Abstract
Magnetic resonance (MR) imaging has demonstrated that variation in brain structure is associated with differences in behavior and disease state. However, it has rarely been practical to prospectively test causal models that link anatomical and functional differences in humans. In the present study we have combined classical mouse genetics with high-field MR to systematically explore and test such structure-functional relations across multiple brain regions. We segmented 33 regions in two parental strains—C57BL/6J (B) and DBA/2J (D)—and in nine BXD recombinant inbred strains. All strains have been studied extensively for more than 20 years using a battery of genetic, functional, anatomical, and behavioral assays. We compared levels of variation within and between strains and sexes, by region, and by system. Average within-strain variation had a coefficient of variation (CV) of 1.6% for the whole brain; while the CV ranged from 2.3–3.6% for olfactory bulbs, cortex and cerebellum, and up to ~18% for septum and laterodorsal thalamic nucleus. Variation among strains averages ranged from 6.7% for cerebellum, 7.6% for whole brain, 9.0% for cortex, up to ~26% for the ventricles, laterodorsal thalamic nucleus, and the interpeduncular nucleus. Heritabilities averaged 0.60 ± 0.18. Sex differences were not significant with the possible (and unexpected) exception of the pons (~20% larger in males). A correlation matrix of regional volumes revealed high correlations among functionally related parts of the CNS (e.g., components of the limbic system), and several high correlations between regions that are not anatomically connected, but that may nonetheless be functionally or genetically coupled.
Introduction
Both the absolute and relative size of brain regions are highly variable within populations. For example, the surface area of primary visual cortex varies nearly three-fold among normal humans (Stensaas, Eddington et al. 1974);(Horton and Hoyt 1991) (Andrews, Halpern et al. 1997) and this variation may well relate to the wide range in variation in performance (Halpern, Andrews et al. 1999). To what extent does neuroanatomical variation in one structure match that in other related structures—does the three-fold variation in striate cortex match differences in the subcortical visual system, such as the lateral geniculate nucleus and the optic tract? Purves and colleagues used classical postmortem neuroanatomical methods and showed impressively tight covariation among several components of the visual systems. Pearson product-moment correlations ranged from 0.5 to 0.8 (Andrews, Halpern et al. 1997). This is an important finding both with respect to CNS function and possible genetic, developmental, and environmental mechanisms that produce the covariation. Many questions remain unaddressed. Is covariation restricted to known components of specific neuronal circuits? Is covariation generated by hardwired genetic control of cell division and cell growth or is covariation the result of functional plasticity of connected CNS regions? To what extent can covariation itself be used to map putative or known functional relations among CNS compartments? And finally, can covariation across systems be used to predict functional capacity and disease progression?
To begin to answer these questions we need to know much more about the covariance structure of CNS compartments in humans and common experimental animal models. What makes this work particularly daunting is the need to obtain accurate estimates of volume from large numbers of subjects and large numbers of regions (Wright, Sham et al. 2002). Ideally, one should be able to control both the genetics and the environment of all subjects and to resample subjects at different stages and across different treatments. To a limited extent this can be done in humans by exploiting groups of related individuals, for example entire families or sets of monozygotic and dizygotic twins (Pennington, Filipek et al. 2000); (White, Andreasen et al. 2002); (Wright, Sham et al. 2002)). For example, Wright and colleagues imaged 20 pairs of twins to compute heritability for multiple brain regions and to parse the covariation matrix of these brain regions into principal components to statistically define regions with shared patterns of variation.
Corresponding work in experimentally tractable rodents with small brains is now finally practical. Over the last few years, we have developed high-field strength magnetic resonance microscopy (MRM) protocols, contrast enhancement methods, and semi-automatic segmentation procedures that now enable us to quantify 30–40 well defined CNS regions in mouse with unprecedented precision, despite its small brain size (Johnson et al., 2007, Sharief et al., 2008). In a recent study we used this suite of methods to quantify the level of variation of 33 regions within a set of six isogenic males (essentially a clone of identical twins) all raised under identical laboratory conditions (Badea et al., 2007). For this analysis we chose the C57BL/6J strain that has been used so widely in genetic and genomic studies and for which we currently have the most complete sequence data and CNS gene expression data (Lein, Hawrylycz et al. 2007). The analysis of one genotype raised in a uniform environment provides a way to study the limits of precision with which developmental mechanisms can control CNS structure (Pearson and Goodman 1979), (Williams, Strom et al. 1996). These baseline data are also essential in order to understand how much genetic, developmental, and environmental factors contribute to structural and functional variation.
In the present study we have extended our analysis of C57BL/6J in several ways. First, we explore variation across different genotypes using identical procedures. This genetic extension includes the original set of C57BL/6J animals and a sample of ten other strains. We have added one of the oldest common inbred strains of mice, DBA/2J. We also added nine BXD-type recombinant inbred strains derived from an intercross between C57BL/6J and DBA/2J. We have used this set of strains as an initial resource to compare variation within and among strains when environmental variation is minimized. One can think of this as an analysis, with replication, of a family of humans—mother, father, and nine offspring. Second, we have segmented a sufficiently large number of regions that we have been able to produce a neuroanatomical covariance matrix similar to those computed recently using cohorts of monozygotic and dizygotic human twins. The dataset that we have generated is still modest in size and limited in the extent of replication within strain, but it nonetheless comprises the most comprehensive dataset on variation and covariation among multiple CNS regions in any rodent species. The data provide a useful proof of principle for future large-scale analyses of more genotypes, more replicates, and systematic environmental perturbations. All image data and segmented volumes are available as part of the Biomedical Informatics Research Network (BIRN) at www.nbirn.net/bdr/mouse_civm/index.shtm and www.civm.duhs.duke.edu/bxd/index.html. Similarly all of the morphometric data are integrated in GeneNetwork (www.genenetwork), along with more than 1000 other classic phenotypes for these same strains of mice.
Methods
Animal Preparation
We studied age-matched pairs (male and female) of mice belonging to 11 inbred strains (56–64 day of age) that were obtained directly from the Jackson Laboratory (www.jax.org): C57BL/6J (B6), DBA/2J (D2), and the following nine BXD recombinant inbred strains—BXD1, BXD6, BXD15, BXD16, BXD24, BXD28, BXD29, BXD34, and BXD40. We intentionally studied age-matched male-female pairs from different litters. This is essential in order to ensure that the low levels of within-strain variance is not simply due to a common litter effects. (This is analogous to the situation of monozygotic human twins raised in different environments.)
In previous companion studies we have developed semi-automated segmentation procedures and quantified 33 brain regions within a single isogenic strain of mouse (Badea et al., 2007, Sharief et al., 2008). For this previous work we used six young adult (~9 weeks old) male B6 mice. B6 is the most widely used inbred strain and is also the genetic background for most mutations and knockouts. B6 is also the maternal parent of the large set of BXD recombinant inbred strains used in the present study.
The BXD RI strains used in this study are also fully inbred, but each of the BXD strains is a unique genetic “mosaic” of the genomes of the maternal and paternal parental strains, B6 and D2, respectively. The BXDs are a family of isogenic but highly diverse strains that can be used to study genetic factors that contribute to differences in brain structure and function. They can also be used to study covariance among traits, while minimizing the effects of sampling error and environmental confound (Chesler et al., 2003). A wide range of phenotypes for as few as 8 to as many as 80 of these BXD strains are available online in GeneNetwork (GN, www.genenetwork.org). Phenotypes include morphometric and stereological datasets that are closely related to the MRM volumetric data we describe here. GN also includes brain gene expression data for adult BXD strains, including data for the whole brain and the following four major CNS regions that we have also segmented: cerebellum, hippocampus, striatum, and the neocortex.
All experiments were conducted in accordance with NIH guidelines, using protocols approved by the Duke University Institutional Animal Care and Use Committee. Mice were anesthetized with 100 mg/kg pentobarbital (i.p.) and then fixed by transcardial perfusion, first with a flush of a mixture of 0.9% saline and gadoteridol contrast agent—ProHance (Bracco Diagnostics, Princeton, NJ) (10:1, v:v), followed by a mixture of 10% formalin and ProHance (10:1, v:v), (Johnson, Cofer et al. 2002). Whole heads were stored overnight in formalin, and then trimmed to remove the lower jaw and muscle. Brains were scanned within the cranial vault to a avoid distortions or damage to the tissue during excision from the cranium.
Image Acquisition
The fixed specimens were imaged using a 9.4 T (400 MHz) vertical bore Oxford magnet with a GE EXCITE console (Epic 11.0). A 14-mm diameter solenoid RF coil was used for the ex-vivo, in-situ mouse brains. We used a 3D spin warp sequence with the readout gradient applied along the long (anterior-posterior) axis of the brain. Two different acquisitions were used to provide multispectral data. A T1-weighted sequence was acquired with the following parameters: echo time (TE) 5.1 ms, repetition time (TR) 50 ms, 62.5 kHz bandwidth, field of view (FOV) of 11×11×22 mm. A T2 multiecho sequence was acquired with a Carr Purcell Meiboom Gill sequence using the same FOV and bandwidth, with TR of 400 ms and echo spacing of 7 ms (16 echoes). Post-processing included a Fourier transform along the echo time line to produce data heavily dependent on T2 differences (Sharief and Johnson 2006). Asymmetric sampling of k-space with dynamic adjustment of receiver gain and partial zero filling of k-space were used to achieve an image matrix size of 1024×512×512, resulting in an isotropic 21.5 μm resolution, in 2 hours 7 minutes for the T1 dataset. A matrix of 512×512×256 with isotropic resolution of 43 μm was generated for the T2-weighted data with total acquisition time of 4 hours, 20 minutes.
Brain Segmentation
The B6 mouse brain atlas that we recently constructed as part of a companion study was used as the reference for segmentation of all cases (Badea, Ali-Sharief et al. 2007). The atlas, labeled at a 43 μm resolution, consists of a T1 and a T2 image, and has 33 labeled structures. The use of two MR image contrasts allowed the identification of several regions with better accuracy than what would have been possible by the use of a single imaging protocol. For example the geniculate nuclei have a contrast-to-noise ratio (CNR) on the order of 0.89 in the T1-weighted images, but 3.74 in the T2-weighted image. Similarly, the pontine nuclei have a CNR on the order of 0.82 in the T1-weighted images, but 5.75 in the T2-weighted image.
To match the atlas resolution, the more recent T1 MRM datasets (BXD, as well as additional B6 and D2 brains) were down-sampled to 43 μm. The datasets were skull-stripped applying a sequence of morphological operations in ImageJ (http://rsbweb.nih.gov/ij/). The algorithm (Badea, Ali-Sharief et al. 2007) starts by Gaussian smoothing, followed by thresholding and erosions. Region growing started from a seed point located in the center of the volume. Voxels with values between fixed thresholds and connected to the initial region were added to the brain mask. Dilation was applied at the end on the brain mask, for the same number of times erosion has been applied. This process removed most of the skull surrounding the brain. The remaining skull areas were removed manually. Following skull stripping, the voxel values were intensity normalized. The atlas brain was then registered using an affine transform, and then a non-rigid transform (Rueckert, Sonoda et al. 1999) to each of the other brains. The transform needed to map the atlas onto the particular brain was also applied to the atlas labels. Following automated segmentation some manual corrections was performed using SHIVA (Shattuck and Leahy 2002) by a single investigator (AB).
The segmentation accuracy was tested by comparing the results of automated with those of manual segmentation of the hippocampus, for a subsample consisting of the male brains from BXD6, BXD16, BXD24, BXD29, BXD28, and BXD40. The sample was selected based on a large variability in hippocampal weight and brain size. First, volumes of automatically segmented hippocampi were compared with those of manually traced hippocampi, by means of the percent volume difference:
Second, segmentation accuracy was measured by means of percentage voxel overlap (Rijsbergen 1979) between automatically and manually segmented hippocampi using the equation:
Data Analysis
Absolute volumes for individual segmented structures and the whole brain were calculated using the number of voxels, and multiplying this number by the voxel volume. Composite volume structures were calculated as follows: brain - as the sum of all individual structures; thalamus - as the sum of ventral thalamic nuclei, laterodorsal thalamic nuclei, geniculate bodies and rest of thalamus; medulla and midbrain - as the sum of medulla, unlabeled regions of midbrain plus periaqueductal gray, substantia nigra, interpeduncular nucleus, cochlear nucleus, mesencephalic reticular nucleus (plus red nucleus), anterior pretectal nucleus, and trigeminal tract; brainstem - as the sum of medulla and midbrain and pons. The coefficient of variation (CV) of volume (the ratio between the standard deviation and the mean volume) was used to compare variability of structures.
MATLAB (The MathWorks, Natick, MA www.mathworks.com) was used for statistical analysis. Sex differences were tested using a two-tailed paired t-test (within strain, male-female pairs) with 10 degrees of freedom, at a confidence level of 95%. One-way analysis of variance (ANOVA) was conducted with the volumes adjusted for sex differences as the dependent variable and strain as the independent variable. When appropriate, we corrected for multiple tests using Benjamini and Hochberg’s method (Benjamini, Drai et al. 2001) to estimate false discovery rates. Estimates of heritability (h2) were calculated from the ANOVA tables as h2 = SSB/SST, where SSB is the sum of squares between subjects and SST is the sum of squares total. This estimated the proportion of variability that is attributable to genetic differences.
Correlation between volumes was measured using the Pearson product-moment coefficients, and we performed principal component analysis (PCA) in order to nominate functional and regional covariate systems. Correlation and PCA were carried out at two levels: that of all individual cases, neglecting strain as a variable, and that of strain means. The former method has approximately twice the sample size (n = 22) and will provide better power to detect true correlations. The latter method (n = 11) increases the fraction of genetic covariance that contributes to correlations by averaging two samples per strain. In both cases the volumes were adjusted for overall brain size differences.
The strain averages were entered into GN (http://www.genenetwork.org, and set search parameters as follows: Choose Species = Mouse, Group = BXD, Type = Phenotypes, and then enter the text string badea in the ANY field). This allowed us to study the correlation of the all MRM-based CNS volumetric phenotypes (36 were entered) with other morphometric, behavioral, and even molecular datasets in GN.
Results
We imaged brains of nine BXD RI strains (BXD1, BXD6, BXD15, BXD16, BXD24, BXD28, BXD29, BXD34, BXD40), as well as their parental strains—B6 and D2—using a well-characterized MRM protocol (Johnson, Ali-Sharief et al. 2007). We analyzed anatomical variability and patterns of covariance between structures using Pearson product-moment correlation coefficients and principal component analysis. The results complement and extend our previous studies on normal variability within a single isogenic strain. We segmented the same 33 structures as was done previously for B6 (Badea, Ali-Sharief et al. 2007) (Sharief, Badea et al. 2008) and added age-matched males and females for each of 10 strains.
The average within-strain variation measured by the coefficient of variation of volume (CV) for the whole panel of 11 strains across all structures was 7.5 ± 3.5%. This is very close to the within-strain variation measured in our companion study of B6 mice of 7.9 ± 4.0%. Estimates of within-strain sample of B6 mice therefore provide a relatively accurate approximation of within-strain variability for the BXD progeny strains. However, the magnitude of variations across the different BXD strains is naturally much higher due to the segregation and random assortment of allelic variants, and the average between-strain CV for volumes of structures, was almost double, at 15.2 ± 5.8%.
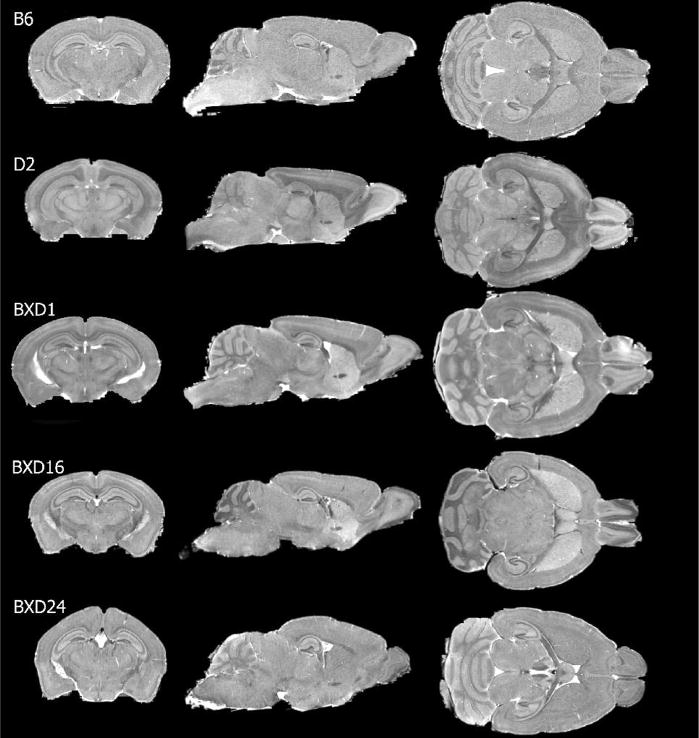
Not only does overall brain size differ among strains (from 406.0±6.3 mm3 for BXD6 to 511.5±5.8 mm3 for B6), but regions also differ appreciably in shape (Figure 1). For example, in comparison to other strains the dorsal aspect of brains of both BXD24 cases (the only BXD strain with retinal degeneration) appear rounded, a preliminary finding supported in part by comparison with data for other BXD24 cases in the Mouse Brain Library (MBL, http://www.mbl.org/).
Figure 1.
Cross-sections through C57BL/6 (B6), DBA2 (D2), BXD1, BXD16 and BXD24 mouse brain illustrate differences in size and shape among these strains. Note the rounded dorsal surface of the BXD24 mouse brain.
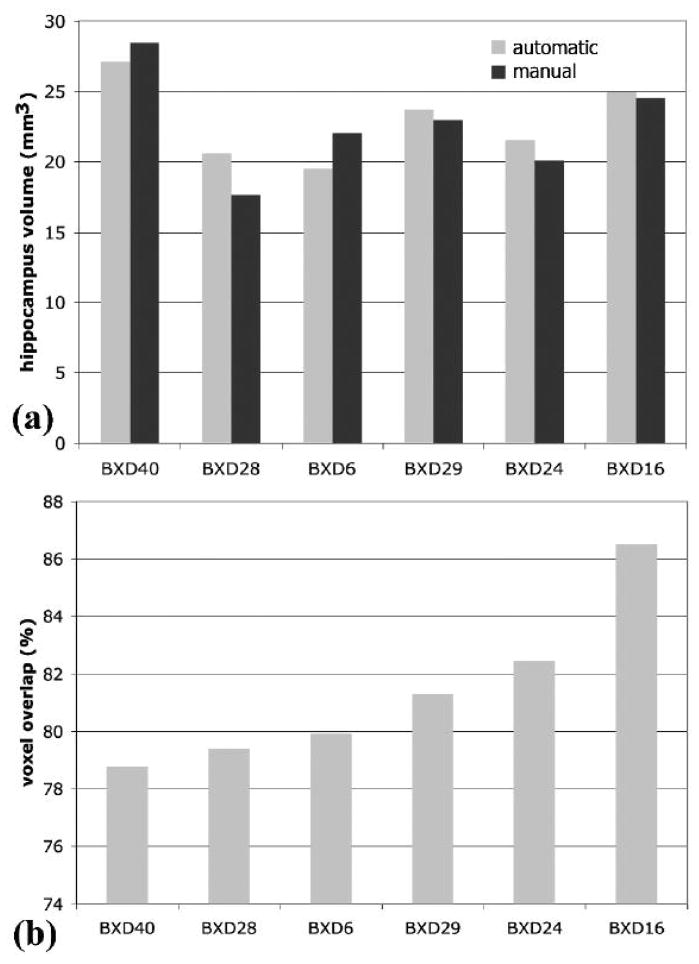
A prerequisite for a quantitative structural dissection of the brain is accurate segmentation (Figure 2). The accuracy of segmentation was previously reported for B6 based on absolute volume differences and local voxel overlap between automated and manual segmentation(Sharief, Badea et al. 2008). In the present study, we reevaluated the accuracy of automated segmentation (Figure 3), when dealing with the added challenge of differences in shape and size. We chose to analyze a single structure—the hippocampus—because this brain region is known to have large variation in volume (Lu, Airey et al. 2001) and shape (Wimer, Wimer et al. 1976) across this group of strains. The average volume difference between the automated and manual segmentation was 7.5 ± 5.4% (Figure 3a), while the average volume overlap was 81.4 ± 2.8% (Figure 3b).
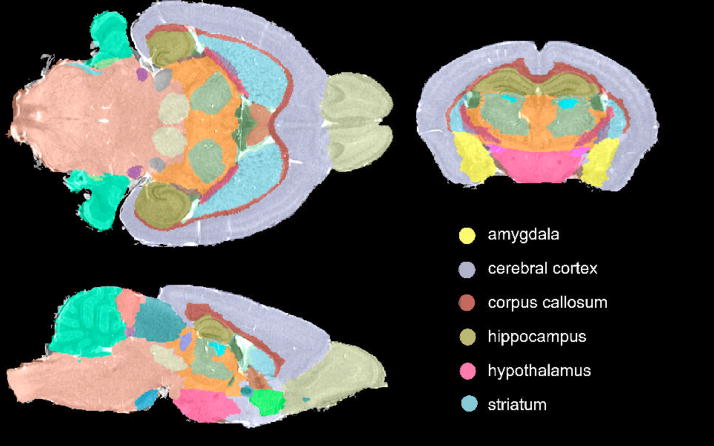
Figure 2.
Visualization of segmentation results, shown in the background of the anatomy, allows a qualitative evaluation of the segmentation accuracy, as well as preliminary comparisons among strains.
Figure 3.
Segmentation validation. (a) The percentage volume difference between the estimates of hippocampal volume based on manual and automated segmentation was 7.45±5.44%. (b) The percent voxel overlap between manual and automated labels in the case of hippocampus averaged 81.38±2.83%.
Volume Estimates
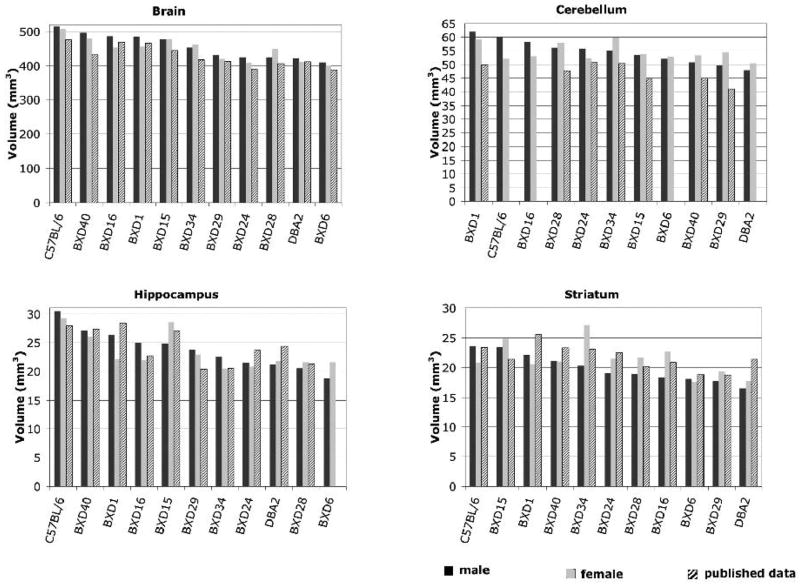
There was substantial variation in volumes of brain regions across strains. The range of variation amounted to about ±12.5% of the mean for total brain volume (CV = 7.7%). The volume of the hippocampus ranged from 20.2±2.0 mm3 in BXD6 to 29.9±0.8 mm3 in B6, a remarkable variation of about ±25% of the mean (Figure 4). For most structures, B6 had the largest volume (whole brain, hippocampus, striatum). BXD1, BXD40, BXD15, and BXD16 had large values as well. BXD6 and D2 were at the other end of the spectrum, with small volumes for all structures. BXD27 had the smallest brain and cerebellum, while BXD6 had the smallest hippocampus among strains.
Figure 4.
Volumes of selected brain structures from 9 BXD mouse strains and their parental strains (male –shown as black bars, and female – gray bars) compare well with published data shown as hash-patterned bars (Zhou et al., 1999 for brain, Airey et al., 2001 for cerebellum, Peirce et al, 2003 for hippocampus, Rosen et al., 2005 for striatum).
A comparison among several pairs of estimates based on our MRM method and comparable data in GN (Figure 4) yielded significant correlations for brain size (r =0.88, p = 0.000043 (Zhou and Williams 1999)), and hippocampusdata, (r = 0.75, p = 0.0057, (Peirce, Chesler et al. 2003)). For cerebellum (Airey, Lu et al. 2001) and striatum (Rosen, Pung et al. 2008) the correlations were more modest: for cerebellum (r =0.72, p = 0.04), and for striatum (r = 0.44, p = 0.15). The percentage volume difference, which assumes systematic bias, was 5.4% for brain and 6.9% for hippocampus, with MRM estimates being higher. The volume difference was 8.1% for striatum, and as high as 16.2% for cerebellum. Part of this difference may be due to different boundary selection, differential shrinkage, and statistical sampling error.
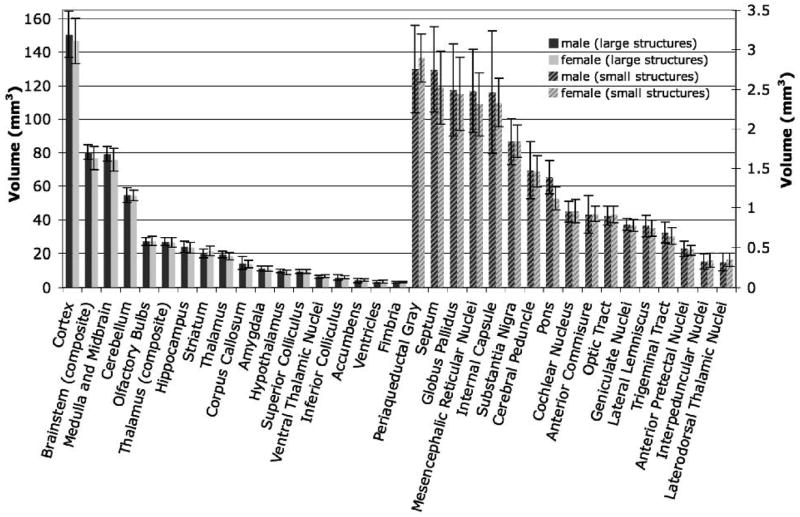
Volumes of the 33 segmented structures (Figure 5) across 11 strains spanned a wide range of sizes, from small structures such as the interpeduncular nucleus and the laterodorsal thalamic nuclei (~ 0.33 ± 0.09 mm3) to large structures such as the neocortex volume (148.6 ± 34.6 mm3). This variation in size has repercussions on magnitude of error and therefore on heritability estimates and the ability to detect covariation among structures.
Figure 5.
Volumes of segmented brain structures, and estimates of variability. Smaller structures are represented on the right axis.
Sex Difference
An analysis of sex differences (males and females from 11 strains) using a two-tailed paired t test, and a 95% significance value detected very modest differences. Without correction for multiple tests, the medulla and midbrain (p <0.02), brainstem (p <0.009), hypothalamus (p <0.02), and pons (p <0.0012) reached nominal significance thresholds. The hypothalamus averaged 9.9 ± 1.2 mm3 in the male cohort and 8.9 ± 1.2 mm3 in the female cohort (10% effect). However, after the appropriate adjustment for multiple tests and using a false discovery rate of 0.05 (Benjamini, Drai et al. 2001), only the volume of pons differed significantly between sexes.
Strain Difference
There was a significant difference in total brain volume among strains (F(10, 21)= 6.3, p <0.003). Strain differences accounted for ~85% of the variance in total brain volume, with or without correction for sex effects. Twenty-three out of the 33 segmented structures also had significant strain differences in volume, even with our modest within-strain replication. After adjustment for total brain volume as a cofactor, 10 out of the 33 structures retained significant strain differences, illustrating impressive regional variation among strains. The mean heritability of brain structure volumes, after removing effects of differences in total brain volume was 0.60 ± 0.18. Heritabilities for hippocampus and amygdala were 0.64 and 0.72. Heritabilities for olfactory bulbs, inferior colliculus, ventricles were somewhat higher, averaging about 0.80. The heritability was highest for the globus pallidus at 0.87. At the opposite end of the spectrum the heritability of the periaqueductal gray and ventral thalamic nuclei were only ~0.15. The correlation between the size of a region and its heritability was not significant (r = +0.01). This is of interest because a systematic increase in measurement error for small regions would deflate their apparent heritability relative to large regions. The lack of a strong positive covariation between size and heritability suggests that our measurement error is not unduly biased against small structures (see below).
Variability Measures
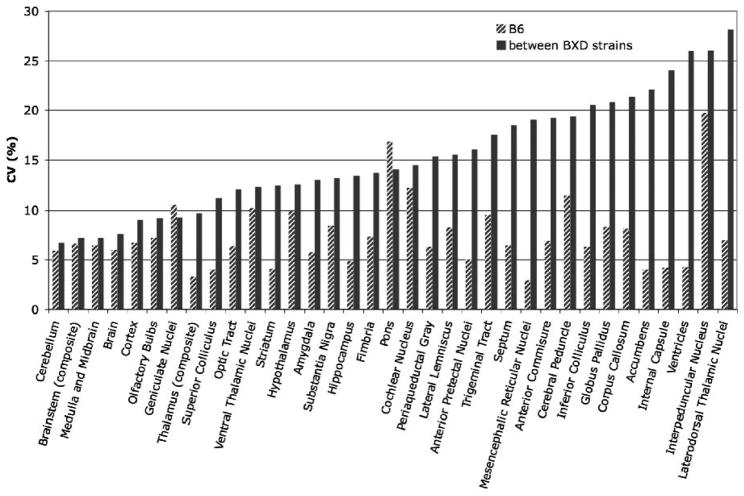
We analyzed the variability of mean volumes for 33 brain structures in 11 strains and compared these data with previous estimates (Badea, Ali-Sharief et al. 2007) based on a sample of B6 brains only (Figure 6). Between-strain variation for all segmented structures averaged 15.2 ± 5.8%, compared to an average 8.0 ± 4.0% within-strain variation for the BXD sample, and 7.4 ± 3.5% within the B6 sample. The hippocampus was characterized by 13.4% between strains variation, compared to the 4.9% estimate based on the B6 sample. Minimal variability was obtained for several large structures such as the cerebellum (6.7% between strains, 5.9% for B6), brainstem (7.2% between strain, 6.6% for B6), and cortex (9.0% between strain, 6.8% for B6), but also for small nuclei such as the geniculate bodies (9.3% between strains, 10.5% for B6). Large CV values were obtained for white matter structures including the internal capsule (26.1% between strain, 4.2% for B6), for the ventricles (26.0% between strain, 4.2% for B6), and for small nuclei such as the thalamic nuclei (28.2% between strains, 7.0% for B6) and the interpeduncular nucleus (26.1% between strains, 19.8% for B6).
Figure 6.
The coefficient of variation of the volume for segmented anatomical structures presents a means of comparing variability regardless of the absolute volumes. Small CV values were characteristic to large structures such as the cerebellum (5.93% for B6, 6.73% between strains), and brainstem (6.6% for B6, 7.19% between strains). Large values were obtained for white matter structures such as corpus callosum (8.16% for B6, 21.35% between strain), for the ventricles (4.24% for B6, 25.96% between strain%), and for small nuclei such as laterodorsal thalamic nuclei (6.98% for B6, and 28.18% between strains).
Small CV values were a characteristic to volumes of large structures (percentage of brain volume) such as cortex (2.8%), midbrain (4.7%), brainstem (4.7%), and thalamus (4.8%), and for some small structures such as thalamic geniculate (lateral and medial combined) nuclei (7.3%). A number of small nuclei had large CVs, including the laterodorsal thalamic nuclei (25.5%) and interpeduncular nucleus (25.9%). As mentioned above in relation to heritability estimates, some of this variation is undoubtedly due to segmenting errors. But several large CNS compartments that were comparatively easy to segment and nonetheless had high CVs including the internal capsule (22.9%) and the ventricles (23.0%).
Correlations Between Brain Regions
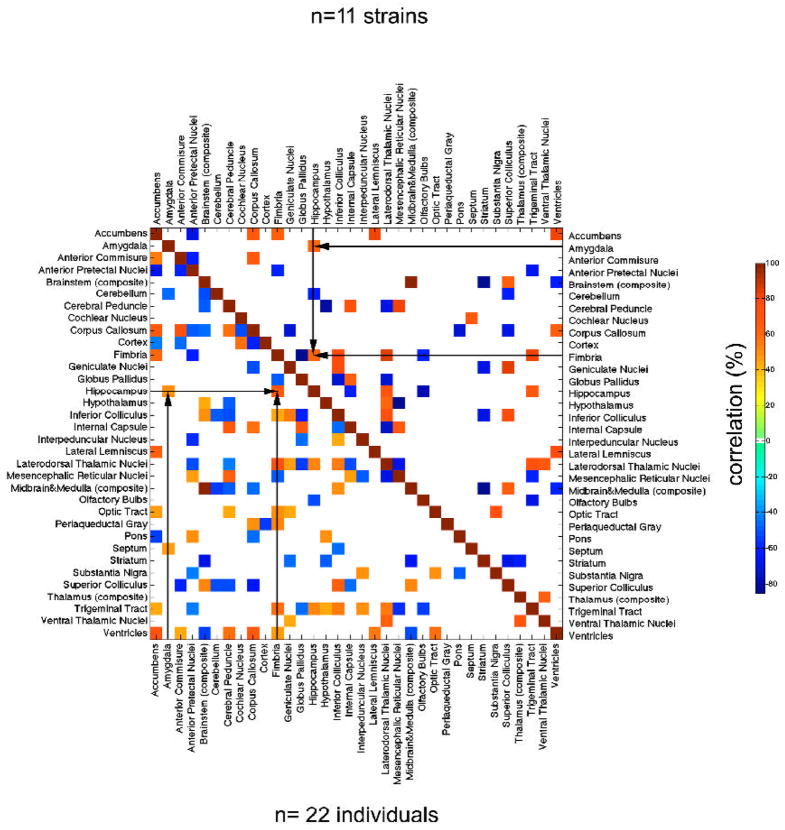
To understand the covariance structure of CNS compartments we computed Pearson correlations (Figure 7) between structures adjusted for differences in brain size. Correlations were computed using all individuals (n = 22, neglecting strain as a variable), and using strain mean volumes (n = 11 male-female pairs). We describe patterns of correlations for only a few select regions, but the entire covariation matrix makes it possible for readers to review any of the 528 [(332 − 33)/2)] correlations and to test their own hypotheses regarding structural covariation. Furthermore, all of the volumetric data can be studied in relation to many behavioral phenotypes in these same strains at www.genenetwork.org.
Figure 7.
Significant correlations (p <0.05) were found between volumes of brain structures in the BXD family of strains. The area above the diagonal illustrates cross-correlations among (N=11) strains mean volumes, while the area under the diagonal illustrates the cross-correlation map for (N=22) individuals. Only significant correlations are shown (p<0.05) and we note those among components of the limbic system such as hippocampus and amygdala, as well as associated fiber bundle – the fimbria.
For the new dataset, the volume of cortex was positively correlated to the anterior pretectal nuclei (r =0.55, p < 0.09 approaching significance), but negatively correlated with thalamus (r = −0.50, approaching significance at p < 0.12). The hippocampus was positively correlated with the fimbria (r = 0.60, p <0.05), amygdala (r = 0.61, p <0.05), and the laterodorsal thalamic nuclei (r=0.69, p < 0.02), but negatively correlated with the cerebellum (r =−0.62, p <0.05) and olfactory bulbs (r = −0.77%, p <0.006). The olfactory bulbs were also correlated with the trigeminal tract (r = −69.12, p < 0.02).
The thalamus was negatively correlated with striatum (r = −0.64, p < 0.04), with which it shares multiple parallel connections, while the correlation with brainstem approached significance (r = 0.50, p < 0.12). The brainstem was positively correlated with superior colliculus (r = 0.52, p <0.02), but negatively with striatum (r = −0.85, p <0.001), and ventricles (r =−0.63, p < 0.04). The superior colliculus was correlated with geniculate nuclei (r = 0.85, p < 0.001), and inferior colliculus (r = 0.77, p <0.006); but negatively with striatum (r= −0.70, p < 0.02), and cerebellum (r= −0.62, p < 0.04).
Using strain means for the regional volumes in the genetically diverse panel generally yielded overall tighter correlations, compared to using individual cases (average of 0.33 versus 0.26). However, the sample size is twice as large for the individual cases and this produced more significant correlation values (p of 0.3 versus 0.4). For example the correlation between hippocampus and amygdala was r = 0.6, p = 0.05 based on strain means, but r = 0.5, p = 0.02 based on 22 individual cases.
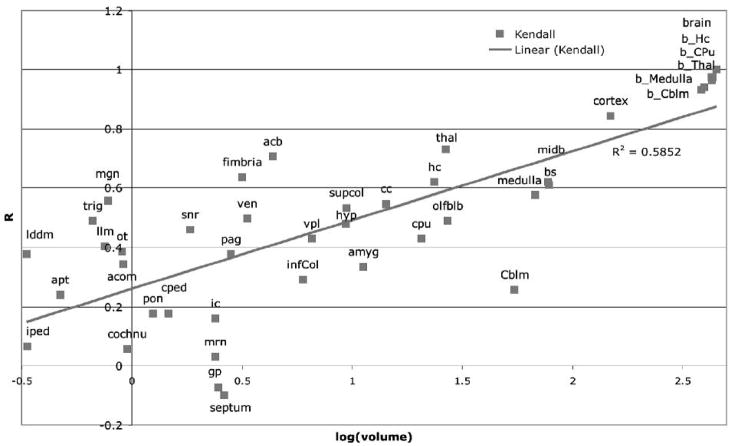
We examined correlations between regional and total brain volume as a function of the regional volumes (Figure 8). Better correlations for larger structures can be attributed to the increased segmentation accuracy compared to small structures. However there are several exceptions. The cerebellum is a large structure (54 mm3, representing 12% of the brain volume) and has good segmentation accuracy but poor correlation with brain volume (25.5%). The fimbria (3.2 mm3 or 0.70% of brain volume) and substantia nigra (1.84 mm3, or 0.41% of brain volume) are smaller structures but have a better correlation with the brain volume (r = 0.63 and 0.46, respectively).
Figure 8.
Correlations between structure volumes and brain volume are in general better for large structures compared to small structures. These results can be attributed to the decreased segmentation accuracy characteristic to small structures. However structures like fimbria and cerebellum constitute exceptions. Cerebellum is a relatively large structure (54.45 mm3, representing 12.04% of the brain volume), and has good segmentation accuracy but poor correlation with brain volume (25.54%). Fimbria (3.16 mm3 or 0.70% of brain volume) and substantia nigra (1.84 mm3, or 0.41% of brain volume) are smaller structures, but have a better correlation with the brain volume (63.64%, and 45.99% respectively).
Correlations With Other Phenotypic Traits
We performed a search for interesting and possibly significant correlations between our MRM volume estimates and other phenotypic traits(Table 1). As expected, other morphometric traits covary positively with MRM data. For example, our hippocampus volume data correlates very well with brain weight (Lu, Airey et al. 2001) (r = 0.87, p <0.00014); hippocampus volume (Peirce, Chesler et al. 2003) (0.7, p <0.02), dentate gyrus volume (Peirce, Chesler et al. 2003) (0.74, p <0.01), and dorsal thalamus volume (Dong, Martin et al. 2007) (0.73, p <0.008).
Table 1.
Correlates between MRM based volume estimates of anatomical structures and other phenotypes.
| Label | Correlated phenotypes | R | p | Correlated phenotypes | R | p |
|---|---|---|---|---|---|---|
| Amygdala | Hippocampus volume | 0.79 | 0.002 | Dopamine transporter expression in ventral midbrain (Jones, Tarantino et al. 1999) | 0.92 | 0.005 |
| Body weight (Williams, Airey et al. 2001) | 0.77 | 0.004 | Saccharin consumption (Belknap, Crabbe et al. 1992) | 0.67 | 0.05 | |
| Forebrain weight (Lu, Williams, unpublished) | 0.72 | 0.01 | Seizure threshold pressure (McCall and Frierson 1981) | 0.71 | 0.046 | |
| Brain weight (Lu, Airey et al. 2001) | 0.73 | 0.009 | Distance traveled in cocaine open field test (Jones, Tarantino et al. 1999) | −0.83 | 0.04 | |
| Ethanol preference (Rodriguez, Plomin et al. 1994) | 0.91 | 0.001 | Acute functional tolerance in ethanol induced ataxia (Kirstein, Davidson et al. 2002) | −0.78 | 0.005 | |
| Consumption of ethanol (Phillips, Crabbe et al. 1994) | 0.90 | 0.004 | Plasma iron concentration (Jones, Beard et al. 2007) | −0.77 | 0.005 | |
|
| ||||||
| Hippocampus | Total brain weight (Lu, Airey et al. 2001) | 0.87 | 0.001 | Ethanol preference (Rodriguez, Plomin et al. 1994) | 0.83 | 0.009 |
| Forebrain weight (Lu and Williams, 2005 unpublished) | 0.86 | 0.001 | Morphine consumption (Phillips, Belknap et al. 1991) | 0.69 | 0.04 | |
| Hippocampus volume (Peirce, Chesler et al. 2003) | 0.70 | 0.02 | Morris water maze – log latency (Milhaud, Halley et al. 2002) | 0.72 | 0.07* | |
| Dentate gyrus volume (Peirce, Chesler et al. 2003) | 0.73 | 0.01 | Seizure susceptibility to high atmospheric pressure (Plomin, McClearn et al. 1991) | 0.67 | 0.08* | |
| Dorsal thalamus volume (Dong, Martin et al. 2007) | 0.73 | 0.01 | LDL cholesterol levels (Colinayo, Qiao et al. 2003) | −0.98 | 0.001 | |
| Lateral geniculate volume (Seecharan, Kulkarni et al. 2003) | 0.67 | 0.03 | Cocaine open field activity (Jones, Tarantino et al. 1999) | −0.94 | 0.004 | |
| Seizure threshold pressure (McCall and Frierson 1981) | 0.73 | 0.04 | Open field habituation (Jones, Tarantino et al. 1999) | −0.85 | 0.04 | |
| Ethanol acceptance – total ethanol intake (Crabbe, Kosobud et al. 1983) | 0.89 | 0.001 | Cocaine stereotypy (Jones, Tarantino et al. 1999) | −0.81 | 0.05 | |
|
| ||||||
| Label | Correlated phenotypes | R | p | Correlated phenotypes | R | p |
|
| ||||||
| Brainstem | Brain weight (Williams, Airey et al. 2001) | 0.86 | 0.001 | Mean water consumption (versus saccharin) (Phillips, Crabbe et al. 1994) | 0.89 | 0.001 |
| Hippocampus weight (Lu, Airey et al. 2001) | 0.81 | 0.001 | HDL cholesterol levels (Colinayo, Qiao et al. 2003) | −0.81 | 0.03 | |
| PPI of the acoustic startle response (McCaughran, Bell et al. 1999) | 0.73 | 0.02 | Plasma corticosterone levels post ethanol administration (Roberts, Phillips et al. 1995) | −0.55 | 0.09* | |
| Ethanol preference (Rodriguez, Plomin et al. 1994) | 0.75 | 0.02 | Drd1 expression in nucleus accumbens (Jones, Tarantino et al. 1999) | −0.93 | 0.004 | |
|
| ||||||
| Cerebellum | Cerebellum weight (Airey, 2001) | 0.73 | 0.03 | Cocaine exploratory activity (Jones, Tarantino et al. 1999) | 0.76 | 0.09* |
| Brain weight (Belknap, Phillips et al. 1992) | 0.76 | 0.02 | Locomotor response to allopregnolone injection (Palmer, 2002) | −0.68 | 0.05 | |
| Femoral cross section area (Klein, Turner et al. 2002) | 0.84 | 0.007 | Retinal ganglion cell number (Williams, Strom et al. 1998) | −0.82 | 0.004 | |
| PPI of the acoustic startle response (McCaughran, Bell et al. 1999) | 0.92 | 0.001 | Tolerance/sensitization to ethanol effects on locomotor activity (Cunningham 1995) | −0.63 | 0.08* | |
|
| ||||||
| Striatum | Gray matter volume (Cheverud, 2005- unpublished) | 0.86 | 0.003 | Response to contextual fear (Owen, Christensen et al. 1997) | 0.79 | 0.02 |
| Forebrain weight (Lu, Williams 2005, unpublished) | 0.73 | 0.01 | High affinity choline uptake in frontal cortex (Tarricone, Hwang et al. 1995) | 0.72 | 0.03 | |
| Brain weight (Belknap, Crabbe et al. 1992) | 0.78 | 0.01 | Plasma corticosterone post ethanol (Roberts, Phillips et al. 1995) | 0.85 | 0.04 | |
| Amygdala neuron number (Mozhui, Hamre et al. 2007) | 0.70 | 0.02 | Cocaine concentration to induce tonic seizure (Hain, Crabbe et al. 2000) | −0.85 | 0.002 | |
| Body mineral density (Klein, Mitchell et al. 1998) | 0.82 | 0.005 | Transferring saturation (Jones, Beard et al. 2007) | −0.69 | 0.02 | |
|
| ||||||
| Pons | Baseline handling induced convulsions (Belknap, Metten et al. 1993) | 0.78 | 0.01 | Vocalization threshold - mild foot shock (Matthews et al, 2006, unpublished) | −0.84 | 0.003 |
|
| ||||||
| Label | Correlated phenotypes | R | p | Correlated phenotypes | R | p |
|
| ||||||
| Thalamus | Brain weight (Williams, Airey et al. 2001) | 0.94 | 0.001 | Hippocampus weight (Lu, Airey et al. 2001) | 0.78 | 0.004 |
| Forebrain weight (Lu, Williams, 2005 – npublished) | 0.93 | 0.001 | Total ethanol intake (Crabbe, Kosobud et al. 1983) | 0.85 | 0.002 | |
| Dorsal thalamic volume (Dong, Martin et al. 2007) | 0.87 | 0.001 | Cocaine open field activity (Jones, Tarantino et al. 1999) | −0.83 | 0.04 | |
| Lateral geniculate cell number (Seecharan, Kulkarni et al. 2003) | 0.75 | 0.02 | Plasma corticosterone levels 6h post 4g/Kg ethanol (Roberts, Phillips et al. 1995) | −0.84 | 0.003 | |
| Lateral geniculate volume (Seecharan, Kulkarni et al. 2003) | 0.70 | 0.03 | Cholesterol levels (Colinayo, Qiao et al. 2003) | −0.90 | 0.011 | |
|
| ||||||
| Inferior colliculus | PPI of acoustic startle reponse (McCaughran, Bell et al. 1999) | 0.78 | 0.02 | Plasma corticosterone levels (Roberts, Phillips et al. 1995) | −0.85 | 0.003 |
| Response to auditory stimulus in contextual fear conditioning (Owen, Christensen et al. 1997) | 0.76 | 0.03 | Rate of formation of aortic lesions following 16 week atherogenig diet (Colinayo, Qiao et al. 2003) | −0.90 | 0.011 | |
| Locomotor activity after ethanol conditioning (Cunningham 1995) | 0.69 | 0.04 | Duration of post ethanol loss of righting reflex (Browman and Crabbe 2000) | −0.72 | 0.03 | |
| Ethanol induced locomotor response (Demarest, Koyner et al. 2001) | 0.69 | 0.04 | Cocaine exploratory activity (Jones, Tarantino et al. 1999) | −0.85 | −0.03 | |
Approaches significance
A total of 37,851 correlations can be computed using the MRM data in concert with 1146 other phenotypes for the BXD strains in listed in GeneNetwork. This obviously raises a multiple testing problem with respect to the significance of the nominal p values listed from the correlations above. An overly stringent Bonferroni correction would require a p of ~0.00005 to reject the null hypothesis. A better alternative is to permute the index phenotype (e.g., striatal volume), recalculate the entire set of 1146 correlations repeatedly, and estimate the empirical p value required to reach a particular alpha level from the distribution of these permuted correlations. Rosen and colleagues (2008, and GD Rosen, personal communication) performed this analysis for striatal volume and found that a correction factor of ~50 was appropriate for their dataset. A nominal p value of 0.001 to 0.0005 will often be close to p = 0.05, even given the numbers of phenotypes that can be tested.
Not only do morphometric phenotypes covary with our volumetric estimates but so do physiological and behavioral phenotypes. Amygdala volume was positively correlated with phenotypes related to addiction such as ethanol preference (Rodriguez, Plomin et al. 1994) (r =0.91, p = 0.0006), but was negatively correlated with functional tolerance on dowel test in ethanol induced ataxia (Kirstein, Davidson et al. 2002) (r =−78.28%, p = 0.005) and plasma iron concentration (Jones, 2005; 0.77, p = 0.0042).
The hippocampal volume was negatively correlated with LDL cholesterol levels (Colinayo, Qiao et al. 2003) (−0.98, p = 0.0001). Correlations with open field habituation (Jones, Tarantino et al. 1999) (−0.85, p = 0.032) and cocaine stereotypy- (Jones, Tarantino et al. 1999) (−0.81, p = 0.05) are nominally significant, but would not be significant given the large numbers of tests.
Midbrain structures are important mediators of prepulse inhibition (PPI) stimuli; among these the inferior colliculus (IC) serves as a relay for acoustic prepulses for PPI (Fendt, Li et al. 2001). We found that the IC volume was correlated with the of the acoustic startle response (0.82, p = 0.005), (McCaughran, Bell et al. 1999). Overall brainstem volume was also correlated with the PPI of the acoustic startle response (McCaughran, Bell et al. 1999) (0.71, p = 0.03).
The pontine nuclei were negatively correlated with vocalization threshold (−0.84, p = 0.0028), but positively with the baseline handling induced convulsions (Belknap, Metten et al. 1993) (0.78, p = 0.009).
The medial cerebellum has been shown to be involved in long-term habituation of the acoustic startle response (Leaton and Supple 1986). Cerebellar volume correlated with the prepulse inhibition of the acoustic startle response (McCaughran J, Bell J, Hitzeman, unpublished) (0.92, p = 0.0002).
An inverse search starting from memory-related traits within GN revealed associations between latencies in the Morris water maze (Milhaud, Halley et al. 2002) with structures known to be involved in spatial learning such as hippocampus (0.68, p = 0.06), but also with accumbens (0.80, p = 0.02), fimbria (0.83, p = 0.008), periaqueductal gray (0.87, p = 0.003), inferior (0.69, p = 0.06) and superior (0.75, p = 0.03) colliculus. While not directly involved in spatial learning, the colliculi are however involved in integration of multiple sensory inputs(Jain and Shore 2006). Also the protein kinase activity for total hippocampus (Wehner, Sleight et al. 1990) was negatively correlated with the hippocampal volume (−0.80, p = 0.06). We have found interesting correlations between anatomical and functional parameters for several structures. However, many of these correlations would not remain significant after such a correction for multiple tests. These results should therefore be considered as hypotheses to be retested on a larger sample.
Brain Subsystems
Brain regions that have anatomical connections or that are directly or indirectly involved in common functions would be expected to covary in size. We explored the correlations between structures using principal component analysis (PCA) on the volumes of segmented structures after adjusting for brain volume differences (Table 2). For the B6 data taken from our previous work (Badea, Ali-Sharief et al. 2007), the first principal component, which explains 79% of the variance, provided contrast between the cerebellum (negative loading of −0.3) and a group of structures including the cortex, hippocampus, medulla and midbrain (positive loadings: 0.61, 0.14, and 0.16, respectively). The second component (14% of the variance) provided contrast between a group including limbic structures (hippocampus [−0.2], amygdala [−0.2], and accumbens [−0.1]) and brainstem structures (medulla and midbrain [−0.3], inferior colliculus [−0.3], pons [−0.2], and a second group including the cortex (0.45), cerebellum (0.61), and thalamus (0.11). The third PCA (5% of variance) provided contrast between olfactory bulbs (0.6) and brainstem structures (medulla, midbrain [−0.5] and pons [−0.1]).
Table 2.
Principal component (PC) analysis of brain structure volumes, only regions characterized by absolute values of eigenvector coefficients equal or larger than 0.1 are shown.
| Regions | B6 (N=6 cases) |
BXD (N=11 strains) |
||||
|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | |
| Accumbens | −0.13 | −0.15 | ||||
| Amygdala | −0.15 | −0.16 | 0.12 | −0.19 | ||
| Cerebellum | −0.28 | 0.61 | 0.10 | −0.64 | 0.43 | |
| Corpus Callosum | −0.68 | −0.35 | −0.21 | −0.56 | ||
| Cortex | 0.61 | 0.45 | −0.22 | 0.35 | −0.76 | 0.31 |
| Hippocampus | 0.14 | −0.18 | 0.23 | −0.07 | −0.22 | |
| Hypothalamus | −0.18 | 0.10 | 0.10 | |||
| Inferior Colliculus | −0.25 | 0.18 | 0.15 | 0.10 | ||
| Midbrain and Medulla (composite) | 0.16 | −0.28 | −0.49 | 0.44 | 0.54 | 0.26 |
| Olfactory Bulbs | 0.59 | −0.16 | 0.13 | 0.40 | ||
| Pons | −0.17 | −0.11 | ||||
| Septum | 0.10 | |||||
| Striatum | −0.29 | −0.23 | −0.15 | |||
| Superior Colliculus | 0.11 | 0.16 | 0.11 | |||
| Thalamus | 0.11 | 0.16 | ||||
| Ventral Thalamic Nuclei | −0.05 | 0.31 | −0.19 | |||
| Ventricles | 0.10 | −0.12 | 0.16 | −0.17 | ||
For the current set of genetically diverse strains the first principal component (~40% of the variance) provided contrast between cerebellum (−0.64) and striatum (−0.29), as the group with largest negative loadings, and cortex (0.35) and brainstem structures (0.44), which share high positive loadings. The second component (~30% of the variance) provided contrast between cortex (−0.76) and striatum (−0.23) as one group and medulla and midbrain (0.54). We found on the third PCA (~15% of variance) contrast between limbic structures and ventricles (strong negative loadings), and olfactory bulbs and cerebellum (strong positive loadings). Another subsystem could be defines on the third PC, consisting of the thalamus-accumbens-striatum, and amygdala, which are parts of a cortico-striato-thalamo-cortical loop. As expected performing the PCA analysis for the new dataset either using 11 strain means or 22 individual cases preserved the same groups of contrasting structures that load strongly on the same principal component.
Discussion
Synopsis
We performed a systematic analysis of covariation among 33 regions of the mouse CNS using a single panel of genetically diverse strains of mice. Each of these normal inbred strains was sampled sparsely, but the full collection of 22 cases provides a good first estimate of how variation among different brains regions is coupled. The data we have generated can be used to answer a large number of simple questions regarding covariation among volumes of CNS compartments, for instance whether the volume of the hippocampus in mouse covaries with that of the fimbria; or that of the lateral geniculate nucleus with that of the optic tract. Much of the positive covariation between these types of CNS compartments is associated with overall differences in brain size, but this general effect can be statistically removed in order to study covariation that is generated by shared genetics effects, known input-output relations, and a system-specific plasticity.
Variation in the Impact of Technical Error
Segmentation accuracy can be better for larger structures compared to small structures, but it also depends on the relative contrast of these regions in the MR images (Sharief, Badea et al. 2008). Manual corrections performed to reduce the amount of variability that can be attributable to technical errors, can however be impeded by the complexity of anatomical shapes. The advantage of MRM data is that it does preserve the integrity of shapes in three dimensions and does not suffer from distortions or shrinkage that often affect histologically processed tissue. Automated segmentation accuracy was estimated through the percentage voxel overlap to be in the range of 80% for hippocampus, but can be less for smaller structures. These systematic errors could in principle be reflected in higher coefficients of variation for smaller structures, lower correlation values among small structures, and higher heritability estimates for larger structures. Heritability estimates for regional volumes were computed based on adjusted values, to remove the effects of heritable variations in overall brain size. There was no linear relation however between the size of structures and their coefficients of variations, or heritability estimates (the correlation was not significant). In some cases, this may be due to counterbalancing types of errors. For example, the geniculate nuclei are small but have good contrast, a simple shape and are clearly defined. In contrast, the hippocampus is large, but has a complex shape and a highly variable intensity distribution.
Volume Variability
The magnitude of variation in the volumes of brain structures in the diverse set of brains that we have studied greatly exceeded the variability within strain. The variation across strains had an average coefficient of variation (CV) of 15.2%, roughly twice that within strain. On average genotype accounted for ~60% of variability. The magnitude of variability differed among structures. The ventricles were the most variable CNS compartment—a CV of 26.0% among strains and 9.7% within strain. While parts of this variability can be attributable to inaccurate segmentation for small structures, our results replicate those found in humans. (Lange, Giedd et al. 1997) found that variability differed significantly across structures in humans; the lateral ventricles having the highest CV, and the putamen the lowest.
Family and twin studies have demonstrated significant familial effects in multiple brain regions. (Wright, Sham et al. 2002) for example, found the brain volume variability to be under substantial genetic control, while the lateral ventricles had a much lower heritability. Among genetically diverse sets of strains heritability of the whole brain volume and that of the ventricles (~0.80) are much more closely matched. This is also true for the ventricular volume of the AXB set of mouse strains(Zygourakis and Rosen 2003). The apparent species difference is mainly technical. Heritabilities computed using a panel of strains of mice are usually based on averages from two or more individuals per strain. This “cheat” increases the correlation between genotype and phenotype. The other factor is that the environment can be well controlled for strains of mice. These factors produce inflated estimates of heritability that are advantageous from the point of view of mapping sequence variants, but that should not be considered particularly relevant to natural populations living in complex environments.
Sex differences in volumes were negligible in this well balanced but small sample. After the appropriate correction for multiple tests (false discovery rate correction (Benjamini, Drai et al. 2001) only the volume of pons retained a significant sex difference (p <0.05). Although surprising to us, this difference is consistent with differences noted in normal humans by Raz and colleagues (Raz, Gunning-Dixon et al. 2001). In contrast we did not discover sex differences in hypothalamus, perhaps because these differences are restricted to a small part of the rostal hypothalamus and preoptic areas, which we did not segment. Improved MR methods and image analysis, in particular more refined segmentation algorithms, may allow us to define smaller regions such as the locus coeruleus and the medial preoptic areas, which might be more relevant to sex differences.
Covariance Among Neuroanatomical Phenotypes
The correlational analysis of CNS compartments after correction for differences in brain volume identified a large number of structures that are positively correlated, indicating possible pleiotropic gene effects and shared influences of the environment and functional plasticity. A limbic component was singled out based on strong correlations among tightly interconnected subcomponents such as the amygdala, hippocampus, and fimbria. Other correlations were identified between interconnected structures such as hippocampus and thalamic nuclei (r = 0.69, p = 0.02), the superior colliculus and the geniculate nuclei (r= 0.85, p = 0.001), both involved in vision: the superior colliculus in oculomotor control, and the lateral group of the geniculate in relaying visual information to visual cortex. Another example is an intriguing and unexpected correlation between the olfactory bulbs and trigeminal tract. These regions do not share any intimate anatomical connections, nor are they parts of the same major CNS developmental compartments. But in rodents, the olfactory bulbs and trigeminal systems are functionally coupled by the close integration of olfactory exploration (sniffing) and whisking of the vibrissae (Cain and Murphy 1980). Besides those correlations between anatomical regions that have anatomical and functional connections, and are involved in similar functions, some correlations might be due to the vicinity of those regions which might share common developmental influences (i.e. inferior colliculus and cerebellum [r = −0.5, p < 0.02, for N=22 cases; r = −0.44, non-significant for N=11 strain means). We do not expect that all correlations will be preserved between and within strains. Paradoxically, covariation within an isogenic strain cannot be generated by shared genetic effects because the genome is a constant and cannot be the source of within-strain variation. The major causes of variation within the B6 panel (Badea et al, 2008) are due to epigenetic and environmental effects. In contrast, an additional major cause of variation across the entire panel is due to the segregation of over a million sequence variants among these strains of mice.
Brain subsystems
The principal component analysis identified morphological subregional systems associated with the largest proportion of total variance. The first component (~40% variance for the diverse set of strains) presented a contrast between cortex and brainstem structures as a group characterized by strong positive loadings, and cerebellum characterized by negative loadings. This kind of contrast reflected in opposite signs for the loading factors on the same principal component for brainstem structures (medulla and midbrain) and cerebellum is also reflected in negative correlations between their volumes (r = −0.55, p = 0.009 for n = 22 cases, r = −0.5; p = 0.14 non-significant for n = 11 strains).
The ventricular system and cortex had loadings of opposite signs on the second and third PC for the B6, and on the third PC for the whole family of strains respectively, and we can interpret this as an inverse relationship between the size of the two regions for a same volume of brain. Components of the same subregional system, defined by strong loadings of the same sign on the same PC, might be under the influence of several common factors, including positively correlated growth patterns, anatomical connectivity, and cooperative functioning (Wright, Sharma et al. 1999).
Correlations With Physiological And Behavioral Phenotypes
Interestingly an exploratory analysis using a public database (www.genenetwork.org) revealed not only correlations among morphometric phenotypes, but also that behavioral phenotypes correlated with anatomical phenotypes. Memory related traits such as the log latency in the Morris water maze test were correlated with volumes of structures associated with the limbic system (hippocampus, accumbens and fimbria), while the activity of PCK in the hippocampus were inversely correlated with the hippocampal volume. Also the cerebellum, whose vermis is part of a neural circuit that modulates PPI and habituation, was correlated to the PPI startle response. PPI is deficient in a number of psychiatric and neurological disorders associated with abnormalities in the limbic and cortico-pallido-striato-thalamic circuitry. Increasing evidence suggests that cholesterol plays a central role in the pathophysiology of Alzheimer’s disease (Gaudreault, Dea et al. 2004; van Helmond, Miners et al. 2007). In this context, it is of interest that we found a negative correlation between LDL cholesterol level and hippocampal size.
New hypotheses can be generated efficiently by correlation analysis, even using a small dataset. But of more interest, this approach opens up the long-term possibility of discovering gene variants and sets of polymorphism that generate correlations and that contribute to networks of covarying phenotypes. Our current sample size are likely to generate interesting hypotheses but with considerable risk of false discovery because of our small sample size. The inclusion of more strains and more cases per strain will add to the power and enable more in depth exploration and analysis of the genetic basis of variation in brain structure, and functional and causal covariates of differences in brain structure.
Acknowledgments
We are grateful to Gary Cofer and Boma Fubara for technical support. This work was performed at the Duke Center for In Vivo Microscopy, an NCRR/NCI National Biomedical Technology Resource Center supported by grants (P41 RR005959/U24 CA092656). The Mouse Bioinformatics Research Network (MBIRN) (U24 RR021760) provided major support for this study. GeneNetwork is also supported by NIAAA U24AA13513 and U01AA13499, a Human Brain Project funded jointly by NIDA, NIMH, and NIAAA (P20-DA21131), and NCI U01CA105417.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Airey DC, Lu L, et al. Genetic control of the mouse cerebellum: identification of quantitative trait loci modulating size and architecture. J Neurosci. 2001;21(14):5099–109. doi: 10.1523/JNEUROSCI.21-14-05099.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TJ, Halpern SD, et al. Correlated size variations in human visual cortex, lateral geniculate nucleus, and optic tract. J Neurosci. 1997;17(8):2859–68. doi: 10.1523/JNEUROSCI.17-08-02859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea A, Ali-Sharief AA, et al. Morphometric analysis of the C57BL/6J mouse brain. Neuroimage. 2007;37(3):683–93. doi: 10.1016/j.neuroimage.2007.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, et al. Single-locus control of saccharin intake in BXD/Ty recombinant inbred (RI) mice: some methodological implications for RI strain analysis. Behav Genet. 1992;22(1):81–100. doi: 10.1007/BF01066794. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Metten P, et al. Quantitative trait loci (QTL) applications to substances of abuse: physical dependence studies with nitrous oxide and ethanol in BXD mice. Behav Genet. 1993;23(2):213–22. doi: 10.1007/BF01067426. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Phillips TJ, et al. Quantitative trait loci associated with brain weight in the BXD/Ty recombinant inbred mouse strains. Brain Res Bull. 1992;29(3–4):337–44. doi: 10.1016/0361-9230(92)90065-6. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1–2):279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- Browman KE, Crabbe JC. Quantitative trait loci affecting ethanol sensitivity in BXD recombinant inbred mice. Alcohol Clin Exp Res. 2000;24(1):17–23. [PubMed] [Google Scholar]
- Cain WS, Murphy CL. Interaction between chemoreceptive modalities of odour and irritation. Nature. 1980;284(5753):255–7. doi: 10.1038/284255a0. [DOI] [PubMed] [Google Scholar]
- Colinayo VV, Qiao JH, et al. Genetic loci for diet-induced atherosclerotic lesions and plasma lipids in mice. Mamm Genome. 2003;14(7):464–71. doi: 10.1007/s00335-002-2187-2. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Kosobud A, et al. Polygenic and single-gene determination of responses to ethanol in BXD/Ty recombinant inbred mouse strains. Neurobehav Toxicol Teratol. 1983;5(2):181–7. [PubMed] [Google Scholar]
- Cunningham CL. Localization of genes influencing ethanol-induced conditioned place preference and locomotor activity in BXD recombinant inbred mice. Psychopharmacology (Berl) 1995;120(1):28–41. doi: 10.1007/BF02246142. [DOI] [PubMed] [Google Scholar]
- Demarest K, Koyner J, et al. Further characterization and high-resolution mapping of quantitative trait loci for ethanol-induced locomotor activity. Behav Genet. 2001;31(1):79–91. doi: 10.1023/a:1010261909853. [DOI] [PubMed] [Google Scholar]
- Dong H, Martin MV, et al. Quantitative trait loci linked to thalamus and cortex gray matter volumes in BXD recombinant inbred mice. Heredity. 2007;99(1):62–9. doi: 10.1038/sj.hdy.6800965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt M, Li L, et al. Brain stem circuits mediating prepulse inhibition of the startle reflex. Psychopharmacology (Berl) 2001;156(2–3):216–24. doi: 10.1007/s002130100794. [DOI] [PubMed] [Google Scholar]
- Gaudreault SB, Dea D, et al. Increased caveolin-1 expression in Alzheimer’s disease brain. Neurobiol Aging. 2004;25(6):753–9. doi: 10.1016/j.neurobiolaging.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Hain HS, Crabbe JC, et al. Cocaine-induced seizure thresholds: quantitative trait loci detection and mapping in two populations derived from the C57BL/6 and DBA/2 mouse strains. J Pharmacol Exp Ther. 2000;293(1):180–7. [PubMed] [Google Scholar]
- Halpern SD, Andrews TJ, et al. Interindividual variation in human visual performance. J Cogn Neurosci. 1999;11(5):521–34. doi: 10.1162/089892999563580. [DOI] [PubMed] [Google Scholar]
- Horton JC, Hoyt WF. The representation of the visual field in human striate cortex. A revision of the classic Holmes map. Arch Ophthalmol. 1991;109(6):816–24. doi: 10.1001/archopht.1991.01080060080030. [DOI] [PubMed] [Google Scholar]
- Jain R, Shore S. External inferior colliculus integrates trigeminal and acoustic information: unit responses to trigeminal nucleus and acoustic stimulation in the guinea pig. Neurosci Lett. 2006;395(1):71–5. doi: 10.1016/j.neulet.2005.10.077. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Ali-Sharief A, et al. High-throughput morphologic phenotyping of the mouse brain with magnetic resonance histology. Neuroimage. 2007;37(1):82–9. doi: 10.1016/j.neuroimage.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, et al. Morphologic phenotyping with magnetic resonance microscopy: the visible mouse. Radiology. 2002;222(3):789–793. doi: 10.1148/radiol.2223010531. [DOI] [PubMed] [Google Scholar]
- Jones BC, Beard JL, et al. Systems genetic analysis of peripheral iron parameters in the mouse. Am J Physiol Regul Integr Comp Physiol. 2007;293(1):R116–24. doi: 10.1152/ajpregu.00608.2006. [DOI] [PubMed] [Google Scholar]
- Jones BC, Tarantino LM, et al. Quantitative-trait loci analysis of cocaine-related behaviours and neurochemistry. Pharmacogenetics. 1999;9(5):607–17. [PubMed] [Google Scholar]
- Kirstein SL, Davidson KL, et al. Quantitative trait loci affecting initial sensitivity and acute functional tolerance to ethanol-induced ataxia and brain cAMP signaling in BXD recombinant inbred mice. J Pharmacol Exp Ther. 2002;302(3):1238–45. doi: 10.1124/jpet.302.3.1238. [DOI] [PubMed] [Google Scholar]
- Klein RF, Mitchell SR, et al. Quantitative trait loci affecting peak bone mineral density in mice. J Bone Miner Res. 1998;13(11):1648–56. doi: 10.1359/jbmr.1998.13.11.1648. [DOI] [PubMed] [Google Scholar]
- Klein RF, Turner RJ, et al. Mapping quantitative trait loci that influence femoral cross-sectional area in mice. J Bone Miner Res. 2002;17(10):1752–60. doi: 10.1359/jbmr.2002.17.10.1752. [DOI] [PubMed] [Google Scholar]
- Lange N, Giedd JN, et al. Variability of human brain structure size: ages 4–20 years. Psychiatry Res. 1997;74(1):1–12. doi: 10.1016/s0925-4927(96)03054-5. [DOI] [PubMed] [Google Scholar]
- Leaton RN, Supple WF., Jr Cerebellar vermis: essential for long-term habituation of the acoustic startle response. Science. 1986;232(4749):513–5. doi: 10.1126/science.3961494. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lu L, Airey DC, et al. Complex trait analysis of the hippocampus: mapping and biometric analysis of two novel gene loci with specific effects on hippocampal structure in mice. J Neurosci. 2001;21(10):3503–14. doi: 10.1523/JNEUROSCI.21-10-03503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall RD, Frierson D., Jr Evidence that two loci predominantly determine the difference in susceptibility to the high pressure neurologic syndrome type I seizure in mice. Genetics. 1981;99(2):285–307. doi: 10.1093/genetics/99.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughran J, Jr, Bell J, et al. On the relationships of high-frequency hearing loss and cochlear pathology to the acoustic startle response (ASR) and prepulse inhibition of the ASR in the BXD recombinant inbred series. Behav Genet. 1999;29(1):21–30. doi: 10.1023/a:1021433705004. [DOI] [PubMed] [Google Scholar]
- Milhaud JM, Halley H, et al. Two QTLs located on chromosomes 1 and 5 modulate different aspects of the performance of mice of the B x D Ty RI strain series in the Morris navigation task. Behav Genet. 2002;32(1):69–78. doi: 10.1023/a:1014412029774. [DOI] [PubMed] [Google Scholar]
- Mozhui K, Hamre KM, et al. Genetic and structural analysis of the basolateral amygdala complex in BXD recombinant inbred mice. Behav Genet. 2007;37(1):223–43. doi: 10.1007/s10519-006-9122-3. [DOI] [PubMed] [Google Scholar]
- Owen EH, Christensen SC, et al. Identification of quantitative trait loci involved in contextual and auditory-cued fear conditioning in BXD recombinant inbred strains. Behav Neurosci. 1997;111(2):292–300. doi: 10.1037//0735-7044.111.2.292. [DOI] [PubMed] [Google Scholar]
- Pearson KG, Goodman CS. Correlation of variability in structure with variability in synaptic connections of an identified interneuron in locusts. J Comp Neurol. 1979;184(1):141–66. doi: 10.1002/cne.901840109. [DOI] [PubMed] [Google Scholar]
- Peirce JL, Chesler EJ, et al. Genetic architecture of the mouse hippocampus: identification of gene loci with selective regional effects. Genes Brain Behav. 2003;2(4):238–52. doi: 10.1034/j.1601-183x.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, et al. A twin MRI study of size variations in human brain. J Cogn Neurosci. 2000;12(1):223–32. doi: 10.1162/089892900561850. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Belknap JK, et al. Use of recombinant inbred strains to assess vulnerability to drug abuse at the genetic level. J Addict Dis. 1991;10(1–2):73–87. doi: 10.1300/J069v10n01_06. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, et al. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res. 1994;18(4):931–41. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- Plomin R, McClearn GE, et al. Use of recombinant inbred strains to detect quantitative trait loci associated with behavior. Behav Genet. 1991;21(2):99–116. doi: 10.1007/BF01066330. [DOI] [PubMed] [Google Scholar]
- Raz N, Gunning-Dixon F, et al. Age and sex differences in the cerebellum and the ventral pons: a prospective MR study of healthy adults. AJNR Am J Neuroradiol. 2001;22(6):1161–7. [PMC free article] [PubMed] [Google Scholar]
- Rijsbergen CJv. Information Retrieval. London: Butterworths; 1979. [Google Scholar]
- Roberts AJ, Phillips TJ, et al. Genetic analysis of the corticosterone response to ethanol in BXD recombinant inbred mice. Behav Neurosci. 1995;109(6):1199–208. doi: 10.1037//0735-7044.109.6.1199. [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, et al. Alcohol acceptance, preference, and sensitivity in mice. I. Quantitative genetic analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res. 1994;18(6):1416–22. doi: 10.1111/j.1530-0277.1994.tb01444.x. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Pung CJ, et al. Genetic modulation of striatal volume in BXD recombinant inbred mice. Genes Brain Behavin press. 2008 doi: 10.1111/j.1601-183X.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueckert D, Sonoda LI, et al. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18(8):712–21. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Seecharan DJ, Kulkarni AL, et al. Genetic control of interconnected neuronal populations in the mouse primary visual system. J Neurosci. 2003;23(35):11178–88. doi: 10.1523/JNEUROSCI.23-35-11178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharief AA, Badea A, et al. Automated segmentation of the actively stained mouse brain using multi-spectral MR microscopy. Neuroimage. 2008;39(1):136–45. doi: 10.1016/j.neuroimage.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharief AA, Johnson GA. Enhanced T2 contrast for MR histology of the mouse brain. Magn Reson Med. 2006;56(4):717–25. doi: 10.1002/mrm.21026. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002;6(2):129–42. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Stensaas SS, Eddington DK, et al. The topography and variability of the primary visual cortex in man. J Neurosurg. 1974;40(6):747–55. doi: 10.3171/jns.1974.40.6.0747. [DOI] [PubMed] [Google Scholar]
- Tarricone BJ, Hwang WG, et al. Identification of a locus on mouse chromosome 17 associated with high-affinity choline uptake using BXD recombinant inbred mice and quantitative trait loci analysis. Genomics. 1995;27(1):161–4. doi: 10.1006/geno.1995.1019. [DOI] [PubMed] [Google Scholar]
- van Helmond ZK, Miners JS, et al. Caveolin-1 and -2 and their relationship to cerebral amyloid angiopathy in Alzheimer’s disease. Neuropathol Appl Neurobiol. 2007;33(3):317–27. doi: 10.1111/j.1365-2990.2006.00815.x. [DOI] [PubMed] [Google Scholar]
- Wehner JM, Sleight S, et al. Hippocampal protein kinase C activity is reduced in poor spatial learners. Brain Res. 1990;523(2):181–7. doi: 10.1016/0006-8993(90)91485-y. [DOI] [PubMed] [Google Scholar]
- White T, Andreasen NC, et al. Brain volumes and surface morphology in monozygotic twins. Cereb Cortex. 2002;12(5):486–93. doi: 10.1093/cercor/12.5.486. [DOI] [PubMed] [Google Scholar]
- Williams RW, Airey DC, et al. Genetic dissection of the olfactory bulbs of mice: QTLs on four chromosomes modulate bulb size. Behav Genet. 2001;31(1):61–77. doi: 10.1023/a:1010209925783. [DOI] [PubMed] [Google Scholar]
- Williams RW, Strom RC, et al. Natural variation in neuron number in mice is linked to a major quantitative trait locus on Chr 11. J Neurosci. 1998;18(1):138–46. doi: 10.1523/JNEUROSCI.18-01-00138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Strom RC, et al. Genetic and environmental control of variation in retinal ganglion cell number in mice. J Neurosci. 1996;16(22):7193–205. doi: 10.1523/JNEUROSCI.16-22-07193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimer RE, Wimer CC, et al. The genetic organization of neuron number in Ammon’s horns of house mice. Brain Res. 1976;118(2):219–43. doi: 10.1016/0006-8993(76)90709-5. [DOI] [PubMed] [Google Scholar]
- Wright IC, Sham P, et al. Genetic contributions to regional variability in human brain structure: methods and preliminary results. Neuroimage. 2002;17(1):256–71. doi: 10.1006/nimg.2002.1163. [DOI] [PubMed] [Google Scholar]
- Wright IC, Sharma T, et al. Supra-regional brain systems and the neuropathology of schizophrenia. Cereb Cortex. 1999;9(4):366–78. doi: 10.1093/cercor/9.4.366. [DOI] [PubMed] [Google Scholar]
- Zhou G, Williams RW. Eye1 and Eye2: gene loci that modulate eye size, lens weight, and retinal area in the mouse. Invest Ophthalmol Vis Sci. 1999;40(5):817–25. [PubMed] [Google Scholar]
- Zygourakis CC, Rosen GD. Quantitative trait loci modulate ventricular size in the mouse brain. J Comp Neurol. 2003;461(3):362–9. doi: 10.1002/cne.10697. [DOI] [PubMed] [Google Scholar]