Abstract
In Saccharomyces cerevisiae, Pph21 and Pph22 are the two catalytic subunits of type 2A phosphatase (PP2Ac), and Sit4 is a major form of 2A-like phosphatase. The function of these phosphatases requires their association with different regulatory subunits. In addition to the conventional regulatory subunits, namely, the A and B subunits for Pph21/22 and the Sap proteins for Sit4, these phosphatases have been found to associate with a protein termed Tap42. In this study, we demonstrated that Sit4 and PP2Ac interact with Tap42 via an N-terminal domain that is conserved in all type 2A and 2A-like phosphatases. We found that the Sit4 phosphatase in the sit4-102 strain contains a reverse-of-charge amino acid substitution within its Tap42 binding domain and is defective for formation of the Tap42-Sit4 complex. Our results suggest that the interaction with Tap42 is required for the activity as well as for the essential function of Sit4 and PP2Ac. In addition, we showed that Tap42 is able to interact with two other 2A-like phosphatases, Pph3 and Ppg1.
INTRODUCTION
In S. cerevisiae, there are two forms of PP2A catalytic subunits (PP2Ac) encoded by PPH21 and PPH22 and three 2A-like phosphatases encoded by PPH3, PPG1, and SIT4 (Sneddon et al., 1990; Ronne et al., 1991; Sutton et al., 1991; Posas et al., 1993). The products of PPH21 and PPH22 genes, Pph21 and Pph22, are two highly identical and functionally redundant phosphatases, which exist primarily in cells as a heterotrimeric complex with two regulatory subunits, designated A and B. The TPD3 gene encodes the only A subunit in yeast, which serves as a scaffolding protein to accommodate one catalytic subunit and one of the two alternative B subunits, encoded by CDC55 and RTS1 (Healy et al., 1991; van Zyl et al., 1992; Shu et al., 1997). Depletion of both PPH21 and PPH22 eliminates 80-90% of the cellular PP2A activity and drastically reduces cell growth (Sneddon et al., 1990). Pph3 phosphatase activity is believed to be responsible for sustaining cell growth in the absence of PP2A. Deletion of PPH3 in combination with pph21Δ pph22Δ is lethal (Ronne et al., 1991). Despite this, the Pph3 phosphatase differs from PP2A in its enzymatic properties and subunit composition, and inactivation of Pph3 alone is without any effect on cell growth (Hoffmann et al., 1994). Yeast cells contain a second 2A-like phosphatase catalytic subunit, encoded by SIT4, which performs functions in the cell distinct from those of Pph21/22 (Sutton et al., 1991). Sit4 normally associates with a family of related proteins termed Sap proteins, including Sap155, Sap185, Sap190, and possibly Sap4 (Luke et al., 1996). Sit4 in complex with any of the Sap proteins promotes progression through G1, via regulation of G1 cyclin production (Fernandez-Sarabia et al., 1992; Sutton et al., 1991). In addition, yeast cells have another 2A-related phosphatase encoded by PPG1. The function of Ppg1 is unclear. Inactivation of Ppg1 reduces glycogen accumulation in yeast cells but does not affect cell growth (Posas et al., 1993).
In addition to the association with the conventional regulatory subunits, PP2Ac and Sit4 have been found to form complexes with a phosphatase-associated protein termed Tap42 (Di Como and Arndt, 1996). Formation of the Tap42-phosphatase complex is independent of the other regulatory subunits of the phosphatases. For instance, PP2Ac is able to associate with Tap42 in the absence of Tpd3, and Sit4 is able to bind to Tap42 without the Sap proteins (Di Como and Arndt, 1996). In fact, it has been found that the association with Tap42 prevents PP2Ac from interacting with Tpd3 and Cdc55, suggesting that Tap42 competes with Tpd3 and Cdc55 for PP2Ac binding (Jiang and Broach, 1999).
The interaction of Tap42 with phosphatases is regulated by the Tor proteins, the targets of rapamycin in yeast cells. It has been shown that Tor signaling activity promotes the interaction of Tap42 with phosphatases (Di Como and Arndt, 1996). Tap42 is phosphorylated by the Tor proteins (Jiang and Broach, 1999). The Tor-dependent phosphorylation seems to be important for the interaction of Tap42 with phosphatases, because inactivation of the Tor proteins prevents formation of the Tap42-phosphatase complexes (Di Como and Arndt, 1996). The Tap42-phosphatase complexes have been demonstrated to play a major role in Tor-dependent phosphorylation of many factors downstream of the Tor proteins. Rapid dephosphorylation of these factors is found to accompany the dissociation of Tap42 from phosphatases (Schmidt et al., 1998; Bertram et al., 2000). This observation has led to the suggestion that Tap42 acts as a phosphatase inhibitor (Jacinto et al., 2001). To better understand the role of Tap42 in phosphatase regulation, we defined the domains in the Sit4 and Pph21 phosphatases that are required for their interaction with Tap42. We demonstrated that the ability to interact with Tap42 is required for the activity of the phosphatases to which it associates. In addition, we found that Tap42 interacts with Pph3 and Ppg1, two 2A-like phosphatases in yeast.
MATERIALS AND METHODS
Yeast Strains and Reagents
The strains used in this study are listed in Table 1. Yeast cells were normally grown in YP or synthetic complete (SC) medium lacking appropriate amino acid(s) for selection. All media contained 2% glucose as a carbon source. Standard methods were used for yeast transformation and other manipulations (Guthrie and Fink, 1991). Table 2 lists all the plasmids used in this study. 5-Fluoroorotic acid (5-FOA) (Toronto Research Chemicals, North York, Ontario, Canada) was used at a concentration of 1 mg/ml for the drug sensitivity assay. Rapamycin (Sigma-Aldrich, St. Louis, MO) was stored in 10% Tween 20 and 90% ethanol at a concentration of 1 mg/ml and was added to growth medium to a final concentration of 300 ng/ml. Anti-Tpd3 antibody has been described previously (Jiang and Broach, 1999). Anti-Tap42 and anti-Sit4 antibodies were raised in rabbits against recombinant six-histidine tagged Tap42 and Sit4, respectively. Purified anti-GST antibody and anti-hemagglutinin (HA) (12CA5) antibodies were purchased from Zymed Laboratories (South San Francisco, CA) and were used according to manufacturer's instructions.
Table 1.
Strains used in this study
| All strains are derivatives of W303 (ade2-1 can1-100 his3-11,15 leu2-3, 112 trp1-1 ura3-1 GAL) except those indicated (Thomas and Rothstein, 1989). | ||
|---|---|---|
| Strain | Genotypea | Source |
| Y661 | MATa W.T. (W303-1A) | Laboratory stock |
| Y253b | MATα trp1-289 leu2-2,112 ura2-52 prb1 pep4 | Laboratory stock |
| Y397b | MATa sit4::HIS3 SSD1-vl ura3 leu2-3 his3 trp1-1 | Laboratory stock |
| Y398 | MATa sit4::HIS3 ssd1-d1 [Ycp50-sit4-102 (URA3 CEN)] | Laboratory stock |
| Y421 | MATa sit4::HIS3 ssd1-d1 [pRS416-SIT4 (URA3 CEN)] | Laboratory stock |
| Y531 | MATα pph22::HIS3, pph21::LEU2 | Laboratory stock |
| Y729b | MATa sit4::HIS3 SSD1-vl ura3 leu2-3 his3 trp1-1 [pRS316-GLN3(myc)13 (URA3 CEN)] | This study |
| Y732 | MATa W303-1A [pRS314-(HA)2PPH3 (TRP1 CEN)] | This study |
| Y733 | MATa W303-1A [pRS314-(HA)2PPG1 (TRP1 CEN)] | This study |
Plasmids are indicated in square brackets. Yeast markers carried on each plasmid are listed in parentheses after the plasmid designation
Derivatives of S288C
Table 2.
Plasmids used in this study
| Plasmids | Genes | Vector | Source |
|---|---|---|---|
| pRS314 | CEN TRP1 | Sikorski and Hieter (1989) | |
| pRS416 | CEN URA3 | Sikorski and Hieter (1989) | |
| pRS425 | 2μ LEU2 | Sikorski and Hieter (1989) | |
| pRS416-GLN3(myc)13 | GLN3(myc)13 | CEN URA3 | This study |
| pRS314-SIT4 | SIT4 | CEN TRP1 | This study |
| pRS314-(HA)3PPH21 | (HA)3PPH21 | CEN TRP1 | Jiang and Broach (1999) |
| pRS425-TAP42 | TAP42 | 2μ LEU2 | This study |
| pRS314-(HA)2PPH3 | (HA)2PPH3 | CEN TRP1 | This study |
| pRS314-(HA)2PPG1 | (HA)2PPG1 | CEN TRP1 | This study |
| pJY635 | GST | CEN URA3 | This study |
| Ycp50-sit4-102 | sit4-102 | CEN URA3 | Sutton et al. (1991) |
Plasmid Construction
Cloning and Epitope Tagging of GLN3. The GLN3 gene was amplified from yeast genomic DNA by polymerase chain reaction (PCR) by using highfidelity Taq polymerase (Roche Diagnostics, Indianapolis, IN). The 5′ primer for the PCR was placed ∼500 bp upstream of the initial codon of the gene and the 3′ primer was placed immediately before the stop codon. The PCR product was digested with SacI and BamHI, which were incorporated into the product via the two primers and cloned into pRS416 (CEN URA3) at the corresponding sites. The resulting plasmid was then digested with BamHI and ligated with a 0.8-kb BamHI-BglII DNA fragment containing sequences for 13 tandem copies of Myc epitope followed by an ADH1 termination sequence that was excised from pFA6a-13myc-TRP1 (Longtine et al., 1998). The BamHI site in the plasmid was engineered so that after the ligation, the myc epitope sequence in the insert was fused in-frame with the GLN3 gene. The resulting plasmid [pRS416-GLN3(myc)13] was then used to express the Myc epitope tagged Gln3.
pRS425-TAP42 (2μ LEU2), a high-copy plasmid (2μ) containing a TAP42 gene under control of its native promoter, was constructed by cloning into pRS425 a 2.3-kb PvuII DNA fragment containing the TAP42 gene, which was excised from a previously generated plasmid pRS424-TAP42 (Jiang and Broach, 1999).
Cloning and Epitope Tagging of PPH3 and PPG1. A 1.5-kb DNA fragment containing PPH3 was amplified from yeast genomic DNA by PCR. The PCR product was digested with BamHI and SacII that were introduced via the two PCR primers used for amplification and cloned into pRS314 (CEN TRP1) to generate plasmid pRS314-PPH3. The double HA-tagged PPH3 gene was created by site-directed mutagenesis as described previously (Kunkel et al., 1991). The resultant HA-PPH3 gene contained two tandem copies of HA epitope sequence in place of the second codon of the gene. The HA-PPG1 was generated in a similar way. A 1.6-kb PCR product containing the PPG1 gene was cloned into pRS314 after digestion with XbaI and XhoI, whose sites were introduced into the PCR product via the two primers used for PCR. The HA-PPG1 gene contained a double HA epitope sequence in place of the second codon of the gene.
Mutagenesis of SIT4 and PPH21. Single-strand DNA templates were generated from plasmid pRS314-SIT4 (CEN TRP1) and pRS314-(HA)3PPH21 (CEN TRP1) and used for oligo-directed mutagenesis to replace codons of appropriate amino acids in both phosphatases with that of alanine (Kunkel et al., 1991). The (HA)3PPH21 gene was constructed previously (Jiang and Broach, 1999). The expression of the SIT4 and PPH21 genes was under control of their native promoter.
Glutathione S-Transferase (GST)-Fusion Constructs and Pull-Down Assay
Plasmid pJY635 used for making GST-fusion constructs of SIT4 and PPH21 was derived from pXZ134-L1, a gift from Dr. Martin Schmidt (University of Pittsburgh, Pittsburgh, PA). It is a single copy plasmid (CEN URA3) containing a 5′ portion of the GST gene under control of an ADH1 promoter and ADH1 terminator. The truncated versions of SIT4 or PPH21 were generated by PCR and cloned into the ClaI site of pJY635 at the end of the GST open reading frame. The 5′ end of the PCR generated fusion cassettes was engineered so that the truncated genes were fused in-frame with GST. Plasmids containing the GST-fused sit4 and pph21 constructs were transformed into a protease deficient strain (Y253). The resulting strains were grown to mid-log phase. Cells were collected, washed twice with ice-cold lysis buffer (50 mM Tris-Cl, pH 7.4, 50 mM NaF, 5 mM EDTA, 1 mM dithiothreitol [DTT] and 5% glycerol), resuspended in the same buffer containing protease inhibitor cocktails (Roche Diagnostics), and lysed by vortexing with glass beads. Cell lysates were diluted fourfold with wash buffer containing 50 mM Tris-Cl, pH 7.4, 50 mM NaF, 200 mM NaCl, 1 mM DTT, 1% Triton X-100, and protease inhibitors. Insoluble cell debris was removed by centrifugation at 12,000 × g for 15 min. Protein concentration of the lysates was determined using Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA). An aliquot of supernatant (1 mg of protein) was incubated with 15 μl of glutathione-conjugated agarose beads (Pharmacia, Peapack, NJ) at 4°C for 1 h. After the incubation, beads were washed three times with wash buffer and once with 20 mM Tris-Cl, pH 7.4, and boiled for 5 min in 70 μl of 2× SDS sample buffer. An aliquot of 20 μl of sample was fractionated with a 10% SDS polyacrylamide gel and transferred to nitrocellulose membrane, which was then immunoblotted with either anti-GST (1:2000) or anti-Tap42 antibody (1:1500).
Cross-Linking of Anti-Tap42 Antibody to Protein A Beads
An aliquot of anti-Tap42 serum (1 ml) was diluted 10-fold with phosphate-buffered saline and incubated with 1 ml of protein A-conjugated Sepharose beads (Zymed Laboratories) at 4°C for 2 h with gentle shaking. The beads were washed three times with phosphate-buffered saline buffer and two times with 0.2 M sodium borate, pH 9.0. After the final wash, the beads were resuspended in 10 ml of the sodium borate buffer. The cross-linking reaction was carried out using dimethylpimelimidate (Pierce Chemical, Rockford, IL) as described previously (Harlow and Lane, 1988).
Coimmunoprecipitation
Yeast cells were grown in appropriate medium to early log phase. Cells were collected and lysed as described in the preceding section. To precipitate Tap42, an aliquot of cell extract (1 mg of protein) was incubated with 15 μl of anti-Tap42 antibody conjugated protein A beads at 4°C for 3 h. After the incubation, beads were washed three times with wash buffer (50 mM Tris-Cl, pH 7.4, 50 mM NaF, 200 mM NaCl, 1 mM DTT, 1% Triton X-100), once with 20 mM Tris-Cl, pH 7.4, and boiled for 5 min in 70 μl of 2× SDS sample buffer. An aliquot (20 μl) of sample was fractionated with a 10% SDS polyacrylamide gel and transferred to nitrocellulose membrane, which was then immunoblotted with anti-Tap42 (1:1500 dilution), anti-Sit4 (1:1000 dilution), or anti-HA (12CA5 at 1:1000 dilution) antibody. To precipitate Tpd3, an aliquot of extract containing 1 mg of protein was incubated with 2 μl of anti-Tpd3 antibody at 4°C for 2 h. The Tpd3 protein-antibody complexes were then precipitated with protein A beads during a 90-min incubation at 4°C. Beads were washed three times with wash buffer and once with 20 mM Tris-Cl, pH 7.4, and boiled for 5 min in 70 μl of 2× SDS sample buffer. Western blot analysis was then performed as described above by using anti-Tpd3 (1:2000) and anti-HA antibodies.
RESULTS
Sit4 and PP2A Interact with Tap42 via a Conserved Region Located at Their N Termini
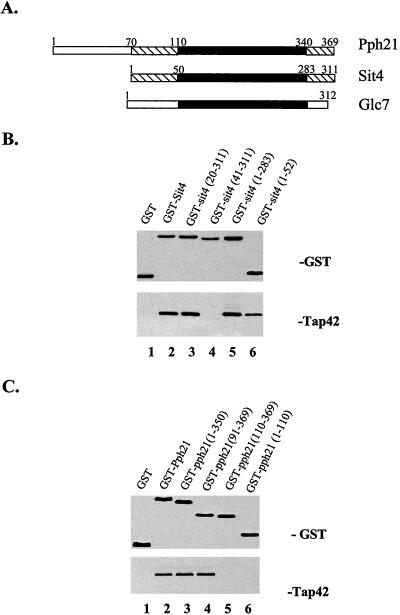
In yeast, two major PP2Ac, Pph21 and Pph22, and a 2A-like phosphatase, Sit4, have been found to associate with Tap42 in a Tor-dependent manner. However, Glc7, the yeast protein phosphatase 1 (PP1), lacks Tap42 binding activity (Di Como and Arndt, 1996). A sequence alignment between Pph21 and Glc7 reveals that these two phosphatases are highly identical in their central catalytic domain but differ significantly at their N- and C-terminal regions. In contrast, both the N and C termini of Pph21, specially the N terminus, are closely related to the same regions in Sit4 (Figure 1A). Thus, it is conceivable that the Tap42 binding domain of Sit4 and Pph21 is located at their N or C terminal region. Accordingly, we made a series of N- and C-terminal deletions of Sit4 and Pph21 and expressed them in yeast as GST fusion proteins in which the mutant phosphatases were fused to the C terminus of the GST protein. The interaction between the GST tagged mutant phosphatases and the endogenous Tap42 protein was then examined using GST pull-down assay.
Figure 1.
An N-terminal domain in Sit4 and Pph21 is required for their interaction with Tap42. (A) Schematic comparison of the sequences of Pph21, Sit4, and Glc7. The conserved regions are marked by hatched bar, divergent regions by open bar, and catalytic domain by solid bar. Yeast cells expressing a variety of GST-sit4 (B) and GST-pph21 (C) fusion constructs were grown to exponential log phase and lysed. The interaction of the expressed GST-fusion proteins with endogenous Tap42 was determined by GST-pull down followed by Western blot analysis of the GST precipitates by using anti-GST and anti-Tap42. Lane 1, GST vector alone; lane 2, GST with full length of Sit4 (B) or of Pph21 (C); and lanes 3-6, GST fused with a variety of truncated Sit4 (B) and Pph21 (C) proteins. The numbers in the parentheses represent the corresponding positions in either Sit4 or Pph21 that were fused with GST.
As shown in Figure 1B, deletion of the entire C-terminal region outside of the catalytic domain of Sit4 did not affect its binding to Tap42; however, deletion of its first 40 amino acids completely abolished the activity (Figure 1B, compare lanes 4 and 5). A smaller deletion, which removed the first 20 amino acids of Sit4, yielded a protein that was as capable of binding Tap42 as the full-length protein (Figure 1B, compare lanes 2 and 3). Furthermore, a GST-fusion construct containing the first 52 amino acids of Sit4 was found to be able to bind to Tap42 (lane 6). Together, these results suggest that the Tap42-interacting domain of Sit4 is located between positions 20-52. Similar results were obtained for Pph21 (Figure 1C), in which case we observed that a truncated Pph21 missing the first 90 amino acids (91-369) was able to interact with Tap42, whereas a protein missing additional 19 amino acids (110-369) was devoid for the activity (Figure 1C, compare lanes 4 and 5). This result suggests that the region between positions 90-110 is critical for Tap42 binding. Because Pph21 contains a unique N-terminal extension of ∼70 amino acids, the position of the Tap42-interacting domain in Pph21 is analogous to that of Sit4. Together, these results demonstrate that PP2Ac and Sit4 interact with Tap42 via a region located at their N terminus. However, we were unable to demonstrate that the region containing the first 110 amino acids of Pph21 was capable of Tap42 binding (Figure 1C, lane 6). It is possible that the unique N-terminal extension in Pph21 interferes with the interaction of the fusion protein with Tap42.
A detailed comparison of the N-terminal region in Sit4 with that in Glc7 reveals a domain between positions 29-46 in Sit4 that differs significantly to the corresponding region in Glc7 (Figure 2A). Furthermore, a database search reveals that the putative Tap42-interacting domain is present in virtually all 2A and 2A-like phosphatases (our unpublished data) but is absent in PP1 from different organisms (Figure 2B). Among the phosphatases containing this domain, many of them, including Pph21, Pph22, and Sit4 in yeast, PP2Ac, PP4, and PP6 in mammalian cells, have been shown to associate with Tap42 or α-4 protein (Di Como and Arndt, 1996; Murata et al., 1997; Chen et al., 1998; Nanahoshi et al., 1998), supporting the notion that this domain mediates the interaction of a phosphatase with Tap42 or a Tap42 counterpart.
Figure 2.
Putative Tap42 interacting domain is conserved in PP2A and 2A-like phosphatases. (A) Sequence alignment between the N-terminal regions of Sit4 and that of yeast PP1, Glc7, by using Bestfit program in the GCG sequence analysis package. (B) Sequence alignment of the putative Tap42 interacting domains in all the phosphatases that have been shown to interact with Tap42 in yeast or its counterpart in mammalian cells. Letters in bold represent invariant amino acids.
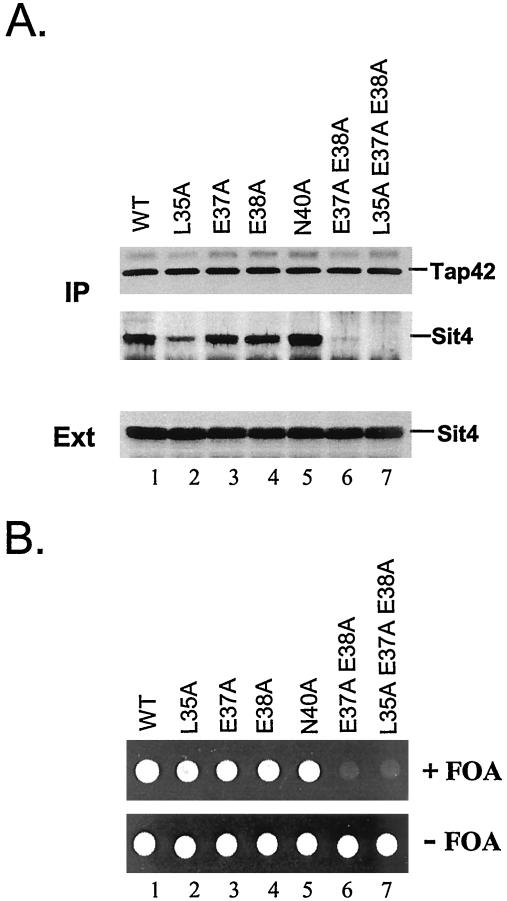
Three invariant amino acids were identified within the putative Tap42 binding domain of Sit4 that exist exclusively in PP2A and 2A-like phosphatases but are absent from PP1 (Figure 2B). We thus determined the role of these residues in Sit4 for its Tap42 binding. Accordingly, we substituted the invariant amino acids in Sit4 (35L, 38E, and 40N) with alanine by site-directed mutagenesis and expressed these mutant proteins in a sit4 deletion strain (sit4Δ) (Y397, a derivative of S288C strain in which SIT4 was not essential because of the presence of the SSD1-v1 allele) (Sutton et al., 1991). The interaction between the mutant Sit4 proteins and the endogenous Tap42 was determined by coimmunoprecipitation. As shown in Figure 3A, a single alanine substitution at position 35 of Sit4 significantly reduced its Tap42 binding (compare lanes 1 and 2), whereas the substitution at position 38 or 40 was without major effect (compare lane 1 with 4 and 5). A 35A 38A double and a 35A 38A 40A triple substitution yielded proteins with a Tap42 binding capacity similar to that of a 35A single substitution (our unpublished data). Because positions 37 and 38 both contain glutamic acid, a negatively charged residue, we further examined the role of 37E for Tap42 binding. We found that although a Sit4 protein containing a single alanine substitution at position 37 was able to interact with Tap42 as effective as the wild-type one, a protein containing double substitution at positions 37 and 38 (Sit437A38A) lost most Tap42 binding activity. Furthermore, a triple substitution at positions 35, 37, and 38 (Sit435A37A38A) virtually abolished the ability of Sit4 for Tap42 binding. The absence of Tap42 binding in the last case was not due to the absence of Sit4 expression, because the amount of the mutant Sit4 protein was comparable with that of wild-type one (Figure 3A, Ext). Together, these findings suggest that 35L and the negatively charged residues at positions 37 and 38 in Sit4 are critical in mediating its interaction with Tap42.
Figure 3.
The interaction with Tap42 is essential for Sit4 function. (A) The mutant Sit4 proteins containing alanine substitutions at different sites in the putative Tap42 interacting domain were expressed from a centromeric-based TRP1 vector (pRS314) in Y397 (sit4Δ). The interaction between the mutant Sit4 proteins and endogenous Tap42 was determined by coimmunoprecipitation by using anti-Tap42 antibody that was conjugated to protein A beads. The presence of Tap42 (A, top) and Sit4 (A, middle) in the precipitates was determined by Western blotting. The amount of the Sit4 proteins in the cell extracts used for coimmunoprecipitation was also examined by Western blotting (A, bottom). (B) The same set of mutant Sit4 proteins was expressed in Y421 [sit4Δ with plasmid pRS316-SIT4 (CEN URA3)] in which SIT4 was essential. The cells were grown in SC- Trp medium to mid-log phase and spotted on either SC- Trp plates (B, bottom) or SC medium containing 5-FOA (top). The plates were photographed after incubated at 23°C for 2 d.
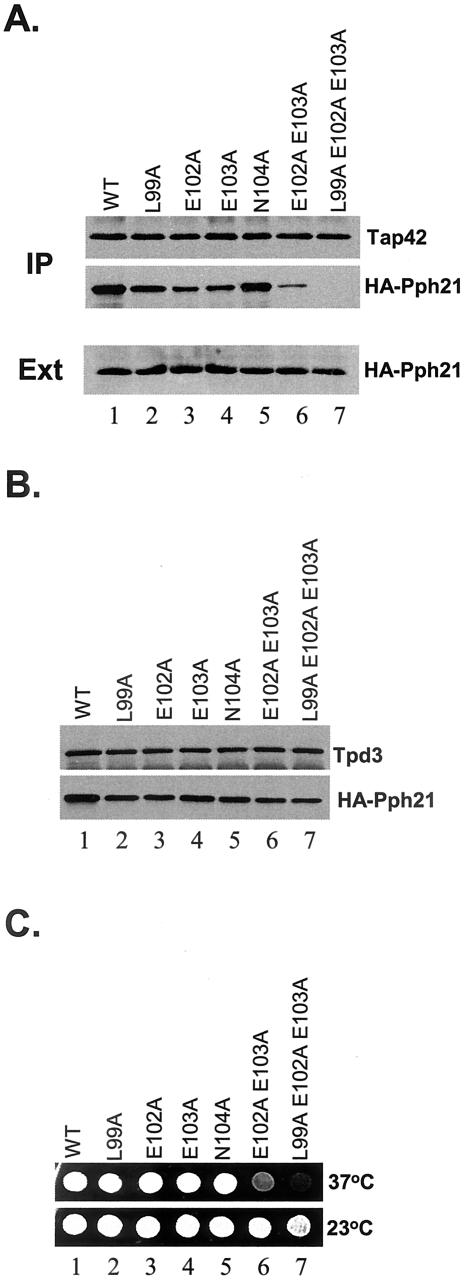
To confirm the results from Sit4, we further examined the role of the equivalent residues, including 99L, 102E, and 104N in Pph21 for its Tap42 binding. In this case, the genes encoding HA epitope-tagged mutant Pph21 proteins were expressed in a pph21Δ pph22Δ strain (Y531). The expression levels for all the mutant Pph21 proteins were found to be comparable with those of wild-type (Figure 4A, Ext). The interaction between the HA tagged Pph21 proteins and endogenous Tap42 was determined by coimmunoprecipitation. The results, as shown in Figure 4A, were similar to those for Sit4 (Figure 3A), which indicated that 99L and the two negatively charges residues, 102E and 103E, were critical for Tap42 binding. It is worth noting, however, that 103E in Pph21 is not at the equivalent position to 37E in Sit4, because the former is C-terminal to the invariant glutamic acid, whereas the latter is N-terminal. This may suggest that the negative charge of the second glutamic acid is more critical than its position.
Figure 4.
Interaction with Tap42 is essential for Pph21 function. The mutant HA-Pph21 proteins containing different alanine substitutions in the Tap42 interacting domain were expressed from a centromeric-based vector (CEN TRP1) in Y531 (pph21Δ pph22Δ). (A) Interaction of the expressed Pph21 proteins with endogenous Tap42 was determined by coimmunoprecipitation by using anti-Tap42 antibody that was conjugated with protein A beads. The presence of HA-Pph21 and Tap42 in the precipitates was assessed by Western blot analysis with anti-Tap42 (top) and anti-HA (middle) antibodies. The expression levels of the mutant HA-Pph21 proteins were determined by Western blotting (bottom). (B) Interaction of the expressed mutant HA-Pph21 proteins with Tpd3 was determined by coimmunoprecipitation by using anti-Tpd3 antibody. The presence of Tpd3 and HA-Pph21 in the precipitates was examined with anti-Tpd3 (top) and anti-HA antibodies, respectively. (C) Same cells used in A were grown to mid-log phase in SC- Trp medium and spotted on plates containing the same medium. The plates were photographed after incubated at either 23°C (bottom) or 37°C (top) for 2 d.
Because Pph21 exists mainly as a heterotrimeric complex containing, in addition to Pph21, Tpd3 and one of the alternative B subunits, Cdc55 or Rts1, we asked whether the mutations in Pph21 affecting its Tap42 binding would also affect its association with Tpd3. Accordingly, we examined the interaction of the mutant Pph21 proteins with Tpd3 in the same cells used above. As shown in Figure 4B, all the mutant Pph21 proteins used in the experiment showed a slight reduction in their binding to Tpd3 in comparison with the wild-type one. However, the mutant proteins containing double (E102A E103A) or triple (L99A E102A E103A) substitution did not show binding defects more severe than those with single substitution, suggesting that these residues do not play a critical role for Tpd3 binding as they do for Tap42 binding.
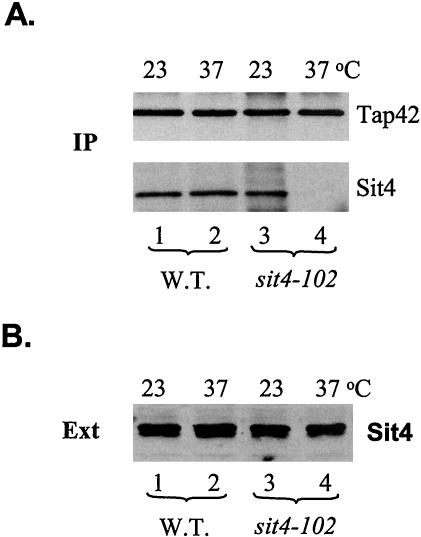
The sit4-102 Alleles Is Deficient for Formation of the Tap42-Sit4 Complex
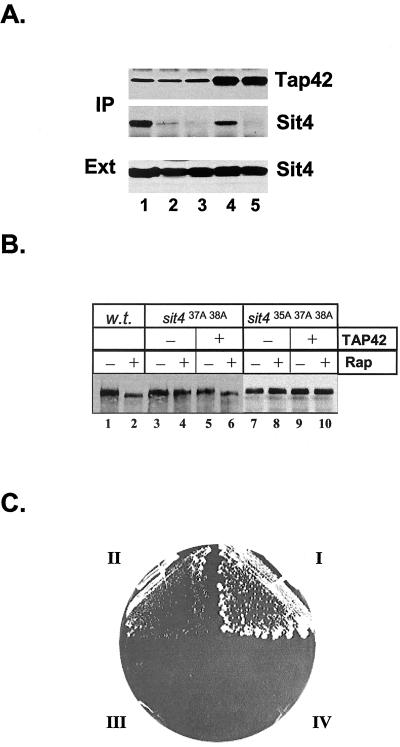
The above-mentioned results demonstrate that the two glutamic acids at positions 37 and 38 are critical for Sit4 to bind to Tap42. Interestingly, Sutton et al. (1991) have isolated a ts- allele of SIT4, sit4-102, in which the mutant Sit4 protein contains a reverse-of-charge amino acid substitution (E to K) at position 38 (Figure 2B). This raised the possibility that the sit4-102 mutant allele, which was used extensively for studying Sit4 function, was defective in the interaction of Sit4 with Tap42. The fact that TAP42 was originally isolated as a high copy suppressor of the sit4-102 mutant supports this possibility (Di Como and Arndt, 1996). Therefore, we examined the interaction between Sit4 and Tap42 in the sit4-102 strain. The mutant cells were grown to early log phase at 23°C and the shifted to 37°C for 2 h. The interaction between Sit4 and Tap42 in cells before and after the shift was examined by coimmunoprecipitation. In this case, we immunoprecipitated Tap42 from cell extracts made from both wild-type and mutant cells and detected the presence of Sit4 in the precipitates. As shown in Figure 5A, we found that the mutant Sit4 protein bound to Tap42 to the same extent as the wild-type Sit4 protein at 23°C (compare lanes 1 and 3). However, it failed to interact with Tap42 after incubation at 37°C for 2 h (compare lanes 2 and 4). The absence of Sit4 in the Tap42 precipitate from the mutant cells grown at 37°C was not due to loss of expression of the Sit4 mutant protein, since we found that the expression levels of Sit4 in cells before and after the shift were essentially the same (Figure 5B). Thus, the sit4-102 strain is specifically defective in the interaction between Sit4 and Tap42.
Figure 5.
Mutant Sit4 protein in the sit4-102 strain is defective for Tap42 interaction. Wild-type (lanes 1 and 2) and the sit4-102 mutant cells (Y398) (lanes 3 and 4) were grown to early log phase at 23°C (lanes 1 and 3) and shifted to 37°C for 2 h (lanes 2 and 4). The cells before and after the shift were lysed. The cell extracts were precipitated with anti-Tap42 antibody conjugated to protein A beads. (A) Presence of Tap42 and Sit4 was determined by Western blotting by using anti-Tap42 (top) and anti-Sit4 antibodies (bottom), respectively. (B) Expression levels of Sit4 in both wild-type (lanes 1 and 2) and sit4-102 cells (lanes 3 and 4) at 23 and 37°C were determined by Western blotting by using anti-Sit4 antibody.
The Ability to Interact with Tap42 Is Required for the Essential Function of Sit4 and Pph21
Because both Sit4 and Pph21 play important roles for cell growth in yeast, we determined whether the interaction with Tap42 was required for their function. To test the ability of the above-mentioned mutant Sit4 phosphatases to support cell growth, genes for these mutant proteins were expressed in a strain (Y421) in which SIT4 was essential (SIT4 is essential in strains containing an ssd1-d allele). This strain contains a chromosomal deletion of SIT4, whose essential function is provided by a SIT4 gene expressed from a URA3 plasmid (sit4Δ with pRS416-SIT4). On transforming the strain with the mutant sit4 genes expressed from a centromeric based plasmid (CEN TRP1), the transformants were tested for growth on plates containing 5-FOA, which is toxic to URA3+ cells. If an introduced mutant sit4 gene were functional, it would be able to replace the wild-type SIT4 gene on the URA3 plasmid and thus render the URA3 plasmid dispensable and confer 5-FOA resistance to the cells expressing it. We found that genes encoding Sit4 proteins with any of the single substitution were able to support cell growth (Figure 3B, lanes 2-5); however, genes encoding Sit4 proteins with multiple substitutions (Sit437A38A and Sit435A37A38A) failed to do so (Figure 3B, lanes 6 and 7). Because the Sit4 proteins with multiple substitutions cannot interact with Tap42, this result suggests that the ability to interact with Tap42 is essential for Sit4 function. Similar results were obtained by the functional analysis of the Pph21 mutant phosphatases. In this case, the mutant pph21 genes were expressed in a pph21Δ pph22Δ strain, which was temperature sensitive in that it grew at 23°C but not at 37°C (our unpublished data). The function of the mutant pph21 genes was tested by their ability to restore the cell growth at the nonpermissive temperature. As shown in Figure 4C, the two mutant genes encoding the Pph21 phosphatases that were unable to interact with Tap42 failed to support cell growth at 37°C (lanes 6 and 7), whereas the rest of the mutant genes did. Together, our findings demonstrate that the ability to interact with Tap42 is essential for Sit4 and Pph21 function.
Formation of the Tap42-Sit4 Complex Is Required for Dephosphorylation of Gln3
Gln3 is a GATA transcription factor required for expressing genes whose products are involved in nitrogen catabolism in yeast (Cooper, 2002). The Tap42-Sit4 complex has been shown to regulate its phosphorylation levels in response to Tor signaling. Inactivation of the Tor signaling pathway induces dephosphorylation of Gln3, which in turn depends on the Sit4 phosphatase (Bertram et al., 2000). To further examine the role of Tap42 in regulation of the Sit4 phosphatase, we asked whether the interaction with Tap42 is required for Sit4-dependent dephosphorylation of Gln3. Accordingly, we expressed the mutant Sit4 proteins that were defective for interaction with Tap42 in a sit4 deletion strain carrying a GLN3(myc)13 plasmid (Y729), and examined the ability of the mutant phosphatases to dephosphorylate Gln3 in response to rapamycin treatment. As shown in Figure 6, we found that rapamycin treatment failed to induce dephosphorylation of Gln3 in the sit4 deletion cells expressing a control vector (compare lanes 7 and 8), which was consistent with the notion that SIT4 is required for Gln3 dephosphorylation (Bertram et al., 2000). Rapamycin-induced dephosphorylation of Gln3 in the sit4 deletion cells was restored by the expression of either the wild-type (compare lanes 1 and 2) or the mutant Sit4 phosphatases that were able to interact with Tap42 (our unpublished data) but not by the expression of the phosphatases that were unable to interact with Tap42 (lanes 3-6). These results demonstrated a straight correlation between the ability of the Sit4 phosphatase to interact with Tap42 and its ability to dephosphorylate Gln3, suggesting that the ability to interact with Tap42 is required for Sit4 activity.
Figure 6.
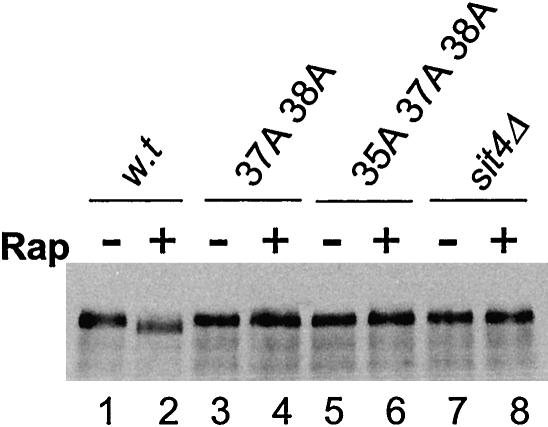
Interaction with Tap42 is required for rapamycin-induced dephosphorylation of Gln3. The sit4Δ cells (Y729) expressing a Myc tagged Gln3 were transformed with a single-copy plasmid (pRS314) containing a wild-type SIT4 (lanes 1 and 2), sit437A38A (lanes 3 and 4), sit435A37A38A (lanes 5 and 6), and vector alone (lanes7 and 8). The cells of the transformants were grown to early log phase at 23°C and split. One-half was treated with rapamycin (300 nM) for 30 min (+), the other half without (-), and lysed. Cell extracts were fractionated with gel electrophoresis and blotted with anti-Myc antibody. Rapamycin-induced dephosphorylation of Gln3 was characterized by the appearance of the fast migrating band (lane 2).
The above-mentioned results could also be explained if the sit4 mutant phosphatases that were unable to interact with Tap42 were catalytically inactive. To rule out this possibility, we further tested whether restoration of the interaction of the mutant phosphatases with Tap42 would enable the mutant Sit4 phosphatases to dephosphorylate Gln3. As indicated in Figure 3A, the Sit437A 38A mutant displayed a residual Tap42 interacting activity despite being functionally inactive. We thus asked whether overexpression of TAP42 would be able to enhance the interaction of the mutant Sit437A 38A phosphatase with Tap42 and whether the enhanced interaction with Tap42 would restore its function. Accordingly, we overexpressed TAP42 together with the sit437A 38A gene in the sit4Δ cells (Y397), in which SIT4 was not essential, and examined the interaction of the mutant phosphatase with Tap42. As shown in Figure 7A, we found that more Sit437A 38A was copurified with Tap42 in cells overexpressing TAP42 than in those expressing a control vector (compare lanes 2 and 4), suggesting that overexpression of TAP42 allowed more mutant Sit4 phosphatase to be associated with Tap42. In contrast, overexpression of TAP42 did not restore the interaction of Sit435A 37A 38A with Tap42 (compare lanes 3 and 5). To determine whether the enhanced interaction with Tap42 restored Sit4 activity, we examined whether rapamycin treatment could induce Gln3 dephosphorylation in the sit4Δ cells (Y729) overexpressing TAP42. As shown in Figure 7B, rapamycin induced dephosphorylation of Gln3 in cells coexpressing TAP42 and sit437A 38A (lanes 5 and 6) but not in cells coexpressing a control vector and sit437A 38A (lanes 3 and 4). This finding, together with that shown in Figure 7A, suggests that the enhanced interaction of Tap42 with Sit437A 38A enables the mutant phosphatase to dephosphorylate Gln3. It also indicates that Sit437A 38A is catalytically active. In contrast, rapamycin failed to induce dephosphorylation of Gln3 in cells coexpressing TAP42 and Sit435A 37A 38A. Given the fact that overexpression of TAP42 is unable to enhance the interaction of Sit435A 37A 38A with Tap42 (Figure 7A), this finding indicates that dephosphorylation of Gln3 requires Sit4 to interact with Tap42. To further demonstrate that restoration of Tap42 interaction reestablish the essential function of Sit437A 38A, we expressed TAP42 in the cells used for the experiment shown in Figure 3 [Y421, sit4Δ +pSIT4(CEN URA3)] and examined 5-FOA sensitivity of the cells. As indicated in Figure 7C, although cells expressing sit437A 38A alone were 5-FOA sensitive (IV), the same cells became 5-FOA resistant when sit437A 38A was coexpressed with TAP42 (II). Conversely, overexpression TAP42 failed to confer 5-FOA resistance to cells expressing sit435A 37A 38A (III). These results, together with those shown in 7A, project a straight correlation between the essential function of Sit4 and its ability to interact with Tap42, indicating that overexpression of Tap42 restores Sit437A 38A function by enhancing its interaction with Tap42 but not by bypassing its function.
Figure 7.
Enhancement in Tap42 interaction restores the function of the mutant Sit4 protein. (A) Overexpression of TAP42 enhances the interaction of sit437A38A with Tap42. The sit4Δ cells (Y729) expressing wild-type SIT4 (lane 1), or sit437A38A (lanes 2 and 4), or sit435A37A38A (lanes 3 and 5) together with either a high copy vector containing TAP42 (lanes 4 and 5) or the vector alone (lanes 1, 2, and 3) were grown to early log phase. The interaction between Sit4 and Tap42 in these cells was determined by coimmunoprecipitation by using anti-Tap42 antibody conjugated protein A beads. The presence of Tap42 (top) and Sit4 (middle) in the precipitates was examined by Western blot analysis. The expression levels of Sit4 in the cells were also determined by Western blot analysis (bottom). (B) Overexpressing Tap42 enables the Sit437A38A phosphatase to dephosphorylate Gln3 in response to rapamycin. sit4Δ cells expressing GLN3(myc)13 (Y729) were transformed with wild-type SIT4 (lanes 1 and 2) or sit437A38A (lanes 3-6) or sit435A37A38A (lanes 7-10) together with a high copy vector (-TAP42) or the same vector containing TAP42 (+TAP42). The cells of the transformants were grown to early log phase at 23°C and treated with (+Rap) or without (-Rap) rapamycin for 30 min. Cells were lysed and the phosphorylation levels of Gln3 in the lysates were determined by Western blotting analysis. (C) Overexpression of TAP42 confers the essential function of SIT4 to the sit437A38A mutant. Y421 (sit4Δ + pRS416-SIT4) cells were cotransformed with two plasmids in the following combinations: I, pRS314-SIT4 and pRS425; II, pRS314-sit437A38A and pRS425-TAP42; III, pRS314-sit435A37A38A and pRS425-TAP42; and IV, pRS314-sit437A38A and pRS425. Transformants were streaked on a plate containing 5-FOA. The plate was photographed after incubation at 30°C for 3 d.
Tap42 Associates with Pph3 and Ppg1
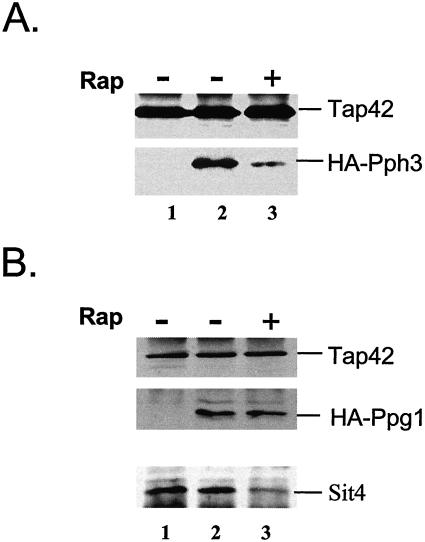
In addition to Sit4, yeast also contains two other type 2A-like protein phosphatases encoded by PPH3 and PPG1 (Ronne et al., 1991; Posas et al., 1993). The subunit composition of these two phosphatases is unknown, despite the fact that PPH3 is required for survival of yeast cells in the absence of PPH21 and PPH22 (Ronne et al., 1991). Because there is no indication that Pph3 is able to form a complex with the A and B subunits of PP2A, Pph3 is unlikely to be the third PP2A catalytic subunit in yeast. However, a sequence comparison reveals that both Pph3 and Ppg1 contain a putative Tap42 binding domain (our unpublished data), indicating that these two phosphatases may associate with Tap42. To confirm this notion, we constructed HA-tagged version of the PPH3 and PPG1 genes, and expressed them individually in wild-type yeast (Y661). The interaction between the epitope tagged phosphatases and Tap42 was then examined by coimmunoprecipitation. As shown in Figure 8A, we found that HA-Pph3 effectively copurified with Tap42 from extracts made from actively growing yeast cells, suggesting that like Sit4 and PP2Ac, the Pph3 phosphatase is also able to interact with Tap42. On the other hand, when the cell extracts were prepared from cells treated with rapamycin, less HA-Pph3 was copurified with Tap42, indicating the interaction of Pph3 with Tap42 is, at least partially, sensitive to rapamycin. Similarly, we found that HA-tagged Ppg1 copurified with Tap42 in extracts prepared from actively growing yeast cells (Figure 8B, lane 2), suggesting the existence of the Tap42-Ppg1 complex in the cells. These results, together with those from previous studies (Di Como and Arndt, 1996; Jiang and Broach, 1999), demonstrate that all the 2A and 2A-related phosphatases in yeast are able to interact with Tap42. However, in the case of Ppg1, it seemed that the interaction of this phosphatase with Tap42 was insensitive to rapamycin treatment, because rapamycin failed to disrupt the interaction (Figure 8B, lane 3). As a control, we further examined the interaction of Sit4 with Tap42 in the same cell extract used for coimmunopurifying Ppg1 and Tap42. We found that rapamycin treatment significantly reduced the interaction between Sit4 and Tap42 (Figure 8B, compare lanes 2 and 3), suggesting that the interaction between Ppg1 and Tap42 was less sensitive to rapamycin than that between Sit4 and Tap42. Intriguingly, in our strain background, we have never observed a complete disassociation of Tap42 from Sit4, Pph21, and Pph22 regardless the concentration of rapamycin used to treat the cells.
Figure 8.
Pph3 and Ppg1 are able to interact with Tap42. Cells expressing either HA-PPH3 (A) or HA-PPG1 (B) were grown to early log phase and treated with rapamycin (300 nM) for 1 h. Cells extracts were made from cells before (Rap-) and after (Rap+) the drug treatment and precipitated with anti-Tap42 antibody conjugated protein A beads. The presence of Tap42, HA-Pph3, and HA-Ppg1 in the precipitates was determined by Western blot analysis by using anti-Tap42 and anti-HA antibodies, respectively. Wild-type cells expressing neither of the tagged genes were also treated in parallel as a control (A and B, lane 1). Western blot analysis was repeated to detect the presence of Sit4 in the same samples used for Ppg1 detection (C).
DISCUSSION
All Type 2A and 2A-like Phosphatases Associate with Tap42 in Yeast
Formation of the Tap42-phosphatase complexes is a critical step in the Tor signaling pathway; Tap42 interacts with phosphatases when the Tor proteins are active and disassociates from phosphatases when the Tor proteins are inhibited (Di Como and Arndt, 1996). In this study, we determined the structural basis for a phosphatase to interact with Tap42. Our results demonstrate that Sit4 and Pph21 interact with Tap42 via a conserved region located at their N termini. Consistent with this notion, Chen et al. (1998) have shown that a fragment containing the first 50 amino acids of PP6, a mammalian phosphatase that is closely related to Sit4, is able to interact with α-4 protein in a two-hybrid assay. In addition, Mann et al. (1993) have found that a chimeric protein containing the N-terminal region of PPV, the Drosophila counterpart of Sit4, fused to the catalytic domain of Drosophila PP1, is able to function as Sit4 in yeast, suggesting that the N-terminal region of PPV contains all the elements essential for converting a heterogeneous phosphatase to act as Sit4, in which Tap42 binding is expected to be included, because the interaction with Tap42 is essential for Sit4 function.
A database search reveals that the Tap42 interacting domain is present in all type 2A and 2A-like phosphatases identified so far (our unpublished data), suggesting that Tap42 is a phosphatase associating protein that is able to associate with all 2A and 2A-like phosphatases. In support of this notion, we demonstrate that the other two 2A-like phosphatases in yeast, Pph3 and Ppg1, are capable of associating with Tap42. This result, together with findings from previous studies that Tap42 associates with Pph21/22 and Sit4, establishes that Tap42 is able to interact with all type 2A and 2A-like phosphatases in yeast. In accordance with this conclusion, studies in mammalian cells have demonstrated that α-4 protein, the mammalian counterpart of Tap42, were able to interact with, in addition to PP2Ac, PP4, and PP6, two 2A-like protein phosphatases (Murata et al., 1997; Chen et al., 1998).
Our results indicate that a glutamic acid residue (38E in Sit4 and 102E in Pph21) within the Tap42 binding domain is critical for a phosphatase to interact with Tap42. Although an alanine substitution at this position does not have a significant effect on the interaction of these phosphatases with Tap42, a reverse-of-charge substitution (E to K) at this position renders a conditional binding between these phosphatases and Tap42 (Figure 5) (Wang and Jiang, 2003). Because the glutamic acid residue is invariant in all type 2A and 2A-like phosphatases identified so far, our observation suggests that the equivalent change in other type 2A and 2A-like phosphatases is likely to yield a protein that displays a thermal sensitive interaction with Tap42 or the counterparts of Tap42 in different organisms, in which case, the mutant phosphatase would provide a valuable tool to dissect the role of the complex formed between the phosphatase and Tap42.
Interaction with Tap42 Is Required for the Essential Function of Sit4 and PP2A
Sit4 and PP2A play pleiotropic roles in controlling yeast cell growth. To perform their function, PP2Ac has to be associated with a scaffolding protein encoded by TPD3 and one of the two regulatory components encoded by either CDC55 or RTS1, and Sit4 has to be associated with the Sap proteins (Healy et al., 1991; van Zyl et al., 1992; Luke et al., 1996; Shu et al., 1997). These regulatory components are believed to play a critical role in defining the substrate specificity as well as the subcellular localization of these phosphatases (Stark, 1996). In contrast, the role of Tap42 in phosphatase regulation is poorly understood despite its being found to associate with phosphatases. Our deletion study of Sit4 and Pph21 demonstrate that the Tap42 interacting domain of these phosphatases is located immediately N-terminally adjacent to their catalytic domain. Structural analysis of protein phosphatase 1 revealed that the equivalent region in PP1 is positioned at the back of the catalytic core, away from the activity center (Goldberg et al., 1995). Because the catalytic domain of PP2A is highly similar to that of PP1, it has been suggested that the core structure of the two types of phosphatases are alike (Goldberg et al., 1995). Therefore, it is conceivable that binding of Tap42 does not block the accessibility of the phosphatase to its substrate. Although we cannot rule out the possibility that Tap42 may contact phosphatases via other domains of the proteins in a way that blocks the catalytic center, in vitro studies showing that the α-4 protein-PP2Ac complex possessed phosphatase activity argue against it (Murata et al., 1997). While these findings are in accordance with a notion that Tap42 acts as a regulatory subunit, many studies in yeast have seen a straight correlation between activation of the Sit4 phosphatase and its disassociation from Tap42, indicating that the nature of the Tap42 interaction is inhibitory for phosphatase activity (Schmidt et al., 1998; Cardenas et al., 1999; Bertram et al., 2000). In this study, by using the mutant Sit4 and Pph21 phosphatases that were defective for Tap42 interaction, we demonstrated that the interaction with Tap42 is required for the essential function of both Sit4 and PP2Ac. Furthermore, we found that the interaction of Sit4 with Tap42 is required for rapamycin-induced dephosphorylation of Gln3. Thus, it is clear that despite being negative on phosphatase activity, the interaction with Tap42 is required for a phosphatase to dephosphorylate its substrate. One way to explain this paradox is to suggest that Tap42 is involved in targeting the phosphatase to its substrates and that dephosphorylation does not occur unless the phosphatase is released from Tap42. In this view, Tap42 acts as a regulatory subunit to bring a phosphatase to its substrate, yet the Tap42 interaction restricts the activity of the phosphatase until the interaction is disrupted. Because the interaction of Tap42 with phosphatase is controlled by the Tor signaling pathway, such a mechanism ensures a rapid dephosphorylation of factors downstream of the Tor pathway in response to inactivation of the Tor proteins. This model is in accordance with the notion that the Tor proteins act as a checkpoint protein to control cell growth in response to nutrient conditions (Schmelzle and Hall, 2000). In this way, alterations in nutrient conditions are sensed by the Tor proteins and an acute response is ensured by a rapid activation of phosphatases via the action of Tap42.
Previous studies of the sit4-102 allele have indicated that the mutant cells are defective for accumulation of the mRNAs of the G1 cyclins, including CLN1, CLN2, and HCS26, at late G1 phase, a process that is required for the cells to progress into S phase (Fernandez-Sarabia et al., 1992). Our finding that the mutant Sit4 phosphatase in the sit4-102 strain is specifically defective for Tap42 interaction suggests that the mutant phenotypes associated with this allele are caused by defects in the Tap42-Sit4 complex, providing the first indication that Tap42 is involved in the G1/S transition of the cell cycle. Further analysis the role of Tap42 and of the Tap42-Sit4 complex in controlling G1 cyclin expression will allow us to test this notion.
Acknowledgments
We thank Drs. Lisa Schneper and Jane Wang for critical reading of this manuscript. We are grateful to Dr. Martin Schmidt for providing plasmids. This work was supported by the Competitive Medical Research Fund from University of Pittsburgh School of Medicine and by American Cancer Society (grant RSG0316901-TBE to Y.J.).
References
- Bertram, P.G., Choi, J.H., Carvalho, J., Ai, W., Zeng, C., Chan, T.F., and Zheng, X.F. (2000). Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J. Biol. Chem. 275, 35727-25733. [DOI] [PubMed] [Google Scholar]
- Cardenas, M.E., Cutler, N.S., Lorenz, M.C., Di Como, C.J., and Heitman, J. (1999). The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13, 3271-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Peterson, R.T., and Schreiber, S.L. (1998). Alpha 4 associates with protein phosphatases 2A, 4, and 6. Biochem. Biophys. Res. Commun. 247, 827-832. [DOI] [PubMed] [Google Scholar]
- Cooper, T.G. (2002). Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26, 223-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Como, C.J., and Arndt, K.T. (1996). Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10, 1904-1916. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sarabia, M.J., Sutton, A., Zhong, T., and Arndt, K.T. (1992). SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2, and HCS26 RNAs during late G1. Genes Dev. 6, 2417-2428. [DOI] [PubMed] [Google Scholar]
- Goldberg, J., Huang, H.B., Kwon, Y.G., Greengard, P., Nairn, A.C., and Kuriyan, J. (1995). Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376, 745-753. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. (1991). Guide to Yeast Genetics and Molecular Biology; San Diego, CA; Academic Press.
- Harlow, E.D., and Lane, D. (1988). Preparing antibody affinity columns. In: Antibodies, A Laboratory Manual, ed. E.D. Harlow and D. Lane; Cold Spring Harbor, NY; Cold Spring Harbor Laboratory Press. 519-552.
- Healy, A.M., Zolnierowicz, S., Stapleton, A.E., Goebl, M., DePaoli-Roach, A.A., and Pringle, J.R. (1991). CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11, 5767-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, R., Jung, S., Ehrmann, M., and Hofer, H.W. (1994). The Saccharomyces cerevisiae gene PPH3 encodes a protein phosphatase with properties different from PPX, PP1 and PP2A. Yeast 10, 567-578. [DOI] [PubMed] [Google Scholar]
- Jacinto, E., Guo, B., Arndt, K.T., Schmelzle, T., and Hall, M.N. (2001). TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8, 1017-1026. [DOI] [PubMed] [Google Scholar]
- Jiang, Y., and Broach, J.R. (1999). Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18, 2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, T.A., Bebenek, K., and McClary, J. (1991). Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204, 125-139. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Luke, M.M., Della Seta, F., Di Como, C.J., Sugimoto, H., Kobayashi, R., and Arndt, K.T. (1996). The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell Biol. 16, 2744-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, D.J., Dombradi, V., and Cohen, P.T. (1993). Drosophila protein phosphatase V functionally complements a SIT4 mutant in Saccharomyces cerevisiae and its amino-terminal region can confer this complementation to a heterologous phosphatase catalytic domain. EMBO J. 12, 4833-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, K., Wu, J., and Brautigan, D.L. (1997). B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 94, 10624-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanahoshi, M., Nishiuma, T., Tsujishita, Y., Hara, K., Inui, S., Sakaguchi, N., and Yonezawa, K. (1998). Regulation of protein phosphatase 2A catalytic activity by alpha4 protein and its yeast homolog Tap42. Biochem. Biophys. Res. Commun. 251, 520-526. [DOI] [PubMed] [Google Scholar]
- Posas, F., Clotet, J., Muns, M.T., Corominas, J., Casamayor, A., and Arino, J. (1993). The gene PPG encodes a novel yeast protein phosphatase involved in glycogen accumulation. J. Biol. Chem. 268, 1349-1354. [PubMed] [Google Scholar]
- Ronne, H., Carlberg, M., Hu, G.Z., and Nehlin, J.O. (1991). Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell Biol. 11, 4876-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle, T., and Hall, M.N. (2000). TOR, a central controller of cell growth. Cell 103, 253-262. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., Beck, T., Koller, A., Kunz, J., and Hall, M.N. (1998). The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17, 6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, Y., Yang, H., Hallberg, E., and Hallberg, R. (1997). Molecular genetic analysis of Rts1p, a B'regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell Biol. 17, 3242-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon, A.A., Cohen, P.T., and Stark, M.J. (1990). Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J. 9, 4339-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, M.R. (1996). Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast 12, 1647-1675. [DOI] [PubMed] [Google Scholar]
- Sutton, A., Immanuel, D., and Arndt, K.T. (1991). The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell Biol. 11, 2133-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B.J., and Rothstein, R. (1989). Elevated recombination rates in transcriptionally active DNA. Cell 56, 619-630. [DOI] [PubMed] [Google Scholar]
- van Zyl, W., Huang, W., Sneddon, A.A., Stark, M., Camier, S., Werner, M., Marck, C., Sentenac, A., and Broach, J.R. (1992). Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol. Cell Biol. 12, 4946-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and Jiang, Y. (2003). The Tap42-protein phosphatase type 2A catalytic subunit complex is required for cell-cycle-dependent distribution of actin in yeast. Mol. Cell Biol. 23, 3116-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]