Abstract
In higher plants, the circadian clock controls a wide range of cellular processes such as photosynthesis and stress responses. Understanding metabolic changes in arrhythmic plants and determining output-related function of clock genes would help in elucidating circadian-clock mechanisms underlying plant growth and development. In this work, we investigated physiological relevance of PSEUDO-RESPONSE REGULATORS (PRR 9, 7, and 5) in Arabidopsis thaliana by transcriptomic and metabolomic analyses. Metabolite profiling using gas chromatography–time-of-flight mass spectrometry demonstrated well-differentiated metabolite phenotypes of seven mutants, including two arrhythmic plants with similar morphology, a PRR 9, 7, and 5 triple mutant and a CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1)-overexpressor line. Despite different light and time conditions, the triple mutant exhibited a dramatic increase in intermediates in the tricarboxylic acid cycle. This suggests that proteins PRR 9, 7, and 5 are involved in maintaining mitochondrial homeostasis. Integrated analysis of transcriptomics and metabolomics revealed that PRR 9, 7, and 5 negatively regulate the biosynthetic pathways of chlorophyll, carotenoid and abscisic acid, and α-tocopherol, highlighting them as additional outputs of pseudo-response regulators. These findings indicated that mitochondrial functions are coupled with the circadian system in plants.
Keywords: circadian, mitochondria, metabolomics, GC-MS, tricarboxylic acid cycle
In higher plants, the endogenous circadian clock controls various cellular processes ranging from photosynthesis to stress responses (1–3). The clock provides plants with the ability to adapt to daily changes in environmental conditions, thereby temporally organizing their physiological and metabolic processes. Recent studies, using mainly Arabidopsis, have begun to shed light on the mechanism of circadian clock at a molecular level (4, 5). Three candidate genes, CCA1 (CIRCADIAN CLOCK-ASSOCIATED 1), LHY (LATE ELONGATED HYPOCOTYL), and TOC1 (TIMING OF CAB EXPRESSION 1), are associated with the circadian oscillator. They form the first main part of the interlocked transcriptional/translational feedback loops in the model of circadian oscillation. Although this loop is essential for clock function, the CCA1/LHY-TOC1 feedback circuit alone is not sufficient to account for various aspects of circadian behaviors (6, 7).
TOC1 is a member of the PSEUDO-RESPONSE REGULATOR (PRR) family proteins, which include five elements (PRR9, PRR7, PRR5, PRR3, and TOC1/PRR1) (8, 9). PRR9 and PRR7 are reported to be crucial components of a temperature-sensitive circadian system (10), whereas triple-knockout plants of PRR9, 7, and 5 (d975) show arrhythmic expression of clock-associated genes under continuous light. Furthermore, PRR9/7/5 repress the accumulation of CCA1/LHY mRNA (9), whereas CCA1 and LHY activate transcription of PRR9 and PRR7 by binding with their promoter regions (11). d975 has a pleiotropic phenotype, including developmental abnormalities such as late flowering, long hypocotyls under constant red light, and dark green leaves (9, 12). These phenotypes are substantially similar to those of arrhythmic plants overexpressing the CCA1 gene (CCA1-ox) (13). Using transcriptome analysis, we clarified that d975 is tolerant to abiotic stresses, such as cold and drought, as a result of up-regulation of the dehydration-responsive element-binding protein 1 (DREB1) or C-repeat-binding factor (CBF) gene (14). Other reports on the relationship between circadian-clock regulation and abiotic stress responses (15, 16) suggest that clock genes have unknown output in addition to their clock-related output (17–19). Recent biochemical approaches demonstrated the importance of posttranscriptional and posttranslational control for the circadian mechanism (20–22). In the postgenomics era, metabolomics has been used not only to dissect plant metabolism per se, but also to identify unknown gene functions by comparing profiles of wild-type (WT) and genetically altered plants or during developmental changes (23) and diurnal changes (24). Therefore, characterizing an arrhythmic mutant from the viewpoint of metabolomic changes will help identify unknown output of clock genes.
Such study in Arabidopsis may help us understand the evolutionary relationship between the clock function and metabolism across biological kingdoms and help in development of treatment methods for diseases caused by impaired biological clock and metabolic disorders. For example, recent investigations in mouse showed that clock genes are likely related to lifestyle diseases such as obesity and metabolic syndrome (25, 26). Peroxisome proliferator-activated receptor-γ coactivator 1 (PGC-1), a key component in energy regulation in mammals, integrates the circadian clock with energy or lipid metabolism (27). This component enhances mitochondrial biogenesis and mitochondrial remodeling (28), and although an involvement of mitochondria in circadian rhythmicity in mammals and fungi has been proposed (29, 30), the link between the clock function and mitochondria in plants remains unclear.
In the present work, we investigated Arabidopsis thaliana with abnormal rhythmicity and performed transcriptomic and metabolomic analyses to gain insights into clock function and metabolism. Comparative metabolomics revealed unique elevated levels of tricarboxylic acid (TCA) cycle intermediates in d975. Because the unique pattern remained unchanged under different light conditions, a robust link between circadian-clock function and metabolic homeostasis in the TCA cycle was suggested. Integrated analysis of transcriptomics and metabolomics further revealed outputs of PRR 9/7/5 related to central metabolism, mainly in mitochondria.
Results
Comparative Genomics Suggests Structural and Functional Conservation of the Multigene Family of PRRs Among Plant Species.
Genomic analysis has shown that PRR genes in Arabidopsis are encoded by a multigene family containing five members closely associated with the clock oscillator (8). The sequence analyses based on phylogenetic tree show high conservation of the PRR amino acid sequence (e.g., pseudo-receiver domain and CCT motif) among seven plant species [see also supporting information (SI) Methods]. By assessing syntenic relationships based on the Plant Genome Duplication Database (PGDD) (31), phylogenetic analysis demonstrated that PRR3 and PRR7 in Arabidopsis are paralogous genes (Fig. S1A, solid red arrow). Many cross-genome syntenic relationships among the seven plant species were also indicated (Fig. S1A, black arrows). Expression patterns using the DIURNAL database (see SI Methods) indicate similar diurnal rhythms in the transcript level of the PRRs among three plant species (A. thaliana, Oryza sativa, and Populus trichocarpa) (Fig. S1B), although the function of PRR9/7/5 is largely unknown in these species, except in A. thaliana and O. sativa (32). Thus, our comparative genomics study suggests structural and functional conservation of the multigene family of PRRs across a variety of plant species.
Metabolite Phenotyping Differentiates d975 from CCA1-ox Exhibiting Similar Morphology.
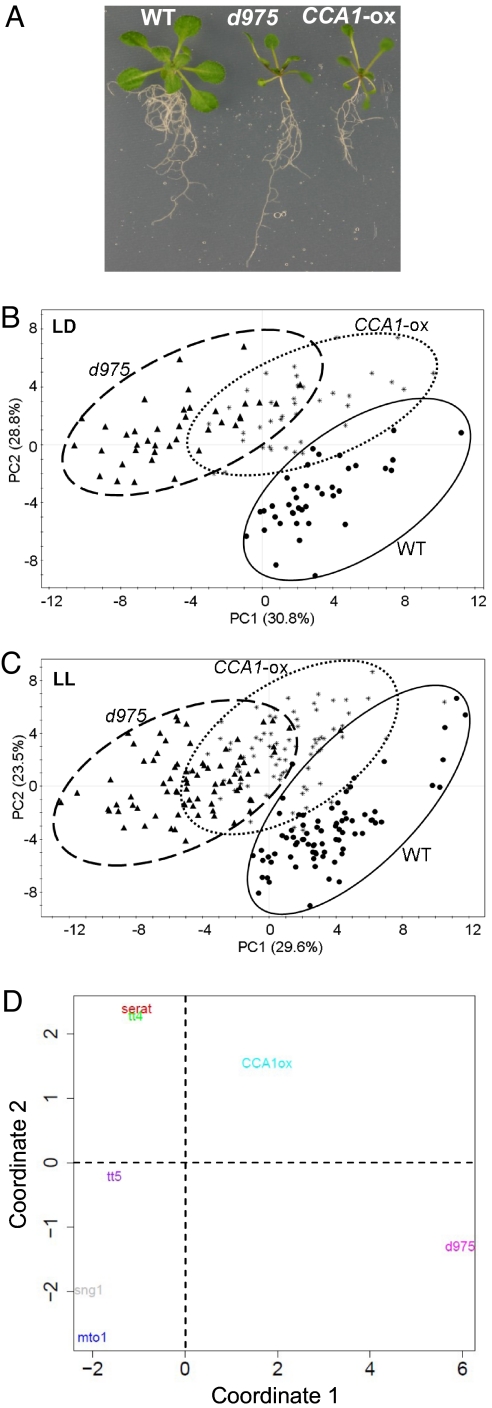
We confirmed that d975 and CCA1-ox were morphologically similar to each other under identical growth conditions (Fig. 1A and Table S1). By partial least-squares discriminate analysis (PLS-DA), a supervised multivariate-regression technique involving a dummy variable for classification, metabolite phenotypes were investigated for WT, d975, and CCA1-ox under light/dark (LD) and continuous light (LL) conditions (see Methods and Fig. S2). The results displayed clear separations between genotypes under both conditions despite the morphological similarity of d975 and CCA1-ox (Fig. 1 B and C). To identify the metabolites that contributed to these separations, we examined the first principal component loadings (Dataset S1) of each genotype in each PLS-DA model. The number of metabolites that changed in d975 was larger than that in CCA1-ox, suggesting more pronounced metabolic changes induced by PRR9/7/5 mutation than by overexpression of CCA1.
Fig. 1.
Morphologic change and metabolite phenotyping of two arrhythmic A. thaliana mutants. (A) Phenotypes of Arabidopsis WT, d975, and CCA1-ox grown under long days (16-h light and 8-h dark). (B and C) Metabolite phenotyping of the three genotypes under both LD (B) and LL (C). At each time, samples (biological replicates, n = 5; see also Dataset S2) were harvested from the whole plant of each plant both under LD (sample n values: WT, 39; d975, 39; and CCA1-ox, 39) and LL (sample n values: WT, 77; d975, 79; and CCA1-ox, 80). Note that the numbers of symbols are equal to Ng * Nt, plot points, where Ng represents the number of biological replicates for a genotype and Nt represents the number of times measured. LD data were sampled 8 times: ZT (0, 3, 6, 9, 12, 15, 18, and 21). LL data were sampled 16 times: ZT (1, 4, 7, 10, 13, 16, 19, 22, 26, 29, 32, 35, 38, 41, 44, and 47). Circles, WT; triangles, d975; stars, CCA1-ox. The PLS-DA model in both conditions showed three significant components according to cross-validation (see Methods). The arrhythmic mutants exhibited distinct metabolic constituents under both light conditions. (D) MDS plot of seven mutants based on 52 commonly detected metabolites at ZT 12 (see Methods). Genotypes of d975, CCA1-ox, mto1, sng1, double mutant serat2;1 serat2;2, tt4, and tt5 were compared. Log2 ratio values compared with the WT plants and Euclidean distance were used (see Methods and SI Methods). Note that d975 exhibited the greatest change in primary metabolism among the mutants examined.
d975 Exhibited the Most Pronounced Changes in Primary Metabolism Among the Examined Mutants.
To characterize d975 further, we compared metabolite profiles among seven mutants: d975, CCA1-ox, mto1 (33), sng1 (34), tt4, tt5 (35), and double mutant serat2;1 serat2;2 (36). The mutants mto1, sng1, and serat2;1 serat2;2, possess mutations related to primary metabolism, whereas mutants tt4 and tt5 possess mutations related to flavonoid metabolism. Profile clustering, using MDS (multidimensional scaling), was based on 52 metabolites that were commonly detected in the seven mutants. A 2D MDS plot (Fig. 1D), representing similarity among the seven mutants in metabolite profiles, clearly separated d975 from the other mutants in coordinate 1. Thus, d975 showed the most pronounced and distinct changes in the metabolite levels among the mutants examined.
d975 Exhibits Considerably Increased TCA Cycle Intermediates, Whereas CCA1-ox Shows Less Change in Primary Metabolism.
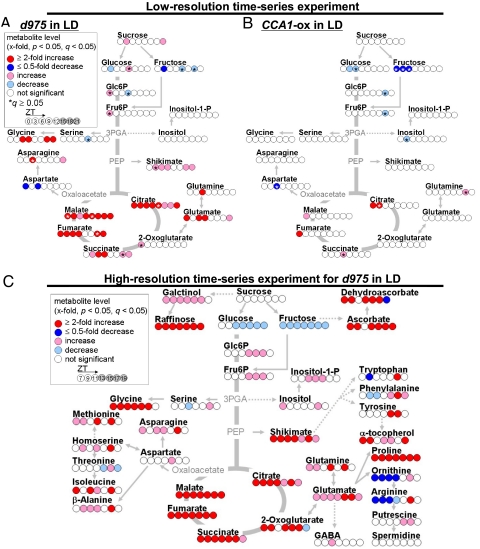
To differentiate metabolite profiles of the two arrhythmic plants, significant metabolite changes (FDR, q < 0.05) observed in d975 or CCA1-ox were compared against WT on a metabolic map (Fig. 2 A and B). A comprehensive list of all measured peaks and level changes is shown in Dataset S2. We found that the increase in TCA cycle intermediates in d975 was remarkable and significant (Fisher's exact test, P = 0.002) under LD conditions. Similar changes in d975 were also found under LL conditions (Fig. S3), suggesting that overaccumulation of TCA cycle intermediates occurred regardless of light condition. In particular, elevated levels of citrate and malate were observed at all time points (Fig. 2A). The level of shikimate was significantly higher in d975 than in WT, implying that the change in shikimate-derived secondary metabolism may affect stress tolerance in the triple mutant (14–16). In contrast, less pronounced changes were observed in CCA1-ox than in d975 (Fig. 2B). Taken together, these results suggest the presence of a PRR9/7/5-dependent control in the central metabolism (e.g., TCA cycle) of Arabidopsis. Because the level of CCA1/LHY mRNA was increased in both d975 and CCA1-ox (9), it is possible that a PRR/9/7/5-specific regulatory mechanism independent of CCA1 exists.
Fig. 2.
Changes in metabolite levels. (A and B) Lower-resolution time series experiments in d975 (A)and CCA1-ox plants (B) Changes in metabolite contents were calculated by dividing metabolite level in the mutant with that in WT. The level of significance was set at q < 0.05 (see also Dataset S3). Red and blue colors indicate increased and decreased metabolite levels, respectively. The eight circles under each metabolite name indicate the corresponding ZT. The figure indicates that d975 had a dramatic increase in TCA cycle intermediates (Fisher's exact test, P = 0.002), whereas CCA1-ox exhibited less-pronounced changes in primary metabolism. (C) Changes in metabolite levels in d975 created with the higher resolution time-series experiment (n = 15, at 2-h sampling period). In addition to the change in TCA cycle intermediates, d975 exhibited remarkable increases in metabolite levels associated with antioxidant vitamins (e.g., ascorbate and α-tocopherol). 3PGA, 3-phosphoglycerate; PEP, phosphoenolpyruvate; Fru6P, fructose 6-phosphate; Glc6P, glucose 6-phosphate. *, q ≥ 0.05.
In a higher-resolution time series experiment, we further investigated the metabolite profiles of d975 from Zeitgeber time (ZT) 7 to 19 (Fig. 2C and Fig. S2B). Fig. 2C shows the significant increases observed in the levels of TCA cycle intermediates, including 2-oxoglutarate, succinate, and fumarate in addition to citrate and malate. The enhanced levels of glycine, glutamate, and shikimate were consistent with the result of lower-resolution experiments, but more accumulation occurred (compare Fig. 2 A and C). The levels of osmolytes (e.g., proline, galactinol, and raffinose) increased as reported (14). However, arginine and ornithine levels decreased significantly, suggesting a shift in the glutamate-metabolic flux to proline production rather than to ornithine production (Fig. 2C). Dramatic increases in antioxidant vitamins (ascorbate and α-tocopherol) were also observed in d975 (Fig. 2C and Dataset S3).
Transcriptomic and Metabolomic Changes Reveal PRR9/7/5 Function in Regulation of Biosynthetic Pathways Associated with Chlorophyll, Carotenoid and Abscisic Acid (ABA), and α-Tocopherol.
Using MapMan (37), an ontology tool, we examined the patterns of transcriptional change in genes involved in central metabolism (Fig. S4A). We observed the up-regulation of genes encoding synthetic enzymes of osmolytes such as proline, galactinol, and raffinose (e.g., AT2G39800, P5CS1) as reported (14). There were general trends for induction of expression of genes that are involved in starch synthesis and degradation, photorespiration, tetrapyrrole metabolism, and terpene biosynthesis. Particularly, to facilitate the integration of marked changes in gene expression and metabolite accumulation in d975, we focused on TCA cycle (Fig. S4B). The expressions of a gene encoding fumarase (AT2G47510) and putative 2-oxoglutarate dehydrogenase (AT5G65750) in the TCA cycle were down-regulated in d975. These changes might suggest a reason for the overaccumulation of TCA cycle intermediates in the triple mutant (see Discussion).
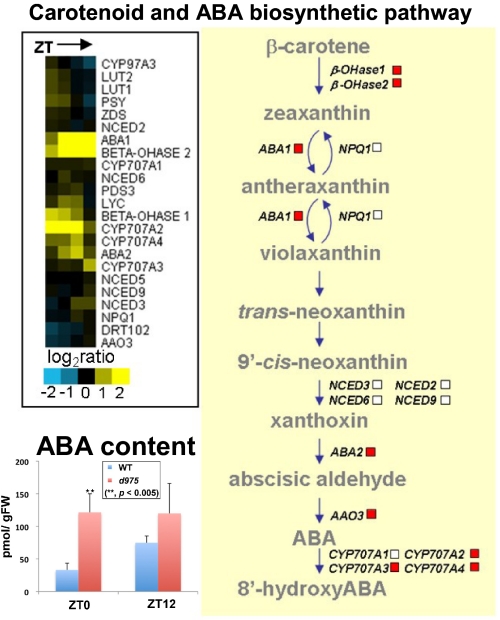
To detect the transcriptional coordination at the metabolic pathway level further, the number of genes with significant changes in expression in d975 at each time point was counted. Within each AraCyc (38) pathway, we compared the proportion of genes with significant changes in expressions (Dataset S4). Marked induction was observed in the metabolic pathways associated with (i) chlorophyll biosynthesis, (ii) carotenoid and ABA, and (iii) α-tocopherol (Fig. 3 and Fig. S4C). Fig. 4C Upper indicates that significant increases in the levels of glutamate (1.3- to 4.7-fold) and phytol (1.2- to 1.4-fold) were associated with the up-regulation of gene expression in tetrapyrrole biosynthetic pathway (Dataset S4). However, the expression of gene encoding PHYTOCHROME-INTERACTING FACTOR 1 (PIF1), which directly and indirectly regulates chlorophyll biosynthesis pathways (39), showed no significant changes in d975 (Dataset S5), suggesting that the regulation of this pathway by PRR9/7/5 is independent of PIF1 (see Discussion). The expression of genes involved in the carotenoid and ABA biosynthetic pathways increased in d975 (Fig. 3), and furthermore, the levels of ABA increased in d975 (see Fig. 3, Left Bottom). Moreover, the enhancement of α-tocopherol production was explained by both the transcript and metabolite levels in α-tocopherol biosynthesis (Fig. S4C and Dataset S4).
Fig. 3.
Schematic view integrating metabolite with transcript data. Pathway-level changes in gene expression involved in biosynthetic pathway of carotenoid and ABA for d975 are shown. To visualize the qualitative differences in transcript levels between d975 and WT, we used a hierarchical cluster analysis with the “Euclidean distance” and the “average linkage” method (see the boxes). ABA content in d975 increased significantly (t test, P < 0.05) (Left Lower). Viewed as a whole, the figure suggests that the biosynthetic pathway of carotenoid and ABA is under the control of PRR9/7/5.
Discussion
A detailed metabolomic analysis using a triple mutant, d975, clearly demonstrated that PRR9/7/5 is involved in maintaining mitochondrial homeostasis in Arabidopsis. Although it has been proposed that abnormalities in mitochondrial function probably affect clock functions in animals and fungi (29, 30), there were no reports indicating such a relationship in plants. Here, a robust link between the function of PRR9/7/5 and mitochondrial metabolism was detected by transcriptomic and metabolomic analyses. First, despite the morphological similarity between the two arrhythmic mutants (d975 and CCA1-ox), we were able to distinguish clearly their metabolite phenotypes under both LD and LL (Fig. 1 B and C). The mutant d975 was characterized by a dramatic increase in TCA cycle intermediates and antioxidant vitamins in addition to osmolytes (14), whereas no drastic changes occurred in these metabolites in CCA1-ox (Fig. 2). These results suggest the presence of a PRR9/7/5-mediated mechanism for cooperative control, through which appropriate metabolite levels may be maintained, especially those in the TCA cycle.
Overaccumulation of TCA cycle intermediates in d975 may be explained, at least partially, by the down-regulation of genes encoding fumarase (AT2G47510) and putative 2-oxoglutarate dehydrogenase (AT5G65750), which are supposed to be localized in mitochondria (Fig. S4). Because of the reduction in expression of these genes, the levels of malate, fumarate, and amino acids, produced via 2-oxo-glutarate, increased in d975. This is in accordance with a report that showed elevated levels of malate, fumarate, and glycine in transgenic tomato with low fumarase activity (40). Sweetlove et al. (41) have proposed an interaction of ascorbate metabolism, respiration, and photosynthesis, and it is known that ascorbate levels in higher plant leaves show a diurnal rhythm (42). Both reactive oxygen species (ROS) production and ROS defense in plants are likely controlled, in part, by a functional circadian clock. Because the last enzyme in the ascorbate biosynthesis pathway, l-galactono-1,4-lactone dehydrogenase (GLDH), is known as an integral membrane protein in mitochondria (43), the clock function involved in cellular redox homeostasis and stress resistance may be closely related with the mitochondrial function.
Here, we observed pathway-level changes in expression of genes encoding enzymes associated with the chlorophyll biosynthetic pathways (Fig. S4C Upper). However, no change in the expression level of PIF1 (39), one of the regulators of chlorophyll biosynthesis, was observed in d975. This implies that regulation of PRR9/7/5 for chlorophyll biosynthetic pathway is independent of PIF1-mediated regulation. It is possible that up-regulation of gene expressions in the chlorophyll biosynthetic pathway resulted in the mature, dark green leaves of d975 (9). This hypothesis is further supported by the significant increase of phytol in d975, as phytol is generated during chlorophyll catabolism by chlorophyllase.
As an output pathway regulated by PRR9/7/5, we also clarified the biosynthesis of carotenoid and ABA (Fig. 3). ABA production is presumably clock-regulated, thereby generating diurnal rhythms of ABA levels in Arabidopsis (44). This raises the possibility that PRR9/7/5-mediated control of ABA may enhance fitness of plants to abiotic stresses.
The significance of ascorbate and α-tocopherol as protectants against light-induced oxidative stress is well established (45, 46), and α-tocopherol content follows a diurnal rhythm in leaves (47). Significant increase of α-tocopherol level in d975, together with up-regulation of gene expressions of the α-tocopherol biosynthetic pathway (Fig. S4C Lower), suggests that PRR9/7/5 may play a role in regulating the α-tocopherol level in the diurnal cycle.
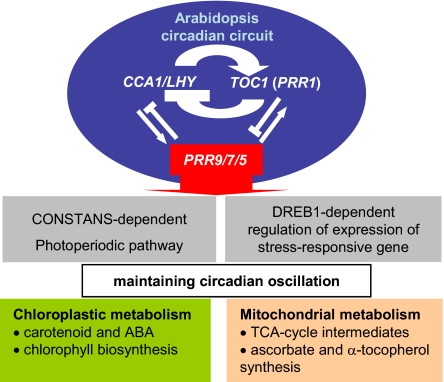
As a conclusion, the function of PRR9/7/5, as elucidated so far, is summarized in Fig. 4. PRR9/7/5, together with CCA1/LHY and TOC1 (PRR1), is integral to the generation of circadian rhythm in Arabidopsis. PRR9/7/5 also plays crucial roles in stress responses controlled by DREB1 (14) and in activation of CO (CONSTANS) expression during daytime (9, 12). The present approach of integrated transcriptomics and metabolomics provided evidence of unique output of PRR9/7/5. Such output coordinately maintains central metabolism, particularly the TCA cycle. The biosynthetic pathways of chlorophyll, carotenoid and ABA, and α-tocopherol were also indicated to be under the control of PRR9/7/5. Our present findings suggest the possibility of universal simultaneous control of clock and mitochondrial function presence in all biological kingdoms as indicted in mammals and fungi (29, 30). General application of such knowledge is also expected to help solve problems related to efficient plant biomass production and plant stress responses.
Fig. 4.
Proposed output functions of PRR9/7/5. The figure suggests that PRR9/7/5, together with CCA1/LHY and TOC1 (PRR1), generates a normal oscillation rhythm. Furthermore, it shows that PRR9/7/5 is involved in DREB1-dependent regulation of stress-responsive gene expression and photoperiodic control of flowering time. The outputs of PRR9/7/5, such as maintaining central metabolism including that of the TCA cycle and antioxidant vitamins, are also shown. In addition, PRR9/7/5 negatively regulates the biosynthetic pathways associated with chlorophyll, carotenoid-ABA, and α-tocopherol biosynthesis in chloroplasts.
Methods
Plant Materials and Growth Conditions.
A. thaliana accession Columbia (Col-0) was used as a WT plant source. Triple-knockout Arabidopsis mutant prr9-10/prr7-11/prr5-11 has been described in ref. 9. Seedling were grown on Murashige and Skoog (392-00591;Wako) with 0.3% gellan gumand 2% sucrose at pH 5.7 under continuous light (LL) or 12-h light/12-h dark (LD) cycles at 22 °C for 18 days. Whole plants were sampled during days 18–19 to reduce possible developmental effects.
Experimental Design.
Two types of experiments were carried out. The lower temporal resolution experiment (Design 1) was designed for metabolite phenotyping of WT, d975, and CCA1-ox (Fig. S2A), over 24 sampling times (3 genotypes were harvested at 3-h intervals). To facilitate LD metabolite phenotyping, we represent three successive sampling times as points ZT 3, 6, and 9 for LD1 and ZT 15, 18, and 21 for LD2 (Fig. S2A Top). Similarly, under LL conditions, sampling times (ZT 4, 7, and 10) were designated as point LL1–1, (ZT 16, 19, and 22) as LL1–2 (Fig. S2A Middle), (ZT 26, 29, and 32) as LL2–1, and (ZT 38, 41, and 44) as LL2–2 (Fig. S2A Bottom). The second experiment (Design 2) was a higher-resolution time series intended to obtain insight into the functional role of PRR 9/7/5 from ZT 7 to ZT 19 with respect to WT and d975 (Fig. S2B). This includes transcript profiling investigated previously (14). The number of time points in transcript profiling was smaller (4 time points, ZT 8 to 14 with 2-h intervals) than that in metabolite profiling (7 time points, ZT 7 to 19 with 2-h intervals).
Metabolite Profiling.
Each sample was extracted, derivatized, and analyzed by using gas chromatography–time-of-flight (GC-TOF/MS) as described in ref. 48. See also SI Methods.
Quantification of ABA.
One hundred mg of 18-day-old plants grown under LD conditions were harvested and ABA contents were measured as reported (49). Three biological replicates were harvested for ABA measurement.
Transcript Profiling.
The data were obtained from a previous study with the permission of authors (14). All raw CEL files have been deposited in the Nottingham Arabidopsis Stock Center microarray database (NASCArrays) under accession number NASCARRAYS-421. Raw CEL files were normalized by robust multiarray average (RMA) (50) with Bioconductor Simpleaffy package (51). See also SI Methods.
Comparative Metabolome Analysis.
Metabolite profiles of d975 and CCA1-ox in Design 1 (Fig. S2A) were compared with those of other metabolome datasets: d975 (9) and CCA1-ox (13); mto1 (methionine-over accumulation 1) (33), tt4 (transparent testa4) (35), and double mutant serat2;1 serat2;2 (36); tt5 (35) and sng1 (sinapoylglucose accumulator 1) (34). All mutants used are in the Col-0 background. To cancel out the effect of different mass spectrometric conditions (e.g., detector responses, detector sensitivity, and column performance) among datasets the log2 ratio of the samples to the control samples in each dataset was used. We used MDS with Euclidean distance as implemented in the statistical R package.
Statistical Data Analysis.
PLS-DA, which is a highly suited regression technique for the analysis of “omics” data that contain typically many more variables (e.g., metabolites) than observations (e.g., replicates), was calculated by using SIMCA-P 11.0 software (Umetrics AB) with log10 transformation and unit variance scaling. The PLS-DA models for Fig. 1 B and C showed three significant components according to cross-validation, respectively. In metabolite phenotyping for each genotype under LD (Fig. 1B), the explained variation in the X matrix (R2X) and the Y matrix (R2Y) is 0.36 and 0.78, and the predictive ability by means of cross-validation (Q2Y) is 0.72. In LL (Fig. 1C), R2X, R2Y, and Q2Y are 0.35, 0.70, and 0.66, respectively. Statistical analysis was performed for genes encoding enzymes included in the TAIR AraCyc database (38). All genes were obtained from AraCyc version 4.1 as text format dump file (aracyc_dump.20071005). Genes without a matching known AGI locus number or with a duplicate number within a given pathway were deleted. Fisher's exact test was used to compare the difference in metabolite levels of intermediates in TCA cycle between d975 and CCA1-ox. The numbers of genes within each AraCyc pathway that were up- or down-regulated significantly (q < 0.05) were counted. Hierarchical cluster analyses, along with visualization for expression level (log2 ratio), were performed by using Cluster 3.0 (52) and Java TreeView software. Heatmap representation and statistical testing were performed by using logarithm transformation. False-discovery rate (FDR) corrections for multiple testing across all transcripts and metabolites were performed with the “qvalue” package (53) in R (www.r-project.org).
Supplementary Material
Acknowledgments.
We thank Thomas Moritz from Umeå Plant Science Centre (Umeå, Sweden) for excellent technical advice on GC-TOF/MS profiling, Pär Jonsson from Umeå University (Umeå, Sweden) for hyphenated data analysis (HDA) assistance, and Hans Stenlund from Umeå University for raw data analysis (RDA) assistance. We thank Elaine Tobin at University of California (Los Angeles, CA) for the CCA1-ox plants. We thank Masanori Arita and Henning Redestig from RIKEN Plant Science Center (Yokohama, Japan) for discussions. This work was supported in part by grants-in-aid from the Ministry of Education, Science, Culture, Sports, and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900952106/DCSupplemental.
References
- 1.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 2.McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–448. doi: 10.1146/annurev.genet.40.110405.090603. [DOI] [PubMed] [Google Scholar]
- 4.Mas P. Circadian-clock function in Arabidopsis thaliana: Time beyond transcription. Trends Cell Biol. 2008;18:273–281. doi: 10.1016/j.tcb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009 doi: 10.1146/annurev.arplant.043008.092054. in press. [DOI] [PubMed] [Google Scholar]
- 6.Locke JC, et al. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol Syst Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeilinger MN, Farre EM, Taylor SR, Kay SA, Doyle FJ., 3rd A novel computational model of the circadian clock in Arabidopsis that incorporates PRR7 and PRR9. Mol Syst Biol. 2006;2:58. doi: 10.1038/msb4100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- 9.Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005;46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 10.Salome PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farre EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 12.Nakamichi N, et al. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7, and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–832. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- 13.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 14.Nakamichi N, et al. Transcript profiling of an Arabidopsis PSEUDO-RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 15.Kant P, et al. Functional-genomics-based identification of genes that regulate Arabidopsis responses to multiple abiotic stresses. Plant Cell Environ. 2008;31:697–714. doi: 10.1111/j.1365-3040.2008.01779.x. [DOI] [PubMed] [Google Scholar]
- 16.Bieniawska Z, et al. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 2008;147:263–279. doi: 10.1104/pp.108.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakin-Thomas PL. Circadian rhythms: New functions for old clock genes. Trends Genet. 2000;16:135–142. doi: 10.1016/s0168-9525(99)01945-9. [DOI] [PubMed] [Google Scholar]
- 18.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez RA, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA. 2008;105:4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugano S, Andronis C, Green RM, Wang ZY, Tobin EM. Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc Natl Acad Sci USA. 1998;95:11020–11025. doi: 10.1073/pnas.95.18.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiba T, Henriques R, Sakakibara H, Chua NH. Targeted degradation of PSEUDO-RESPONSE REGULATOR 5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell. 2007;19:2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara S, et al. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- 23.Oksman-Caldentey KM, Saito K. Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr Opin Biotechnol. 2005;16:174–179. doi: 10.1016/j.copbio.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Gibon Y, et al. Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 2006;7:R76. doi: 10.1186/gb-2006-7-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimba S, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci USA. 2005;102:12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turek FW, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 28.Lin JD. Minireview. The PGC-1 coactivator networks: Chromatin-remodeling and mitochondrial energy metabolism. Mol Endocrinol. 2009;23:2–10. doi: 10.1210/me.2008-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brody S. Circadian rhythms in Neurospora crassa: The role of mitochondria. Chronobiol Int. 1992;9:222–230. doi: 10.3109/07420529209064531. [DOI] [PubMed] [Google Scholar]
- 30.Langmesser S, Albrecht U. Life time circadian clocks, mitochondria and metabolism. Chronobiol Int. 2006;23:151–157. doi: 10.1080/07420520500464437. [DOI] [PubMed] [Google Scholar]
- 31.Tang H, et al. Synteny and collinearity in plant genomes. Science. 2008;320:486–488. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 32.Murakami M, Ashikari M, Miura K, Yamashino T, Mizuno T. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003;44:1229–1236. doi: 10.1093/pcp/pcg135. [DOI] [PubMed] [Google Scholar]
- 33.Inaba K, et al. Isolation of an Arabidopsis thaliana mutant, mto1, that overaccumulates soluble methionine (temporal and spatial patterns of soluble methionine accumulation) Plant Physiol. 1994;104:881–887. doi: 10.1104/pp.104.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorenzen M, Racicot V, Strack D, Chapple C. Sinapic acid ester metabolism in wild type and a sinapoylglucose-accumulating mutant of Arabidopsis. Plant Physiol. 1996;112:1625–1630. doi: 10.1104/pp.112.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe M, et al. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. Plant Cell. 2008;20:2484–2496. doi: 10.1105/tpc.108.060335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thimm O, et al. MapMan: A user-driven tool to display genomics datasets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 38.Mueller LA, Zhang P, Rhee SY. AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huq E, et al. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- 40.Nunes-Nesi A, et al. Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function. Plant J. 2007;50:1093–1106. doi: 10.1111/j.1365-313X.2007.03115.x. [DOI] [PubMed] [Google Scholar]
- 41.Sweetlove LJ, Fait A, Nunes-Nesi A, Williams T, Fernie AR. The mitochondrion: An integration point of cellular metabolism and signalling. Crit Rev Plant Sci. 2007;26:17–43. [Google Scholar]
- 42.Dutilleul C, et al. Leaf mitochondria modulate whole-cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell. 2003;15:1212–1226. doi: 10.1105/tpc.009464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millar AH, et al. Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiol. 2003;133:443–447. doi: 10.1104/pp.103.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee KH, et al. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell. 2006;126:1109–1120. doi: 10.1016/j.cell.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 45.Muller-Moule P, Havaux M, Niyogi KK. Zeaxanthin deficiency enhances the high light sensitivity of an ascorbate-deficient mutant of Arabidopsis. Plant Physiol. 2003;133:748–760. doi: 10.1104/pp.103.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanwischer M, Porfirova S, Bergmuller E, Dormann P. Alterations in tocopherol cyclase activity in transgenic and mutant plants of Arabidopsis affect tocopherol content, tocopherol composition, and oxidative stress. Plant Physiol. 2005;137:713–723. doi: 10.1104/pp.104.054908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munne-Bosch S, Alegre L. The function of tocopherols and tocotrienols in plants. Crit Rev Plant Sci. 2002;21:31–57. [Google Scholar]
- 48.Kusano M, et al. Unbiased characterization of genotype-dependent metabolic regulations by metabolomic approach in Arabidopsis thaliana. BMC Syst Biol. 2007;1:53. doi: 10.1186/1752-0509-1-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naito T, et al. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci Biotechnol Biochem. 2007;71:1269–1278. doi: 10.1271/bbb.60681. [DOI] [PubMed] [Google Scholar]
- 50.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 51.Wilson CL, Miller CJ. Simpleaffy: A bioconductor package for Affymetrix quality control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- 52.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 53.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.