Abstract
Ubiquitin is a small modifier protein that is conjugated to substrates to target them for degradation. Recently, a surprising number of ubiquitin-like proteins have been identified that also can be attached to proteins. Herein, we identify two molecular functions for the posttranslational protein modifier from Saccharomyces cerevisiae, Urm1p. Simultaneous loss of Urm1p and Cla4p, a p21-activated kinase that functions in budding, is lethal. This result suggests a role for the urmylation pathway in budding. Furthermore, loss of the urmylation pathway causes defects in invasive growth and confers sensitivity to rapamycin. Our results indicate that the sensitivity to rapamycin is due to a genetic interaction with the TOR pathway, which is important for regulation of cell growth in response to nutrients. We have found that Urm1p can be attached to a number of proteins. Loss of five genes that are also essential in a cla4Δ strain, NCS2, NCS6, ELP2, ELP6, and URE2, affect the level of at least one Urm1p conjugate. Moreover, these five genes have a role in invasive growth and display genetic interactions with the TOR pathway. In summary, our results suggest the urmylation pathway is involved in nutrient sensing and budding.
INTRODUCTION
A principal feature of morphogenesis in eukaryotes is cell polarization, which involves the asymmetric organization of the cytoskeleton and the attendant asymmetric organization of internal membranes and proteins (Drubin and Nelson, 1996). The Rho-GTPase Cdc42p plays a critical role in regulation of polarized cell growth and in the regulation of other morphogenetic events such as cytokinesis (Adams et al., 1990; Johnson and Pringle, 1990; Ziman et al., 1993; Li et al., 1995; Richman and Johnson, 2000). Cdc42p orchestrates these events via activation of effector proteins. For example, activated Cdc42 binds to and regulates Gic1p and Gic2p, which are important for the polarization of the actin cytoskeleton (Brown et al., 1997; Chen et al., 1997). The p21-activated kinases (PAKs) Ste20p and Cla4p (Cvrckova et al., 1995; Peter et al., 1996; Leberer et al., 1997) are two other Cdc42p effectors. Both Ste20p and Cla4p have poorly characterized roles in polarizing the actin cytoskeleton and in septin function but together these two PAKs are required for viability (Cvrckova et al., 1995).
To gain insight into the physiological events that have been perturbed in a ste20Δ cla4Δ mutant, we used a synthetic lethal mutant screen to identify additional genes that are required in the absence of Cla4p. This effort led to the identification of NCS3 (Needs CLA4 to Survive; identical to UBA4) and a number of other genes as well (Mitchell and Sprague, 2001; Goehring et al., 2003). UBA4 encodes a protein that has 47% sequence similarity to Uba1p (amino acids 416-574), an ubiquitin-activating enzyme (E1). Uba1p forms a thioester linkage between a cysteine within Uba1p and the C-terminal glycine of ubiquitin. Ubiquitin is then transferred to another carrier protein (E2) with the formation of a new thioester linkage and eventually is covalently attached to proteins targeted for destruction. Recently, Uba4p was shown to function in a novel ubiquitin-like conjugation pathway as a protein activation enzyme (E1) of ubiquitin related modifier (Urm1p). In parallel with the ubiquitin system, Urm1p forms a thioester bond with Uba4p (Furukawa et al., 2000). This interaction depends on the carboxy-terminal glycine residue of Urm1p and a cysteine residue in Uba4p (Furukawa et al., 2000).
Ubiquitin-like proteins (Ubl) have recently been discovered in most eukaryotes, suggesting that the covalent attachment of these Ubls to target proteins is a common posttranslational modification (Hochstrasser, 2000). In contrast to ubiquitin, most of these Ubls don't seem to promote proteolysis. For example, small-ubiquitin-related-modifier (SUMO1) is required for the localization of Ran GTPase activating protein (RanGAP1) to the nuclear pore (Mahajan et al., 1997, 1998). Similarly, although related-to-ubiquitin (Rub1p) conjugation to the ubiquitin-ligase (Skp1p/cullin-1/F-box protein) may regulate the ubiquitin conjugation pathways (Lammer et al., 1998; Liakopoulos et al., 1998; Kawakami et al., 2001), this conjugation does not promote proteolysis directly. Although the target(s) or function(s) for the urmylation pathway is unknown, Urm1p seems to have a human counterpart and is suggested to fulfill a basic function in eukaryotic cells (Furukawa et al., 2000).
In this study, we show that the Urm1p-conjugation system in Saccharomyces cerevisiae plays a role in budding and haploid invasive growth. Both Uba4p and Urm1p are required in cells lacking CLA4. In addition, loss of either UBA4 or URM1 leads to a defect in invasion. Using an antibody to Urm1p, we identified several potential urmylated targets. Moreover, we found that loss of a subset of proteins, Ncs2p, Ncs6p, Elp6p, and Ure2p, which are essential in cla4Δ cells, affect the levels of at least one potential urmylated target. Loss of Ncs2p, Ncs6p, Elp2p, Elp6p, or Ure2p causes a defect in invasive growth. Ure2p has a known role in pseudohyphal growth in diploids, is a repressor of genes involved in nitrogen and glucose metabolism, and is part of a signaling cascade downstream of the Tor proteins (Cardenas et al., 1999; Hardwick et al., 1999; Kuruvilla et al., 2002). Together, our results suggest that the Urm1p conjugation pathway is functionally affiliated with vegetative and invasive growth and may play a regulatory role in TOR signaling.
MATERIALS AND METHODS
Strains and Growth Conditions
Yeast strains used in this study are listed in Table 1. Haploid MATa nonessential yeast deletion strains were purchased from Research Genetics (Huntsville, AL). Other gene deletions were constructed by PCR (Baudin et al., 1993) by using either the pRS (Sikorski and Hieter, 1989) or pFA6a (Longtine et al., 1998) plasmid series as templates. In all cases, the entire coding region was replaced with the indicated marker and successful replacement was confirmed by PCR and phenotype when applicable. Yeast strains were propagated using standard methods (Rose et al., 1990). Rich yeast media, containing 2% glucose (YPD), and synthetic yeast media containing either 2% glucose (SCD) or 2% galactose (SCG), were prepared as described (Rose et al., 1990). Strains were cured of URA3-containing plasmids by using 5-fluoroorotic acid (5-FOA; Biovectra, CA). Geneticin (Biovectra) selection was performed as previously described (Longtine et al., 1998). Rapamycin (Sigma-Aldrich, St. Louis, MO) was added to the medium from a concentrated stock solution in 90% ethanol/10% Tween 20. Yeast transformations were performed as described previously (Gietz et al., 1995).
Table 1.
Yeast strains used in this study
| Straina | Relevant genotype | Reference |
|---|---|---|
| SY3357 | MATaleu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1-lacZ | Mitchell and Sprague, 2001 |
| SY3358 | MATα leu2-Δ1 ura3-52 lys2-801 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1::FUS1lacZ | Mitchell and Sprague, 2001 |
| SY3362 | SY3357 except cla4Δ::TRP1 (pRS316ADE8CLA4) | Mitchell and Sprague, 2001 |
| SY3380 | SY3357 except cla4Δ::TRP1 (YCpHIS3cla4-75) | Mitchell and Sprague, 2001 |
| SY3764 | SY3357 except cla4Δ::TRP1 ste20Δ::TRP1 (YCpHIS3cla4-75) | Goehring et al., 2003 |
| SY3754 | SY3358 except cla4Δ::TRP1 ste20Δ::TRP1 (pRS316ADE8CLA4) | This study |
| SY3805 | SY3358 except uba4Δ::KanMX6 | This study |
| SY3806 | SY3358 except urm1Δ::KanMX6 | This study |
| SY3807 | SY3357 except cla4Δ::TRP1 uba4Δ::HIS3 (pRS316ADE8CLA4) | This study |
| SY3808 | SY3357 except cla4Δ::TRP1 urm1Δ::KanMX6 (pRS316ADE8CLA4) | This study |
| SY3809 | SY3357 except cla4Δ::TRP1 uba4Δ::KanMX6 (YCpHIS3cla4-75) | This study |
| SY3810 | SY3357 except cla4Δ::TRP1 urm1Δ::KanMX6 (YCpHIS3cla4-75) | This study |
| SY3815 | SY3358 except ncs2Δ::KanMX6 | This study |
| SY3816 | SY3358 except ncs6Δ::KanMX6 | This study |
| SY3817 | SY3358 except elp2Δ::KanMX6 | This study |
| SY3818 | SY3358 except elp6Δ::KanMX6 | This study |
| SY3819 | SY3358 except ure2Δ::KanMX6 | This study |
| SY3820 | SY3357 except cla4Δ::TRP1 KanMX6 PGAL1-YNL119w (pRS316ADE8CLA4) | This study |
| SY3821 | SY3357 except cla4Δ::TRP1 KanMX6 PGAL1-YNL120c (pRS316ADE8CLA4) | This study |
| SY3822 | SY3357 except cla4Δ::TRP1 ncs2Δ::KanMX6 (YCpHIS3cla4-75) | This study |
| SY3823 | SY3357 except cla4Δ::TRP1 ncs6Δ::KanMX6 (YCpHIS3cla4-75) | This study |
| SY3824 | SY3357 except cla4Δ::TRP1 elp6Δ::KanMX6 (YCpHIS3cla4-75) | This study |
| SY3825 | SY3357 except cla4Δ::TRP1 ure2Δ::KanMX6 (YCpHIS3cla4-75) | This study |
| HYL333 | MATα ura3-52 | G. Fink |
| HYL334 | MATaura3-52 | G. Fink |
| SY3826 | HYL333 except his3Δura3 | This study |
| SY3827 | SY3826 except uba4Δ::KanMX6 | This study |
| SY3828 | SY3826 except urm1Δ::KanMX6 | This study |
| SY3829 | SY3826 except ncs2Δ::KanMX6 | This study |
| SY3830 | SY3826 except ncs6Δ::KanMX6 | This study |
| SY3831 | SY3826 except elp2Δ::KanMX6 | This study |
| SY3832 | SY3826 except elp6Δ::KanMX6 | This study |
| SY3833 | SY3826 except ure2Δ::KanMX6 | This study |
| SY3834 | HYL333 except ste20Δ::URA3 | This study |
| SY4148 | HYL333/HYL334 except leu2ΔURA3 | This study |
| SY4149 | SY4148 except uba4Δ::ura3/uba4Δ::ura3 | This study |
| SY3835 | BY4741 except rrd1Δ::KanMX6 | This study |
| SY3836 | BY4741 except gln3Δ::KanMX6 | This study |
| SY3837 | HYL333 except ste20Δ:: URA3 urm1Δ::ura3 | This study |
| SY3838 | HYL333 except ste20Δ:: ura3 uba4Δ::URA3 | This study |
| BY4741 | MATahis3Δ1 leu2Δ0 metl5Δ0 ura3Δ0 | Research Genetics |
| SY3839 | BY4741 urm1Δ::KanMX4 | Research Genetics |
| SY3840 | BY4741 uba4Δ::KanMX4 | Research Genetics |
| SY3841 | BY4741 ncs2Δ::KanMX4 | Research Genetics |
| SY3842 | BY4741 ncs6Δ::KanMX4 | Research Genetics |
| SY3843 | BY4741 elp2Δ::KanMX4 | Research Genetics |
| SY3844 | BY4741 elp6Δ::KanMX4 | Research Genetics |
| SY3845 | BY4741 ure2Δ::KanMX4 | Research Genetics |
| SY4150 | SY3845 except gln3Δ::NatMX4 | Research Genetics |
| SY4151 | BY4741 except gln3Δ::KanMX4 | Research Genetics |
SY3357 and SY3358 are derivatives S288C
b Strains containing the designation ura3 were made URA3 at the designated chromosomal locus and then ura3 by selection on 5-FOA as described in MATERIALS AND METHODS.
Identification of NCS2
Previously, we described NCS2 as either of two overlapping open reading frames (ORFs), YNL119w/YNL120c. To determine whether loss of YNL119w or YNL120c was lethal in combination with a cla4Δ mutation, the promoter for each gene was replaced separately with a GAL1 promoter in strain SY3358. The GAL1 promoter was amplified by PCR by using the pFA6a-kanMX6-PGAL1 (Longtine et al., 1998) plasmid as a template. Growth of the resulting strains was tested on SCD or SCG supplemented with 5-FOA.
Plasmids
The plasmids used in this study are shown in Table 2. Enzymes used for DNA manipulations were purchased from New England Biolabs (Beverly, MA), or Boehringer Mannheim Biochemicals (Indianapolis, IN). Oligonucleotides were synthesized by Keystone Laboratories (Camarillo, CA). Bacterial transformations and DNA preparations were performed by standard methods (Sambrook et al., 1989). The PGAL1 pRS315 plasmid was created using homologous recombination to replace the URA3 gene of PGAL1 pRS316 with LEU2 (Ma et al., 1987; Baudin et al., 1993). The plasmid for PGAL1-driven expression of Urm1p was produced by inserting polymerase chain reaction (PCR)-derived fragments from yeast genomic URM1 DNA by using Taq polymerase (Promega, Madison, WI) and the oligonucleotides 5′-GCCCGGGATCCGATGGTAAACGTGAAAGTGGAG-3′ (incorporating a BamHI site shown in bold type) and 5′-CTCGAGTGCGGCCGCCTGCTATTTTTAACCACCATG-3′ (incorporating a NotI site shown in bold type; sequence corresponding to URM1 is underlined). This 300-base pair PCR product was cloned into BamHI- and NotI-digested PGAL1pRS315 (pSL2775), thus creating PGAL1-pRS315 URM1. The PGAL1-driven URM1 construct that bears the N-terminal extension MTSHHHHHHMHDYKDDDDKMGS containing His6- and FLAG-tags was generated by inserting PCR-derived fragments into BamHI- and NotI-digested PGAL1 pRS315 (pSL2775). The His6-FLAG URM1 fragments were created by a two-step PCR. The first round of PCR generated FLAG URM1 by using the oligonucleotides 5′-CATGCACGATTACAAGGATGACGACGATAAGATGGGGTCGATGGTAA ACGTGAAAGTGGAG (FLAG tag shown in italics; sequence corresponding to URM1 is underlined) and 5′-CTCGAGTGCGGCCGCCTGCTATT TTTAACCACCATG-3′ (incorporating a NotI site, shown in bold type; sequence corresponding to URM1 is underlined). To generate His6-FLAG URM1, the FLAG URM1 PCR product was used as a template in a reaction with the oligonucleotides 5′-GCCCGGGATCCGCATATGA CCTCGCATCATCACCACCATCACATGCACGATTACAAGGATGAC (incorporating a BamHI site and a NdeI site, both shown in bold type; His6 tag, shown in italics) and 5′-CTCGAGTGCGGCCGCCTGCTATTTTTAACCACCATG-3′ (incorporating a NotI site, shown in bold type; sequence corresponding to URM1 is underlined). This PCR product was digested with BamHI and NotI and inserted into BamHI and NotI digested PGAL1-PRS315, thus creating PGAL1-PRS315 HFURM1. pET-HFURM1 is a pET-22b-derived plasmid (Novagen, Madison, WI) that was generated by digest of PGAL1-PRS315 HFURM1 with NdeI and NotI and followed by insertion into pET-22b. pMBP-URM1 is a pMAL-c2 (New England Biolabs) derived plasmid that was created by subcloning the BamHI/NotI digested PCR product generated by oligonucleotides 5′-GCCGGATCCATGGTAAACGTGA AAGTGGAG-3′ (incorporating a BamHI site, shown in bold type) and 5′-GAGCTCTA ACTGCAGGGTAGTACCCGGTAGCCCTTC-3′ (incorporating a PstI site, shown in bold type) into pMAL-c2. The other plasmids used in this study, YCpHIS3cla4-75, pML40, pML41, pRS316ADE8CLA4, and 2 μ URA3 TAP42 (CB2516) have been described previously (Cvrckova et al., 1995; Lorenz and Heitman, 1995; Di Como and Arndt, 1996; Mitchell and Sprague, 2001).
Table 2.
Plasmids used in this study
| Plasmid | Description | Plasmid number | Reference |
|---|---|---|---|
| YCpHIS3cla4-75 | pSL2679 | Cvrckova et al., 1995; Mitchell and Sprague, 2001 | |
| pRS316ADE8CLA4 | pSL2764 | Mitchell and Sprague, 2001 | |
| pML40 | pRS315 TOR2 | CY3801 | Lorenz and Heitman, 1995 |
| pML41 | pRS315 TOR2-1 | CY3802 | Lorenz and Heitman, 1995 |
| CB2516 | 2μ URA3 TAP42 TAP42 | PSL2826 | Di Como and Arndt, 1996 |
| PGAL1 pRS315 | GAL1 promoter in pRS315 | pSL2775 | This study |
| PGAL1 pRS315 URM1 | URM1 in pSL2772 | pSL2776 | This study |
| PGAL1 pRS315 HFURM1 | N-terminal His6-FLAG tagged URM1 in pSL2776 | pSL2777 | This study |
| pET HFURM1 | N-terminal His6-FLAG tagged URM1 in pET22b | pSL2778 | This study |
| pMBP-URM1 | N-terminal MBP tagged URM1 in pMAL-c2 | pSL2779 | This study |
Antibodies
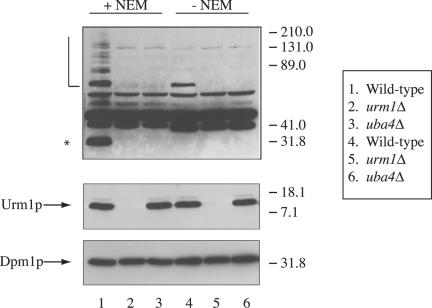
A rabbit polyclonal antibody was raised against His6-FLAG-Urm1p and was affinity purified on a MBP-Urm1p affinity column (Harlow and Lane, 1988). To create the MBP-Urm1p affinity column, 10 ml of a saturated culture of Escherichia coli BL21(DE3) carrying pSL2779 was diluted into 1 liter of LBH supplemented with 0.2% glucose and 50 μg/ml ampicillin, grown at 37°C to an OD600 of 0.6, supplemented with isopropyl-1-thio-B-d-galactopyranoside to 0.3 mM, and incubated at 37°C for another 3 h. Cells were harvested by centrifugation, resuspended in 50 ml of lysis buffer (20 mM Tris pH 7, 200 mM NaCl, 1 mM EDTA), and lysed by French press. Lysates were clarified by centrifugation at 12,000 × g, diluted with 250 ml of lysis buffer, loaded onto 5 ml of amylose resin (New England Biolabs, Beverly, MA), washed with lysis buffer, and maltose binding protein eluted with lysis buffer supplemented with 100 mM maltose. The eluate was dialyzed against 25 mM HEPES pH 7.5 (Sigma-Aldrich) and then 35 mg of MBP-Urm1p was coupled to 1 ml of Affi-gel-15 (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. In a whole cell lysate from a uba4Δ mutant, the affinity-purified anti-Urm1p antibody recognized free unconjugated Urm1p, as well as a many other bands unrelated to Urm1p (our unpublished observations). Therefore, the antibody was further purified by incubation with an acetone pellet of a urm1Δ mutant strain (SY3805) as described previously (Harlow and Lane, 1988) with a few modifications. Before the addition of acetone, the cells were incubated with zymolyase (∼1 mg/1000 ODs; Seikagaku America, Ijamsville, MD) for 20 min at 30°C. After acetone precipitation, the powder was added to the sera at a concentration of 10 mg/ml, incubated overnight at 4°C, and then removed by centrifugation. After the second purification of the anti-Urm1p antibody, the antibody recognized free unconjugated Urm1p, as well as five bands (∼131, 65, 55, 50, and 41 kDa) unrelated to Urm1p in a whole cell lysate of a uba4Δ mutant (Figure 2, lane 2).
Figure 2.
Immunoblot analysis with anti-Urm1p antibodies. Exponential cultures of haploid strains SY3358 (WT), SY3806 (urm1Δ) and SY3805 (uba4Δ), were lysed in the presence of 20 mM NEM (lanes 1-3) or in the absence of NEM (lanes 4 - 6). Lysates were analyzed by SDS-PAGE and immunoblot analysis by using affinity-purified polyclonal anti-Urm1p antibodies and monoclonal antibodies to Dpm1p (to confirm equal protein loading). The band corresponding to unconjugated Urm1p is indicated. A half-open square bracket designates high molecular mass Urm1p modified species, whereas an asterisk denotes the position of a 32-kDa Urm1p-modified species.
A rabbit polyclonal antibody against Cdc3p, a generous gift of J. Pringle (University of North Carolina, Chapel Hill, NC) was affinity purified on a nitrocellulose filter containing MalE-Cdc3p as described previously (Kim et al., 1991).
Microscopy
Indirect immunofluorescence was performed essentially as described previously (Goehring et al., 2003) to visualize the septins by using an α-Cdc3p antibody. For indirect immunofluorescence of Urm1p, cells were grown in YEPD at 30°C to 1 OD600/ml, fixed by the addition of 3% formaldehyde for 1 h, and incubated for 16 h at room temperature in 4% paraformaldehyde, 50 mM KPO4 pH 6.5. Cells were converted to spheroplasts by using zymolyase 100T, permeabilized by treatment with 5% SDS for 5 min, and adhered to poly(l-lysine) multiwell-coated slides. Nonspecific antibody binding was blocked by incubation of the cells in phosphate-buffered saline containing 5 mg/ml bovine serum albumin. All antibodies were diluted in phosphate-buffered saline with 5 mg/ml bovine serum albumin, which was also used for all washes. Antibodies against Urm1p were preabsorbed to yeast proteins (to remove nonspecific binding) by incubation with urm1Δ cells. Antibody incubations were performed at room temperature for 1 h. Alexa (A594)-conjugated goat anti-rabbit antibodies were obtained from Molecular Probes. Images were generated by using an Axioplan 2 fluorescence microscope (Carl Zeiss, Thornwood, NY) fitted with an Orca 100 digital camera (Hamamatsu, Bridgewater, NJ). All assays were performed in triplicate.
Invasive and Pseudohyphal Growth Assays
For the plate-washing invasive growth assay, strains were patched on YPD plates and incubated at 30°C. After 2 d of incubation (unless otherwise indicated), plates were washed under a stream of running water as the surface was gently rubbed with a finger to remove cells not in the agar, and invasive growth was scored (Roberts and Fink, 1994). The single cell invasive growth assay was performed as described previously (Cullen and Sprague, 2000). Cells were grown to stationary phase in synthetic medium, washed twice with water, and plated onto fresh synthetic medium lacking glucose (SC) at a concentration of 104 cells/plate. The plates were incubated at 25°C for 12 h. Cells were scraped from plates by using 4 ml of distilled water, concentrated by centrifugation, resuspended in 20 μl of water and assayed by microscopy for the formation of the filamentous form. All assays were performed in triplicate. For the pseudohypal growth assay cells were plated to SLAD medium, grown for 3 d at 30°C, followed by microscopic examination of pseudohypal formation (Gimeno et al., 1992).
Immunoblot Analysis of Whole Yeast Lysates
Yeast strains were grown to mid-exponential phase (OD600 ∼0.8) in YPD at 30°C. Culture aliquots containing equal numbers of cells, determined by OD600, were collected by centrifugation, washed once in water, and the yeast pellet was frozen. The pellet was resuspended in buffer A (50 mM Tris pH 8.0, 1% NP-40, 50 mM NaCl, 1 mM EDTA) containing a mixture of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml aprotinin [all from Sigma-Aldrich], and 1 tablet of protease inhibitor mixture Complete/25 ml [Roche Diagnostics, Indianapolis, IN]). The lysis buffer also contained 20 mM N-ethylmaleimide (NEM; Sigma-Aldrich) unless otherwise noted. The cells were broken by vortexing with glass beads, and the resulting extracts were centrifuged at 2000 × g to remove glass beads and unbroken cells. SDS- and dithiothreitol-containing loading buffer was added to the extracts, which were loaded onto 10% SDS-PAGE, subjected to electrophoresis, and electroblotted to nitrocellulose. Urm1p was detected using polyclonal antibody to Urm1p, a secondary anti-rabbit horse-radish peroxidase-conjugated antibody (Bio-Rad), and chemiluminescence (Pierce Chemical, Rockford, IL). A monoclonal antibody against Dpm1p (a kind gift from T. Stevens, University of Oregon, Eugene, OR) and a secondary anti-mouse horseradish peroxidase-conjugated antibody (Bio-Rad) were used to confirm that equal amounts of protein were loaded in each lane.
RESULTS
The Urmylation Pathway Is Essential in cla4Δ Cells
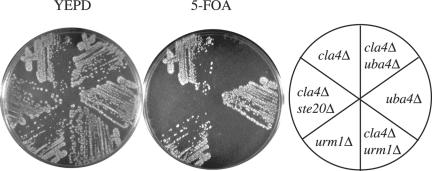
Previously, we identified Uba4p in a screen for proteins that were essential in a cla4Δ mutant (Goehring et al., 2003). Because Urm1p had been shown to form a thioester linkage with Uba4p (Furukawa et al., 2000), we tested whether the loss of Urm1p was essential in a cla4Δ background. Like uba4 mutations, mutations in URM1 were synthetically lethal with cla4Δ (Figure 1).
Figure 1.
Loss of Urm1p and Uba4p is synthetically lethal with cla4Δ. Strains SY3362 (cla4Δ), SY3805 (uba4Δ), SY3806 (urm1Δ), SY3807 (uba4Δ cla4Δ), SY3808 (urm1Δ cla4Δ), and SY3754 (ste20Δ cla4Δ) harboring pRS316ADE8CLA4 were streaked on YEPD plates and incubated at 30°C for 2 d. Plates were then replica-plated to 5-FOA and incubated at 30°C for 3 d.
Identification of Potential Urmylated Products
To detect free Urm1p and possible Urm1p-protein conjugates, we generated polyclonal antibodies to Urm1p (see MATERIALS AND METHODS). One difficulty in identifying targets of the ubiquitin-like modifiers has been that these proteins are rapidly deconjugated by isopepetidases upon cell lysis in nondenaturing buffers (Li and Hochstrasser, 2000). We therefore prepared lysates with and without NEM, an isopeptidase inhibitor. In Western analysis with the polyclonal Urm1p antibody, lysates prepared either in the presence or absence of NEM contained a polypeptide of ∼13 kDa (free Urm1p; Figure 2, lanes 1, 3, 4, and 6) and a polypeptide ∼75 kDa, although the latter was less abundant in the absence of NEM (Figure 2, lane 3). Intriguingly, Urm1p itself may undergo posttranslational modification because it forms as a doublet (Figure 2, lanes 1, 3, 4, and 6). In the presence of NEM, the Urm1p antibody also recognized an ∼32-kDa protein and many other polypeptides of higher molecular weight. Each of these bands presumably represents an urmylated protein because they are not detected in extracts of urm1Δ or uba4Δ cells.
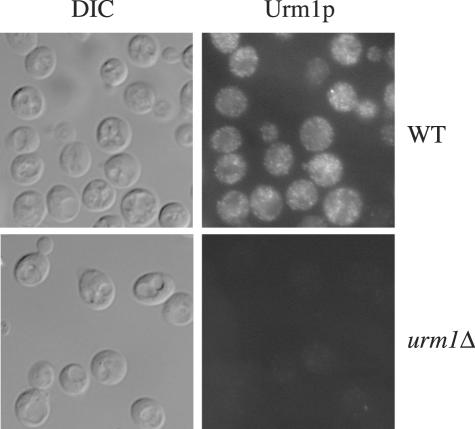
To determine the intracellular localization of Urm1p and the urmylated products, we used the antibody to Urm1p. Urm1p seemed to localize to the cytoplasm and to punctuate spots within the cytoplasm (Figure 3). To gain additional information about the location of Urm1p and the possible Urm1p-protein conjugates, wild-type and uba4Δ cell lysates were fractionated by centrifugation into a 13,000 × g (P13), a 100,000 × g pellet (P100), and a 1000,000 × g supernatant solution (S100). Essentially all of the Urm1p and Urm1p-protein conjugates were found in the cytosol (our unpublished observations).
Figure 3.
Immunolocalization of Urm1p. Immunofluorescence microscopy of strains SY3358 (WT) and SY3806 (urm1Δ) by using a polyclonal antibody against Urm1p. Cells were also visualized by differential interference contrast.
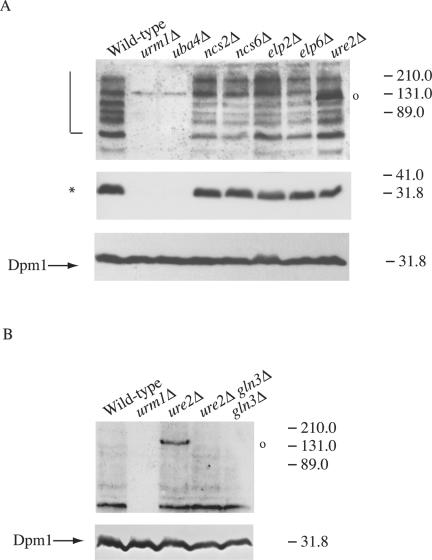
Loss of Ncs2, Ncs6p, Elp2, Elp6p, and Ure2p Affects the Presence of Potential Urmylated Products
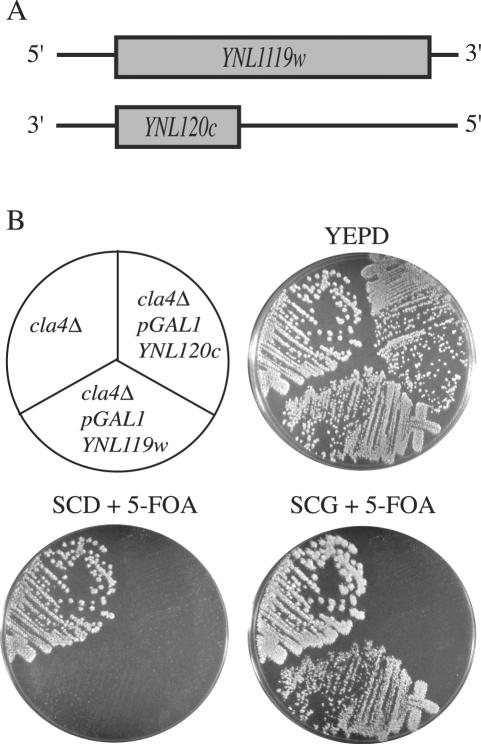
Previously, we identified a large number of proteins that were essential in a cla4Δ mutant (Goehring et al., 2003). To determine whether any of these proteins are components or targets of the urmylation pathway, we performed Western analysis with the anti-Urm1p antibody on strains lacking all 65 genes identified as NCSs. Except for UBA4, loss of the identified NCS genes did not completely eliminate any of the potential urmylated targets (our unpublished observations). However, loss of four genes (NCS2, NCS6, ELP6, and ELP2) did decrease the abundance of the ∼75-, 80-, 89-, and 125-kDa polypeptides (Figure 4A, lanes 4-7, respectively). Elp2p and Elp6p are components of the elongator RNA polymerase II holoenzyme. They stimulate the transcription elongation and are important for normal histone acytelation levels (Fellows et al., 2000; Krogan and Greenblatt, 2001; Winkler et al., 2001, 2002). NCS6 is an open reading frame (ORF), YGL211w, encoding for a protein of unknown function; it has homology to other proteins, also of unknown function (Goehring et al., 2003). NCS2 was identified as one of two overlapping, uncharacterized ORFs, YNL119w or YNL120c (Goehring et al., 2003; Figure 5A). To identify the bona fide NCS2 gene, a GAL1 promoter was inserted in place of the promoter for each ORF and the resulting constructs introduced into a cla4Δ mutant strain. On medium containing 5-FOA and glucose (SCD + 5-FOA), neither PGAL1YNL119w cla4Δ nor PGAL1YNL120c cla4Δ was able to survive. Only GAL1-driven expression of YNL119w allowed for growth on 5-FOA galactose media (Figure 5B), indicating that YNL119w is NCS2.
Figure 4.
Loss of Ncs2p, Ncs6p, Elp2p, Elp6p, and Ure2p alter the pattern of Urm1p-conjugates. (A) Strains SY3358 (wild-type) SY3806 (urm1Δ), SY3805 (uba4Δ), SY3815 (ncs2Δ), SY3816 (ncs6Δ), SY3817 (elp2Δ), SY3818 (elp6Δ), and SY3819 (ure2Δ) were grown to mid-log in YEPD at 30°C. Lysates were analyzed by SDS-PAGE and immunoblot analysis by using affinity-purified polyclonal anti-Urm1p antibodies and monoclonal antibodies to Dpm1p (to confirm equal protein loading). A half-open square bracket designates high molecular mass Urm1p-modified species, whereas an asterisk designates the positions of a 32-kDa Urm1p-modified species. A 131-kDa Urm1p-modified species that occurs in a ure2Δ is indicated with an open circle. (B) Urm1p immunoblot analysis was performed on strains BY4741 (wild-type) SY3839 (urm1Δ), SY3845 (ure2Δ), SY4150 (ure2Δ gln3Δ), and SY4151 (gln3Δ) as in part A with the exception that the immunoblots were exposed for a shorter time.
Figure 5.
Identification of NCS2 as YNL119w. (A) A small yeast ORF of unknown function, YNL120c, overlaps YNL119w on the other DNA strand. (B) Strains SY3362 (cla4Δ), SY3820 (PGAL1-YNL119w cla4Δ), and SY3821 (PGAL1-YNL120c cla4Δ) harboring pRS316ADE8CLA4 were streaked on YEPD plates and incubated at 30°C for 2 d. Plates were then replica-plated to SCD + 5-FOA or SCG + 5-FOA and incubated at 30°C for 3 d.
Loss of a fifth gene, URE2, did not affect the ∼75-, 80-, 89-, and 125-kDa polypeptides, but instead, in this mutant background, a new band recognized by the Urm1p antibody occurred at ∼131 kDa (Figure 4A, lane 8). URE2 encodes for a protein that is part of a signaling cascade downstream of the TOR proteins and is a transcriptional repressor for genes involved in nitrogen and glucose signaling (Cardenas et al., 1999; Hardwick et al., 1999; Kuruvilla et al., 2002). Both Ure2p and the TOR pathway function in diploids to promote the transition from a vegetative form to a pseudohyphal form in response to nutrient limitations. One protein regulated by Ure2p is Gln3p, a GATA-type transcription factor that functions downstream of the TOR kinases to regulate the expression of genes involved in nutrient sensing (Beck and Hall, 1999; Bertram et al., 2000; Rempola et al., 2000). To determine whether Gln3p also affects the presence of urmylated products, we tested whether the 131-kDa band recognized by the Urm1p antibody is present in a gln3Δ ure2Δ mutant. We found that in the absence of Gln3p, the 131-kDa product was not detected (Figure 4B), suggesting that the urmylation of this protein requires Gln3p.
Urmylation Pathway Has a Role in Invasive and Pseudohyphal Growth
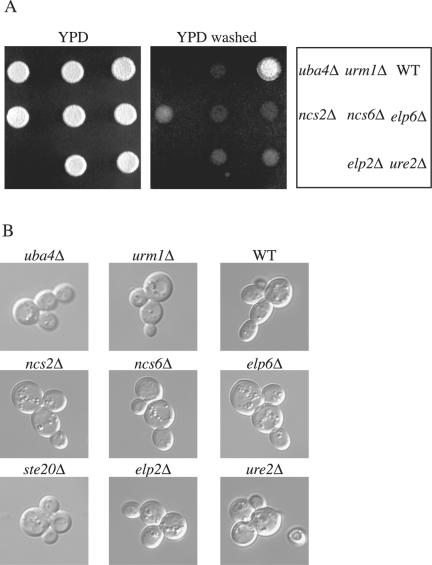
When haploid cells are starved for nutrients, the cells undergo a switch from a vegetative form to a filamentous form. During vegetative growth, the cells are round and bud in an axial manner (buds are formed adjacent to the previous bud site). When nutrients are limiting, the cells become elongated, bud primarily from one pole, and can invade the agar substratum. Because Ste20p was shown to have a role in invasive growth and, like the urmylation pathway components, is essential in a cla4Δ mutant background, we tested to see whether loss of the urmylation pathway components and the genes that affect the potential Urm1p-protein conjugates cause a defect in invasion. Loss of UBA4, URM1, ELP2, ELP6, NCS2, and NCS6 renders cells unable to invade agar under starvation conditions (Figure 6A). To investigate whether loss of these genes causes defects in other aspects of invasive growth (change in bud pattern and formation of elongated cells), we used the single cell invasive growth assay (Cullen and Sprague, 2000). This assay takes advantage of the fact that cells undergo invasive growth in response to glucose depletion. As previously reported, ste20 mutants had a defect in bud pattern (unipolar budding) and in cell elongation by using this assay (Cullen and Sprague, 2000). We found that uba4Δ, urm1Δ, elp2Δ, elp6Δ, ncs2Δ, and ncs6Δ mutants had a defect in cell elongation, but not bud pattern, in response to glucose depletion (Figure 6B). However, ure2Δ mutants not only had a defect in elongation but also had a defect in bud pattern.
Figure 6.
Role of the urmylation pathway in invasive growth. (A) Equal concentrations of strains SY3826 (WT), SY3828 (urm1Δ), SY3827 (uba4Δ), SY3832 (elp6Δ), SY3830 (ncs6Δ), SY3829 (ncs2Δ), SY3833 (ure2Δ), and SY3831 (elp2Δ) were spotted on YEPD plates and incubated at 30°C for 2 d. Plates were photographed, washed, and then photographed again. (B) Equal concentrations of strains SY3826 (WT), SY3828 (urm1Δ), SY3827 (uba4Δ), SY3832 (elp6Δ), SY3830 (ncs6Δ), SY3829 (ncs2Δ), SY3833 (ure2Δ), and SY3831 (elp2Δ) were spread onto SC medium, incubated for 16 h at 25°C, and photographed.
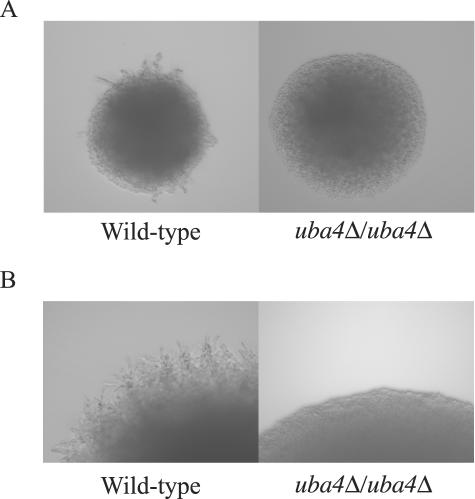
We next asked whether the urmylation pathway has a role in pseudohyphal development by diploid cells. We found that uba4Δ/uba4Δ mutants failed to form pseudohypae under nitrogen limiting conditions (Figure 7A). Hence, we conclude that the urmylation pathway and the other proteins affecting urmylation have a role in both haploid invasive growth and in diploid pseudohyphal development.
Figure 7.
UBA4 is required for diploid pseudohypal development. (A) Wild-type (SY4148) and uba4Δ/uba4Δ (SY4149) diploids were plated to SLAD medium for 3 d at 30°C. Colonies were then visualized with a 20× objective for pseudohypal formation. (B) Wild-type (SY4148) and uba4Δ/uba4Δ (SY4149) diploids containing the TAP42 high copy plasmid (CB2516) were plated to SLAD medium for 3 d at 30 degrees. Colonies were then visualized with a 20× objective for pseudohypal filament formation.
Ste20p and the Urmylation Pathway May Function in Parallel Pathways during Invasive Growth
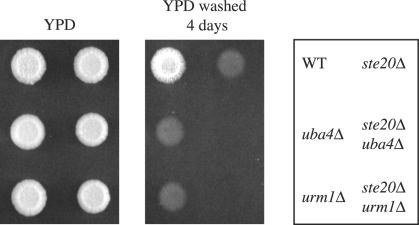
Previously, it has been shown that there are several pathways that contribute to invasive growth, one of which includes Ste20p. As shown above, the urmylation pathway also seems to play a role in invasive growth. By using the single cell assay, we showed that ste20Δ mutants have a defect in both bud pattern and in cell elongation, whereas urm1Δ mutants only have a defect in cell elongation (Figure 6). To determine whether the urmylation pathway is functioning with Ste20p in invasive growth, we compared the phenotype of the double mutants to that of the single mutants. We found ste20Δ urm1Δ and ste20Δ ncs3Δ double mutants have a more severe invasive growth defect than either single mutant (Figure 8). One interpretation of these results is that URM1 and STE20 function independently in the invasive growth program. Alternatively, URM1 and STE20 may have overlapping roles in orchestration of invasive growth, for example, sharing a role in cell elongation but having distinct roles as well.
Figure 8.
Invasive growth phenotype of ste20Δ urm1Δ double mutants. Equal concentrations of strains SY3826 (WT), SY3828 (urm1Δ), SY3827 (uba4Δ), SY3834 (ste20Δ), SY3837 (urm1Δ ste20Δ), and SY3838 (uba4Δ ste20Δ) were spotted on YEPD plates and incubated at 30°C for 4 d. Plates were photographed, washed, and then photographed again.
The Urmylation Pathway Is Implicated in TOR Pathway Signaling
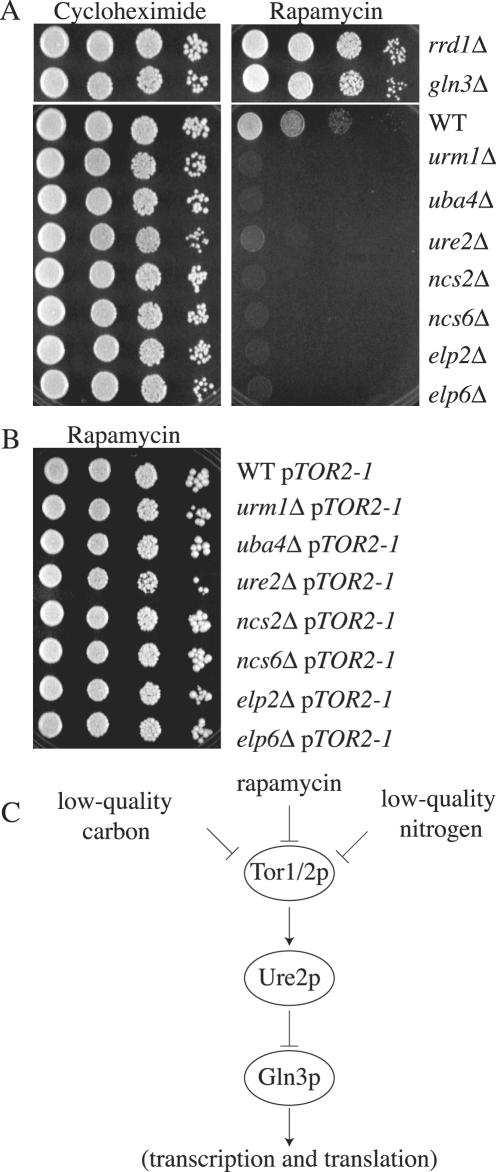
As this work was ongoing, another group completed a large-scale genome-wide screen and identified ure2Δ, ncs6Δ, and uba4Δ mutants as being hypersensitive to rapamycin, a macrocyclic antibiotic (Chan et al., 2000). Rapamycin inhibits Tor1p and Tor2p, ultimately resulting in cellular responses characteristic of nutrient deprivation through a mechanism involving translational and transcriptional arrest (Kunz et al., 1993; Helliwell et al., 1994; Barbet et al., 1996; Komeili et al., 2000) (see model in Figure 9C). Moreover, the TOR proteins have been shown to directly modulate a glucose sensitive signaling pathway (Hardwick et al., 1999), suggesting that the TOR pathway may regulate invasive growth. Although URM1, ELP2, ELP6, and NCS2 were not identified in screens for TOR pathway mutants, they could have been missed because an incomplete set of gene deletion strains was used for the analysis. We therefore tested whether loss of these genes conferred sensitivity to rapamycin and whether the mutations exhibited genetic interaction with TOR pathway mutations. A rrd1Δ/ncs1Δ mutant, which is defective for a protein phosphatase type 2A essential in cla4Δ cells, and a gln3Δ mutant, which is defective for a GATA-type transcription factor regulated by the TOR kinases and by the Ure2p repressor (Beck and Hall, 1999; Bertram et al., 2000; Rempola et al., 2000; Mitchell and Sprague, 2001), served as rapamycin-resistant control strains. A ure2Δ mutant served as a control for rapamycin sensitivity. We found that null mutants lacking URM1, ELP2, ELP6, or NCS2 were unable to grow after 5 d on medium containing rapamycin (Figure 9A). Furthermore, loss of the urmylation pathway, or ELP2, ELP6, NCS2, or NCS6, conferred greater sensitivity to rapamycin than did loss of URE2. None of the mutants was sensitive to cycloheximide, suggesting that sensitivity to rapamycin did not reflect general drug sensitivity and was not due to a reduction of protein synthesis (Figure 9A).
Figure 9.
Interaction of the urmylation pathway with TOR signaling. Strains SY3835 (ncs1Δ), SY3836 (gln3Δ), BY4741 (wild type), SY3839 (urm1Δ), SY3840 (uba4Δ), SY3841 (ncs2Δ), SY3842 (ncs6Δ), SY3843 (elp2Δ), SY3844 (elp6Δ), and SY3845 (ure2Δ) were grown to mid-log in YEPD at 30°C. A serial dilution (1/10) was performed starting with 10,000 cells. Cells were spotted onto either YEPD + 100 nM cycloheximide (left) or YEPD + 25 nM rapamycin (right) and grown 3 d at 30°C. (B) Strains SY3835 (ncs1Δ), SY3836 (gln3Δ), BY4741 (wild type), SY3839 (urm1Δ), SY3840 (uba4Δ), SY3841 (ncs2Δ), SY3842 (ncs6Δ), SY3843 (elp2Δ), SY3844 (elp6Δ), and SY3845 (ure2Δ) carrying pTOR2-1 (pML41) were grown to mid-log in YEPD at 30°C. A serial dilution (1/10) was performed starting with 10,000 cells. Cells were spotted YEPD + 25 nM rapamycin (right) and grown 3 d at 30°C. (C) A model for genetic interaction between TOR and other components in the rapamycin-sensitive pathway.
To verify that the sensitivity of these mutants to rapamycin reflected inactivity of the TOR pathway, we measured the rapamycin sensitivity of urm1Δ, elp2Δ, elp6Δ, and ncs2Δ, mutants carrying a plasmid borne TOR2S1972I (TOR2-1) allele (Lorenz and Heitman, 1995). This allele of TOR2 has previously been shown to confer rapamycin resistance to many sensitive mutants. We found that all the double mutants were no longer sensitive to rapamycin (Figure 9B). In contrast, the urm1Δ, elp2Δ, elp6Δ, or ncs2Δ mutants carrying a wild-type version of TOR2 on a plasmid were still sensitive to rapamycin (our unpublished observations). These results suggest that mutants defective for the urmylation pathway are hypersensitive to rapamycin because TOR pathway signaling is abrogated.
We also investigated the relationship between the TOR and urmylation pathways in diploid pseudohyphal development. Previous studies have shown that overexpression of TAP42, which encodes a protein phosphatase, exacerbates pseudohypal filament formation in wild-type diploids. However, we observed that uba4Δ/uba4Δ homozygous diploids overexpressing TAP42 failed to form filaments (Figure 7B). This result further implicates the urmylation pathway in diploid pseudohyphal growth and suggests that the pathway acts downstream of the Tap42p phosphatase.
Terminal Phenotype of Cells lacking Ncs2p, Ncs6p, Elp6p, and Ure2p in a cla4Δ Mutant Strain
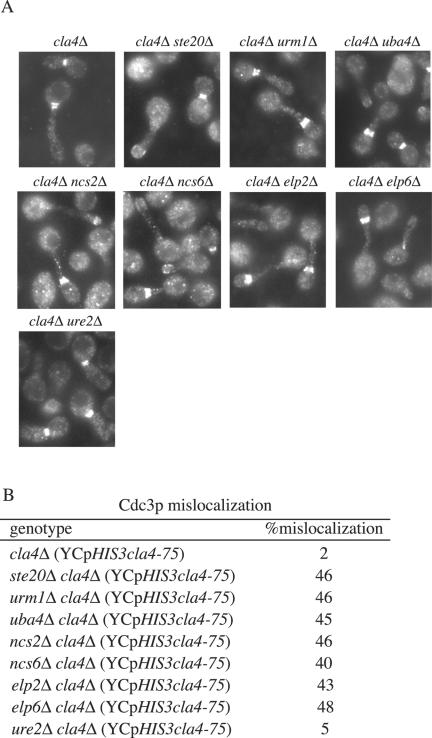
Previously, the ste20Δ cla4Δ YCpHIS3cla4-75 mutants were shown to have a terminal phenotype that included a failure to maintain the septin ring at the bud neck (Cvrckova et al., 1995). We therefore examined septin localization in urm1Δ cla4Δ YCpHIS3cla4-75 and in uba4Δ cla4Δ YCpHIS3cla4-75 double mutants. The phenotype of urm1Δ cla4Δ YCpHIS3cla4-75 and uba4Δ cla4Δ YCpHIS3cla4-75 mutants at the restrictive temperature resembled that of ste20Δ cla4Δ YCpHIS3cla4-75 mutants: the cells were elongated with mislocalized septins (Figure 10). To test further the phenotypic parallels between urm1Δ cla4Δ and cla4Δ strains lacking ELP2, ELP6, NCS2, NCS6, and URE2, we compared the terminal phenotypes of the double mutants carrying a plasmidborne thermosensitive allele of CLA4 (YCpHIS3cla4-75). We found that the loss of ELP2, ELP6, NCS2, or NCS6 in a cla4Δ mutant conferred a terminal phenotype indistinguishable from the loss of the urmylation components in a cla4Δ mutant (Figure 10). However, loss of URE2 in a cla4Δ strain did not result in a defect in septin ring localization. ure2Δ cla4Δ YCpHIS3cla4-75 cells arrest with elongated buds in which the septin ring is localized normally at the motherbud junction.
Figure 10.
Terminal phenotype of cells lacking Ncs2p, Ncs6p, Elp6p, and Ure2p in a cla4Δ mutant strain. Exponential cultures of SY3380 (cla4Δ), SY3810 (urm1Δ cla4Δ), SY3809 (uba4Δ cla4Δ), SY3822 (ncs2Δ cla4Δ), SY3823 (ncs6Δ cla4Δ), SY3824 (elp6Δ cla4Δ), and SY3825 (ure2Δ cla4Δ) carrying YCpHIS3cla4-75 were grown at 25°C in YEPD, shifted to 37°C for 4 h, fixed, and stained for Cdc3p. (B) Quantitation of Cdc3p mislocalization. For each strain, 250 cells were counted in three independent experiments.
In previous studies, we showed that loss of Swe1p, a protein tyrosine kinase that regulates Cdc28p, can suppress some facets of the synthetic lethal phenotype exhibited by ncs cla4 double mutants. In particular, swe1Δ suppressed completely the lethality of ncs1Δ cla4Δ double mutants, suppressed weakly the lethal phenotype of uba4Δ cla4Δ, ncs6Δ cla4Δ, and elp2Δ cla4Δ double mutants and did not suppress at all the lethal phenotype of the remaining NCS genes that were tested, including ste20, bud6, and bni1. We have extended this analysis and shown that swe1Δ also suppresses weakly the lethal phenotype of urm1Δ cla4Δ double mutants. Hence, the subgroup of NCS genes that affect the urmylation pattern also share this genetic suppression phenotype.
DISCUSSION
In this study, we identified roles for the urmylation pathway in both vegetative and invasive growth. Components of this novel ubiquitin-like pathway, Ncs3p/Uba4p and Urm1p, are essential in a cla4Δ mutant. The simultaneous loss of Cla4p and urmylation pathway components cause cells to arrest with a severe defect in budding, and we therefore suggest that the urmylation pathway has a role in vegetative growth. Using an antibody to the modifier protein, Urm1p, we identified several potential urmylated targets. Moreover, we found that loss of a subset of proteins that is essential in cla4Δ-Ncs2p, Ncs6p, Elp2p, Elp6p, and Ure2p, affects the level of at least one potential urmylated target. In addition to its role in vegetative growth, we found the urmylation pathway plays a role in both haploid invasive growth and diploid pseudohyphal development. Finally, loss of the urmylation pathway, as well as Ncs2p, Ncs6p, Elp2p, Elp6p, and Ure2p, causes cells to be sensitive to rapamycin suggesting a connection between these proteins and the TOR pathway. These results identify the first process in which urmylation and other proteins that affect the urmylation pathway play a role.
Urmylation of Targets
The identity of the Urm1p conjugates remains to be determined. The pattern of Urm1p conjugates is consistent with Urm1p being attached to at least seven distinct proteins in the molecular mass range of 62-122 kDa. The most prominent band was at ∼32 kDa, suggesting a substrate of ∼19 kDa. A second major band at ∼75 kDa suggests a substrate of ∼62 kDa. We tested deletion mutants of each NCS gene for the presence of these urmylated products. In no case did one of the bands disappear, implying that none of these urmylated proteins corresponds to the product of an NCS gene.
In addition to modifying other proteins, Urm1p might itself be posttranslationally modified. When immunoblot analysis was performed on wild-type lysates, Urm1p occurs as a doublet (Figure 2, lanes 1, 3, 4, and 6), but only a small amount of the higher molecular mass band is detected, suggesting that only a fraction Urm1p is modified. Another ubiquitin-like protein, Apg12p also seems to be modified: Apg12 was shown to be conjugated to phosphatidylethanolamine thorough an amide bond between the C-terminal glycine and the amino group of phosphatidyletanolamine (Ichimura et al., 2000). This lipidation event is mediated by a ubiquitin-like system required for autophagy. However, the Urm1p modification does not does not require Uba4p (Figure 2, lane 3) and hence is not analogous to the Apg12p modification.
The majority of Urm1p seems to be unconjuagted. This observation is similar to ubiquitin, where 20-70% is found unconjugated (Haas and Bright, 1988; Haas et al., 1988) but is in contrast to Smt3p/SUMO where very little protein is unconjugated (Johnson et al., 1997).
Interaction of Elp2p, Elp6p, Ncs2p, Ncs6p, and Ure2p with the Urmylation Pathway
Loss of Elp2p, Elp6p, Ncs2p, and Ncs6p caused a decrease in the amount of the higher molecular mass urmylated products. Presumably this decrease reflects less urmylation rather than a reduction in the amount of these proteins, but this presumption cannot be tested until the identity of the proteins is known. The biochemical functions of Ncs2p and Ncs6p are unknown. Elp2p and Elp6p are components of a RNA polymerase II elongator complex important for the regulated expression of a subset of genes (Krogan and Greenblatt, 2001). The reduced urmylation observed in these mutants could be due to a decrease in the synthesis of urmylation enzymes or targets. Alternatively, loss of Elp2p, Elp6p, Ncs2p, and Ncs6p could affect the localization of the enzymes or targets. It is tempting to suggest that the loss of these genes is synthetically lethal with cla4Δ because of the decrease in abundance of the Urm1p conjugates. Identification of these products will help clarify why loss of these genes is lethal in combination with cla4Δ.
The pattern of Urm1p conjugates in an ure2Δ mutant was distinct from that in ncs2Δ, ncs6Δ, elp2Δ, and elp6Δ mutants. First, the abundance of the 32-kDa and 75- to 210-kDa conjugates did not change. More strikingly, a new conjugate (131 kDa) was detected. Ure2p is known to regulate transcription of genes involved in nitrogen and glucose signaling (Cardenas et al., 1999; Hardwick et al., 1999; Kuruvilla et al., 2002). It is possible that this 131-kDa gene product is involved in invasive growth. Perhaps the loss of Ure2p results in misregulation of this gene, which in turn leads to a defect in invasion. In addition to the difference in the pattern of Urm1p conjugates, loss of Ure2p also has different physiological consequences from loss of Elp2, Elp6p, Ncs2p, and Ncs6p. The ure2Δ mutants are defective in both bud pattern (unipolar budding) and in cell elongation during invasive growth, whereas elp2Δ, elp6Δ, ncs2Δ, and ncs6Δ mutants were defective only in elongation. Finally, ure2Δ cla4Δ mutants arrest with a terminal phenotype in which the cells have elongated buds but properly localized septin rings. The elp2Δ cla4Δ, elp6Δ cla4Δ, ncs2Δ cla4Δ, and ncs6Δ cla4Δ mutants arrest as cells with elongated buds and mislocalized septins. Identification of the 131-kDa conjugate will help clarify why Ure2p is essential in a cla4Δ mutant, its role in invasion, and how Ure2p interacts with the urmylation pathway.
Connection between Urmylation, TOR Signaling, Invasive, and Pseudohypal Growth
We have shown that loss of URM1, UBA4, URE2, ELP2, ELP6, NCS2, and NCS6 confers sensitivity to rapamycin. Perhaps these gene products serve as enhancers of TOR signaling or of a cellular process regulated by TOR. In addition, we found the urmylation pathway, as well as URE2, ELP2, ELP6, NCS2, and NCS6, plays a role in starvation response in haploids (invasive growth) and diploids (pseudohyphal growth). Given that glucose depletion results in invasive growth and pseudohyphal growth (Cullen and Sprague, 2000, 2002) and given that the TOR proteins directly modulate a glucose sensitive signaling pathway (Hardwick et al., 1999), perhaps the urmylation pathway links TOR signaling and these morphogenetic transitions.
Implications for Urmylation and TOR Signaling in Mammals
The TOR proteins are conserved from yeast to mammals and play a role in nutrient sensing. The mammalian target of rapamycin, mTOR, has been shown to link growth factor signaling and proteins involved in cell cycle progression (Brown et al., 1994; Chiu et al., 1994). In response to nitrogen availability, TOR regulates transcription and translation. Recent clinical advances suggest rapamycin may be a novel chemotherapy agent for rapamycin-sensitive tumors such as glioblastoma and prostate carcinoma (Shi et al., 1995; Hosoi et al., 1999; Louro et al., 1999). Moreover, addition of rapamycin has been shown to impair TOR-dependent cell proliferation in leukemic cells (Iiboshi et al., 1999). Intriguingly, based on homology, there seems to be a functional counterpart to Urm1p in mammals. Perhaps in mammals the urmylation pathway also functions in nutrient sensing. Clearly, much still remains to be learned about the connection between the urmylation pathway and the TOR regulation of nutrient signaling, but studies in yeast could provide insights into conserved pathways as targets for therapy.
Acknowledgments
We thank for J. Pringle, J. Heitman, K. Arndt, and T. Stevens for providing plasmids and reagents. Thanks also to Paul Cullen, Hilary Kemp, Megan Keniry, Dave Mitchell, and Greg Smith for helpful comments and suggestions. This work was supported by a research grant (GM-30027 to G.F.S.) and training grants (HD-07348 to D.M.R. and GM-07759 to A.S.G) from the National Institutes of Health.
References
- Adams, A.E., Johnson, D.I., Longnecker, R.M., Sloat, B.F., and Pringle, J.R. (1990). CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111, 131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet, N.C., Schneider, U., Helliwell, S.B., Stansfield, I., Tuite, M.F., and Hall, M.N. (1996). TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7, 25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F., and Cullin, C. (1993). A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T., and Hall, M.N. (1999). The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689-692. [DOI] [PubMed] [Google Scholar]
- Bertram, P.G., Choi, J.H., Carvalho, J., Ai, W., Zeng, C., Chan, T.F., and Zheng, X.F. (2000). Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275, 35727-35733. [DOI] [PubMed] [Google Scholar]
- Brown, E.J., Albers, M.W., Shin, T.B., Ichikawa, K., Keith, C.T., Lane, W.S., and Schreiber, S.L. (1994). A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756-758. [DOI] [PubMed] [Google Scholar]
- Brown, J.L., Jaquenoud, M., Gulli, M.P., Chant, J., and Peter, M. (1997). Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 11, 2972-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas, M.E., Cutler, N.S., Lorenz, M.C., Di Como, C.J., and Heitman, J. (1999). The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13, 3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, T.F., Carvalho, J., Riles, L., and Zheng, X.F. (2000). A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc. Natl. Acad. Sci. USA 97, 13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.C., Kim, Y.J., and Chan, C.S. (1997). The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 11, 2958-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, M.I., Katz, H., and Berlin, V. (1994). RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 91, 12574-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., and Sprague, G.F., Jr. (2000). Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97, 13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, P.J., and Sprague, G.F., Jr. (2002). The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol. Biol. Cell 13, 2990-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova, F., De Virgilio, C., Manser, E., Pringle, J.R., and Nasmyth, K. (1995). Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9, 1817-1830. [DOI] [PubMed] [Google Scholar]
- Di Como, C.J., and Arndt, K.T. (1996). Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10, 1904-1916. [DOI] [PubMed] [Google Scholar]
- Drubin, D.G., and Nelson, W.J. (1996). Origins of cell polarity. Cell 84, 335-344. [DOI] [PubMed] [Google Scholar]
- Fellows, J., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (2000). The Elp2 subunit of elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. J. Biol. Chem. 275, 12896-12899. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., Mizushima, N., Noda, T., and Ohsumi, Y. (2000). A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J. Biol. Chem. 275, 7462-7465. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., Schiestl, R.H., Willems, A.R., and Woods, R.A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355-360. [DOI] [PubMed] [Google Scholar]
- Gimeno, C.J., Ljungdahl, P.O., Styles, C.A., and Fink, G.R. (1992). Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and. RAS. Cell 68, 1077-1090. [DOI] [PubMed] [Google Scholar]
- Goehring, A.S., Mitchell, D.A., Tong, A.H., Keniry, M.E., Boone, C., and Sprague, G.F., Jr. (2003). Synthetic lethal analysis implicates Ste20p, a p21-activated protein kinase, in polarisome activation. Mol. Biol. Cell 14, 1501-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, A.L., and Bright, P.M. (1988). The resolution and characterization of putative ubiquitin carrier protein isozymes from rabbit reticulocytes. J. Biol. Chem. 263, 13258-13267. [PubMed] [Google Scholar]
- Haas, A.L., Bright, P.M., and Jackson, V.E. (1988). Functional diversity among putative E2 isozymes in the mechanism of ubiquitin-histone ligation. J. Biol. Chem. 263, 13268-13275. [PubMed] [Google Scholar]
- Hardwick, J.S., Kuruvilla, F.G., Tong, J.K., Shamji, A.F., and Schreiber, S.L. (1999). Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96, 14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Helliwell, S.B., Wagner, P., Kunz, J., Deuter-Reinhard, M., Henriquez, R., and Hall, M.N. (1994). TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5, 105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, M. (2000). Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2, E153-157. [DOI] [PubMed] [Google Scholar]
- Hosoi, H., Dilling, M.B., Shikata, T., Liu, L.N., Shu, L., Ashmun, R.A., Germain, G.S., Abraham, R.T., and Houghton, P.J. (1999). Rapamycin causes poorly reversible inhibition of mTOR and induces p53-independent apoptosis in human rhabdomyosarcoma cells. Cancer Res. 59, 886-894. [PubMed] [Google Scholar]
- Ichimura, Y., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488-492. [DOI] [PubMed] [Google Scholar]
- Iiboshi, Y., Papst, P.J., Hunger, S.P., and Terada, N. (1999). L-Asparaginase inhibits the rapamycin-targeted signaling pathway. Biochem. Biophys. Res. Commun. 260, 534-539. [DOI] [PubMed] [Google Scholar]
- Johnson, D.I., and Pringle, J.R. (1990). Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111, 143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.S., Schwienhorst, I., Dohmen, R.J., and Blobel, G. (1997). The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, T., et al. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.B., Haarer, B.K., Pringle, J.R. (1991). Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell. Biol. 112, 535-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili, A., Wedaman, K.P., O'Shea, E.K., and Powers, T. (2000). Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151, 863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., and Greenblatt, J.F. (2001). Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 8203-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, J., Henriquez, R., Schneider, U., Deuter-Reinhard, M., Movva, N.R., and Hall, M.N. (1993). Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73, 585-596. [DOI] [PubMed] [Google Scholar]
- Kuruvilla, F.G., Shamji, A.F., Sternson, S.M., Hergenrother, P.J., and Schreiber, S.L. (2002). Dissecting glucose signalling with diversity-oriented synthesis and small-molecule microarrays. Nature 416, 653-657. [DOI] [PubMed] [Google Scholar]
- Lammer, D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer, E., Wu, C., Leeuw, T., Fourest-Lieuvin, A., Segall, J.E., and Thomas, D.Y. (1997). Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16, 83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Zheng, Y., and Drubin, D.G. (1995). Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J. Cell Biol. 128, 599-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (2000). The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell Biol. 20, 2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos, D., Doenges, G., Matuschewski, K., and Jentsch, S. (1998). A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Lorenz, M.C., and Heitman, J. (1995). TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 270, 27531-27537. [DOI] [PubMed] [Google Scholar]
- Louro, I.D., McKie-Bell, P., Gosnell, H., Brindley, B.C., Bucy, R.P., and Ruppert, J.M. (1999). The zinc finger protein GLI induces cellular sensitivity to the mTOR inhibitor rapamycin. Cell Growth Differ. 10, 503-516. [PubMed] [Google Scholar]
- Ma, H., Kunes, S., Schatz, P.J., and Botstein, D. (1987). Plasmid construction by homologous recombination in yeast. Gene 58, 201-216. [DOI] [PubMed] [Google Scholar]
- Mahajan, R., Delphin, C., Guan, T., Gerace, L., and Melchior, F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97-107. [DOI] [PubMed] [Google Scholar]
- Mahajan, R., Gerace, L., and Melchior, F. (1998). Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140, 259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.A., and Sprague, G.F. (2001). The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 488-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, M., Neiman, A.M., Park, H.O., van Lohuizen, M., and Herskowitz, I. (1996). Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15, 7046-7059. [PMC free article] [PubMed] [Google Scholar]
- Rempola, B., Kaniak, A., Migdalski, A., Rytka, J., Slonimski, P.P., and di Rago, J.P. (2000). Functional analysis of RRD1 (YIL153w) and RRD2 (YPL152w), which encode two putative activators of the phosphotyrosyl phosphatase activity of PP2A in Saccharomyces cerevisiae. Mol. Gen. Genet. 262, 1081-1092. [DOI] [PubMed] [Google Scholar]
- Richman, T.J., and Johnson, D.I. (2000). Saccharomyces cerevisiae Cdc42p GTPase is involved in preventing the recurrence of bud emergence during the cell cycle. Mol. Cell Biol. 20, 8548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R.L., and Fink, G.R. (1994). Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8, 2974-2985. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (ed) (1990). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Shi, Y., Frankel, A., Radvanyi, L.G., Penn, L.Z., Miller, R.G., and Mills, G.B. (1995). Rapamycin enhances apoptosis and increases sensitivity to cisplatin in vitro. Cancer Res. 55, 1982-1988. [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G.S., Kristjuhan, A., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (2002). Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 99, 3517-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G.S., Petrakis, T.G., Ethelberg, S., Tokunaga, M., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (2001). RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276, 32743-32749. [DOI] [PubMed] [Google Scholar]
- Ziman, M., Preuss, D., Mulholland, J., O'Brien, J.M., Botstein, D., and Johnson, D.I. (1993). Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Biol. Cell 4, 1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]