Abstract
Identifying peptides from mass spectrometric fragmentation data (MS/MS spectra) using search strategies that map protein sequences to spectra is computationally expensive. An alternative strategy uses direct spectrum-to-spectrum matching against a reference library of previously observed MS/MS that has the advantage of evaluating matches using fragment ion intensities and other ion types than the simple set normally used. However, this approach is limited by the small sizes of the available peptide MS/MS libraries and the inability to evaluate the rate of false assignments. In this study, we observed good performance of simulated spectra generated by the kinetic model implemented in MassAnalyzer (Zhang, Z. (2004) Prediction of low-energy collision-induced dissociation spectra of peptides. Anal. Chem. 76, 3908–3922; Zhang, Z. (2005) Prediction of low-energy collision-induced dissociation spectra of peptides with three or more charges. Anal. Chem. 77, 6364–6373) as a substitute for the reference libraries used by the spectrum-to-spectrum search programs X!Hunter and BiblioSpec and similar results in comparison with the spectrum-to-sequence program Mascot. We also demonstrate the use of simulated spectra for searching against decoy sequences to estimate false discovery rates. Although we found lower score discrimination with spectrum-to-spectrum searches than with Mascot, particularly for higher charge forms, comparable peptide assignments with low false discovery rate were achieved by examining consensus between X!Hunter and Mascot, filtering results by mass accuracy, and ignoring score thresholds. Protein identification results are comparable to those achieved when evaluating consensus between Sequest and Mascot. Run times with large scale data sets using X!Hunter with the simulated spectral library are 7 times faster than Mascot and 80 times faster than Sequest with the human International Protein Index (IPI) database. We conclude that simulated spectral libraries greatly expand the search space available for spectrum-to-spectrum searching while enabling principled analyses and that the approach can be used in consensus strategies for large scale studies while reducing search times.
Identification of proteins in complex samples is a major new area in bioinformatics. The most successful method currently available is shotgun proteomics where proteins are proteolyzed into peptides (usually by trypsin) followed by large scale sequencing of peptides by on-line chromatographic separation and fragmentation in a mass spectrometer (LC-MS/MS). The fragmentation process generates spectra (referred to as MS/MS spectra) from which peptide sequences consistent with the observed fragment ions can be identified (1). A common computational strategy for matching an MS/MS spectrum to a peptide sequence involves interconverting spectral and sequence information (2). Spectrum-to-sequence database search programs match peptide sequences to spectra in one of two ways: by 1) extracting sequence information from an observed spectrum and matching the sequences against peptides contained in a protein database or 2) converting peptide sequences from the protein database into simple spectra (e.g. predicting a subset of possible b and y fragment ions generated by peptide bond cleavage) and matching the predicted fragment ions to those observed. Various scoring methods are then used to evaluate overlap between observed and predicted fragments, including use of probability functions or spectral similarity metrics (2).
An alternative strategy involves direct spectrum-to-spectrum matching of experimental spectra against reference MS/MS in a spectral library (3). Programs that search sequence databases and spectral libraries are similar in many ways. Both match an experimental spectrum by selecting candidates from a reference database, use preprocessing and filtering functions to simplify the matching, and rank candidates using scores that evaluate the ability of the candidates to account for the observed fragment ions. However, spectrum-to-spectrum matching more easily allows use of fragment ion intensities as well as information on fragments other than the major b and y ions. Furthermore spectral library searching is simpler conceptually and faster to execute (4) because it is unnecessary to interconvert between spectra and sequences during the scoring process. For this reason, reference libraries of peptide MS/MS derived from experimental data sets are actively under development with libraries of human proteins now available from the National Institute of Standard and Technology (223,793 spectra), the MacCoss laboratory (320,658 spectra) (5), and the Beavis laboratory (297,519 spectra) (4). All contain mainly tryptic peptides but also include a significant number of non-tryptic and covalently modified peptides.
One limitation of spectrum-to-spectrum matching using MS/MS libraries derived from observed spectra is that all possible peptide sequences are not represented. In the human database, tryptic products alone predict ∼3,300,000 spectra in the mass range detectable by MS when different charge forms are included.1 Thus, the available reference libraries contain only a small fraction of all potentially observable spectra for tryptic peptides. Spectra are absent because they are rarely sampled experimentally, are derived from proteins of low abundance, are found in rare forms (e.g. alternative splice products), and/or ionize with low efficiency. Thus, only a few spectra might be available in a library to identify a protein, which is problematic due to the fact that shotgun proteomics depends on peptide sampling. Furthermore it is difficult to compare performances of spectrum-to-spectrum matching with spectrum-to-sequence strategies because of large differences in the sizes of their database search spaces and the unpredictable representation of a protein in the libraries.
In this study, we hypothesized that spectrum-to-spectrum searching can be improved by using a spectral library composed of simulated spectra for all tryptic peptides in the human database. Simulated spectra were generated using a kinetic model, which simulates fragment ion intensities based on known mechanisms for peptide fragmentation (implemented in the MassAnalyzer program developed by Z. Zhang) (6, 7). The MassAnalyzer MS/MS simulator was generated by fitting kinetic parameters to known peptide gas phase chemistries based on similarity scores to a large library of previously identified MS/MS spectra. The simulator satisfactorily predicts most cleavage patterns noted in manual analysis, such as enhanced fragmentation near Pro, Asp, Glu, His, and Ile/Leu/Val and near proton-donating side chains for peptide ions when proton mobility is limited. It also models common neutral losses, internal fragment ions, and losses of the C-terminal residue and provides a simple modeling of charge distribution between the various products, which is an important factor in simulating the spectra of MH+, MH22+, and MH33+ charge states. Results from Zhang (6) showed that the simulated spectra show good discrimination when scored by similarity to peptide standards. We have further shown that the spectra can be used to evaluate chemical plausibility in a program designed to mimic manual analysis of MS/MS spectra (8) because it provides a way to use fragment ion intensities in evaluating candidate sequences. Rescoring Mascot or Sequest assignments based on similarity to simulated spectra yielded improved discrimination with large scale data sets and enabled validation of hits with low scores from the search program, thus identifying a large class of correct assignments that were normally rejected using conventional score thresholds (8).
Here we develop methods for using a library of MassAnalyzer-generated simulated spectra of peptides from human proteins for spectrum-to-spectrum matching. The simulated spectral library is 10 times larger than the current reference libraries, providing a search space comparable to that used by spectrum-to-sequence searching of protein databases. Methods were developed to partition this library to accommodate the memory limits of the computer and operating system and to manage searches of multiple files. This approach also allows generation of randomized or inverted sequence libraries for target-decoy searches (9) to apply principled methods to evaluate the significance of peptide matches. Use of simulated spectra for spectrum-to-spectrum searching improves performance over available reference libraries and provides more rapid searching of large scale data sets.
EXPERIMENTAL PROCEDURES
Data Sets—
LC-MS/MS sequencing of proteins was performed on a Thermo LCQ Classic mass spectrometer interfaced with an Agilent Cap1100 HPLC instrument (15 cm × 250-μm inner diameter, Jupiter C18, Phenomenex) (10) or a Thermo LTQ-Orbitrap mass spectrometer interfaced with an Eksigent NanoLC-2D HPLC instrument (10 cm × 75-μm inner diameter, Zorbax C18, Agilent). Three data sets were used in this study. 1) The “LCQ data set” contained 845 manually curated and validated tryptic peptide assignments derived from a data set of 4,051 MS/MS spectra collected on a trypsinized soluble protein extract from human K562 erythroleukemia cells using an LCQ Classic ion trap mass spectrometer as described previously (10). 2) The “ABRF2 data set” contained 5,854 MS/MS spectra collected on a tryptic digest of the ABRF standard mixture of 49 proteins (Sigma-Aldrich, UPS1) collected on an LTQ-Orbitrap. A digest of 200 fmol of total protein was loaded, and peptides were eluted with a gradient of 2–40% acetonitrile in 0.1% formic acid, water over 150 min. MS/MS were collected enabling monoisotopic precursor and charge selection settings. Each MS scan was followed by five LTQ MS/MS scans targeting the top five most intense ions with a dynamic exclusion of 180 s and a repeat count of 2. The maximum injection time for Orbitrap parent scans was 500 ms with two microscans and an automatic gain control of 1 × 106. The maximum injection time for the LTQ MS/MS was 250 ms with three microscans and an automatic gain control of 1 × 104. The normalized collision energy was 35% with activation Q of 0.25 for 30 ms. 3) The “large scale data set” contained 90,411 MS/MS spectra collected on tryptic peptides derived from cytosolic protein extracts of WM115 human melanoma cells fractionated by quaternary aminoethyl anion exchange (Mono Q) fast protein liquid chromatography. LC-MS/MS data collection was carried out on each fraction using an LTQ-Orbitrap as above but with reverse phase elution from 2 to 40% acetonitrile (0.1% formic acid) in 120 min, running each sample three times and scanning different mass ranges (350–708, 700–1108, and 1100–1600 Da).
Search Programs—
Spectrum-to-spectrum search programs used in this study were X!Hunter, which was developed by Beavis and co-workers (4), and BiblioSpec, which was developed by MacCoss and co-workers (5). X!Hunter v.Win32 July 1, 2007 was tested on an Intel Pentium 4 3.2-GHz CPU (hyperthread) with 2-GB RAM using Windows XP Professional, and BiblioSpec v.1.0 was tested on a Dual Intel Xeon 2.4-GHz CPU with 2-GB RAM using Linux (Fedora Core 4). Results were compared with two spectrum-to-sequence search programs: Mascot v.2.2 (11), run on a Dual Intel Xeon 2.8-GHz CPU (hyperthread) with 8-GB RAM and Windows 2003, and TurboSequest (v.27 revision 12) (12) run on an Intel Pentium 4 3.0-GHz CPU (hyperthread) with 1-GB RAM using Windows XP Professional.
Error tolerances were ±1.2 Da for the parent ion using Mascot or X!Hunter and ±0.8 Da for fragment ions using Mascot (optimized for sensitivity and discrimination) or ±0.5 Da for fragment ions using X!Hunter (this default value is in the code and is not controlled by the user setting in the parameter file). In BiblioSpec, the fragment ion mass tolerance is controlled by fragment ion binning during spectral preprocessing, producing mass tolerances varying with m/z that cannot be altered by the user. BiblioSpec also bases parent ion mass tolerances on m/z value, and tolerances used for simulated spectral library searching were ±1.2, ±0.6, and ±0.4 Da, respectively, for MH+, MH22+, and MH33+ ions. Mascot and X!Hunter convert experimentally observed parent ion m/z to MH+ (M is the uncharged peptide mass) and then bases mass tolerance on that value, which produces varying tolerances for different charge forms. Therefore, we partitioned the simulated spectral library by charge and set different tolerances for each charge form to compare results between these programs. In addition to mass tolerances, we allowed up to two missed cleavages and fixed carbamidomethyl-Cys modification for protein database searches. For spectral library searches, there is no parameter to exclude other modifications; therefore, those MS/MS cases are removed by postfiltering.
Scoring methods utilized default functions: Mascot reports a Mowse score (also called ion score), BiblioSpec reports a dot product similarity score (Σ(Ii × Ij)/[(ΣIi2)1/2 × (ΣIj2)1/2]) between reference library and experimental spectra, and X!Hunter converts the weighted dot product score into an expectation score and then adds probability-based scoring to report a modified expectation score. To make comparisons equivalent, some functions in the expectation scoring were modified or turned off. In the X!Hunter reference library derived from observed spectra, each entry is assigned an initial expectation value as part of the final expectation score calculation. Because this information was not available for the simulated spectra, a default value of 0.001 was used for searches with both the observed and simulated spectral libraries. In addition, the partitioning of the input data sets (described below) required turning off a scoring correction for the number of total MS/MS analyzed. These changes had no effect on the search results (not shown), although the use of 0.001 as the initial expectation value shifted the range of the final expectation scores below a cutoff value, which varied between data sets.
Construction of a Human Simulated Spectral (SS) Library—
Reference libraries of observed spectra for X!Hunter and BiblioSpec were downloaded from the developers’ Web sites. The simulated spectral libraries were based on the human IPI protein database v.3.29 (13) or a “decoy sequence database” where each protein sequence was read in reverse. A database of sequences was generated for tryptic peptides with mass between 900 and 4500 Da, allowing up to two missed cleavages and removing unlikely missed cleavage products (14). Charge forms up to MH33+ were included for each sequence provided that a sufficient number of basic residues were present. For this in silico generated peptide database, there are a total of 2,918,714 peptide sequences that were at least 9 amino acids long. Of these, 1,452,058 passed the missed cleavage rules of Yen et al. (14). Simulation of spectra utilized the program MassAnalyzer (v.2.1), which generates the simulated spectra as DTA files. To convert the simulated spectra into a spectral library, DTA files for each simulated spectra were converted to the appropriate text file format (extensible markup language (XML) for X!Hunter and spectrum-sequence list (SSL) + MS2 for BiblioSpec) and then converted into the required binary format for each program. The number of stored ions had to be specified for the X!Hunter library format and was set to the default of 20 ions. Table I shows versions and sizes of the public domain libraries and the SS library used in this study.
Table I.
Spectral libraries of human peptide sequences used in this study
| SS library | X!Hunter library (xRef) | BiblioSpec library | |
|---|---|---|---|
| Version | IPI v.3.29 | 9/28/2007 | v.23.2, 11/5/2007 |
| Number of spectra | 3,306,625a | 320,658b | 297,519 |
| Number of unique sequences | 1,452,058 | 122,314 | 292,337 |
| Number of ions per spectrum | No limit | Up to 20 | Unknownc |
The in silico digested peptide sequences are limited by mass within 900–4500 Da, number of missed cleavages up to 2, charge state up to 3, allowing fixed carbamidomethyl-Cys, and passing missed cleavage rules (14).
xRef is managed by protein entry, so the number of spectra may include duplications.
The number of ions per spectrum is spectrum-dependent and unknown for entries in the BiblioSpec library.
Input experimental MS/MS spectra were extracted by extract_msn.exe (distributed with Bioworks 3.2) using the parameters -M1.4 -B85 -T4500 -S5 -G1 -I35 -C0 for LCQ data and the parameters -M0.2 -B85 -T4500 -S0 -G1 -I35 -C0 -P2 for LTQ data. Files were then formatted as MGF files for both X!Hunter and Mascot, and a converter was developed to change MGF-formatted experimental data to the MS2 format (15) used by BiblioSpec. X!Hunter XML and BiblioSpec SQT (15) output files were converted into an in-house MSPlus format (10) consisting of a comma-separated value (.csv) file with designated columns used in our regular work flow. DTAs for charge forms ≥4+ were excluded (less than 12% of spectra in data sets used in this study).
Spectral library search applications require libraries that are preloaded into system memory. Because it is difficult to accommodate large libraries using 32-bit operating systems, searches against the simulated spectral library were performed by partitioning the input data set and library as described previously for peptidecentric database searching (14). MGF files of input experimental data and simulated spectral libraries were partitioned by charge and by MH+ with masses overlapping between adjacent partitions to accommodate the parent mass tolerance. For searching with data sets containing multiple files, we developed an automated graphical user interface tool for X!Hunter that executes the searches and generates a single output file. This tool divides input MGF files by parent m/z and charge criteria, makes X!Hunter parameter files for the divided MGF files, invokes X!Hunter searches pairing the divided MGF files to the corresponding partition of the spectral library, and generates an MSPlus-formatted output by extracting information from the X!Hunter output files.
The simulated library is available for download from the X!Hunter Web site along with the graphical user interface tool and documentation. Other results files can be obtained by request from the corresponding author.
RESULTS
Testing Input/Output Conversions—
The experiments to evaluate the use of the MassAnalyzer-generated simulated spectra for spectrum-to-spectrum matching were carried out using two spectral library search programs, X!Hunter and BiblioSpec. These programs, the library-generating utilities, and the X!Hunter reference spectral library of observed MS/MS (referred to as “xRef”) were obtained from developer Web sites and implemented as described under “Experimental Procedures.” The different input and output formats required several complex conversions; therefore, we devised a simple experiment to test the generation of spectral libraries, preprocessing and filtering functions of the search programs, and conversion of output files into an in-house format (“MSPlus”) (10). In this experiment, a reference library was constructed from 845 high quality experimental MS/MS spectra taken from a larger, curated data set (LCQ data set; see “Experimental Procedures”) where manual analysis had confirmed each peptide assignment. The same spectra were then used as the input data set for a search.
Ideally every MS/MS input as experimental data should match to its corresponding entry in the reference library generated from the same spectra. BiblioSpec correctly matched all 845 MS/MS spectra to their corresponding reference library entries. X!Hunter correctly matched 777 spectra to their reference entries with three mismatches and 65 cases showing no assignment. Experiments revealed that the lower number of matches was caused by removal of spectra by eight data preprocessing filters in X!Hunter (supplemental Table 1). For example, of the cases with no assignment, 55 were never evaluated because they were rejected by a preprocessing filter designed to remove noisy or weak spectra; these were correctly assigned when the filter was turned off. When only the “clean isotopes” filter was used, X!Hunter correctly matched 841 MS/MS with three mismatches and one case where no match was reported. We attributed the mismatches to the fact that only 20 peaks were saved in each reference library entry, whereas 50 peaks were considered for input data files. For LTQ-Orbitrap data, preprocessing of the spectra by X!Hunter had a different effect: the noise processing eliminated less than 0.5% of the MS/MS, but the other filters appeared to be helpful (supplemental Table 2), so default settings were used. Despite this complication, the experiment showed that input/output conversions were carried out correctly.
Performance of Simulated Spectra against Observed Spectra—
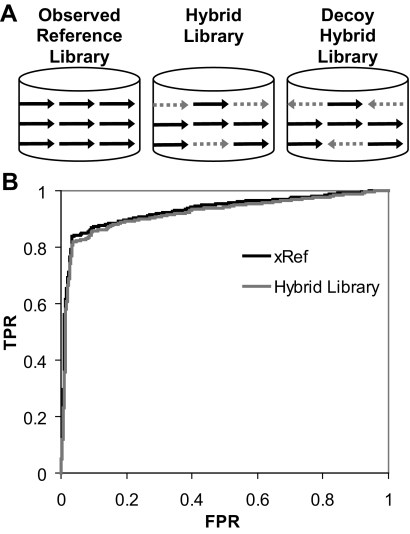
We next asked whether the simulated spectra generated by MassAnalyzer could substitute for reference library spectra in spectrum-to-spectrum searching. To address this, we compared xRef, generated entirely from observed MS/MS, with a “hybrid library” where a subset of the spectra was replaced by simulated spectra as shown in Fig. 1A. The replacement spectra were derived from 49 standard proteins used to generate an experimental data set (ABRF data set; see “Experimental Procedures”); the ABRF data set was then searched using xRef or the hybrid library. The purpose of this experiment was to test the ability to identify standard peptides when holding the background of other spectra constant. The subset comprised the same peptide sequences and charge states found in xRef, replacing only the tryptic peptide sequences (allowing carbamidomethyl-Cys) and ignoring peptides with non-tryptic cleavages or other modifications. The resulting hybrid library replaced 8,682 of 320,658 spectra in xRef with simulated spectra, maintaining the same database size and background MS/MS. A “decoy hybrid library” was also generated, replacing the same 8,682 spectra with simulated spectra generated from inverted sequences of the same peptides where the C-terminal residue was held constant to maintain tryptic specificity.
Fig. 1.
Performance of simulated MS/MS spectra within a reference library. A, strategy for generating spectral libraries to test simulated spectra. Spectra in the reference library (xRef) that corresponded to peptides in 49 standard proteins were replaced by simulated spectra of the same peptide ions generated by MassAnalyzer, producing the hybrid library. A decoy hybrid library was created by replacing xRef spectra with simulated spectra generated against peptide sequences reversed to read from C to N terminus, holding the C-terminal amino acid constant to maintain the tryptic nature of the sequence. Only the simulated spectra correspond to inverted sequences and thus represent decoy targets. B, ROC plots of results using xRef or the hybrid library to search an LC-MS/MS data set collected for a tryptic digest of the 49 standard proteins (ABRF data set). Each plotted point represents the true positive rate (TPR) as the proportion of the MS/MS assigned correctly (as ABRF proteins), when scoring above the threshold at each plotted point, divided by the total number of MS/MS assigned correctly in this data set. The false positive rate (FPR; (FP/(TN + FP))) is the proportion of incorrect assignments (not assigned to any ABRF protein) above the threshold at each plotted point divided by the total number of incorrect assignments. The algorithm for calculating these values is given in the supplemental data.
Of the 5,854 MS/MS spectra in the ABRF data set, 4,881 spectra passed the data preprocessing and filtering functions of X!Hunter and were carried forward in the search to generate a peptide identification. Peptide sequences were assigned to 3,521 and 3,531 spectra for xRef and hybrid library searches, respectively. These included assignments to both ABRF proteins or other proteins; we refer to the ABRF assignments as “correct assignments” and the others as “incorrect assignments,” distinguishing these from true and false positives, which we determined by other criteria (see below). The xRef search yielded 994 correct assignments to standard proteins, whereas the hybrid library search yielded 986 correct assignments. We wanted to know how many of the correct assignments might be matched by chance. Searching the ABRF data set against the decoy hybrid library revealed a total of 33 matches to inverted ABRF sequences, or a 0.9% chance for a random match for this data set. Using manual analysis of the MS/MS, we evaluated whether the correct assignments were valid or due to a chance match. All 941 cases where both xRef and hybrid library searches made the same assignment were classified as either valid or plausible (the plausible class was 3% of the total and represented low intensity spectra that were more difficult to validate by manual analysis). Manual analysis of the 53 matches unique to the xRef search classified 30 as invalid/ambiguous and 23 as valid/plausible. Of the 45 matches unique to the hybrid library search, 31 were classified as invalid/ambiguous, and 14 were valid/plausible. The 30 or 31 manually confirmed invalid assignments were consistent with the 33 assignments to ABRF proteins by chance alone as predicted from the decoy hybrid library search.
The number of invalid assignments by X!Hunter was larger than that observed by Mascot where searches against a decoy IPI human sequence database yielded only seven MS/MS files corresponding to inverted sequences of the ABRF proteins (i.e. only 0.15% of 4,792 MS/MS with a reported assignment). The difference between the X!Hunter and Mascot results could be attributed to overrepresentation of peptides for the ABRF proteins in xRef. Whereas xRef contained 8,682 spectra derived from the 49 standard proteins (2.9% of the 297,519 spectra in the library), the IPI human protein database (v.3.27) used by Mascot contained 3,682 unique tryptic peptides from the standard proteins, equaling 0.25% of all 1,452,058 tryptic peptides. Thus, the chance of randomly matching an ABRF standard protein should be ∼10-fold higher for xRef than for the IPI human database, consistent with the observed 0.9 versus 0.15%. The fact that the observed false positive assignments to ABRF proteins in each of these two situations were lower than calculated suggests that the library entries are not equally probable, most likely because their mass or charge distributions do not match those of the experimental data set.
Receiver-Operator Characteristic (ROC) Analysis of Results with xRef Versus the Hybrid Library—
To provide a more comprehensive comparison of the performance of xRef versus the hybrid library in this search, a ROC analysis was carried out (Fig. 1B) using an algorithm described in the supplemental data. Search results were classified as class positives (including true positives (TP) and false negatives) or class negatives (including false positives (FP) and true negatives (TN)) depending on whether the assignment was to a peptide derived from the ABRF versus non-ABRF proteins. The expectation score was varied to determine the number of each class that was above the acceptance threshold for every plotted value. These classes cannot always be quantified in proteomics experiments; however, use of the ABRF data set facilitated classification of sequence matches as correct versus incorrect based on whether they matched one of the 49 standard proteins without considering any score.
The ROC analysis showed very little difference between the xRef and hybrid library searches. Our confidence in this result is increased because a detailed evaluation showed no systematic bias. The nearly identical numbers of MS/MS assignments in the two searches indicated that any bias introduced by chemical differences between peptides assigned in each search was very small. Overall the total class positive assignments for the hybrid library search were estimated as 953 MS/MS (986 − 33) yielding 4,901 total class negatives (5,854 − 953). The relatively small difference between 986 and 953 makes it unlikely that invalid ABRF assignments have significantly affected the ROC analyses or our assumption that assignments to ABRF peptides are valid. The class positives for the xRef search were 994 − 30 = 964, which is similar to that of the hybrid library search, increasing the confidence in the comparison of the two search methods.
Taken together, the results showed very little error introduced into the ROC analyses by the small number of invalid assignments to ABRF proteins made by chance alone and highlight the potential usefulness of the simulated spectra. It is important to note that the experiment controlled for differences in the sequence and charge forms available to be candidates in the search. The 13 correct assignments unique to the hybrid library search represented cases where reference spectra were apparently from TOF/TOF data as judged by the presence of many low mass ions in the spectrum. The 16 correct assignments unique to the xRef search represented cases where the simulated spectra did not simulate the experimental spectra very well. The low number of the latter cases (23 compared with 941 MS/MS) provided further support for comparable performances of the simulated and xRef spectra when the library size and spectra for other proteins are held constant. Therefore, we expected that replacing xRef with a library of simulated spectra would gain from increasing the search space to a level comparable to protein databases without a significant loss of correct matches normally obtained with xRef.
Simulated Spectral Libraries—
Next the simulated spectra libraries were constructed, and tools were developed to handle the searches. We used MassAnalyzer to generate the simulated spectra for peptides in the human IPI protein database (“SS library”) as well as a “decoy SS library” generated from inverted protein sequences. Only tryptic peptides with mass 900–4,500 Da and charge states 1+, 2+, or 3+ were considered, and peptides contained up to two missed cleavages, eliminating unlikely missed cleavage products as described previously (14). The SS library was then formatted to conform to the requirements of the search programs, producing normal libraries for X!Hunter (“xSS”) and BiblioSpec (“bSS”) and inverted sequence libraries “decoy xSS” and “decoy bSS.” Each SS library contained 3.3 million spectra, a size that was limiting for the memory of the computer and operating system. To deal with this, each library was partitioned by charge and calculated mass (MH+), which enabled simple parallel processing because each partition could be analyzed independently of the others. In addition, software was developed to partition multiple input MGF files tailored to the library partitions, trigger the searching, and extract information from multiple output files into a single file summarizing the search results.
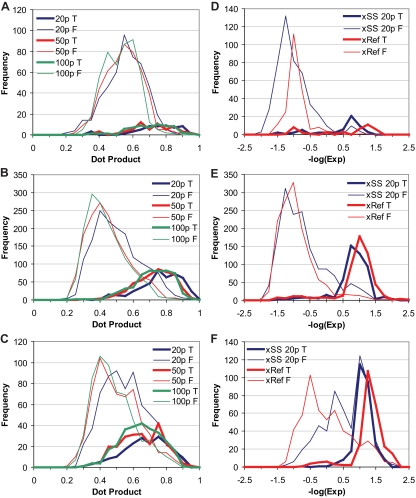
A major difference between X!Hunter and BiblioSpec libraries is the number of fragment ions saved (which we refer to as “peaks”) and the number of fragment ions in the experimental spectra that are utilized in the search. For X!Hunter, the library-generating software saves 20 peaks from each spectrum by default, whereas BiblioSpec saves the most or all of the ions in the spectrum and allows the user to vary the number of peaks during the analysis. The effect of varying peak number was tested in searches of the ABRF data set against the SS library using BiblioSpec to analyze the top 20, 50, or 100 fragment ions and compared with X!Hunter, which compares 50 fragment ions3 from the experimental spectra with 20 peaks in the reference spectra. In BiblioSpec searches against bSS, the number of assignments to standard proteins increased with the number of peaks in each library entry (Table II) with little change in the number of assignments from a decoy bSS search between peak numbers. Small effects on discrimination were observed as assessed by the separation in dot product score distributions for correct assignments to standard proteins (i.e. those most likely valid) versus other proteins (incorrect assignments, which are all invalid). The incorrect assignments showed broad distributions and large overlap with correct assignments. Discrimination varied with charge state (Fig. 2, A–C); for MH+ ions it increased as the number of peaks varied from 20 to 100, whereas smaller effects were observed with MH22+ or MH33+ ions.
Table II.
Results of searches using the ABRF data set
| Search program and database
|
|||||
|---|---|---|---|---|---|
| X!Hunter
|
BiblioSpec
|
||||
| xRefa 20pb | xSSa 20pb | bSSa 20pb | bSSa 50pb | bSSa 100pb | |
| Number of assigned DTAsc | 3342 | 3407 | 3436 | 3436 | 3436 |
| Number of correct assignmentsd | 994 | NDe | ND | ND | ND |
| Number of correct tryptic assignmentsd,f | 717 | 831 | 697 | 799 | 844 |
| Number of decoy assignmentsd | ND | 6 | 7 | 6 | 9 |
xRef, xSS, and bSS are libraries used by X!Hunter and BiblioSpec. xRef is the reference library generated from observed spectra; xSS and bSS are simulated spectral libraries generated by MassAnalyzer for X!Hunter and BiblioSpec, respectively. The SS library is based on tryptic peptides in the IPI human database (see text).
“p” indicates the number of peaks considered for similarity scoring in the library entries.
Total number of MS/MS (out of 5,854) assigned a sequence by the search program.
d Correctly identified MS/MS are taken as those assigned to 49 standard proteins in the ARBF sample considering only tryptic peptides regardless of score. Assignments to other proteins are considered incorrect. For the decoy assignments, if an assignment maps to 49 reversed standard proteins in the ABRF sample, it will be counted as a correct decoy assignment.
ND, not determined.
179 MS/MS were removed from the ABRF data set that represent non-tryptic peptide assignments and unlikely missed cleavage products in xRef search. This allows direct comparison between results using xRef and xSS.
Fig. 2.
Score distributions for searches of the ABRF data set. A–C show dot product score distributions of BiblioSpec assignments for ions with charge state MH+, MH22+, and MH33+. D–F show expectation score distributions for X!Hunter assignments for ions with charge state MH+, MH22+, and MH33+. Assignments are called true (T) when the identified peptide sequence corresponds to one of the 49 standard proteins and called false (F) when it does not. p indicates the number of peaks considered for similarity scoring in the library entries.
X!Hunter searches of the ABRF data set against xSS showed greater sensitivity compared with BiblioSpec, yielding higher numbers of correct assignments to standard proteins when 20 ions were considered (Table II). In addition, distributions of expectation scores showed greater separation between correct and incorrect assignments at all charge states compared with BiblioSpec (Fig. 2, D–F). The difference may be due to additional score processing by X!Hunter, which calculates a probability assessment for each assignment in addition to the expectation score. Overall the analyses suggested better results with X!Hunter; this led us to use X!Hunter in subsequent studies.
Comparison of xRef and xSS Search Results—
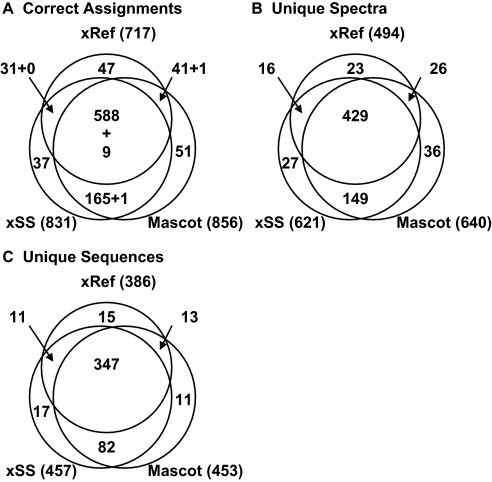
We next compared the performances of searches against xRef and xSS. Searching the ABRF data set against xRef assigned 994 spectra of which 277 were correctly assigned to non-tryptic peptide sequences (155), modified peptide sequences (98; mostly unalkylated Cys-containing peptides), and unlikely missed cleavage products (24). Because these cases were excluded from xSS, we modified the ABRF data set to remove these MS/MS files to directly compare searches using the two libraries. Searches of the modified ABRF data set against xRef gave 717 tryptic assignments to standard proteins, and searches against xSS gave 831 tryptic assignments (Fig. 3A). Searches against decoy xSS gave only six assignments (Table II), indicating very low numbers of invalid assignments to ABRF proteins in the xSS searches. The estimated number of invalid assignments was lower than the 33 observed with the decoy hybrid library because the ABRF proteins were not as overrepresented in the SS as in the hybrid library. Furthermore the number of correct ABRF assignments achieved with xSS was comparable to that achieved in Mascot searches of the IPI database (Fig. 3A).
Fig. 3.
Search results for the ABRF data set using X!Hunter or Mascot. Searches with X!Hunter used a reference library of observed spectra (xRef) or the full simulated spectral library (xSS), whereas searches with Mascot used the IPI protein database (“Mascot”). A, summary of all identified MS/MS. B, summary of the identified unique spectra, which count peptide ions with distinct sequence and/or charge state. C, summary of the identified unique peptide sequences. The total numbers of assignments to ABRF proteins for each search strategy are given in parentheses. In a few cases, more than one ABRF assignment was made to the same spectrum, and these are indicated as a second number (e.g. “+ 9” in A, region showing overlap between all three searches). Manual validation confirmed that both assignments were correct because the MS/MS was a chimera of two different parent ions captured in one MS/MS isolation event. To compare xRef and xSS, MS/MS derived from non-tryptic sequences were removed from the ABRF data set. Only peptides with m/z >900 Da and that were derived from singly, doubly, or triply charged parent ions were included.
The 114 additional MS/MS assigned to ABRF proteins using xSS versus xRef supported the usefulness of the simulated spectral library for extracting more information from LC-MS/MS data sets. Additional unique assignments to xSS were revealed by Venn analysis. Only 597 MS/MS were assigned the same peptide sequence by both libraries (Fig. 3A). These included nine cases where the spectra were chimeras derived from fragmentation of two different parent ions with nearly identical m/z and reverse phase elution time. In each of these chimeras, the two top scoring candidates were derived from ABRF proteins and could be matched by each of the two searches although in reversed rank order. Because many of these assignments resampled the same peptide ions, it was important to evaluate the number of the unique sequence assignments and unique spectra (i.e. including different charge forms). Of the xSS identifications, 203 matches (including one chimera) were found only in the xSS search, which represented 176 unique spectra (Fig. 3B) and 99 unique peptide sequences (Fig. 3C). In 174 cases (representing 147 unique spectra and 127 unique peptide sequences), the differences with xRef could be explained by the absence of spectra for the peptide sequences or charge forms in xRef. The other 39 sequences uniquely assigned by xSS searches were present but not identified in the search against xRef. These may represent situations where the reference library spectrum was incorrectly identified or was weak and thus difficult to match to our experimental data. This provided an unexpected role for the simulated spectra in delineating possible errors in xRef.
We also expected that xRef would generate some assignments where observed spectra provided better matches than simulated spectra. In fact, 89 matches were observed only in xRef searches, representing 49 unique charge forms (unique spectra) and 28 unique peptide sequences. All spectra for these sequence and charge forms were present in xSS. We refer to these as distracted cases where other spectra in xSS scored higher than the correct assignment. They usually represented cases where the simulated spectra show poor fragmentation, and the small number of fragment ions creates ambiguity against a large search space, or the distribution between b and y ions was incorrectly modeled in a richly fragmenting peptide, and the use of 20 peaks for the reference spectrum was inadequate. Overall 28 unique peptide sequences were lost by distraction, whereas 127 unique peptide sequences were gained from improved sequence and spectrum coverage of xSS.
Next we asked whether we could validate the correct assignments using the expectation scores; this would predict their performances in shotgun proteomics experiments where correct versus incorrect assignments cannot be assessed as for ABRF proteins. To do this we evaluated a histogram plotting the distribution of the expectation values (Fig. 2, D–F). This showed that score distributions for MS/MS matched to standard proteins (most likely valid assignments) were comparable between xSS versus xRef searches but with the xRef distribution shifted to slightly better values (bold lines). Discrimination was evaluated by the separation between the correct and incorrect assignments (bold versus thin lines). In general, although correct assignments were clustered in a narrow peak within the high scoring region, MH+ and MH22+ ions showed greater separation between correct and incorrect assignments, whereas MH33+ ions showed less separation. Nevertheless substantial overlap was seen between incorrect and correct assignments in all cases. The poor discrimination meant that it would be difficult to achieve high sensitivity and selectivity based on expectation score alone and prompted investigation of alternative approaches to validate search results with complex samples.
Consensus between X!Hunter and Mascot—
We and others have shown that evaluating consensus between independent search programs significantly improves sensitivity between correct and incorrect assignments in situations where discrimination is problematic (10, 16). This approach works best when the scoring methods are significantly different from each other. In an earlier study, we evaluated consensus between Mascot and Sequest searches, which produced a significant false discovery rate (FDR = FP/(FP + TP)); several filters were developed to remove the false positives, achieving an FDR of ∼2% for LCQ data sets. We therefore asked whether a similar consensus approach could be applied to improve confidence with spectrum-to-spectrum searching.
An important criterion for success in a consensus approach is whether good overlap is achieved for sequences identified by the different search programs or scoring methods. To test this, we compared the total number of correct assignments achieved with Mascot searches against the IPI human database versus X!Hunter searches against xSS regardless of scores. X!Hunter and Mascot searches of the ABRF data set both assigned 763 MS/MS to peptide sequences in the standard proteins and 68 and 93 unique peptides, respectively (Fig. 3A). Importantly the overlap was better when only unique sequences were considered. Here there were 429 unique peptides identified in common with 28 and 24 unique sequences identified only in xSS versus Mascot searches, respectively (Fig. 3C). This agreement suggested that a consensus approach between X!Hunter and Mascot would work well without using the low discrimination scoring methods.
Results comparing consensus between Mascot and X!Hunter (“XM”) with consensus between Mascot and Sequest (“SM”) are detailed in Table III. The Mascot-only results in this table report all the sequence assignments for all the MS/MS; it is shown for the purpose of illustrating the function of the mass accuracy filter. The Sequest results are similar to those of Mascot. XM consensus yielded 771 matches to standard proteins (433 unique peptide sequences), whereas SM consensus gave 823 matches (440 unique peptide sequences). In this experiment, a false positive is defined as a consensus assignment that is invalid. False positives were estimated in parallel searches using the decoy IPI protein sequence database (for Mascot and Sequest) or decoy xSS (for X!Hunter). These yielded 93 decoy matches obtained by XM consensus and 278 by SM consensus. The trend agreed with the number of incorrect assignments for non-ABRF proteins in the normal searches, which equaled 65 and 161 for XM and SM, respectively. The results demonstrated that consensus between X!Hunter and Mascot yielded an FDR of 8–12%, which is significantly lower than the 19–34% achieved by Sequest and Mascot consensus.
Table III.
Searches of the ABRF data set using Mascot with and without consensus with Sequest or X!Hunter
| Search methoda | Identifications
|
Protein identifications | |
|---|---|---|---|
| No mass filter | Mass filter appliedb | ||
| Mascotc | |||
| ABRF | 873 (459 unique)d | 765 (433 unique)d | 49 |
| Non-ABRF | 1,761 | 22 | |
| Decoy | 2,391 | 63 | |
| FDR (%) normal/decoy | 67/91e | 2.8/8.0e | |
| Sequest | |||
| ABRF | 893 (466 unique)d | 763 (435 unique)d | 48 |
| Non-ABRF | 1,758 | 39 | |
| Decoy | 2,421 | 63 | |
| FDR (%) normal/decoy | 66/91e | 4.9/7.9e | |
| SM consensus | |||
| ABRF | 823 (440 unique)d | 729 (419 unique)d | 48 |
| Non-ABRF | 161 | 3 | |
| Decoy | 278 | 13 | |
| FDR (%) normal/decoy | 16/28e | 0.4/1.8e | |
| XM consensus | |||
| ABRF | 771 (433 unique)d | 688 (412 unique)d | 48 |
| Non-ABRF | 65 | 1 | |
| Decoy | 93 | 4 | |
| FDR (%) normal/decoy | 66/91e | 4.9/7.9e | |
In each case, the results of the normal search are broken out into correct versus incorrect assignments, and total decoy hits for both are shown. No score thresholds were used in accepting the assignments. Results are filtered to remove unlikely missed cleavage products, peptides with fewer than 9 amino acids, and ions with charge >3+.
Results are postfiltered by parent ion mass accuracy within −2 to +7 ppm.
Criteria applied to Mascot-only result allow spectra with charge up to 3 and sequences with at least 9 amino acids that pass the missed cleavage rules (14). The difference in total number for the decoy search (2,391) versus the normal search (2,634) is due to the fact that the cases assigned as an unlikely missed cleavage product have a higher probability of occurring in the decoy search.
Number of MS/MS identified; number of unique peptide sequences in parentheses.
FDR values are shown for the ABRF protein assignments and for the estimated FDR from the decoy search. A high FDR is obtained for the Mascot- and Sequest-only searches because assignments for all MS/MS are considered correct to show the effect of consensus before the mass accuracy filter is applied.
We next examined the effect of postfiltering assignments based on mass accuracy (17) where only assignments with observed parent mass within −2 to +7 ppm of the predicted mass were accepted. Applying this filter decreased the number of MS/MS matches by 11–12% with a 5–6% decrease in the numbers of unique peptide sequences (Table III). Some of the MS/MS removed were valid assignments where either the second isotope peak was chosen for sequencing or an adjacent peak within a 2-Da window was activated to yield a spectral chimera (not shown). Importantly the postfilter decreased the numbers of decoy database or decoy library searches to four and 13 by XM and SM consensus, respectively (Table III). After accounting for invalid consensus identifications (one by XM and three by SM), we calculated three FP assignments (FDR = 0.4%; 3 ÷ 688) for XM and 10 FP assignments (FDR = 1.4%; 10 ÷ 729) for SM. We conclude that evaluating consensus between X!Hunter and Mascot, together with a postfilter for mass accuracy, reduced FDR to a level acceptable for large scale data sets with minor impact on the numbers of unique peptide sequences.
We also asked whether high mass accuracy was more useful as a mass tolerance window during the search process rather than in postfiltering. Searches performed using mass tolerance of 10 ppm yielded fewer MS/MS assignments with XM or SM consensus as expected because of the reduction in search space (data not shown). In contrast, decoy database or decoy library searches conducted using mass tolerances of 1.2 Da or 10 ppm, respectively, yielded similar numbers of false identifications by either consensus method. We interpret this to mean that consensus methods yield the same number of FP matches against two random lists of sequences regardless of the search strategy or search space. It is therefore more effective to combine consensus assignments with a postfilter based on mass accuracy of the parent.
Analysis of Large Scale Data Sets—
We next applied the consensus approach with a mass accuracy postfilter to a large scale data set of a complex mixture of cytosolic proteins from human melanoma cell lysates fractionated by quaternary aminoethyl chromatography, proteolyzed, and then analyzed by LC-MS/MS (see “Experimental Procedures”). These results were compared with Mascot searches where 46,603 MS/MS (7,717 unique peptide sequences) were validated by using physicochemical filters and high mass accuracy on the parent ion, yielding an FDR of 4%. The Mascot result yielded 1,363 nonredundant proteins. By comparison, XM consensus identified 37,589 MS/MS (6,640 unique sequences) with an FDR of 0.5%, yielding 1,356 nonredundant proteins, and SM consensus identified 44,303 MS/MS (7,227 unique sequences) with an FDR of 1%, yielding 1,436 nonredundant proteins (Table IV). The number of protein identifications supported by only one peptide is considered a measure of the quality of the profile because false positives are higher in these cases. In the XM and SM searches, 31–32% proteins were identified by one peptide, an acceptably low value.
Table IV.
Search results of a large scale data set filtered by mass accuracy of the parent ion, the peptide length, missed cleavage rules, and charge
| Search methoda | Peptide IDsb | MH+ | MH22+ | MH33+ | Unique peptides | Protein IDs (1-pep cases)c |
|---|---|---|---|---|---|---|
| Mascot | ||||||
| Normal | 46,603 | 4,179 | 23,745 | 18,679 | 7,717 | 1,760 (732) |
| Decoy | 1,881 | 346 | 987 | 548 | 1,069 | 1,017 (971) |
| FDR (%) | 4.0 | 8.3 | 4.2 | 2.9 | 13.9 | 58 |
| Sequest | ||||||
| Normal | 47,344 | 4,115 | 24,368 | 18,861 | 7,861 | 1,730 (689) |
| Decoy | 1,754 | 242 | 975 | 537 | 931 | 873 (824) |
| FDR (%) | 3.7 | 5.9 | 4.0 | 2.9 | 12 | 50 |
| SM consensus | ||||||
| Normal | 44,303 | 3,877 | 23,111 | 17,315 | 7,227 | 1,441 (449) |
| Decoy | 427 | 60 | 254 | 113 | 216 | 186 (183) |
| FDR (%) | 1.0 | 1.6 | 1.1 | 0.7 | 3.0 | 13 |
| XM consensus | ||||||
| Normal | 37,589 | 3,008 | 19,836 | 14,745 | 6,640 | 1,359 (438) |
| Decoy | 185 | 37 | 134 | 14 | 81 | 80 (79) |
| FDR (%) | 0.5 | 1.2 | 0.7 | 0.1 | 1.2 | 5.9 |
Results are shown for Mascot search and consensus between Sequest and Mascot using the IPI human database or decoy database and consensus between X!Hunter and Mascot where X!Hunter searches xSS and decoy xSS. There were 90,411 total MS/MS in this data set. For all search results, the assignments were filtered to allow charge up to 3, at least 9 amino acids, following missed cleavage rules, and parent ion mass accuracy within −2 to +7 ppm.
Total MS/MS identified are shown along with the breakdown of MH+, MH22+, and MH33+ cases and total unique peptide sequences.
The total number of protein assignments accepted by these filters is shown, and in parentheses the number of cases that are supported by only one peptide (1-pep cases) is shown. For example, there were 1,760 proteins found in the normal Mascot search of which 732 were protein identifications (IDs) supported by only one peptide.
Overall trends observed for the large scale data set were similar to the prior analyses of ABRF data sets where the false discovery rate was lower with XM than SM consensus and higher for MH+ ions (Table IV). Analyses of overlaps (supplemental Table 3) showed 97 assignments uniquely identified by SM consensus (89 supported by one peptide and eight supported by two) and 24 assignments unique to XM consensus (23 supported by one peptide and one supported by two). Proteomics investigations often relax FDR thresholds but maintain stringency by requiring two charge forms of a peptide or two unique peptide sequences to validate a protein identification (18). Analysis of the Mascot searches using these criteria and setting FDR to 5% identified 997 proteins (supplemental Table 2). Using a Mascot search of an inverted sequence database yielded an FDR for the proteins of 5.8%, which is significantly higher than that of the XM consensus search. Furthermore the XM search identified 359 more proteins (supported by one peptide but two programs) that were rejected by Mascot (supported by two peptides). This is consistent with previous studies showing that greater sensitivity is achieved by evaluating consensus between two search programs, which provides the advantage of minimizing or removing scoring thresholds.
In summary, although spectrum-to-spectrum searching against SS libraries was alone insufficient to achieve high confidence assignments, XM consensus yielded results comparable to the SM spectrum-to-sequence consensus strategy, greatly improving confidence and sensitivity compared with single programs. An important advantage was noted with XM over SM when evaluating the search run time for each of the consensus strategies. Whereas run times with the large scale data set lasted 24 and 2.0 h in searches against the human IPI database by Sequest and Mascot, respectively, searches against xSS by X!Hunter required 18 min to complete without enabling parallel processing of the partitions. Because the comparison of running times is based on the whole protein database versus filtered and partitioned spectra libraries, one can question the results due to the unequivalent search spaces. However, Yen et al. (14) showed that the running time of a Mascot search is not very sensitive to the size of the database. Thus, spectrum-to-spectrum searching against the SS library yields an impressive reduction in search time for the human proteome, providing an important advantage over Sequest in a consensus search strategy.
DISCUSSION
This study demonstrates that good results can be obtained with spectrum-to-spectrum searching using a novel simulated spectral library based on MS/MS spectra simulated by the kinetic modeling program MassAnalyzer. This approach provided three important advantages. First, spectra were generated for all plausible tryptic peptides predicted from protein sequences, which increased the search space by nearly 10-fold over the sizes of current reference libraries, making it comparable to search spaces of protein databases typically used in spectrum-to-sequence searching. This improved peptide identification in a controlled test using 49 standard proteins. Furthermore the approach reduced biases toward high abundance proteins, for example by decreasing representation of proteins from the ABRF standard mixture compared with xRef. Second, X!Hunter searches were much faster than Sequest, enabling a consensus approach with Mascot to validate assignments. Third, decoy spectral libraries could be created that corresponded to inverted protein sequences. This provided a key resource for estimating the numbers of invalid assignments in normal searches and allowed estimation of false discovery rates for spectral matching algorithms in the same manner used by spectrum-to-sequence programs.
To allow a principled evaluation of performances, two approaches were used: 1) replacing selected spectra in the reference libraries of observed MS/MS spectra with the corresponding simulated spectra and 2) generating the library of simulated spectra using the same protein database used for Mascot and Sequest searches. The replacement strategy showed almost no difference between the simulated and observed spectra by analyses of ROC plots and false positive assignments. However, these spectra are likely dominated by easily fragmented peptide ions that can be readily identified by any method. The SS library contained a broader representation of peptide ions. Comparison of searches against xRef and xSS with the ABRF data set showed a large net increase in valid assignments (127 unique peptides gained versus 28 peptide sequences lost). Thus, a net gain by using xSS was observed even though the ABRF proteins are more highly represented in xRef. In addition, the fact that valid assignments were found in xSS, but not the xRef result even though the spectra for those sequences were present in xRef, suggested that simulated spectra can assist in delineating possible errors in xRef. Most importantly, a comparable number of correct assignments were achieved with Mascot searching the protein database and with X!Hunter searching xSS. Together these studies indicate that a library of the MassAnalyzer-simulated MS/MS spectra can be used successfully in spectrum-to-spectrum searching.
On the other hand, we observed relatively poor discrimination between correct and incorrect assignments based on scoring methods of X!Hunter and BiblioSpec. This makes it difficult to validate hits at a low false discovery rate using score thresholds as required for complex samples where the protein composition is unknown. Because X!Hunter and Mascot identified nearly the same set of peptides and each program used a very different scoring method, we tested the use of a consensus strategy. Although slightly better results were achieved with Sequest/Mascot consensus, the X!Hunter/Mascot approach had a significant advantage in the ∼80-fold faster run time for X!Hunter versus Sequest. The new instruments rapidly generate large amounts of data; this faster run time now makes it feasible to use a consensus approach for validation of assignments. The study with the complex sample also showed that X!Hunter/Mascot consensus with an accurate parent mass filter would provide a more sensitive alternative to the often used method allowing a high FDR but accepting only the protein identifications supported by two unique peptide MS/MS spectra (data not shown).
We were initially surprised that similar sensitivity is achieved in comparing Mascot and X!Hunter despite the low discrimination for the X!Hunter searches. The fact that sensitivity was good shows that this approach is capable of bringing the correct assignment to the top score in comparison with other candidates for each MS/MS. Some of the poor discrimination may be due to presence of simulated spectra that are not well simulated, but discrimination with xRef was also relatively poor, indicating that the chemical nature of the peptides is the major contributing factor. This suggests that there are different classes of peptides that have different error models and is probably a major reason why use of score thresholds as acceptance criteria is also difficult in other methods. A major advantage of a consensus approach is that score thresholds need not be considered.
An important factor in using the large simulated database was the previous experience with managing the peptide sequences to minimize the search space along with targeting of a specific subset of MS/MS in a data set to a corresponding subset of the protein database (14). This partitioning approach proved essential in handling the large simulated spectra library and also could be used to enhance search speed by simple parallel processing. We and others also showed that by combining results from independent search programs (e.g. Sequest and Mascot) low false discovery rates could be achieved with reduced score thresholds, allowing validation of a larger percentage of the MS/MS in a data set (10, 16). Finally we developed an early version of the simulated spectral library in a study showing that the MassAnalyzer-generated spectra could be used in rescoring search results to improve validation of search results.
Taken together, our work with search spaces, similarity scoring, and the simulated spectra reveals several areas where improvements could be made. The 15% fewer MS/MS identified by X!Hunter/Mascot consensus compared with Sequest/Mascot consensus in part reflects differences in spectrum-to-spectrum scoring functions of X!Hunter. Data sets of highly complex samples contain many spectral chimeras and weak spectra, which produce lower similarity scores. More sophisticated preprocessing will likely improve the underlying similarity scoring of these cases. A major problem is the use of different processing for the input library and the experimental spectra. This issue is complex because isotope peaks and correlated ions (for example, dehydrated forms) are included, and the number of amino acids in the peptide will influence the number of ions in the spectra. However, our goal was to determine whether the simulated spectra could be used in this application and not to optimize X!Hunter.
It is also likely that the MassAnalyzer simulation of MS/MS spectra can be improved. We have previously determined that a subset of about 20% of the simulated spectra is known to have problems with simulation (8). In particular, the modeling of the chemistry of higher charge forms and of distribution of charge in the fragment ions is an area that would benefit from more research. Work in progress has found that the large size of the model required for MS/MS simulation will require better methods for global fitting of the parameters. Nevertheless it is remarkable that the simulated spectra provide improved sensitivity over current libraries of observed MS/MS given our relatively limited understanding of the gas phase chemistry of peptides. Because of the faster search speed with X!Hunter and the ability to produce a library for any type of peptide, our results show that a modest investment in improving the simulated spectra and scoring functions will likely make this a very competitive search method for shotgun proteomics.
Supplementary Material
Acknowledgments
We are indebted to Z. Zhang for assistance with MassAnalyzer, particularly in providing versions adapted to the needs of our studies, as well as many helpful discussions of the issues involved in simulating these spectra. In addition, we thank R. Beavis for assistance with X!Hunter (University of British Columbia, Vancouver, Canada), B. E. Frewen and M. MacCoss (University of Washington, Seattle, WA) for assistance with BiblioSpec, and M. V. Mannino (University of Colorado, Denver, CO) for helpful discussions on data analysis.
Footnotes
Published, MCP Papers in Press, December 22, 2008, DOI 10.1074/mcp.M800384-MCP200
The number was based on an in silico generated peptide database. There are a total of 2,918,714 peptide sequences that were at least 9 amino acids long. Of these, 1,452,058 passed the missed cleavage rules of Yen et al. (14). The simulated spectra are then generated for charges 1–3.
The abbreviations used are: ABRF, Association of Biomolecular Resource Facilities; FDR, false discovery rate (FP/(TP + FP)); FP, false positive; TP, true positive; TN, true negative; SS, simulated spectra; xRef, spectral library provided by X!Hunter; xSS, simulated spectral library for X!Hunter; bSS, simulated spectral library for BiblioSpec; SM, Sequest and Mascot consensus; XM, X!Hunter and Mascot consensus; CPU, central processing unit; RAM, random access memory; IPI, International Protein Index; MGF, Mascot generic format; ROC, receiver-operator characteristic.
If the parameter “spectrum, total peaks” is set to 0, then all ions that survive the filters will be considered. If another number is input, then peaks surviving the filters will be used, but only up to the most intense 50 peaks will be considered regardless of the input number.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-CA126240 (to K. A. R.).
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 2.Sadygov, R. G., Cociorva, D., and Yates, J. R., III ( 2004) Large-scale database searching using tandem mass spectra: looking up the answer in the back of the book. Nat. Methods 1, 195–202 [DOI] [PubMed] [Google Scholar]
- 3.Stein, S. E., and Scott, D. R. ( 1994) Optimization and testing of mass spectra library search algorithms for compound identification. J. Am. Soc. Mass Spectrom. 5, 859–866 [DOI] [PubMed] [Google Scholar]
- 4.Craig, R., Cortens, J. C., Fenyo, D., and Beavis, R. C. ( 2006) Using annotated peptide mass spectrum libraries for protein identification. J. Proteome Res. 5, 1843–1849 [DOI] [PubMed] [Google Scholar]
- 5.Frewen, B. E., Merrihew, G. E., Wu, C. C., Noble, W. S., and MacCoss, M. J. ( 2006) Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 78, 5678–5684 [DOI] [PubMed] [Google Scholar]
- 6.Zhang, Z. ( 2004) Prediction of low-energy collision-induced dissociation spectra of peptides. Anal. Chem. 76, 3908–3922 [DOI] [PubMed] [Google Scholar]
- 7.Zhang, Z. ( 2005) Prediction of low-energy collision-induced dissociation spectra of peptides with three or more charges. Anal. Chem. 77, 6364–6373 [DOI] [PubMed] [Google Scholar]
- 8.Sun, S., Meyer-Arendt, K., Eichelberger, B., Brown, R., Yen, C.-Y., Old, W. M., Pierce, K., Cios, K. J., Ahn, N. G., and Resing, K. A. ( 2007) Improved validation of peptide MS/MS assignments using spectral intensity prediction. Mol. Cell. Proteomics 6, 1–17 [DOI] [PubMed] [Google Scholar]
- 9.Elias, J. E., and Gygi, S. P. ( 2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 10.Resing, K. A., Meyer-Arendt, K., Mendoza, A. M., Aveline-Wolf, L. D., Jonscher, K. R., Pierce, K. G., Old, W. M., Cheung, H. T., Russel, S., Wattawa, J. L., Goehle, G. R., Knight, R. D., and Ahn, N. G. ( 2004) Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal. Chem. 76, 3556–3568 [DOI] [PubMed] [Google Scholar]
- 11.Papping, D. J. C., Horjup, P., and Bleasby, A. J. ( 1993) Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3, 327–332 [DOI] [PubMed] [Google Scholar]
- 12.Eng, J. K., McCormack, A. L., and Yates, J. R., III ( 1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 13.Kersey, P. J., Durate, J., Williams, A., Karavidopoulou, Y., Birney, E., and Apweiler, R. ( 2004) The international protein index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 [DOI] [PubMed] [Google Scholar]
- 14.Yen, C.-Y., Russell, S., Mendoza, A. M., Meyer-Arendt, K., Sun, S., Cios, K. J., Ahn, N. G., and Resing, K. A. ( 2006) Improving sensitivity in shotgun proteomics using a peptide-centric database with reduced complexity: protease cleavage and SCX elution rules from data mining of MS/MS spectra. Anal. Chem. 78, 1071–1084 [DOI] [PubMed] [Google Scholar]
- 15.McDonald, W. H., Tabb, D. L., Sadygov, R. G., MacCoss, M. J., Venable, J., Graumann, J., Johnson, J. R., Cociorva, D., Yates, J. R., III ( 2004) MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18, 2162–2168 [DOI] [PubMed] [Google Scholar]
- 16.Searle, B. C., Turner, M., and Nesvizhskii, A. I. ( 2008) Improving sensitivity by probabilistically combining results from multiple MS/MS search methodologies. J. Proteome Res. 7, 245–253 [DOI] [PubMed] [Google Scholar]
- 17.Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. ( 2007) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 18.Shi, R., Kumar, C., Zougman, A., Zhang, Y., Podtelejnikov, A., Cox, J., Wisniewski, J. R., and Mann, M. ( 2007) Analysis of the mouse liver proteome using advanced mass spectrometry. J. Proteome Res. 6, 2963–2972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.