Abstract
The CXC subfamily of chemokines plays an important role in diverse processes, including inflammation, wound healing, growth regulation, angiogenesis, and tumorigenesis. The ELR-CXC chemokine, CXCL1 or MGSA/GROα, is traditionally considered to attract neutrophils to sites of inflammation. The non-ELR-CXC chemokine, CXCL10 or IP-10, is chemotactic for monocytes, B cells, and activated T lymphocytes. In addition to its role in leukocyte migration, CXCL10 inhibits the angiogenic functions of the ELR-CXC chemokines as well as bFGF and VEGF. Heparan sulfate proteoglycans (HSPGs) are required for the interaction of bFGF and vEGF ligands and their receptors. However, the role of HSPGs in regulating the ELR-chemokines signaling and biological functions is poorly understood. We show here that the CXCL1 maximal binding to CXCR2 expressed on HEK293 and CHO-K1 cells is dependent on the presence of cell surface HSPGs. The cell surface HSPGs on cells are required for CXCL1-induced PAK1 activation. Moreover, CXCL10 can inhibit CXCL1-induced PAK1 and ERK activation as well as the CXCL1-induced chemotaxis through decreasing CXCL1 binding to cell surface heparan sulfate. These data indicate that HSPGs are involved in modulating CXCL1-induced PAK1 activation and chemotaxis through regulating CXCL1 binding activity to CXCR2 receptor. CXCL10 inhibits CXCL1-induced PAK1 activation and chemotaxis by interfering with appropriate binding of CXCL1 to CXCR2 receptor.
CXC1 chemokines are crucial for timely recruiting of specific populations of the leukocyte to sites of the tissue damage during the inflammatory responses. In addition, these chemokines are also important in angiogenesis, tumor formation, and tumor metastasis (1). In this subfamily, CXC chemokines are further divided into two groups depending on the presence or absence of the amino acid sequence Glu–Leu–Arg (the ELR motif) at the N-terminal domain of the ligands. ELR-CXC chemokines, such as CXCL1 (melanoma growth stimulatory activity/growth regulated protein, MGSA/GRO), CXCL5 (epithelial derived neutrophil-activating peptide 78, ENA-78), CXCL6 (granulocyte chemotactic protein-2, GCP-2), and CXCL8 (interleukin-8), are all neutrophil-activating CXC chemokines, which bind to the CXCR1 or CXCR2 (CXC chemokine receptor 1 or 2) (1). These ELR-CXC chemokines not only attract neutrophils during inflammation, but also induce angiogenesis and tumor development (2–6). The non-ELR-CXC chemokines, such as CXCL10 (interferon-inducible protein-10, IP-10), CXCL4 (Platelet factor 4, PF4), CXCL11 (interferon-inducible T cell alpha chemoattractant, I-TAC), and CXCL9 (monokine induced by IFN-γ, MIG), do not bind the CXCR1 or CXCR2 receptors (7–8) and fail to induce endothelial cell chemotaxis, proliferation, and angiogenesis (9–11). CXCL9, CXCL10, and CXCL11 bind to the CXCR3 receptor, while CXCL12 (stromal cell-derived factor, SDF-1α) only binds the CXCR4 receptor (1). In general, these non-ELR CXC chemokines are able to induce chemotaxis in monocytes B and activated T lymphocytes during inflammation.
The heparan sulfate proteoglycan (HSPGs) are required for bFGF-induced endothelial cell proliferation and angiogenesis by modulating the interaction of ligands and receptors (13–15). When the FGF binding to HSPGs is prevented by treating the cells with heparinase, the FGF binding to its receptor is also reduced together with its ability to stimulate cell proliferation (14). In general, an increasing body of evidence suggests that HSPGs capture cytokine and chemokine ligands such as CXCL10 and CXCL4 at the cell surface and facilitate polymerization of chemokines by immobilizing and enhancing local concentrations of the ligands (16–17), as is the case for FGF and FGF receptor (18). However, the significance of multimerization for chemokine functions remains controversial. It has been reported that multimerization of the MCP-1 ligand is not required for glycosaminoglycan-dependent transendothelial chemotaxis (19). To explore the role of HSPGs in regulating the ELR-CXC chemokine-induced signaling and biologic functions, we investigated whether HSPGs regulate CXCL1 binding to its receptor and effect the PAK1 activation and chemotaxis in response to these ligands.
CXCL10 induces intracellular signals in various cell types through binding to its receptor, CXCR3. For example, Ras, ERK, Src, and PI3-K/Akt are activated when CXCL10 induces cell migration and proliferation in human vascular pericytes (20). Interestingly, the non-ELR-CXC chemokines have been shown to antagonize the angiogenic functions of the ELR-CXC chemokines as well as basic fibroblast growth factor (bFGF) and vascular endothelial growth factor (VEGF) (3–4, 21). The angiostatic properties of the non-ELR-CXC chemokines can inhibit the growth of subcutaneous tumors (9, 12, 22) and delay wound healing (10). One explanation for the CXCL10 inhibition is that CXCL10 binds to cell surface heparan sulfate motif shared with CXCL4 to inhibit endothelial cell proliferation (23). Luster’s group reported that the CXCL10 binds a specific endothelial cell surface heparan sulfate (23). This binding site has a Kd of 25 nM. However, CXCL4 inhibits EGF-induced endothelial cell proliferation and angiogenesis by interference with cell cycle machinery. CXCL4 inhibition is independent of interaction with cell surface glycosaminoglycans (GAGs), such as heparan sulfate (11). Marco Presta, et al reported that CXCL8 inhibits bFGF-induced endothelial cell proliferation by down-regulating FGF receptors. In their systems, CXCL8 did not prevent the binding of bFGF to its receptors nor to cell surface heparan sulfate proteoglycans (HSPGs) (24).
Our earlier studies showed that CXCL1 induces the activation of transcription factors, NF-κB and AP-1, through a Ras-MEKK1-MEK4/6-p38 MAP kinase pathway in melanocytes (25). This pathway is involved in the CXCL1-induced melanocyte transformation (6). CXCL8, another member of the ELR-CXC chemokines, has been demonstrated to activate the PI3-kinase/Ras/Raf cascade in neutrophils (26). This activation may be required for the IL-8-induced chemotaxis. In addition, CXCL1 can induce cdc42-PAK1 activation, which is required for chemotaxis (27), and MAP kinase ERK1/2 (27–28). PAKs (p21-activated kinases) play an important role in diverse cellular processes, including cytoskeletal rearrangements (29–33), growth, and apoptosis (34–36). PAKs are Ser/Thr protein kinases that undergo autophosphorylation and activation upon interacting with the active forms of the small GTPase (p21) Rac or Cdc42 (37). PAK activation is regulated by a variety of external stimuli that act through cell surface receptors, including G protein-coupled receptors (38), growth factor receptor tyrosine kinases (39), proinflammatory cytokine receptors (40), Fc receptors (41), and integrins (42–43).
The role of cell surface HSPGs on the CXCL1-induced signaling and biological function is unknown, and the mechanism by which CXCL10 inhibits CXCL1-induced angiogenesis is not clear. Therefore, the aim of this work was to investigate the biologic role of HSPGs for CXCL1-induced signaling and chemotaxis and to explore whether CXCL10 blocks CXCL1-induced signal pathways, which lead to induction of chemotaxis. Our findings demonstrate that a cell surface HSPG is required for CXCL1 maximal binding to CXCR2 and for the CXCL1-induced signaling and chemotaxis. CXCL10 inhibits CXCL1-induced signaling and significantly affects CXCL1-induced chemotaxis by interrupting the CXCL1 binding to cell surface heparan sulfate.
EXPERIMENTAL PROCEDURES
Cell Culture
293 human embryonic kidney cells (HEK293) were cultured in DMEM supplemented with 50 units/mL penicillin, 50 μg/mL streptomycin, 3 mM glutamine, 5% heat-inactivated fetal bovine serum (GIBCO BRL, Rockville, MD). The CXCR2-expressing HEK293 cells (pooled clones) were cultured in the same media supplemented with 800 μg/mL G418 (Sigma, St. Louis, MO) as previously described (44). The expression level of CXCR2 receptor in the HEK293 cells has been previously verified (44). Wild-type CHO-K1 (Chinese hamster ovary) and the mutant GAG-deficient CHO cells (pgsA-745) (glycosaminoglycan-deficient CHO) cells obtained from ATCC (Manassas, VA) were cultured in F12K supplemented with 50 units/ml penicillin, 50 μg/mL streptomycin, 3mM glutamine, 10% heat-inactivated fetal bovine serum (GIBCO BRL, Rockville, MD). Purified recombinant human CXCL1 (a kind gift of Repligen Corp., Needham, MA) was used at 50 ng/mL. CXCL10 was purchased from R & D Systems (Minneapolis, MN).
Whole Cell Extracts and Western Blot
Whole cell extracts were prepared from CXCR2-expressing HEK293 cells treated with CXCL1 for the indicated time after serum starvation for 14 h. Western blots were performed following protocols provided by Santa Cruz Biotechnology Inc. The cells were washed at 4 °C with 1X PBS and lysed in 0.6 mL of RIPA buffer (1X PBS, 1%NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with protease inhibitor cocktail tablets (Boehringer Mannheim Corp., Indianapolis, IN) and 0.2 mM sodium orthovanadate. A 50 μg sample of soluble protein was boiled, loaded, and electrophoretically separated on a 10% SDS–PAGE reducing gel, then electrophoretically transferred to a 0.45 μm nitrocellulose membrane (BIO-RAD, Hercules, CA). The membrane was blocked with 5% dry milk in TBS-T buffer (10mM Tris-HCl, pH 8.0; 150mM NaCl; 0.05% Tween-20) for 1 h and then incubated for 12–16 h at 4 °C in a 1:1000 dilution (0.2 μg/mL) of the anti-PAK1 antibody (Santa Cruz Biotechnology, Inc.) or the anti-phospho-ERK1/2 (Santa Cruz Biotechnology, Inc.). After washing three times with TBS-T buffer, the membrane was incubated in a 1:3000 dilution of the appropriate anti-mouse or anti-rabbit immunoglobulin conjugated with horseradish peroxidase (Boehringer Mannheim Corp.) in TBS-T buffer with 5% dry milk for 1 h at room temperature. After washing three times with TBS-T buffer, the protein bands were detected with the ECL western blotting detection reagents (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer’s instructions. The blots were stripped and re-probed with anti-ERK2.
Immune Complex Kinase Assays
Whole cell extracts were prepared from CXCR2-expressing HEK293 cells treated with CXCL1 after serum starvation for 14 h. PAK1 kinase assays were performed as described in the manufacturer’s protocol (Upstate Biotechnology, Lake Placid, NY). A 400 μg sample of protein of each whole cell extract was immunoprecipitated with 1μg PAK1 antibody. Immunoprecipitated PAK1 activity was assayed using the PAK1 substrate, myelin basic protein (MBP) (Sigma). Kinase reactions were initiated by addition of 2 μg MBP and kinase buffer containing 500 μM cold ATP and 10 μCi of [γ-32P]ATP. Reactions were incubated for 30 min at 30 °C and terminated by the addition of an equal volume of 2XSDS loading buffer followed by boiling for 5 min. Phosphorylated proteins were resolved on a 10% SDS–PAGE reducing gel and transferred to a 0.45 μm nitro-cellulose membrane (BIO-RAD). The phosphorylated bands were visualized by auto-radiography. The blot was probed with PAK1 antibody to monitor equal loading of PAK1.
Chemotaxis Assay
Chemotaxis assays were performed on the CXCR2-expressing HEK293 cells, or wild-type CHO-K1 and the GAG-deficient CHO (pgsA-745) cells transiently transfected with either empty vector or hCXCR2 plasmid, as described previously (44). In short, a 96-well modified Boyden chamber (Neuroprobe Inc) was used, and the lower compartment of the chamber was loaded with 450 mL chemotaxis buffer (1 mg/mL ovalbumin/DMEM) containing CXCL1 diluted at indicated concentration in the chemotaxis buffer. Polycarbonate membranes (pore size: 10 μm) were coated on both sides with 20 μg/mL human collagen type IV (Sigma), incubated for 2 h at 37 °C, and then stored at 4 °C overnight. The cells were removed from the plate by trypsinization and incubated for restoration of receptors in 5% FBS/DMEM for 2 h at 37 °C. The cells were washed with chemotaxis buffer and then loaded into the upper chamber in 250 μL chemotaxis buffer at 5 × 106 cells/ml. The chamber was incubated for 4 h at 37 °C with 5% CO2; then the membrane was removed, washed, fixed, and stained with a Diff-Quik kit. Cell chemotaxis was quantified by counting cells that crossed the membrane in five high-power microscope fields (X 40). The relative chemotactic index represented the mean number of cells migrating with ligand stimulation versus without the stimulation.
Ligand Binding Assay
Binding assays were performed on the CXCR2-expressing HEK293 cells, parental HEK293 cells. For wild-type CHO-K1 and GAG-deficient CHO (pgsA-745) cells, the cells were transiently transfected with hCXCR2. The CXCR2 expression level on the cell surface was monitored by FACS analysis. In short, the cells were cultured to confluence on 24-well plates. For the CXCR2-stably expressing HEK293 cells, the 24-well plates were precoated with 0.1 mg/mL poly-L-lysine (Sigma, MW 30 000–70 000) for 1 h and washed once with distilled water. For the CHO cells, cells were cultured in complete media for 24 h after transfection. Cells were then incubated in 0.5 mL of serum-free DMEM medium containing 0.025 μCi/ml 125I-CXCL1 for 1 h at 4 °C with or without the indicated concentration of the cold competitors. After the incubation, the cells were washed four times with 1 mL of wash buffer (25 mM HEPES, pH 7.2, 0.5M NaCl) and lysed in 1 mL of 1% SDS with 0.1 N NaOH. The radioactive cell lysates were counted in a γ-counter (Gamma 5500, Beckman). The specific cell surface binding was calculated by subtracting the nonspecific binding from the total binding. Nonspecific binding was determined by performing the binding assay in the presence of excess unlabeled CXCL1 (250 ng/mL).
Heparinase Treatment
The cells were washed once with serum free DMEM and then incubated in same media with the indicated concentration of heparinase (Calbiochem) at 37 °C with 5% CO2 for 1 h. After treatment, the cells were washed three times with binding buffer for binding assays or with serum free DMEM for PAK1 kinase assays as described as above.
RESULTS
CXCL1 Binds to Heparan Sulfate
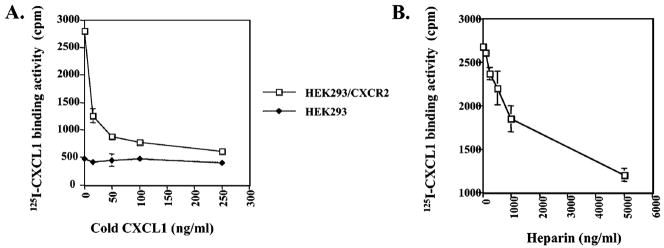
125I-CXCL1 binding was inhibited by unlabeled CXCL1 in a dose-dependent manner on CXCR2-expressing HEK-293 cells (Figure 1A). However, the unlabeled CXCL1 failed to compete for 125I-CXCL1 binding to parental HEK293 cells (Figure 1A). These data demonstrate that CXCL1 specifically binds to CXCR2 receptors on these cells (Figure 1A). CXCL1 has a HSPG-like binding site, which indicates that this chemokine has the capacity to bind to heparan sulfate (HS) (24). We performed a dose-inhibition experiment to determine whether CXCL1 has the capacity to bind soluble heparan sulfate. In this experiment, soluble heparan sulfate could compete for 125I-CXCL1 binding to cells in a concentration-dependent manner (Figure 1B). These data demonstrate that CXCL1 can bind to the soluble heparan sulfate.
Figure 1.
CXCL1 binds to soluble heparan sulfate. CXCL1 binding is specific and competed by heparan sulfate. Binding assays were performed as described in Experimental Procedures. CXCL1 (panel A) or soluble heparan sulfate (panel B) was tested for their ability to compete for 125I-labeled CXCL1 binding to CXCR2-expressing HEK293 cells and parental HEK293 cells. The starting point was taken in the presence of 2.5 ng/mL 125I-CXCL1, without any cold CXCL1. Data are presented as the mean of the absolute binding activity from three independent experiments. The data were analyzed using the Student’s paired t test (p < 0.05).
CXCL1 Binding to Cells Is Dependent on Surface HSPGs
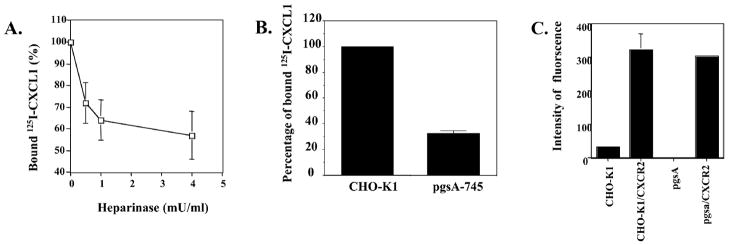
It has been reported that HSPGs capture cytokine and chemokine ligands on the cell surface and facilitate polymerization of chemokines by immobilizing and enhancing local concentrations of the ligands (16, 17). To determine whether HSPGs will affect the specific binding capacity of CXCL1 to its receptor, we next examined whether this binding activity of CXCL1 to CXCR2 is heparinase sensitive in HEK293 cells. Figure 2A showed that CXCR2-expressing HEK293 cells pretreated with different concentrations of heparinase exhibited a 28–43% decrease in the specific binding activity to CXCL1. These data suggest that the cell surface HSPGs are involved in the CXCL1 maximal binding to CXCR2-expressing HEK293 cells. To further confirm the role of cell-surface heparan sulfate in CXCL1 binding to CXCR2, we compared the CXCL1 binding properties of wild type and GAG-deficient CHO cells (pgsA-745) that have been transiently transfected with CXCR2. The data in Figure 2B show that the specific binding of 125I-CXCL1 to the mutant cells is 3-fold less than that to the wild-type CHO cells. The data of FACS analysis demonstrated that both cells express similar level of CXCR2 after transient transfection (Figure 2C). Since we used high salt wash buffer (0.5M NaCl) to inhibit the polymerization of 125I-CXCL1, our results suggest that the cell surface heparan sulfate-enhanced ligand binding to CXCR2-expressing cells is not due to polymerization of ligands. Taken together, these data demonstrate that cell surface heparan sulfate is required for CXCL1 maximal binding to CXCR2 on these cells because CXCL1 failed to specifically bind to parental wild-type CHO and GAG-deficient CHO cells under our experimental conditions (data not shown).
Figure 2.
(A) The role of cell surface HSPGs in facilitating CXCL1 binding to its receptors. The CXCL1 binding sites are heparinase-sensitive. The CXCR2-expressing HEK293 cells were treated with indicated concentration of heparinase in serum free medium for 2 h at 37 °C and then washed with binding buffer two times before the ligand binding assay. Nonspecific binding was determined by performing the binding assay with 125I-labeled CXCL1 in the presence of excess 250ng/mL unlabeled CXCL1. Data are presented as the mean of the percentage of binding where specific binding in the presence of heparinase is divided by specific binding in the absence of heparinase from three independent experiments. The data were analyzed using Student’s paired t test (p < 0.05). (B) The comparison of 125I-labeled CXCL1 binding to wild-type and HSPG-deficient CHO cells. After wild-type and HSPG-deficient CHO cells were transient transfected with hCXCR2, binding assays were performed as in Figure 1. The CXCL1 specific binding to wild-type CXCR2-expressing CHO cells was set as 100%. The results represent mean of the percentage of CXCL1 binding to HSPG-deficient CXCR2-expressing CHO cells, as compared to wild-type CXCR2-expressing CHO cells from three independent experiments. The data were analyzed using Student’s paired t test (p < 0.05). (C) The expression of hCXCR2 in wild-type and HSPG-deficient CHO cells. After wild-type and HSPG-deficient CHO cells were transient transfected with hCXCR2, the cells were stained with anti-PE-coupled CXCR2 antibody and analyzed by FACS. The data are presented as the mean of the intensity of PE fluorescence from three independent experiments. The data were analyzed using the Student’s paired t test (p < 0.05).
Blocking of CXCL1 Binding to the HSPGs Inhibits the CXCL1-Induced Signaling
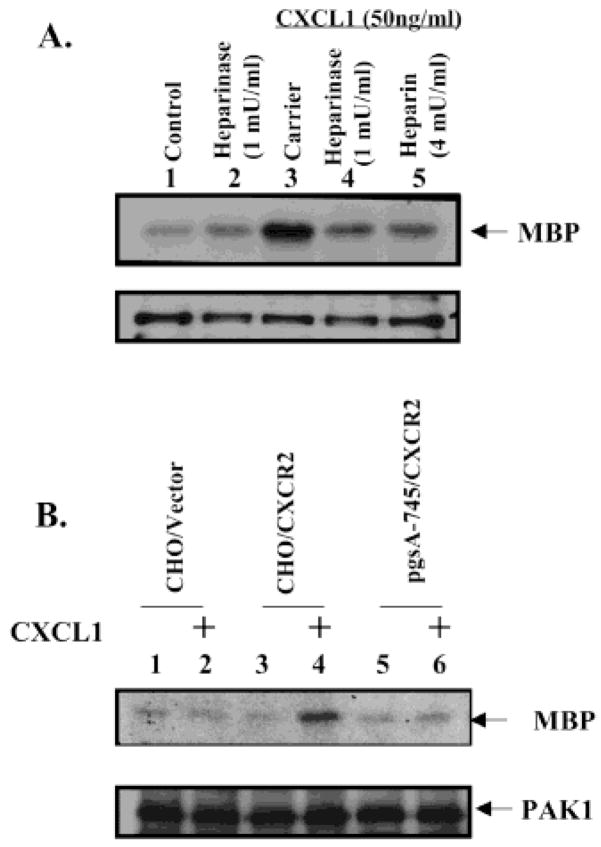
To investigate the role of the cell surface HSPGs in CXCL1-stimulated CXCR2-mediated signaling and biological function, we examined the effects of the blocking of CXCL1 binding to the HSPGs on the CXCL1-induced PAK1 activation. As shown in Figure 3A, the CXCR2-expressing HEK293 cells treated with heparinase exhibited a significant decrease in the CXCL1-induced PAK1 activation, while the treatment with heparinase did not affect the expression level of PAK1. These data indicate that the cell surface HSPGs are involved in the CXCL1-induced signaling. A further demonstration that CXCL1-induced PAK1 activation is dependent on cell surface heparan sulfate comes from studies using GAG-deficient CHO cells CXCL1 induced PAK1 activation in wild-type CHO line K1 but did not induce PAK1 activation in GAG-deficient CHO (Figure 3B).
Figure 3.
Cell surface heparan sulfate is required for CXCL1-induced PAK1 activation. (A) Depletion of heparan sulfate by heparinase blocks CXCL-induced PAK1 activation in CXCR2-expressing HEK293 cells. The CXCR2-expressing HEK293 cells were treated with heparinase as in Figure 2A before the stimulation with CXCL1 for 10 min. PAK1 kinase assays were performed as described in Experimental Procedures. Endogenous PAK1 activity was determined by an amount of MBP phosphorylation (top panel). The blot was reprobed with PAK1 antibody to monitor equal loading of PAK1 (lower panel). This figure is representative of three different experiments with similar results. (B) Absence of heparan sulfate in HSPG-deficient CHO cells inhibits CXCL1-induced PAK1 activation. CXCR2-expressing wild-type and HSPG-deficient CHO cells were either untreated or treated with 50 ng/mL CXCL1 for the 10 min after serum starvation for 14 h. PAK1 kinase assays were performed as in panel A. This figure is representative of three different experiments with similar results.
CXCL10 Inhibits CXCL1-Induced Signaling
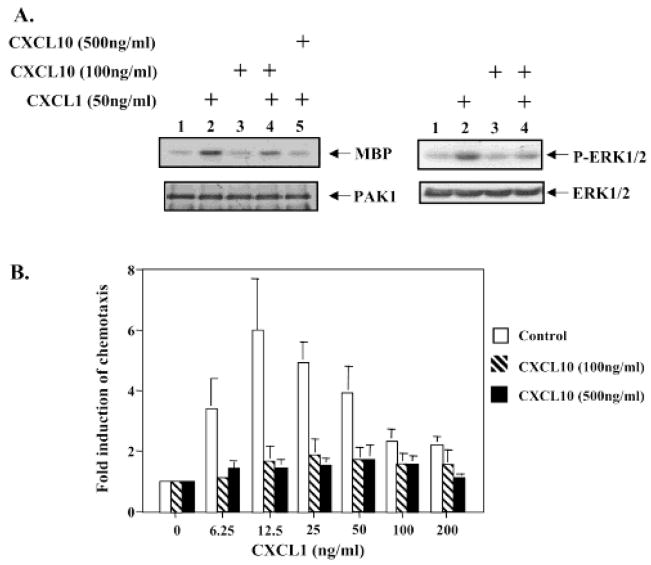
Previous studies demonstrated that CXCL10 blocks the angiogenesis induced by ELR-CXC chemokines such as CXCL8 and CXCL1 as well as growth factors such as bFGF and VEGF in vivo. To better understand the molecular mechanism of the angiostatic properties of CXCL10, we examined whether CXCL10 would block CXCL1-induced signaling pathways. Our previous data showed that CXCL1 induces PAK1 and ERK activation (27). Therefore, we first tested whether CXCL10 blocks CXCL1-induced PAK1 and ERK activation. Immune-complex kinase assays were performed to evaluate PAK1 activity induced by CXCL1 in the CXCR2-expressing HEK293 cells pretreated with CXCL10 at the indicated concentration for 10 min. CXCL10 decreased CXCL1-induced PAK1 kinase activity at a concentration of 100 ng/mL (Figure 4A, upper-left panel, lane 4 vs lane 2) and totally blocked CXCL1 stimulation of PAK1 kinase activity at 500 ng/mL (Figure 4A, upper-left panel, lane 5 vs lane 2). The expression level of PAK1 was not affected by the treatment of CXCL10 (Figure 4A, lower-left panel). Similarly, CXCL10 inhibited CXCL1-induced phosphorylation of ERK at Tyr 204 based upon Western blot using the antibody that specifically detects the Tyr 204-phosphorylated form of ERK (Figure 4A, upper-right panel). The lower-right panel showed the equal loading of ERK on this gel. These data demonstrate that CXCL10 can block CXCL1-induced signaling.
Figure 4.
CXCL10 inhibits CXCL1-induced signaling, which is required for the chemotaxis. (A) CXCL10 inhibits CXCL1-induced PAK1 and ERK activation. CXCR2-expressing cells were either untreated or treated with CXCL10 at indicated concentrations for 10 min before stimulation with 50 ng/mL CXCL1 for 10 min as in Figure 3A. Endogenous PAK1 activity was determined by an immunocomplex kinase assay as in Figure 1A (left panel). The blot was probed with PAK1 antibody to monitor equal loading of PAK1 (left-lower panel). Phosphorylated ERK1/2 was detected by Western Blot (right-upper panel). The blot was reprobed with ERK antibody monitor equal loading of PAK1 (right-lower panel). These figures are representative of three different experiments with similar results. (B) The effect of the CXCL10 on CXCL1-stimulated CXCR2-mediated chemotaxis in HEK293 cells. CXCR2-expressing HEK293 cells were treated with the carrier buffer for control (empty bars), CXCL10 at 100 ng/mL (striped bars) or CXCL10 at 500 ng/mL (solid bars), and then the cells were loaded into the upper chamber. For the CXCL10 treated cells, CXCL10 was added to the lower chambers at same concentration as the upper chamber. Chemotactic response to CXCL1 stimulation was compared as described under Experimental Procedures. Values represent the means ± S.E. of three independent experiments. The data were analyzed using the Student’s paired t test (p < 0.05).
CXCL10 Inhibits CXCL1-Induced Chemotaxis
Our early results demonstrated that PAK1 activation is required for CXCL1-induced chemotaxis (27). The above observation that CXCL10 blocks PAK1 activation indicates that CXCL10 may inhibit CXCL1-stimulated CXCR2-mediated chemotaxis. A CXCL1 concentration-dependent chemotactic response was observed in the CXCR2-expressing HEK293 cells (control), but not in the same cells treated with the indicated concentrations of CXCL10 (100 and 500 ng/mL), where CXCL10 is present in both upper and lower chamber (Figure 4B). These data demonstrated that CXCL10 inhibits CXCL1-stimulated CXCR2-mediated chemotaxis.
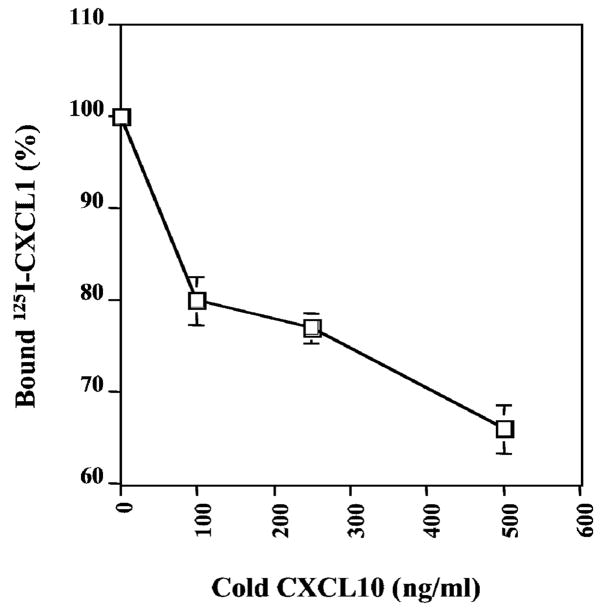
CXCL10 Partially Blocks CXCL1 Binding Activity to CXCR2
The above results showed that the CXCL1 maximal binding activity requires cell surface HSPGs. Since CXCL10 can bind to the same cell surface HSPGs, we postulated that the inhibitory activity of CXCL10 on CXCL1-induced signaling could be explained by blocking CXCL1 binding to HSPGs. We explored the ability of unlabeled CXCL10 to block 125I-CXCL1 binding to the CXCR2-expressing HEK293 cells. As shown as Figure 5, CXCL10 partially blocks CXCL1 binding to the CXCR2-expressing HEK293 (Figure 5). Unlabeled CXCL10 competed much less effectively than unlabeled CXCL1 because CXCL10 could not directly bind to the CXCR2 on the cell surface. Like heparinase, CXCL10 can partially decrease the CXCL1 binding capacity to the cell surface receptor by occupying the heparan sulfate, which is required for the CXCL1 maximal binding to CXCR2-expressing cells.
Figure 5.
The effects of CXCL10 on the CXCL1 binding to CXCR2-expressing cells. CXCR2-expressing HEK293 (panel A) and CXCR2-expressing CHO cells (panel B) were incubated for 1 h at 4 °C with 125I-CXCL1 in the presence of varying concentrations of CXCL10. Data are presented as the percentage of binding where the specific binding in the presence of competitor is divided by specific binding in the absence of competitor × 100 in three independent experiments. Values represent the mean ± S.E. of three independent experiments performed in duplicate. The data were analyzed using the Student’s paired t test (p < 0.05).
DISCUSSION
For the first time, we demonstrate here that the ability of CXCL1 binding to CXCR2 expressed on HEK293 and CHO-K1 cells is dependent on the presence of cell surface heparan sulfate, on the basis of the observations that CXCL1 binding to CXCR2 is (a) inhibited by soluble heparin sulfate, (b) is sensitive to treatment with heparinase, and (c) is absent in GAG-deficient CHO cells (pgsA-745). The cell surface heparan sulfate is also required for CXCL1-induced PAK1 activation, on the basis of the observation that CXCL1 failed to induce PAK1 activation in the GAG-deficient CXCR2-expressing CHO cells (pgsA-745), although CXCL1 can induce PAK1 activation in wild-type CXCR2-expressing CHO cells (CHO-K1). Moreover, CXCL10 can inhibit CXCL1-induced PAK1 and ERK activation as well as the CXCL1-induced chemotaxis. These data indicate that CXCL10 inhibits CXCL1-induced signaling and chemotaxis by interrupting CXCL1 binding to cell surface heparan sulfate, since CXCL10 also binds to cell surface heparan sulfate and effectively competes with CXCL1 for the heparan sulfate binding site (23).
Our data show that the depletion of the heparan sulfate by heparinase treatment decreases CXCL1 binding to the CXCR2-expressing HEK293 cells by 30–40% (Figure 2A), suggesting that cell surface heparan sulfate might be required for the ligand maximal binding to its receptor. Further evidence was provided by the demonstration that the CXCL1-binding activity to CXCR2 in GAG-deficient CHO cells (pgsA-745) is 3-fold less than in wild-type CHO cells (Figure 2B). The GAG-deficient CHO cells have a specific deficiency in xylosyl transferase, which leads to failure to express heparan sulfate (45). Both wild-type and GAG-deficient CHO cells showed a similar morphology and growth rate, an observation consistent with previous studies (45). Wild-type and the GAG-deficient CHO cells have been extensively used to examine the role of cell surface heparan sulfate in regulating ligand functions. On the basis of the above results, we postulated that two facts contribute the residual 60% CXCL1 binding after haparanase treatment. First, there is a residual 33% CXCL1 binding to CXCR2 in the GAG-deficient CHO (Figure 2B). CXCL1 still binds CXCR2 with low binding activity in the lack of the HSPGs. Second, heparanase treatment may not totally cut HS from HSPGs. A combination only 80% heparanase digestion and the residual 33% CXCL1 binding may causes the residual 50% CXCL1 binding after heparanase treatment.
Our results demonstrate that the presence of cell surface heparan sulfate facilitates the activation of ligand-bound receptor, on the basis of our observation that CXCR2 activation of PAK1 was blocked when heparan sulfate was depleted by heparinase treatment in HEK293 cells or when GAG-deficient CHO cells expressing CXCR2 were assayed (Figure 3). Therefore, it is most likely that the absence of CXCL1-induced signaling in the CXCR2-expressing GAG-deficient CHO cells is due to the failure of appropriate presentation of CXCL1 to CXCR2 and/or the formation of stable complex of ligand and its receptor, indicating that the heparan sulfate-rich microenviroment surrounding the cell-surface receptor complex plays an important role in regulating CXCL1 activation of its receptor. One possible interpretation of these results is that cell surface heparan sulfate facilitates ligand binding to high-affinity binding site in the receptor because the CXCR2 receptor has two ligand binding sites with different affinity (46). In the absence of cell surface heparan sulfate, the CXCL1 can still bind to CXCR2 with low binding activity (Figure 2B). The low binding activity of CXCL1 to CXCL2 results in the failure of receptor activation, which could not fully induce signaling and biological functions (47–49).
CXCL8 activation of the PI3-kinase/Ras/Raf pathway is required for human neutrophil migration. The overall mechanism(s) responsible for the CXCL8 activation of the PI3-kinase pathway is likely to be the same for CXCL1 activation. PI3-kinase can regulate PAK activation through Rac/cdc42 (50). PAKs have been shown to regulate the MAP kinases ERK, JNK and p38 in response to stimuli from cytokines, chemoattractants, and various stresses in certain type of the cell (51). In CXCR2-expressing HEK293 cells, ERK is not a downstream target of PAK1 (27). Recently, accumulating data indicate that PAKs phosphorylate key components such as paxillin (52), myosin light-chain kinase (33), and LIM kinase (32), all of which are involved in regulation of the cytoskeletal organization. We have not, however, determined the exact downstream targets for PAK in CXCR2-expressing HEK293 and CHO cells. Future studies will address these unsolved issues.
Our data show that CXCL10 partially blocks CXCL1 binding to its receptors by about 35% and almost completely inhibits CXCL1-induced signaling in the CXCR2-expressing HEK293 cells. These data are consistent with the observation that depletion of heparan sulfate by heparinase only decreases the binding activity of CXCL1 to CXCR2 by about 35%, but almost blocks CXCL1-induced PAK1 activation. Since CXCL10 can bind to the cell surface HSPGs, which are required for the maximal CXCL1 binding to CXCR2, we postulate that CXCL10 decreases the CXCL1 binding effectiveness of the CXCR2 by occupying cell surface heparan sulfate. The lower binding activity of CXCL1 to CXCR2 by CXCL10 leads to complete inhibition of ligand-induced signaling and function. The observation that CXCL10 only reduced 30% of CXCL1 binding to CXCR2 is not a surprise. When cell surface heparan sulfate is totally absent, there is only a 67% reduction of CXCL1 binding to CXCR2-expressing cells. These observations suggest that cell surface HSPGs participate in CXCL10 inhibition on the CXCL1-induced signaling. The mechanism of CXCL10 inhibition of CXCL1 binding has not been examined. It is not known whether CXCL10 destabilizes the complex of CXCL1, HSPGs, and CXCR2 or competes with CXCL1 binding to HSPGs by binding to HSPGs. However, the finding that the non-ELR-CXC chemokine, CXCL10, blocks an ELR-CXC chemokine-induced signaling in a CXCR3 independent manner is novel and potentially important. Previous observations have shown that ELR-positive and non-ELR-CXC chemokines exert opposite effects on angiogenesis in vitro and in vivo (3). The shift in the balance of the ELR-CXC chemokines versus non-ELR-CXC chemokines has been observed in many inflammatory diseases, such as chronic pancreatitis, inflammatory bowel disease and psoriasis (53–54), and tumor development (4–5). Our data provide a potential mechanism for the different biological functions of these two subsets of CXC chemokines.
Footnotes
We are indebted to Grants CA34590 (A.R.) and CA56704 (A.R.) from the NIH for support, Vanderbilt Ingram Cancer Center Grant CA68485, and awards from the Department of Veterans Affairs: the Merit Award (A.R.), the Career Scientist Award (A.R.), and the GRECC Pilot Project (A.R.).
Abbreviations: CXC chemokine, chemokine with the first two conserved cysteine residues separated by an intervening amino acid; DMEM, Dulbecco’s modified Eagle’s medium; CXCL1 or MGSA/GRO, melanoma growth-stimulatory activity/growth-regulated protein; CXCL10 or IP-10, interferon-inducible protein-10; GAGs, glycosaminoglycans; HSPG, heparan sulfate proteoglycans; PAKs, p21-activated kinases; MBP, myelin basic protein.
References
- 1.Rossi D, Zlotnik A. Annu Rev Immunol. 2000;18:217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. J Biol Chem. 1991;266:23128–23134. [PubMed] [Google Scholar]
- 3.Strieter RM, Polverini PJ, Kunkel SL, Arenberg DA, Burdick MD, Kasper J, Dzuiba J, Van Damme J, Walz A, Marriott D, Chan SY, Roczniak S, Shanafelt AB. J Biol Chem. 1995;270:348–357. doi: 10.1074/jbc.270.45.27348. [DOI] [PubMed] [Google Scholar]
- 4.Arenberg DA, Polverini PJ, Kunkel S, Shanafelt A, Hesselgesser JR, Strieter M. J Leukocyte Biol. 1997;62:554–562. doi: 10.1002/jlb.62.5.554. [DOI] [PubMed] [Google Scholar]
- 5.Luan J, Shattuck-Brandt R, Haghnegahdar H, Owen JD, Strieter R, Burdick M, Nirodi C, Beauchamp D, Johnson KN, Richmond A. J Leukococyte Biol. 1997;62:588–97. doi: 10.1002/jlb.62.5.588. [DOI] [PubMed] [Google Scholar]
- 6.Wang D, Yang W, Du J, Devalaraja MN, Liang P, Matsumoto K, Tsubakimoto K, Endo T, Richmond A. Oncogene. 2000;19:4647–4659. doi: 10.1038/sj.onc.1203820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark-Lewis I, Dewald B, Geiser T, Moser B, Baggiolini M. Proc Natl Acad Sci USA. 1993;90:3574–3577. doi: 10.1073/pnas.90.8.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark-Lewis I, Dewald B, Loetscher M, Moser B, Baggiolini M. J Biol Chem. 1994;269:16075–16081. [PubMed] [Google Scholar]
- 9.Sharpe RJ, Byers HR, Scott CF, Bauer SI, Maione TE. J Natl Cancer Inst. 1990;82:848–853. doi: 10.1093/jnci/82.10.848. [DOI] [PubMed] [Google Scholar]
- 10.Luster AD, Cardiff RD, MacLean JA, Crowe K, Granstein RD. Proc Assoc Am Physicians. 1998;110:183–196. [PubMed] [Google Scholar]
- 11.Gentilini G, Kirschbaum NE, Augustine JA, Aster RH, Visentin GP. Blood. 1999;93:25–33. [PubMed] [Google Scholar]
- 12.Sgadari C, Angiolillo AL, Tosato G. Blood. 1996;87:3877–3882. [PubMed] [Google Scholar]
- 13.Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- 14.Bikfalvi A, Klein S, Pintucci G, Rifkin DB. Endocrinol Rev. 1997;18:26–45. doi: 10.1210/edrv.18.1.0292. [DOI] [PubMed] [Google Scholar]
- 15.Rapraeger AC, Krufka A, Olwin BB. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka Y, Adams DH, Shaw S. Immunol Today. 1993;14:111–115. doi: 10.1016/0167-5699(93)90209-4. [DOI] [PubMed] [Google Scholar]
- 17.Hoogewerf AJ, Kuschert GS, Proudfoot AE, Borlat F, Clark-Lewis I, Power CA, Wells TN. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 18.Spivak-Kroizman T, Lemmon MA, Dikic I, Ladbury JE, Pinchasi D, Huang J, Jaye M, Crumley G, Schlessinger J, Lax I. Cell. 1994;79:1015–1024. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 19.Ali A, Palmer ACV, Fritchley SJ, Maley Y, Kirby JA. Biochem J. 2001;358:737–745. doi: 10.1042/0264-6021:3580737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonacchi A, Romagnani P, Romanelli GR, Efsen E, Annunziato F, Lasagni L, Francalanci M, Serio M, Laffi G, Pinzani M, Gentilini P, Marra F. J Biol Chem. 2001;276:9945–9954. doi: 10.1074/jbc.M010303200. [DOI] [PubMed] [Google Scholar]
- 21.Romagnani P, Annunziato F, Lasagni L, Lazzeri E, Beltrame C, Francalanci M, Uguccioni M, Galli G, Cosmi L, Maurenzig L, Baggiolini M, Maggi E, Romagnani S, Serio M. J Clin Invest. 2001;107:53–63. doi: 10.1172/JCI9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao L, Pike SE, Setsuda J, Parekh J, Gupta G, Raffeld M, Jaffe ES, Tosato G. Blood. 2000;96:1900–1905. [PubMed] [Google Scholar]
- 23.Luster AD, Greenberg MS, Leder P. J Exp Med. 1995;182:219–231. doi: 10.1084/jem.182.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Presta M, Belleri M, Vecchi A, Hesselgesser J, Mantovani A, Horuk R. J Biol Chem. 1998;273:7911–7919. doi: 10.1074/jbc.273.14.7911. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Richmond A. J Biol Chem. 2001;276:3650–3659. doi: 10.1074/jbc.M006115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knall C, Worthen GS, Johnson GL. Proc Natl Acad Sci USA. 1997;94:3052–3057. doi: 10.1073/pnas.94.7.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Sai J, Carter G, Sachpatzidis A, Lolis L, Richmond A. Biochemistry. 2002;41:7100–7107. doi: 10.1021/bi025902m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shyamala V, Khoja H. Biochemistry. 1998;37:15918–15924. doi: 10.1021/bi9811415. [DOI] [PubMed] [Google Scholar]
- 29.Sells MA, Knaus UG, Bagrodia S, Ambrose DM, Bokoch GM, Chernoff J. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 30.Manser E, Huang HY, Loo TH, Chen XQ, Dong JM, Leung T, Lim L. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sells MA, Boyd TJ, Chernoff J. J Cell Biol. 1999;145:837–849. doi: 10.1083/jcb.145.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards CD, Sanders CL, Gill NG, Bokoch MG. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 33.Sanders CL, Matsumura F, Bokoch MG, de Lanerolle P. Science. 1999;283:2083–2085. doi: 10.1126/science.283.5410.2083. [DOI] [PubMed] [Google Scholar]
- 34.Qu J, Cammarano MS, Shi Q, Ha KC, de Lanerolle P, Minden A. Mol Cell Biol. 2001;21:3523–3533. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gnesutta N, Qu J, Minden A. J Biol Chem. 2001;276:14414–12219. doi: 10.1074/jbc.M011046200. [DOI] [PubMed] [Google Scholar]
- 36.Jakobi R, Moertl E, Koeppel MA. J Biol Chem. 2001;276:16624–16634. doi: 10.1074/jbc.M007753200. [DOI] [PubMed] [Google Scholar]
- 37.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 38.Knaus UG, Morris S, Dong HJ, Chernoff J, Bockoch GM. Science. 1995;269:221–223. doi: 10.1126/science.7618083. [DOI] [PubMed] [Google Scholar]
- 39.Dharmawardhane S, Sanders LC, Martin SS, Daniels RH, Bokoch GM. J Cell Biol. 1997;138:1265–1278. doi: 10.1083/jcb.138.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S, Han J, Sells MA, Cheeroff J, Knaus UG, Ulevitch RJ, Bokoch GM. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 41.Jones SL, Knaus UG, Bokoch GM, Brown EJ. J Biol Chem. 1998;273:10556–10566. doi: 10.1074/jbc.273.17.10556. [DOI] [PubMed] [Google Scholar]
- 42.Price LS, Leng J, Schwartz MA, Bokoch GM. Mol Cell Biol. 1998;9:1863–1871. doi: 10.1091/mbc.9.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiosses WB, Daniels RH, Otey C, Bokoch GM, Schwartz MA. J Cell Biol. 1999;147:831–844. doi: 10.1083/jcb.147.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W, Schraw PW, Mueller GS, Richmond A. Biochemistry. 1997;36:15193–15200. doi: 10.1021/bi971594u. [DOI] [PubMed] [Google Scholar]
- 45.Esko JD, Stewart TE, Taylor WH. Proc Natl Acad Sci USA. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammond M, Shyamala V, Siani MA, Gallegos CA, Feucht PH, Abbott J, Gena Lapointe R, Moghadam M, Khoja H, Zakel J, Tekamp-Olson P. J Biol Chem. 1996;271:8228–8235. doi: 10.1074/jbc.271.14.8228. [DOI] [PubMed] [Google Scholar]
- 47.Krasilnikov MA. Biochemistry (Mosc) 2000;65:59–67. [PubMed] [Google Scholar]
- 48.Lim L, Manser E, Leung T, Hall C. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 49.Hashimoto S, Tsubouchi A, Mazaki Y, Sabe H. J Biol Chem. 2001;276:6037–6045. doi: 10.1074/jbc.M005854200. [DOI] [PubMed] [Google Scholar]
- 50.Moser B, Dewald B, Barella L, Schumacher C, Baggiolini M, Clark-Lewis I. J Biol Chem. 1993;268:7125–7128. [PubMed] [Google Scholar]
- 51.Jones SA, Dewald B, Clark-Lewis I, Baggiolini M. J Biol Chem. 1997;272:16166–16169. doi: 10.1074/jbc.272.26.16166. [DOI] [PubMed] [Google Scholar]
- 52.Hesselgesser J, Chitnis CE, Miller LH, Yansura DG, Simmons LC, Fairbrother WJ, Kotts C, Wirth C, Gillece-Castro BL, Horuk R. J Biol Chem. 1995;270:11472–11476. doi: 10.1074/jbc.270.19.11472. [DOI] [PubMed] [Google Scholar]
- 53.Walz A, Schmutz P, Mueller C, Schnyder-Candrian S. J Leukocyte Biol. 1997;62:604–611. doi: 10.1002/jlb.62.5.604. [DOI] [PubMed] [Google Scholar]
- 54.Nickoloff BJ, Mitra RS, Varani J, Dixit VM, Polverini PJ. Am J Pathol. 1994;144:820–828. [PMC free article] [PubMed] [Google Scholar]