” was altered at all stages except 6 weeks (Table X). In summary the changes observed may be due to a disturbance of the delicate equilibrium during development because of the large amount of changes naturally occurring at this time (Tables IV and V), or it is conceivable that the early changes act as a compensatory mechanism for early transgene Htt expression already in the absence of symptoms.
When considering the base-line level of false positives of 39 proteins per stage (see “Experimental Procedures” for more information) it becomes obvious that the protein alterations found at 4 and 6 weeks that are just above this level (Table I) should be treated with caution, whereas the other three time points that are more than 4-fold above this threshold are far more reliable. When considering this and the number of pathways altered at 2, 8, and 12 weeks in addition to confirmatory literature data the results of this study provide a valuable contribution to unraveling the dynamic protein changes in HD.
Changes Are Stage-specific on the Individual Protein Level but Overlapping in Terms of Metabolic Pathways—
Unexpectedly we found that most of the changes identified at each stage were stage-specific. The degree of specificity ranged from 71 to 100% (Table VII). One important conclusion of this finding is that a disease is a complex process that is not represented by linear changes in protein expression starting early at low levels, e.g. 2 weeks, increasing steadily to high levels close to terminal disease (12 weeks; Fig. 1). However, the groups of metabolic pathways altered are relatively constant, especially glycolysis/gluconeogenesis and those involved in proteasome function. It is interesting that the number of changes varies considerably between stages ranging from 18 non-redundant proteins at 6 weeks to 158 at 12 weeks (Table III).
Proteasomal Alterations Dominate Late Changes—
Alterations in the expression levels of proteins involved in proteasome function dominated at 12 weeks of age. Although proteasomal changes have already occurred at 2 weeks these were down-regulated. At 8 weeks they were up-regulated but less numerous, and at 12 weeks the following proteins represented by their gene names were up-regulated: PSMA3, PSMA5, PSMB1, PSMB3, PSMB4, PSMB5, PSMC3, PSMC5, PSMD4, and PSMD7. PSMA3 and PSMA5 are members of the 20 S proteasome α-subunits, whereas PSMB1, PSMB3, PSMB4, and PSMB5 are 20 S proteasome β-subunits (58). The β-subunits are catalytically active and responsible for proteasome specificity (58). So far 11 β-subunits have been identified in mice of which four were up-regulated in our study. In addition, all of the dysregulated subunits are essential subunits in yeast that are functionally conserved in humans (58). PSMC3, PSMC5, PSMD4, and PSMD7 belong to the regulatory or 19 S proteasome, which together with the 20 S complex forms the 26 S proteasome in eukaryotes (58–60). PSMA3, PSMB4, and PSMD4 were already up-regulated at 8 weeks of age. In contrast, PSMA5, PSMA6, and PSMB6 were down-regulated at 2 weeks. Only PSMA5 was later up-regulated at 12 weeks. Therefore, proteasomal changes are specific to certain stages of disease. Global changes to the ubiquitin proteasome system have already been reported in HD at intermediate and late stages of disease at the functional level (61). In this context it is interesting that UBE1X (UBA1) was up-regulated in R6/2 mice at 8 and 12 weeks of age. UBE1X is currently thought to be the sole E1 for charging E2s with ubiquitin in mammals (62, 63). E1, E2, and E3 are enzymes that conjugate ubiquitin to cellular proteins in a multistep pathway prior to their degradation (64). In addition UCHL1, which has already been shown to be involved in the pathology of Parkinson disease and exhibits a ligase as well as a ubiquitin hydrolase activity, was down-regulated at 2 weeks of age but up-regulated at 8 and 12 weeks (supplemental Table 1). In summary components of the ubiquitin/proteasome pathway are up-regulated in symptomatic disease. This may reflect efforts to remove protein aggregates that are a hallmark of HD and other neurodegenerative diseases (65–67).
Relatively High Degree of Overlap between Early and Late Changes—
When comparing changes at 2 and 12 weeks of age many of the protein changes can be observed at both stages (Table VIIB). It is interesting that this overlap exists at the level of pathways as well as at the level of individual proteins (Tables VIII and IX). However, the regulatory pattern of alterations is different at all stages (Tables VIIIB and IX). This may be caused by different types of perturbation in early and late stage disease that is reflected by the differential changes. This is supported by the changes observed in glycolysis/gluconeogenesis that were strictly up-regulated at 2 weeks of age but that were divided (five versus five proteins) at 12 weeks. An early effort to compensate for increased energy requirement early in disease (2 weeks) may be followed by a general deterioration of energy metabolism later prior to death.
No Protein Was Altered in All and Only Two Proteins Were Changed at Four Stages—
After comparing the proteins changed at each stage it became obvious that only two proteins, Psd3 and Gapdh, were changed at four of the five stages investigated, and none were altered at all five stages. Interestingly Gapdh has not only been reported to be involved in the pathology of HD (68, 69) but also in Alzheimer and Parkinson diseases (68). The number of proteins changed is therefore of more interest than the protein identities. The number of proteins investigated by a 2-DE proteomics approach is limited to up to 10,000 protein isoform spots depending on the application (18). The total number of proteins of the mouse proteome affected by HD is considerably larger than the number investigated in our study, but the ratio of changed proteins compared to unchanged proteins may remain approximately the same; that is the relative number of protein changes for each time point remains constant between our study and all proteins in the proteome. Therefore this study argues against a model in which there is a gradual increase in the number and magnitude of protein changes during disease progression and suggests a more dynamic regulatory pattern. We suggest that early changes affect late stage disease by changing processes in the mouse brain irreversibly. Our group has already shown that acute and long term proteome changes can be induced by oxidative stress in the developing brain (70). Oxidative stress has been proposed as a pathogenic mechanism for neurodegenerative disease in general. Early energy deficits or structural changes (Table VIII) may act in the same way.
High Correlation between Altered mRNA and Protein Expression on the Pathway Level—
In the past, a large number of mRNA expression studies were done for HD mouse models (14, 21, 22, 43, 47, 71) as well as human tissue (26, 47) and blood samples (27). All studies so far used intermediate or late stage disease samples starting for the R6/2 mouse model used in our study at 6 weeks of age (22, 24). This means that the two earliest time points, 2 and 4 weeks, used in our study could not be correlated to mRNA expression, and it was not possible to investigate whether the early rise in differential protein expression was mirrored by a rise in altered mRNA expression. Still we found that the correlation between mRNA and protein expression changes was very low in our study on the level of individual proteins and did not exceed 30%, although 88% of the differentially expressed proteins were represented by mRNAs. This is consistent with previous studies in which mRNA data did not necessarily correlate well with protein data (72–76), although these studies did not investigate changes due to disease. In a recent study using brain cortex tissue, we found a 60–70% overlap between mRNA and protein changes during embryonic development where processes are highly predetermined and may be regulated to a large degree by mRNA expression with subsequent protein changes (35). Changes from three time points in embryonic development, embryonic days (ED) 9.5, 11.5, and 13.5, were compared with each other. Unfortunately the study investigates only changes up to ED 13.5, ending about 7 days before birth at ED 21 after gestation. Still during disease progression in contrast to development a cell or tissue may have to react to complex, unforeseen, and highly dynamic protein changes that are not predetermined by altered mRNA expression as they are in embryonic development. mRNA changes may be largely the result of pathological processes at the protein level such as protein aggregation or aberrant interactions of Htt that perturb transcription (77, 78).
After studying the overlap on the level of individual proteins we tested the pathway overlap between protein and mRNA data. We compared altered pathways at 8 and 12 weeks of the protein data set with 9 and 12 weeks of the mRNA data set (Tables X and XI). We found that the overlap of the pathways involved was actually very high. Therefore it is important to consider functional units such as pathways to measure the overlap between mRNA and protein data sets and not individual proteins or mRNAs.
In addition, a recent mRNA expression study of total brain extracts from R6/2 mice at 12 weeks of age revealed an alteration of 42 transcripts (see Fig. 1A of Ref. 79) that were all down-regulated in expression (79). When comparing the gene names from this study with our protein expression results we found no overlap on the single gene/protein or pathway level. Using the KEGG pathway analysis tool we found that the five most prominently altered pathways in the mRNA data set (79) were all related to neuronal signaling (“neuroactive ligand-receptor interaction” (six mRNAs), MAPK signaling pathway (six mRNAs), “cytokine-cytokine receptor interaction” (six mRNAs), “long term potentiation” (five mRNAs), and “GnRH signaling pathway” (five mRNAs)). Interestingly when considering the mRNA data investigated in this study (Table XI) we found that the MAPK signaling pathway was altered in both mRNA studies. Upon a more detailed inspection of the KEGG analysis of mRNA data for 12 weeks (data not shown), neuroactive ligand-receptor interaction (39 mRNAs), cytokine-cytokine receptor interaction (34 mRNAs), long term potentiation (22 mRNAs), and GnRH signaling pathway (25 mRNAs) were also altered on the mRNA level in our study. The most likely explanation for the discrepancy between mRNA and protein data is that most altered signaling mRNAs found in the study of Luthi-Carter et al. (79) are proteins that are either hydrophobic (receptors) and/or low in abundance (ligands) and can therefore not be detected by 2-DE. Importantly it would be of tremendous value to study mRNA and protein expression in the same tissue and time points in parallel because only then will we be able to determine the true correlation between differential mRNA and protein expression overlap without confounding influences such as different time points investigated (8 versus 9 weeks) or brain regions (total brain versus striatum).
In summary, we used a large 2-D gel/mass spectrometry-based proteomics approach to investigate HD-induced protein expression alterations and their kinetics prior to the onset of phenotypes and during the course of disease. Unexpectedly we found that protein changes were largely stage-specific (71–100%), and a drastic alteration (almost 6% of the proteome) in protein expression that occurred as early as 2 weeks of age predominantly included up-regulation of glycolysis/gluconeogenesis and down-regulation of the actin skeleton. This suggests a period of highly variable protein expression that precedes the visible HD phenotype. Although an up-regulation of glycolysis/gluconeogenesis-related protein alterations remained dominant during HD progression, late stage alterations at 12 weeks showed an up-regulation of proteins having proteasomal function. Our observations suggest that HD is characterized by a highly dynamic disease pathology not represented by linear protein concentration alterations over the course of the disease. Detailed time course studies for disease progression with emphasis on early and very early stages are important to understand disease pathology and determine the time for intervention when considering HD therapy.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance by Janine Stuwe. We are indebted to Dr. Fanny Mochel for critically reading the manuscript and the valuable suggestions for improving the discussion of the data.
Footnotes
Published, MCP Papers in Press, November 30, 2008, DOI 10.1074/mcp.M800277-MCP200
The abbreviations used are: HD, Huntington disease; 2-D, two-dimensional; 2-DE, two-dimensional gel electrophoresis; Htt, Huntingtin; P, postnatal day; WEBGESTALT, Web-based gene set analysis toolkit; NCBInr, National Center for Biotechnology Information non-redundant; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; E, embryonic day; GnRH, gonadotropin-releasing hormone; Gapdh, glyceraldehyde-3-phosphate dehydrogenase.
This work was supported by the German Ministry for Education and Research (BMBF) within the National Genome Research Network (NGFN2) program.
This work is dedicated to the memory of our valued colleague and friend Maik A. Wacker who died too early in an accident.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Wexler, N. S., Rose, E. A., and Housman, D. E. ( 1991) Molecular approaches to hereditary diseases of the nervous system: Huntington's disease as a paradigm. Annu. Rev. Neurosci. 14, 503–529 [DOI] [PubMed] [Google Scholar]
- 2.The Huntington's Disease Collaborative Research Group ( 1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 3.McMurray, S. E., and McMurray, C. T. ( 2001) Huntington's disease. A sports star and a cook. Lancet 358, (suppl.) S38. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein, D. C., Leggo, J., Coles, R., Almqvist, E., Biancalana, V., Cassiman, J. J., Chotai, K., Connarty, M., Crauford, D., Curtis, A., Curtis, D., Davidson, M. J., Differ, A. M., Dode, C., Dodge, A., Frontali, M., Ranen, N. G., Stine, O. C., Sherr, M., Abbott, M. H., Franz, M. L., Graham, C. A., Harper, P. S., Hedreen, J. C., Jackson, A., Kaplan, J.-C., Losekoot, M., MacMillan, J. C., Morrison, P., Trottier, Y., Novelletto, A., Simpson, S. A., Theilmann, J., Whittaker, J. L., Folstein, S. E., Ross, C. A., and Hayden, M. R. ( 1996) Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am. J. Hum. Genet. 59, 16–22 [PMC free article] [PubMed] [Google Scholar]
- 5.Bates, G. ( 2003) Huntingtin aggregation and toxicity in Huntington's disease. Lancet 361, 1642–1644 [DOI] [PubMed] [Google Scholar]
- 6.Telenius, H., Kremer, H. P., Theilmann, J., Andrew, S. E., Almqvist, E., Anvret, M., Greenberg, C., Greenberg, J., Lucotte, G., Squitieri, F., Starr, A., Goldberg, Y. P., and Hayden, M. R. ( 1993) Molecular analysis of juvenile Huntington disease: the major influence on (CAG)n repeat length is the sex of the affected parent. Hum. Mol. Genet. 2, 1535–1540 [DOI] [PubMed] [Google Scholar]
- 7.Waelter, S., Scherzinger, E., Hasenbank, R., Nordhoff, E., Lurz, R., Goehler, H., Gauss, C., Sathasivam, K., Bates, G. P., Lehrach, H., and Wanker, E. E. ( 2001) The huntingtin interacting protein HIP1 is a clathrin and α-adaptin-binding protein involved in receptor-mediated endocytosis. Hum. Mol. Genet. 10, 1807–1817 [DOI] [PubMed] [Google Scholar]
- 8.Engelender, S., Sharp, A. H., Colomer, V., Tokito, M. K., Lanahan, A., Worley, P., Holzbaur, E. L., and Ross, C. A. ( 1997) Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum. Mol. Genet. 6, 2205–2212 [DOI] [PubMed] [Google Scholar]
- 9.Modregger, J., DiProspero, N. A., Charles, V., Tagle, D. A., and Plomann, M. ( 2002) PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington's disease brains. Hum. Mol. Genet. 11, 2547–2558 [DOI] [PubMed] [Google Scholar]
- 10.Gervais, F. G., Singaraja, R., Xanthoudakis, S., Gutekunst, C. A., Leavitt, B. R., Metzler, M., Hackam, A. S., Tam, J., Vaillancourt, J. P., Houtzager, V., Rasper, D. M., Roy, S., Hayden, M. R., and Nicholson, D. W. ( 2002) Recruitment and activation of caspase-8 by the Huntingtin-interacting protein Hip-1 and a novel partner Hippi. Nat. Cell Biol. 4, 95–105 [DOI] [PubMed] [Google Scholar]
- 11.Kim, Y. J., Yi, Y., Sapp, E., Wang, Y., Cuiffo, B., Kegel, K. B., Qin, Z. H., Aronin, N., and DiFiglia, M. ( 2001) Caspase 3-cleaved N-terminal fragments of wild-type and mutant huntingtin are present in normal and Huntington's disease brains, associate with membranes, and undergo calpain-dependent proteolysis. Proc. Natl. Acad. Sci. U. S. A. 98, 12784–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turmaine, M., Raza, A., Mahal, A., Mangiarini, L., Bates, G. P., and Davies, S. W. ( 2000) Nonapoptotic neurodegeneration in a transgenic mouse model of Huntington's disease. Proc. Natl. Acad. Sci. U. S. A. 97, 8093–8097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goehler, H., Lalowski, M., Stelzl, U., Waelter, S., Stroedicke, M., Worm, U., Droege, A., Lindenberg, K. S., Knoblich, M., Haenig, C., Herbst, M., Suopanki, J., Scherzinger, E., Abraham, C., Bauer, B., Hasenbank, R., Fritzsche, A., Ludewig, A. H., Bussow, K., Coleman, S. H., Gutekunst, C. A., Landwehrmeyer, B. G., Lehrach, H., and Wanker, E. E. ( 2004) A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell 15, 853–865 [DOI] [PubMed] [Google Scholar]
- 14.Kaltenbach, L. S., Romero, E., Becklin, R. R., Chettier, R., Bell, R., Phansalkar, A., Strand, A., Torcassi, C., Savage, J., Hurlburt, A., Cha, G. H., Ukani, L., Chepanoske, C. L., Zhen, Y., Sahasrabudhe, S., Olson, J., Kurschner, C., Ellerby, L. M., Peltier, J. M., Botas, J., and Hughes, R. E. ( 2007) Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 3, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butte, A. ( 2002) The use and analysis of microarray data. Nat. Rev. Drug Discov. 1, 951–960 [DOI] [PubMed] [Google Scholar]
- 16.Irizarry, R. A., Warren, D., Spencer, F., Kim, I. F., Biswal, S., Frank, B. C., Gabrielson, E., Garcia, J. G., Geoghegan, J., Germino, G., Griffin, C., Hilmer, S. C., Hoffman, E., Jedlicka, A. E., Kawasaki, E., Martinez-Murillo, F., Morsberger, L., Lee, H., Petersen, D., Quackenbush, J., Scott, A., Wilson, M., Yang, Y., Ye, S. Q., and Yu, W. ( 2005) Multiple-laboratory comparison of microarray platforms. Nat. Methods 2, 345–350 [DOI] [PubMed] [Google Scholar]
- 17.Klose, J. ( 1999) Fractionated extraction of total tissue proteins from mouse and human for 2-D electrophoresis. Methods Mol. Biol. 112, 67–85 [DOI] [PubMed] [Google Scholar]
- 18.Klose, J., and Kobalz, U. ( 1995) Two-dimensional electrophoresis of proteins: an updated protocol and implications for a functional analysis of the genome. Electrophoresis 16, 1034–1059 [DOI] [PubMed] [Google Scholar]
- 19.Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 20.Ong, S. E., and Mann, M. ( 2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 21.Chan, E. Y., Luthi-Carter, R., Strand, A., Solano, S. M., Hanson, S. A., DeJohn, M. M., Kooperberg, C., Chase, K. O., DiFiglia, M., Young, A. B., Leavitt, B. R., Cha, J. H., Aronin, N., Hayden, M. R., and Olson, J. M. ( 2002) Increased huntingtin protein length reduces the number of polyglutamine-induced gene expression changes in mouse models of Huntington's disease. Hum. Mol. Genet. 11, 1939–1951 [DOI] [PubMed] [Google Scholar]
- 22.Kuhn, A., Goldstein, D. R., Hodges, A., Strand, A. D., Sengstag, T., Kooperberg, C., Becanovic, K., Pouladi, M. A., Sathasivam, K., Cha, J. H., Hannan, A. J., Hayden, M. R., Leavitt, B. R., Dunnett, S. B., Ferrante, R. J., Albin, R., Shelbourne, P., Delorenzi, M., Augood, S. J., Faull, R. L., Olson, J. M., Bates, G. P., Jones, L., and Luthi-Carter, R. ( 2007) Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum. Mol. Genet. 16, 1845–1861 [DOI] [PubMed] [Google Scholar]
- 23.Luthi-Carter, R., Hanson, S. A., Strand, A. D., Bergstrom, D. A., Chun, W., Peters, N. L., Woods, A. M., Chan, E. Y., Kooperberg, C., Krainc, D., Young, A. B., Tapscott, S. J., and Olson, J. M. ( 2002) Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum. Mol. Genet. 11, 1911–1926 [DOI] [PubMed] [Google Scholar]
- 24.Luthi-Carter, R., Strand, A., Peters, N. L., Solano, S. M., Hollingsworth, Z. R., Menon, A. S., Frey, A. S., Spektor, B. S., Penney, E. B., Schilling, G., Ross, C. A., Borchelt, D. R., Tapscott, S. J., Young, A. B., Cha, J. H., and Olson, J. M. ( 2000) Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum. Mol. Genet. 9, 1259–1271 [DOI] [PubMed] [Google Scholar]
- 25.Luthi-Carter, R., Strand, A. D., Hanson, S. A., Kooperberg, C., Schilling, G., La Spada, A. R., Merry, D. E., Young, A. B., Ross, C. A., Borchelt, D. R., and Olson, J. M. ( 2002) Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington's disease mouse models reveal context-independent effects. Hum. Mol. Genet. 11, 1927–1937 [DOI] [PubMed] [Google Scholar]
- 26.Hodges, A., Strand, A. D., Aragaki, A. K., Kuhn, A., Sengstag, T., Hughes, G., Elliston, L. A., Hartog, C., Goldstein, D. R., Thu, D., Hollingsworth, Z. R., Collin, F., Synek, B., Holmans, P. A., Young, A. B., Wexler, N. S., Delorenzi, M., Kooperberg, C., Augood, S. J., Faull, R. L., Olson, J. M., Jones, L., and Luthi-Carter, R. ( 2006) Regional and cellular gene expression changes in human Huntington's disease brain. Hum. Mol. Genet. 15, 965–977 [DOI] [PubMed] [Google Scholar]
- 27.Runne, H., Kuhn, A., Wild, E. J., Pratyaksha, W., Kristiansen, M., Isaacs, J. D., Regulier, E., Delorenzi, M., Tabrizi, S. J., and Luthi-Carter, R. ( 2007) Analysis of potential transcriptomic biomarkers for Huntington's disease in peripheral blood. Proc. Natl. Acad. Sci. U. S. A. 104, 14424–14429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zabel, C., and Klose, J. ( 2004) Influence of Huntington's disease on the human and mouse proteome. Int. Rev. Neurobiol. 61, 241–283 [DOI] [PubMed] [Google Scholar]
- 29.Zabel, C., Chamrad, D. C., Priller, J., Woodman, B., Meyer, H. E., Bates, G. P., and Klose, J. ( 2002) Alterations in the mouse and human proteome caused by Huntington's disease. Mol. Cell. Proteomics 1, 366–375 [DOI] [PubMed] [Google Scholar]
- 30.Zabel, C., Sagi, D., Kaindl, A. M., Steireif, N., Klare, Y., Mao, L., Peters, H., Wacker, M. A., Kleene, R., and Klose, J. ( 2006) Comparative proteomics in neurodegenerative and non-neurodegenerative diseases suggest nodal point proteins in regulatory networking. J. Proteome Res. 5, 1948–1958 [DOI] [PubMed] [Google Scholar]
- 31.Perluigi, M., Poon, H. F., Maragos, W., Pierce, W. M., Klein, J. B., Calabrese, V., Cini, C., De Marco, C., and Butterfield, D. A. ( 2005) Proteomic analysis of protein expression and oxidative modification in r6/2 transgenic mice: a model of Huntington disease. Mol. Cell. Proteomics 4, 1849–1861 [DOI] [PubMed] [Google Scholar]
- 32.Mangiarini, L., Sathasivam, K., Seller, M., Cozens, B., Harper, A., Hetherington, C., Lawton, M., Trottier, Y., Lehrach, H., Davies, S. W., and Bates, G. P. ( 1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- 33.Hockly, E., Woodman, B., Mahal, A., Lewis, C. M., and Bates, G. ( 2003) Standardization and statistical approaches to therapeutic trials in the R6/2 mouse. Brain Res. Bull. 61, 469–479 [DOI] [PubMed] [Google Scholar]
- 34.Woodman, B., Butler, R., Landles, C., Lupton, M. K., Tse, J., Hockly, E., Moffitt, H., Sathasivam, K., and Bates, G. P. ( 2007) The Hdh(Q150/Q150) knock-in mouse model of HD and the R6/2 exon 1 model develop comparable and widespread molecular phenotypes. Brain Res. Bull. 72, 83–97 [DOI] [PubMed] [Google Scholar]
- 35.Hartl, D., Irmler, M., Romer, I., Mader, M. T., Mao, L., Zabel, C., de Angelis, M. H., Beckers, J., and Klose, J. ( 2008) Transcriptome and proteome analysis of early embryonic mouse brain development. Proteomics 8, 1257–1265 [DOI] [PubMed] [Google Scholar]
- 36.Klose, J. ( 1999) Large-gel 2-D electrophoresis. Methods Mol. Biol. 112, 147–172 [DOI] [PubMed] [Google Scholar]
- 37.Berth, M., Moser, F. M., Kolbe, M., and Bernhardt, J. ( 2007) The state of the art in the analysis of two-dimensional gel electrophoresis images. Appl. Microbiol. Biotechnol. 76, 1223–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartl, D., Rohe, M., Mao, L., Staufenbiehl, M., Zabel, C., and Klose, J. ( 2008) Impairment of adolescent hippocampal plasticity in a mouse model for Alzheimer's disease precedes disease phenotype. PLoS ONE 3, e2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao, L., Hartl, D., Nolden, T., Koppelstatter, A., Klose, J., Himmelbauer, H., and Zabel, C. ( 2008) Pronounced alterations of cellular metabolism and structure due to hyper- or hypo-osmosis. J. Proteome Res. 7, 3968–3983 [DOI] [PubMed] [Google Scholar]
- 40.Mao, L., Zabel, C., Herrmann, M., Nolden, T., Mertes, F., Magnol, L., Chabert, C., Hartl, D., Herault, Y., Delabar, J. M., Manke, T., Himmelbauer, H., and Klose, J. ( 2007) Proteomic shifts in embryonic stem cells with gene dose modifications suggest the presence of balancer proteins in protein regulatory networks. PLoS ONE 2, e1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diedrich, M., Mao, L., Bernreuther, C., Zabel, C., Nebrich, G., Kleene, R., and Klose, J. ( 2008) Proteome analysis of ventral midbrain in MPTP-treated normal and L1cam transgenic mice. Proteomics 8, 1266–1275 [DOI] [PubMed] [Google Scholar]
- 42.Nebrich, G., Herrmann, M., Sagi, D., Klose, J., and Giavalisco, P. ( 2007) High MS-compatibility of silver nitrate-stained protein spots from 2-DE gels using ZipPlates and AnchorChips for successful protein identification. Electrophoresis 28, 1607–1614 [DOI] [PubMed] [Google Scholar]
- 43.Strand, A. D., Baquet, Z. C., Aragaki, A. K., Holmans, P., Yang, L., Cleren, C., Beal, M. F., Jones, L., Kooperberg, C., Olson, J. M., and Jones, K. R. ( 2007) Expression profiling of Huntington's disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J. Neurosci. 27, 11758–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L., and Bates, G. P. ( 1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548 [DOI] [PubMed] [Google Scholar]
- 45.Carter, R. J., Lione, L. A., Humby, T., Mangiarini, L., Mahal, A., Bates, G. P., Dunnett, S. B., and Morton, A. J. ( 1999) Characterization of progressive motor deficits in mice transgenic for the human Huntington's disease mutation. J. Neurosci. 19, 3248–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoerndli, F., David, D. C., and Gotz, J. ( 2005) Functional Genomics meets neurodegenerative disorders. Part II: application and data integration. Prog. Neurobiol. 76, 169–188 [DOI] [PubMed] [Google Scholar]
- 47.Strand, A. D., Aragaki, A. K., Baquet, Z. C., Hodges, A., Cunningham, P., Holmans, P., Jones, K. R., Jones, L., Kooperberg, C., and Olson, J. M. ( 2007) Conservation of regional gene expression in mouse and human brain. PLoS Genet. 3, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Djousse, L., Knowlton, B., Cupples, L. A., Marder, K., Shoulson, I., and Myers, R. H. ( 2002) Weight loss in early stage of Huntington's disease. Neurology 59, 1325–1330 [DOI] [PubMed] [Google Scholar]
- 49.Mochel, F., Charles, P., Seguin, F., Barritault, J., Coussieu, C., Perin, L., Le Bouc, Y., Gervais, C., Carcelain, G., Vassault, A., Feingold, J., Rabier, D., and Durr, A. ( 2007) Early energy deficit in Huntington disease: identification of a plasma biomarker traceable during disease progression. PLoS ONE 2, e647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui, L., Jeong, H., Borovecki, F., Parkhurst, C. N., Tanese, N., and Krainc, D. ( 2006) Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell 127, 59–69 [DOI] [PubMed] [Google Scholar]
- 51.McGill, J. K., and Beal, M. F. ( 2006) PGC-1α, a new therapeutic target in Huntington's disease? Cell 127, 465–468 [DOI] [PubMed] [Google Scholar]
- 52.Magistretti, P. J., Pellerin, L., Rothman, D. L., and Shulman, R. G. ( 1999) Energy on demand. Science 283, 496–497 [DOI] [PubMed] [Google Scholar]
- 53.Rothman, D. L., Behar, K. L., Hyder, F., and Shulman, R. G. ( 2003) In vivo NMR studies of the glutamate neurotransmitter flux and neuroenergetics: implications for brain function. Annu. Rev. Physiol. 65, 401–427 [DOI] [PubMed] [Google Scholar]
- 54.Fan, M. M., and Raymond, L. A. ( 2007) N-Methyl-d-aspartate (NMDA) receptor function and excitotoxicity in Huntington's disease. Prog. Neurobiol. 81, 272–293 [DOI] [PubMed] [Google Scholar]
- 55.Li, J. Y., Plomann, M., and Brundin, P. ( 2003) Huntington's disease: a synaptopathy? Trends Mol. Med. 9, 414–420 [DOI] [PubMed] [Google Scholar]
- 56.Weaver, A. M., Young, M. E., Lee, W. L., and Cooper, J. A. ( 2003) Integration of signals to the Arp2/3 complex. Curr. Opin. Cell Biol. 15, 23–30 [DOI] [PubMed] [Google Scholar]
- 57.Li, Z., and Sheng, M. ( 2003) Some assembly required: the development of neuronal synapses. Nat. Rev. Mol. Cell Biol. 4, 833–841 [DOI] [PubMed] [Google Scholar]
- 58.Coux, O., Tanaka, K., and Goldberg, A. L. ( 1996) Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847 [DOI] [PubMed] [Google Scholar]
- 59.Voges, D., Zwickl, P., and Baumeister, W. ( 1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 [DOI] [PubMed] [Google Scholar]
- 60.Zwickl, P., Voges, D., and Baumeister, W. ( 1999) The proteasome: a macromolecular assembly designed for controlled proteolysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354, 1501–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett, E. J., Shaler, T. A., Woodman, B., Ryu, K. Y., Zaitseva, T. S., Becker, C. H., Bates, G. P., Schulman, H., and Kopito, R. R. ( 2007) Global changes to the ubiquitin system in Huntington's disease. Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 62.Pickart, C. M. ( 2004) Back to the future with ubiquitin. Cell 116, 181–190 [DOI] [PubMed] [Google Scholar]
- 63.Pickart, C. M., and Eddins, M. J. ( 2004) Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 64.Hershko, A., and Ciechanover, A. ( 1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 65.Muchowski, P. J., and Wacker, J. L. ( 2005) Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 6, 11–22 [DOI] [PubMed] [Google Scholar]
- 66.Caughey, B., and Lansbury, P. T. ( 2003) Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26, 267–298 [DOI] [PubMed] [Google Scholar]
- 67.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P., and Aronin, N. ( 1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 68.Zabel, C., Andreew, A., Mao, L., and Hartl, D. ( 2008) Protein expression overlap: more important than which proteins change in expression? Expert Rev. Proteomics 5, 187–205 [DOI] [PubMed] [Google Scholar]
- 69.Burke, J. R., Enghild, J. J., Martin, M. E., Jou, Y. S., Myers, R. M., Roses, A. D., Vance, J. M., and Strittmatter, W. J. ( 1996) Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nat. Med. 2, 347–350 [DOI] [PubMed] [Google Scholar]
- 70.Kaindl, A. M., Sifringer, M., Zabel, C., Nebrich, G., Wacker, M. A., Felderhoff-Mueser, U., Endesfelder, S., von der Hagen, M., Stefovska, V., Klose, J., and Ikonomidou, C. ( 2006) Acute and long-term proteome changes induced by oxidative stress in the developing brain. Cell Death Differ. 13, 1097–1109 [DOI] [PubMed] [Google Scholar]
- 71.Crocker, S. F., Costain, W. J., and Robertson, H. A. ( 2006) DNA microarray analysis of striatal gene expression in symptomatic transgenic Huntington's mice (R6/2) reveals neuroinflammation and insulin associations. Brain Res. 1088, 176–186 [DOI] [PubMed] [Google Scholar]
- 72.Greene, N. D., Leung, K. Y., Wait, R., Begum, S., Dunn, M. J., and Copp, A. J. ( 2002) Differential protein expression at the stage of neural tube closure in the mouse embryo. J. Biol. Chem. 277, 41645–41651 [DOI] [PubMed] [Google Scholar]
- 73.Zheng, P. Z., Wang, K. K., Zhang, Q. Y., Huang, Q. H., Du, Y. Z., Zhang, Q. H., Xiao, D. K., Shen, S. H., Imbeaud, S., Eveno, E., Zhao, C. J., Chen, Y. L., Fan, H. Y., Waxman, S., Auffray, C., Jin, G., Chen, S. J., Chen, Z., and Zhang, J. ( 2005) Systems analysis of transcriptome and proteome in retinoic acid/arsenic trioxide-induced cell differentiation/apoptosis of promyelocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 102, 7653–7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mijalski, T., Harder, A., Halder, T., Kersten, M., Horsch, M., Strom, T. M., Liebscher, H. V., Lottspeich, F., de Angelis, M. H., and Beckers, J. ( 2005) Identification of coexpressed gene clusters in a comparative analysis of transcriptome and proteome in mouse tissues. Proc. Natl. Acad. Sci. U. S. A. 102, 8621–8626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gygi, S. P., Rochon, Y., Franza, B. R., and Aebersold, R. ( 1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anderson, L., and Seilhamer, J. ( 1997) A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18, 533–537 [DOI] [PubMed] [Google Scholar]
- 77.Steffan, J. S., Kazantsev, A., Spasic-Boskovic, O., Greenwald, M., Zhu, Y. Z., Gohler, H., Wanker, E. E., Bates, G. P., Housman, D. E., and Thompson, L. M. ( 2000) The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. U. S. A. 97, 6763–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dunah, A. W., Jeong, H., Griffin, A., Kim, Y. M., Standaert, D. G., Hersch, S. M., Mouradian, M. M., Young, A. B., Tanese, N., and Krainc, D. ( 2002) Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296, 2238–2243 [DOI] [PubMed] [Google Scholar]
- 79.Luthi-Carter, R., Apostol, B. L., Dunah, A. W., DeJohn, M. M., Farrell, L. A., Bates, G. P., Young, A. B., Standaert, D. G., Thompson, L. M., and Cha, J. H. ( 2003) Complex alteration of NMDA receptors in transgenic Huntington's disease mouse brain: analysis of mRNA and protein expression, plasma membrane association, interacting proteins, and phosphorylation. Neurobiol. Dis. 14, 624–636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.