Abstract
Cellular polarity is a fundamental property of every cell. Due to their extremely fast growth rate (≥1 μm/s) and their highly elongated form, filamentous fungi represent a prime example of polarized growth and are an attractive model for the analysis of fundamental mechanisms underlying cellular polarity. To identify the critical components that contribute to polarized growth, we developed a large-scale genetic screen for the isolation of conditional mutants defective in this process in the model fungus Neurospora crassa. Phenotypic analysis and complementation tests of ca. 950 mutants identified more than 100 complementation groups that define 21 distinct morphological classes. The phenotypes include polarity defects over the whole hypha, more specific defects localized to hyphal tips or subapical regions, and defects in branch formation and growth directionality. To begin converting this mutant collection into meaningful biological information, we identified the defective genes in 45 mutants covering all phenotypic classes. These genes encode novel proteins as well as proteins which 1) regulate the actin or microtubule cytoskeleton, 2) are kinases or components of signal transduction pathways, 3) are part of the secretory pathway, or 4) have functions in cell wall formation or membrane biosynthesis. These findings highlight the dynamic nature of a fungal hypha and establish a molecular model for studies of hyphal growth and polarity.
INTRODUCTION
Determining and maintaining cell polarity and other aspects of proper cell shape determination are fundamental prerequisites for proper development of any organism. A variety of model organisms, each having unique advantages, has been used to study these problems. Tip-growing organisms provide examples of highly polarized growth, thus making filamentous fungi an attractive system to study the mechanisms underlying this process. It is likely that the fundamental principles leading to the organization of the actin cytoskeleton and the initial polarization of the cell are conserved among the yeast Saccharomyces cerevisiae, filamentous fungi, and higher eukaryotes (Hall, 1998; Schmidt and Hall, 1998; Pruyne and Bretscher, 2000; Wendland, 2001). Problems of special interest in filamentous fungi include the mechanisms of the high rate of cell elongation, the way cells initiate the formation of new branches, and the regulation of the spatial relationships of branch points. Studies of such issues may help to elucidate the development of other highly elongated cells such as neurons. In addition, understanding the mechanisms of hyphal growth is likely to help in decreasing the detrimental effects of fungi on plants and animals/humans and in enhancing the ability of fungi to produce important products.
Neurospora crassa has a long history as an excellent model for genetic, cellular, and biochemical research (Davis, 2000; Perkins et al., 2001; Davis and Perkins, 2002). Because of its extremely fast rate of tip growth (≥1 μm/s), N. crassa represents a potent system to study cell polarization. As in higher eukaryotic cells, its microtubule cytoskeleton provides the structural basis for long-distance vectorial transport of secretory vesicles (Steinberg and Schliwa, 1993; Seiler et al., 1997, 1999). The fusion of these vesicles with the apical plasma membrane at the hyphal tips is thought to be coordinated by the Spitzenkörper, a specialized organelle, which is localized at the hyphal apex and has been proposed to serve as a vesicle supply center (Girbardt, 1969; Bartnicki-Garcia et al., 1989). Through its microtubule-dependent movement and positioning, this organelle is thought to determine the shape of the hypha (Bartnicki-Garcia et al., 1995; Riquelme et al., 1998). As hyphae elongate, new tips are generated in subapical regions by branching. The signals that establish the sites of new branches and regulate the spacing of branch points along the hyphae are entirely unknown. Indeed, relatively few genes have been defined as important for hyphal growth in various fungal systems. In addition to the cytoskeleton (Plamann et al., 1994; McGoldrick et al., 1995; Seiler et al., 1997; Wendland and Philippsen, 2001; Steinberg et al., 2001; and references therein), both the cAMP and MAP kinase pathways have been shown to be important for hyphal morphogenesis (reviewed in Madhani and Fink, 1998; Borges-Walmsley and Walmsley, 2000). Still, the connections between signal transduction, organization of the cytoskeleton, polarized secretion, and cell wall formation (which are the basis for cellular morphogenesis) are missing.
Recently, N. crassa became the first filamentous fungus with a completely sequenced genome, making it a prime model for genome-wide experimental approaches. Its entire genome is expected to comprise 10,000-13,000 genes (Schulte et al., 2002; Galagan et al., 2003; http://www-genome.wi.mit.edu/annotation/fungi/neurospora), approximately twice as many as in the yeasts (S. cerevisiae ≈ 5600; S. pombe ≈ 4900), and more than half as many as in Drosophila melanogaster (≈14,000) or Caenorhabditis elegans (≈19,000; C. elegans Sequence Consortium, 1998; Adams et al., 2000; Wood et al., 2001, 2002). About 35% of the predicted gene products lack significant matches to any database entry or have matches only to N. crassa expressed sequence tags, while an additional 25% show similarities only to hypothetical proteins. Clearly, there is a need for systematic genetic approaches to define novel genes, as well as the many previously characterized genes, that are involved in hyphal growth and development. Unfortunately, previous screens for such genes have been of limited size, have led mainly to the identification of proteins involved in primary metabolic processes, or were performed in industrial settings and have not allowed full data access (Osherov and May, 2000a, 2000b; DeBacker et al., 2001; Hamer et al., 2001). To overcome these difficulties, we developed a new visual screen designed specifically for the identification of genes involved in cellular polarity and hyphal morphogenesis. Through the analysis of a large collection of temperature-sensitive mutants, we have established a basis for future studies of the establishment and maintenance of polar growth and the regulation of branch formation in a filamentous fungus.
MATERIALS AND METHODS
Isolation and Phenotypic Analysis of Mutants
2 × 109 freshly harvested conidia from wild-type strain 74-OR23-1A (FGSC 987, available from the Fungal Genetic Stock Center, University of Kansas Medical Center, Kansas City, USA) were UV-irradiated to ∼50% survival, transferred to liquid Vogels minimal medium (Davis and de Serres, 1970) containing 2% sucrose, and incubated at 39°C with shaking for ∼18 h. Under these conditions, viable conidia germinated, and the majority exhibited wild-type growth characterized by rapidly extending and branching hyphae. By filtering the liquid medium through two layers of cheesecloth every 2 h, starting at 10 h after inoculation, these actively growing hyphae were removed, thereby enriching the culture for mutants defective in germination or hyphal growth. Following the filtration enrichment step, the remaining conidia and small germlings were suspended in Vogels minimal agar medium containing 0.05% sucrose and 2% sorbose, plated, and incubated at 25°C to allow survivors of the mutagenesis and filtration to form colonies. (2% sorbose restricts hyphal extension, leading to the formation of compact colonies [Davis and de Serres, 1970; Taft et al., 1991].)
After two days at 25°C, the 15 cm plates, containing 300-400 discrete colonies, were overlaid with 2% sucrose agar medium lacking sorbose and transferred to 39°C. When hyphae from the individual colonies entered the sucrose layer, they were no longer inhibited by sorbose and showed rapid extension. However, mutants with temperature-sensitive defects in hyphal growth could be identified by their abnormal growth upon entering the sucrose-medium overlay. The plates were kept at 39°C for 10-20 h and monitored continuously for hyphal growth mutants. To eliminate mutants with defects in primary metabolism, conidia from each isolated mutant were germinated under restrictive conditions, and only conidia producing abnormal germ tubes or showing isotropic growth were selected. In addition, these mutants were stained with DAPI, and only strains with an increase of nuclear number when germinated at restrictive temperature, as compared with ungerminated conidia, were analyzed further. As N. crassa conidia have 1-5 nuclei per conidium, it was not possible to determine the exact increase in nuclear content. Only mutants with an about twofold increase in nuclear number were included in the collection. In addition to mutants with defects in polar growth, mutants that showed alterations in growth directionality were also isolated using the same protocol. Although these mutants were able to produce germ tubes at restrictive temperature, they showed delayed germination rates or the formation of slow-growing, compact mycelia that allowed escape from the filtration procedure.
All mutants of interest were colony purified at least once. Due to frequent differences in the phenotypes seen during growth on an agar surface or in liquid culture compared with growth when embedded in agar, only the phenotypes of embedded hyphae were considered in the classification. Pictures were taken using an Olympus Stereomicroscope and Kodak Technical Pan film (ASA100) that was scanned for further electronic editing using Adobe Photoshop.
Determination of Complementation Groups
Complementation tests were conducted by forming heterokaryons between pairs of mutants and assessing whether the heterokaryons grew as wild-type or mutant at restrictive temperature on Vogels minimal medium containing 2% sucrose. (Hyphal fusion in a growing mycelium occurs frequently and is a normal part of the life cycle of a N. crassa colony [Davis and de Serres, 1970].) For each complementation test, conidia of two strains were inoculated as a dense suspension separately and together on a plate, incubated overnight at 25°C to allow germination as well as fusion of the two coinoculated strains, and then shifted to restrictive temperature to score for complementation. Heterokaryon incompatibility was not a problem because all mutants were derived from the same parental strain. Because pair-wise tests among all ∼950 mutants were impracticable, we analyzed strains within a morphological subclass first, and then used representative alleles to test for complementation between members of different subclasses. Further tests for allelism within a specific subclass included crosses between mutants and strains carrying linked auxotrophic markers (available from the FGSC) and the use of complementing cosmids to analyze the other mutants for complementation. Cross-es for mapping were done using standard procedures (Davis and de Serres, 1970).
Isolation of Genes
Genes were isolated by identifying cosmid clones from the Orbach/Sachs library (available from the FGSC) that rescued the growth defects of specific mutants (Orbach, 1994). Transformation was carried out as described by Vollmer and Yanofsky (1986). To identify the specific genes responsible for complementation, the complementing cosmids were cut independently with several restriction endonucleases, and the digested DNAs were cotransformed with plasmid pMP6 (containing a hygromycin-resistance selectable marker) into the respective mutants to identify enzymes that cut within and outside of the complementing genes. (N. crassa is efficiently transformed with linear DNA [Plamann et al., 1994].) By determining the end sequences of the DNA in various complementing cosmids, we could determine the entire DNA sequence for each cosmid from the N. crassa genome sequence. Given this information and the pattern of inactivating restriction-enzyme cuts, it was possible to identify the open reading frame that was likely to complement a specific mutant. Confirmation was obtained by using PCR to amplify this segment of genomic DNA from a wild-type strain and from the respective mutant. In all cases, the wild-type fragment complemented the mutant upon transformation, whereas the corresponding fragment from the mutant did not. This approach also verified that we had cloned the gene that harbored the original mutation and not a multicopy suppressor. (In contrast to other filamentous fungi, multicopy suppression has not been described in N. crassa.) Finally, representative alleles of the genes identified by cloning were crossed to wild-type and were found to exhibit a 1:1 segregation ratio, establishing that the observed phenotypes were due to single mutations.
RESULTS
Isolation of Mutants Affected in Hyphal Growth
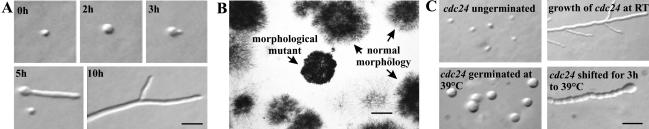
When wild-type N. crassa conidia (asexual spores) are transferred to an appropriate medium, they rehydrate and begin to grow isotropically (Figure 1A; 0-2 h). Growth soon becomes polarized, and a new hyphal tip is generated (Figure 1A; 3 h). Continued polarized growth results in the unidirectional extension of the straight primary hypha into the growth medium (Figure 1A; 5 h). Subsequently, new hyphal tips are generated by branching from subapical compartments (Figure 1A; 10 h). To identify the genes involved in these processes, we isolated temperature-sensitive mutants with defects in morphogenesis. Conditional mutants were isolated because many mutations causing strong defects in morphogenesis were expected to be lethal. Using UV mutagenesis followed by filtration enrichment and a colony-overlay method (see Material and Methods), we visually screened ∼650,000 surviving colonies for temperature-dependent growth defects. Because temperature-sensitive metabolic mutants would presumably also be enriched during the filtration step of this screen (Osherov and May, 2000a, 2000b; DeBacker et al., 2001), and might also resemble morphologenetic mutants during the visual screening (Figure 1B), we performed additional tests, including the visual examination of germinating conidia grown under restrictive conditions and temperature-shift experiments on hyphae (Figure 1C), to identify the bona fide morphologenetic mutants (see MATERIAL AND METHODS). Only mutants displaying one of the following phenotypes were selected: 1) germinating conidia that failed to produce a polar germ tube, but grew in an isotropic manner at high temperature; 2) highly abnormal, bulbous and apolar growing germlings; and 3) strains with defects in growth directionality or defects in branch formation under restrictive conditions. About 900 mutants fulfilled these requirements and were selected for further analysis.
Figure 1.
Wild-type and mutant morphologies. (A) Germination of wild-type conidia. Conidia from a wild-type strain were plated on minimal agar medium, incubated at 30°C, and photographed at various times. (B) Identification of hyphal morphogenesis mutants on the basis of abnormal colony morphology (see MATERIAL AND METHODS for details). (C) Characterization of mutants based on the morphological alterations observed when hyphae or conidia were transferred to restrictive temperature. Bars, 20 μm (A and C) and 500 μm (B).
An important concern was the possibility that certain types of mutants might be excluded by the filtration enrichment technique. For example, rapidly elongating mutants or mutants with branching defects might show sufficient growth to be eliminated during the filtration step. To explore this possibility, conidia were also plated directly after exposure to UV light, and ∼300,000 colonies were screened for interesting phenotypes. An additional ∼70 mutants were isolated that met the requirements mentioned above. Surprisingly, there appeared to be no strong bias of phenotypes due to the filtration procedure. This is probably because many mutants that are able to produce germ tubes at restrictive temperature also show delayed germination rates (e.g., the “growth apolar distal to tip” and “bunches of grapes” mutants; Table 1) or form a slow-growing, compact mycelium that can escape the filtration procedure (e.g., the “compact growth” and “zig-zag growth” mutants). The enrichment of mutants by filtration was estimated to be 5-20 fold depending on the mutant phenotype.
Table 1.
Phenotypic classification of the isolated mutants and analysis of the cloned genes
| Phenotypea
|
Closest homologue inf
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyphae | Conidia | No. of mutants/ complementation groupsb | Cloned genesc | Linkage datad | Complementing cosmide | No. of independent alleles | Animals | S. cerevisiae | S. pombe | |||
| I. Polarity mutants | ||||||||||||
| A. General polarity mutants | ||||||||||||
| Chains of spheres (Figure 2B) | Swollen conidia, rarely bulbous germ tubes | 19/4 | cdc-24 (NCU06067.1)g CDC24 (putative GEF for CDC42) | 51/231 nic-3, Chr VII | X14C6 | 5 | Human (e-11) | Cdc24 (e-32) | Scd1 (e-56) | |||
| bem-1 (NCU06593.1) BEM1 (SH3 domain protein) | 48/364 pyr-1, Chr IV | X5B11 | 4 | Mouse (e-11) | Bem1 (e-53) | Scd2 (e-79) | ||||||
| Irregular chains of spheres (Figure 2C) | Swollen conidia, rarely bulbous germ tubes | 35/3 | mcb* (NCU01166.1) MCB* (regulatory SU of PKA) | 31/291 pyr-6, Chr V | X13C10 | 30 | Mouse (e-55) | Bcy1 (e-67) | SPAC8C9 (e-58) | |||
| Thick, septated, and swollen hyphae (Figure 2D) | Swollen conidia, rarely bulbous germ tubes | 15/6 | ire-1 (NCU02202.1) IRE1 (protein kinase, ER quality control) | Chr VII | X8H8 | 1 | Human Ire2β (e-70) | Ire1 (e-92) | SPAC167 (e-96) | |||
| yif-1 (NCU00624.1) YIF1 (conserved membrane protein) | Chr I | X16D4 | 1 | Human (e-58) | Yif1 (e-25) | SPBC25H2.06 (e-58) | ||||||
| cpc-1* (NCU04050.1) CPC1* (cross pathway control protein) | 15/260 cys-1, Chr VI | X3E6 | 1 | No close homologue | Gen4 (0.007) | No close homologue | ||||||
| Compact growth (heterogeneous group) (Figure 2E) | Swollen conidia or bulbous and compact germlings | 31/14 | sec-5 (NCU07698.1) SEC5 (107-kD exocyst component) | 62/350 pyr-1, Chr IV | X1F8 | 1 | Human (e-12) | Sec5 (e-10) | No close homologue | |||
| Apolar branches (Figure 2, F and G) | Swollen conidia or bulbous germ tubes that lyse | 10/5 | sec-27 (NCU07319.1) SEC27 (β′-COP) | Chr IV | X11D11 | 2 | Human, β′2-COP (0) | Sec27 (0) | β′-COP (0) | |||
| drs-2 (NCU00352.1) DRS2 (Ca2+/P-lipid transporting ATPase) | X14E7 | 2 | Human, ATPase (0) | Drs2 (0) | SPBC887 (0) | |||||||
| gpi-3 (NCU09757.1) GPI3 (GPI anchor biosynthetic protein) | X17F10 | 3 | Mouse, PIG-A (e-108) | Gpi3 (e-128) | GPI biosynthetic protein (e-156) | |||||||
| Highly branched (Figure 2H) | Swollen conidia or bulbous germ tubes that often lyse | 18/8 | stt-4 (NCU09367.1) STT4 (PI-P4-kinase) | Chr I | X3F1 | 2 | Rat, PI-P4-kinase (e-140) | Stt4 (0) | SPBC577 (0) | |||
| mss-4 (NCU02295.1) MSS4 (PI4-P5-kinase) | 41/268 arg-10, Chr VII | X13G3 | 5 | Human, PI4-P5-kinase (e-50) | Mss4 (e-107) | Its1 (e-139) | ||||||
| alg-1 (NCU07261.1) ALG1 (β-1,4 mannosyltransferase) | Chr IV | X6H2 | 1 | D. melanogaster, mannosyltransferase (e-63) | Alg1 (e-58) | SPAC23C4 (e-67) | ||||||
| cop-1 (NCU01002.1) COP1 (ε-COP) | Chr V | X3E4 | 1 | Human ε-COP (e-20) | Sec28 (0.005) | ε-COP (e-22) | ||||||
| sec-21 (NCU01992.1) SEC21 (γ-COP) | Chr I | X7H1 | 2 | Mouse γ 2-COP (0) | Sec21 (e-168) | SPAC57A7 (0) | ||||||
| Highly branched, bulbous and apolar (Figure 2I) | Swollen conidia or bulbous germ tubes | 6/4 | erg-9 (NCU06054.1) ERG9 (squalene synthase) | Chr VII | X5A5 | 2 | Human squalene synthase, (e-76) | Erg9 (e-99) | Squalene synthase (e-109) | |||
| erg-13 (NCU03922.1) ERG13 (HMG CoA synthase) | X24C12 | 1 | D. melanogaster (e-128) | Erg13 (e-155) | HMG CoA synthase (e-163) | |||||||
| hmg-1 (NCU00712.1) HMG1 (HMG CoA reductase) | Chr I | X15D4 | 1 | Human (e-132) | Hmg1 (0) | HMG CoA reductase (0) | ||||||
| Fast lysis, not suppressed by 1 M sorbitol (Figure 2J) | Swollen and lysing conidia | 23/9 | sar-1 (NCU00381.1) SAR1 (GTP binding protein) | X11F9 | 1 | Human, (e-60) | Sar1 (e-65) | Sar1 (e-71) | ||||
| sec-53 (NCU02829.1) SEC53 (Phosphomannomutase) | Chr I | X9C11 | 1 | Human, mannomutase (e-78) | Sec53 (e-86) | Mannomutase (e-101) | ||||||
| apl-4 (NCU04121.1) APL4 (γ-adaptin) | Chr V | X2E12 | 1 | Mouse, AP-1 γ-1 (e-175) | Apl4 (e-95) | SPCP1E11.06 (e-151) | ||||||
| pod-1 (NCU07955.1) POD1 (hypothetical membrane protein) | Chr IV | X7E12 | 2 | No close homologue | Yhr194w (e-121) | SPAC3H1.04c (e-82) | ||||||
| pod-2 (NCU09544.1) POD2 (hypothetical ORF) | X18F8 | 1 | No close homologue | No close homologue | No close homologue | |||||||
| Fast lysis, fully suppressed by 1 M sorbitol (Figure 2K) | Swollen and lysing conidia | 10/6 | gfa-1 (NCU07366.1) GFA1 (glucosamin-fructose-6 phosphate-amidotransferase) | X10E2 | 1 | Mouse, GFAT (0) | Gfa1 (0) | GFAT (0) | ||||
| pod-3 (not annotated, contig3.212, bp113880—117180) POD3 (hypothetical protein) | X24C12 | 1 | No close homologue | No close homologue | No close homologue | |||||||
| B. Mutants affecting primarily regions distal to tip | ||||||||||||
| Apolar growth distal to tip (Figure 2L) | Swollen conidia or germlings growing distally apolar | ≈150/≥2 | cdc-25 (NCU06500.1) CDC25 (putative GEF for Ras) | 29/227 ser-1, Chr III | X3D1 | 112 | Mouse (e-55) | Cdc25 (e-72) | SPBC336 (e-53) | |||
| C. Mutants affecting primarily the tip | ||||||||||||
| Tip branches (Figure 2, M and N) | Compact and tip branching cells, some swollen conidia | 50/8 | act* (NCU04173.1) ACT* (actin) | Chr V | X9H2 | 1 | Human, actin (0) | Act (0) | Actin (0) | |||
| cdc-24 (NCU06067.1)g CDC24 (putative GEF for CDC42) | 51/231 nic-3, Chr VII | X14C6 | 32 | Human (e-11) | Cdc24 (e-32) | Scd1 (e-56) | ||||||
| ypk-1 (NCU07280.1) YPK1 (growth associated kinase) | 38/235 pan-1, Chr IV | X14H12 | 11 | Human, serum/glucocorticoid kinase 2 (e-92) | Ypk1 (e-140) | SPCC4B10 (e-160) | ||||||
| Tips round and swollen (Figure 2O) | Swollen conidia | ≈280/4 | png-1 (NCU00651.1) PNG1 (peptide: N-glycanase) | 25/236 met-6, Chr I | X16C11 | ≈180 | Mouse (e-43) | Png1 (e-44) | SPBC1709.14 (e-58) | |||
| Tips knobby and swollen, slow lysis, reduced branching (Figure 2P) | Swollen conidia, rarely bulbous germ tubes | 25/1 | hsp-70* (NCU09602.1) HSP70* (cytoplasmic chaperone) | 44/322 cys-3, Chr II | X23B12 | 25 | C. elegans HSP 70 (0) | Ssa1 (0) | Hsp70 (0) | |||
| II. Mutants defective in the regulation of polarity and growth directionality | ||||||||||||
| A. Mutants with altered branching patterns and/or defects in tip organization | ||||||||||||
| Branches contorted and one-sided (Figure 2Q) | Wavy and bulbous branched growth | 7/3 | None cloned yeth | |||||||||
| Branches contorted and one-sided (Figure 2Q) | Wavy and bulbous branched growth | 7 /3 | None cloned yeth | |||||||||
| Bunches of grapes (Figure 2, R and S) | Multipolar germination, germination delayed, branches grow contorted | ≈140 /≥5 | pod-4 (NCU02620.1)i POD4 (Kelch-domain protein) | 25/279 arg-1, Chr I | X24B4 | 1 | Leucine zipper transcriptional regulator-1 (e-14) | Kel2 (e-11) | Tea1 (e-21) | |||
| Base of branch bulbous (Figure 2T) | Swollen conidia or bulbous branched germlings | 31 /7 | pod-5 (NCU03628.1) POD5 (conserved WD-repeat protein) | Chr V | X1F6 | 2 | Human, transducin like protein (e-92) | Ylr222c (e-113) | SPCC16A11 (e-135) | |||
| Highly cot-1 like branched (Figure 2, U and V) | Germlings hyperbranched and compact | 30 /7 | cot-1* (NCU07296.1) COT1* (protein kinase) | 18/252 pan-1, Chr IV | Yarden et al., 1992j | 12 | Human Ndr kinase (e-101) | Cbk1 (e-126) | Orb6 (e-136) | |||
| pod-6 (NCU02537.1) POD6k (protein kinase) | 18/169 arg-3, Chr I | X1F7 | 7 | Human, STE20-like kinase (e-45) | Sps1 (e-44) | Nac1 (e-53) | ||||||
| gs-1* (NCU04189.1) GS1* (β-1,3 glucan synthasel) | Chr V | X19H7 | 2 | No close homologue | Smi1 (e-36) | SPBC30D10 (e-60) | ||||||
| gcd-11 (NCU02810.1) GCD11 (γSU of translation initiation factor) | Chr I | X12D3 | 1 | D. melanogaster (0) | Gcd11 (0) | SPBC17G9.09 (0) | ||||||
| Needle-like branch initiations (Figure 2W) | Swollen conidia with multiple initiations of germtubes | 23 /2 | lrg-1 (NCU02689.1) LRG1 (protein with LIM and Rho GAP domains) | 5/59 his-3 Chr I | X8D4 | 22 | D. melanogaster paxillin (e-28) | Lrg1 (e-138) | SPPC3F6 (0) | |||
| B. Mutants affected in growth directionality | ||||||||||||
| Coiled growth (Figure 2X) | Coiled growth | 6 /2 | None cloned yeth | |||||||||
| Zig-zag growth (Figure 2, Y-AA) | Zig-zag growth | 28 /7 | cdc-42 (NCU06454.1) CDC42 (Rho-type GTPase) | Chr III | X1C11 | 5 | Human, CDC42 (e-80) | Cdc42 (e-78) | Cdc42 (e-83) | |||
| mdm-12 (NCU02067.1) MDM12 (mitochondrial protein) | Chr I | X17B2 | 1 | No close homologue | Mdm12 (e-12) | Mdm12 (e-15) | ||||||
| alr-1 (NCU06225.1) ALR1 (conserved membrane protein) | Chr III | X2C3 | 1 | No close homologue | Alr1 (e-79) | SPBC27B12.12 (e-90) | ||||||
| ipp-1 (NCU00951.1) IPP1 (inorganic pyrophosphatase) | Chr I | X2A5 | 1 | D. melanogaster (e-87) | Ipp1 (e-115) | SPAC23C11 (e-121) | ||||||
| Curled hyphae (Figure 2, BB) | Multipolar germination, curled growth | 40 /9 | alf-1 (NCU01713.1) ALF1 (β-tubulin folding cofactor) | X12A10 | 7 | Human, tubulin folding cofactor B (e-27) | Alf1 (e-14) | Alp11 (e-44) | ||||
| ro-1* (NCU06976.1) RO1 (dynein heavy chain) | 20/170 ilv-3 Chr IV | X24F7 | 11 | Mouse, DHC (0) | Dyn1 (0) | DHC (0) | ||||||
| ro-3* (NCU03483.1) RO3* (p150glued, dynactin subunit) | 31/206 thr-2, Chr II | X16C5 | 3 | Human, dynactin-1 (e-58) | No close homologuem | No close homologuem | ||||||
| ro-10* (not annotated in current assembly) RO10* (dynactin subunitn) | X7A6 | 1 | No close homologue | No close homologue | No close homologue | |||||||
| (Continues) | ||||||||||||
Hyphae were grown on Vogel's minimal agar medium containing 2% sucrose, and their phenotype was determined after temperature shift from 25°C to 39°C for 6 h. The phenotypes of conidia were determined after germination at 39°C for 15 h
The exact numbers of isolated mutants and complementation groups are shown except where approximations are indicated
The mutants isolated in this screen were named pod for polarity defective unless cloning of the gene showed that there are well characterized orthologues in other organisms. In these cases, the S. cerevisiae nomenclature has been followed in most cases. Previously described N. crassa genes have kept their original names and are marked with asterisks. The first name in this column refers to the assigned name, and in parentheses the systematic name and a brief functional description of the protein are given
When a gene was closely linked to a given auxotrophic marker (available from the FGSC), the segregation pattern of the progeny was determined. For example, when progeny of a cdc-24xnic-3 cross was plated on minimal medium, among the 231 growing colonies, 180 showed the cdc-24;nic-3+ and 51 the recombinant cdc-24+;nic-3+ phenotype. If only loose linkage with the marker strains was observed, only the chromosome is indicated. These data correspond to the linkage assignments that can be made by locating the identified cosmids in the genome sequence. If no unambiguous linkage was observed, no chromosome is indicated
The cosmid from the Orbach/Sachs library (available from the FGSC) that rescued the mutant phenotype
Shown is the proper or systematic name of the best BLAST hit or (if known) a functional description of the homologue identified, e-values obtained from BLAST searches are indicated
Note that cdc-24 appears in both Class IA and Class IC
In the absence of gene identification by cloning, these mutants carry tentative pod names
The homology of POD4 to Tea1p is restricted to the kelch domains. N. crassa also contains what appears to be a true orthologue of Tea1p (NCU00622.1)
A cosmid described by Yarden et al. (1992) containing the cot-1 gene was used to define the cot-1 complementation group
This protein is in the germinal center kinase group of the STE20 superfamily of protein kinases. An unambiguous assignment to a specific S cerevisiae protein was not possible (Sps1: e-44, Kic1: e-43), so the gene was named pod-6
Enderlin and Selitrennikoff (1994)
S. cerevisiae Nip100p and S. pombe Ssm4p are functional orthologues to D. melanogaster p150glued, but cannot be detected by simple BLAST searches
Minke et al. (1999)
Classification of Mutants and Determination of the Number of Complementation Groups
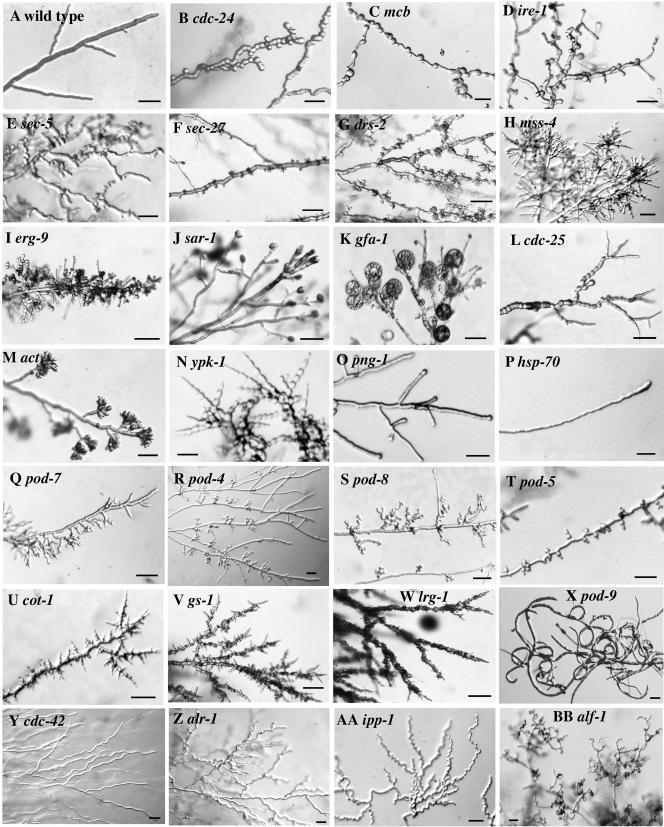
The isolated mutants were divided into five classes and multiple subclasses based on the morphological alterations observed when hyphae or conidia were transferred to restrictive temperature (Table 1, Classes I, A-C and II, A and B, and their subclasses). Classes I, A-C, have (A) general polarity defects (Figure 2, B-K), (B) polarity defects only in regions of hyphae distal to the tips (Figure 2L), and (C) polarity defects restricted to hyphal tips (Figure 2, M-P), respectively. Mutants of all three classes produce conidia that grow in an apolar manner when germinated at elevated temperature. Class IIA mutants have various defects in the establishment and maintenance of hyphal tips and branches (Figure 2, Q-W), whereas class IIB mutants are defective in the directionality of growth (Figure 2, X-BB). Mutants in Classes IIA and IIB can form polar germ tubes at restrictive temperature, although germination is often delayed. Most of the isolated mutants display wild-type morphology when grown at permissive temperature with the exception of members of the subclasses compact growth (class IA), and zig-zag growth and “curled hyphae” (both class IIB). Members of class IIB were isolated as both conditional and nonconditional mutants, suggesting that the corresponding genes might not be essential for viability. Many of the mutants showed aberrant hyphal morphologies or growth patterns shortly after the temperature shift (30-60 min), and all showed their respective phenotypes within 3-6 h after temperature shift. (Note that in these temperature-shift experiments, the agar plate acts as a temperature buffer.) Surprisingly, although most mutants could be unambiguously classified into specific subclasses, most complementation groups within a subclass displayed subtle but distinct differences in phenotype. This is in contrast to findings with budding and fission yeast, where large numbers of mutants display essentially identical morphological phenotypes (e.g., Hartwell et al., 1974; Novick et al., 1980; Verde et al., 1995). It is possible that the more complex and dynamic structure of fungal hyphae allows the visual detection of morphological changes that are too subtle to detect in the unicellular fungi. Alternatively, the increased complexity of growth in filamentous fungi relative to the yeasts may be accompanied by more dynamic regulatory systems, whose actions may result in the generation of distinct morphologies for the various hyphal growth mutants.
Figure 2.
Representative hyphal morphology mutants. The wild-type parent (A) and various mutants (B-BB) were grown at 25°C on minimal agar medium without added osmotic stabilizer and then shifted to restrictive temperature (39°C) for 10 h. Pictures were taken of hyphae growing embedded in agar with the exception of panel X, which shows hyphae growing on an agar surface. (The pod-9 mutant phenotype was similar, but more difficult to photograph, when the hyphae were growing embedded in agar.) All mutants shown here displayed wild-type or nearly wild-type growth at 25°C. Other mutants had phenotypes more-or-less similar to those shown here (see Table 1). Bars, 20 μm.
Complementation tests were conducted by forming heterokaryons between pairs of mutants and assessing whether the heterokaryons grow as wild type or mutant at restrictive temperature (see MATERIALS AND METHODS). The numbers of complementation groups within some subclasses (e.g., “apolar growth distal to tip,” class IB; bunches of grapes, class IIA; and zig-zag growth, class IIB) was difficult to define by heterokaryon formation because of partial dominance of some of the mutations. Nonallelic noncomplementation (in subclasses “highly branched, bulbous and apolar,” class IA and “curled hyphae,” class IIB) and intragenic complementation (in subclass “tips round and swollen,” class IC) were also observed and obscured the determination of complementation groups. Therefore, in the ambiguous cases, we identified complementing cosmids for specific mutants and tested the remaining mutants of the same morphological class for complementation with the appropriate cosmid fragment. In addition, once a gene was defined, we determined whether the remaining mutations within a given morphological class were also linked to an appropriate, closely linked auxotropic marker. Our analyses showed that the ≈950 mutants isolated defined more than 100 genes (Table 1). Only ∼5% of the mutations were found to be completely dominant.
Identification of Genes
To begin converting this mutant collection into meaningful biological information, we identified the genes defined by 45 mutants covering all phenotypic classes (see MATERIALS AND METHODS) and correlated their morphological phenotypes with the proposed functions of the gene products (Table 1). Many of the genes identified encode proteins that are known to have central functions in polar growth in other organisms, such as proteins that function in the organization of actin filaments or microtubules or are parts of signaling pathways. Other major groups of proteins are involved in intracellular vesicle trafficking or have cell wall-related functions. Other genes encode novel proteins lacking known homologues, uncharacterized proteins that are highly conserved between kingdoms, or uncharacterized but fungal-specific proteins. The identified proteins were typically most similar to those from other filamentous fungi, generally followed by fission yeast and often metazoans. Interestingly, 11 of the 45 identified genes (bem-1, ire-1, drs-2, sec-53, apl-4, pod-1, gfa-1, png-1, cot-1, lrg-1, and mdm-12) appear to be essential for polar growth in N. crassa, whereas the corresponding genes have viable deletion phenotypes in S. cerevisiae, at least at room temperature. (Mutants affecting proteins of known multigene families in either N. crassa or S. cerevisiae, where redundancy might obscure an essential function of an individual member, were excluded from this comparison [mcb, hmg-1, ypk-1, hsp-70, and pod-6]).
Many mutants showed rapid lysis at actively growing hyphal tips and branch points when shifted to restrictive temperature (sar-1, sec-53, apl-4, pod-1, pod-2, gfa-1, and pod-3; Figure 2, J and K), and most of the corresponding proteins function in the secretory pathway (Table 1). Mutants with apolar or bulbous branched phenotypes that lysed at slightly later times also have defects in secretory genes (sec-27, drs-2, gpi-3, alg-1, cop-1, and sec-21; Figure 2, F-H). This range in phenotypes may be due at least in part to the varying severity of the respective alleles or the importance of the various proteins in the secretory pathway. Interestingly, drs-2, gpi-3, and sar-1 were not isolated as typical secretion mutants in S. cerevisiae. Addition of osmotic stabilizers could fully suppress the growth defects of two mutants (gfa-1 and pod-3) that show a fast lysis phenotype. gfa-1 encodes a cell wall biosynthetic enzyme, and POD3 is a hypothetical protein with no similarities in the current databases. The generation of protoplasts observed with the gfa-1 mutant (Figure 2K) probably reflects the support provided by the agar medium as the weakened hyphal wall breaks down. In contrast, lysis in the sar-1 mutant (Figure 2J) appears to be the result of rapid rupture at the weakest point at the hyphal apex (the junction between mature wall and the newly synthesized wall), similar to that observed when wild-type hyphae are flooded with distilled water. Some mutants with a hyperbranching phenotype (Figure 2V) contain alleles of gs-1, which encodes the cell wall biosynthetic enzyme β-1,3 glucan synthase. Interestingly, mutations in two genes encoding protein kinases (cot-1 and pod-6) and a gene encoding a component of the translational apparatus (gcd-11) produce similar hyperbranching phenotypes (Figure 2U).
As expected, mutants affected in actin organization showed varying degrees of defective polarized growth. The one actin mutant isolated (act) showed tip-branched hyphae (Figure 2M). Other proteins involved in regulation of actin organization include Rho-type GTPases such as Cdc42 and their regulators including guanine-nucleotide-exchange factors (GEFs) such as Cdc24, GTPase-activating proteins (GAPs) such as Lrg1, and scaffold proteins such as Bem1 (Hall and Nobes, 2000). The isolation of corresponding mutants in N. crassa illustrates the importance of these proteins for hyphal morphogenesis. Several cdc-24 and bem-1 mutants generated chains of spherically growing cells (Figure 2B). Interestingly, other cdc-24 mutants showed a proliferation of tip branches like the act mutant described above, and each of the five cdc-42 mutants grew in an uncoordinated manner (Figure 2Y). The lrg-1 mutants isolated were defective in tip elongation and produced needle-like branch initiations (Figure 2W).
Mutants in the regulatory subunit of PKA (mcb) and in the ras GEF CDC25 also generated chains of spherical cells after temperature shift, a phenotype reminiscent of those resulting from defects in actin organization. However, in the mcb mutants, the chains are much more irregular than those in cdc-24 cells (Figure 2C), suggesting that increased septation, which is a prerequisite for the generation of these chains of spherical cells, is less coordinated. The cdc-25 mutants generated chains of spheres only in subapical regions of hyphae (Figure 2L), although shortly after shift (<1 h), the hyphal tips also became more pointed than wild-type tips. Hyphae of both the mcb and the cdc-25 mutants reverted to more normal phenotypes after prolonged incubation at restrictive temperature (>12 h), suggesting that regulatory mechanisms controlling hyphal growth are able to compensate at least partially for these genetic defects.
The mutants affected in microtubule function (alf-1, ro-1, ro-3, and ro-10) showed multipolar germination and curled hyphae (Figure 2BB), a phenotype observed previously in wild-type conidia that are germinated in the presence of antimicrotubule drugs (That et al., 1988) and in mutants defective in microtubule-dependent motor proteins (Plamann et al., 1994; Bruno et al., 1996a; Seiler et al., 1997, 1999). Mutants defective in several other genes (the bunches of grapes mutants) shared this phenotype, but differed in that their defects appeared specific to the growth of new branches; the hyphal tips initially grew in a wild-type manner, but the branches generated subapically displayed strongly contorted growth after producing a normal-looking stalk of several μm in length (Figure 2, R and S). This unique phenotype suggests that these mutants are defective in forming a mature hyphal tip that is capable of rapid extension.
Our screen also identified several conserved genes without clearly assigned functions (yif-1, pod-1, pod-5, pod-6, and alr-1) as well as several genes encoding proteins not previously implicated in cellular morphogenesis (ire-1, cpc-1, ypk-1, png-1, and ipp-1). YIF1, POD1, and ALR1 are putative membrane proteins, whereas POD5 is a putative WD-repeat protein and POD6 is a putative serine/threonine kinase. IRE1 is a protein kinase with important functions in the ER quality control pathway for newly synthesized proteins. The polarity defect observed with the ire-1 mutant (Figure 2D) is consistent with the finding that many mutations affecting the secretory pathway cause altered polarity. CPC1 is a well-studied regulator of amino acid biosynthesis in N. crassa (Paluh et al., 1988), and its isolation as a morphological mutant was surprising. Interestingly, it was recently shown that the disruption of its homologue gcn-4 led to pseudohyphal growth in Candida maltosa (Takaku et al., 2002; Tripathi et al., 2002), arguing for a possible connection between regulation of amino acid biosynthesis and morphological transitions. YPK1 has homology to vertebrate serum/glucocorticoid-induced kinases. The ypk-1 mutants initially produced tip-branched hyphae that resembled those of the actin mutant (Figure 2M); after prolonged times at restrictive temperature (>6 h), the ypk-1 hyphae showed strong similarity to the proconidial chains associated with asexual reproduction (Figure 2N; see Springer and Yanofsky, 1989); thus, this kinase might be involved in the negative regulation of conidiation. The png-1 gene represented the largest complementation group (∼180 alleles). It encodes a putative peptide:N-glycanase that has been connected to the proteasome-dependent degradation pathway of glycosylated proteins in yeast (Suzuki et al., 2000); however, the yeast mutant shows no apparent growth defect. The N. crassa png-1 mutants show a very pronounced swollen-tip phenotype without additional subapical alterations (Figure 2O), making secondary effects unlikely and suggesting a link between cellular polarity and protein degradation. Interestingly, the phenotypes both of png-1 mutants and of various mutants clearly affected in polar growth (cdc-24, bem-1, mcb, cdc-25, ypk-1, cot-1, pod-6, and lrg-1) were even more pronounced when the mutants were grown in a rich medium (0.5% yeast extract, 0.5% peptone, and 2% sucrose in Vogel's minimal medium) than when they were grown in Vogel's minimal medium containing 2% sucrose alone. IPP1 is an inorganic pyrophosphatase that provides the cell with a fast, accessible energy source, and the isolation of an ipp-1 zig-zag growth mutant could indicate the high energy requirement that is necessary to coordinate hyphal growth. Finally, mutations affecting mevalonate biosynthesis (erg-9, erg-13, and hmg-1) or the two phosphatidylinositol kinases (sst-4 and mss-4) cause very similar apolar growth phenotypes (Figure 2, H and I). Further work is needed to correlate the morphological phenotypes of all these mutants with their potential functions.
DISCUSSION
To initiate genome-wide functional analyses of fungal growth, we combined the recently released N. crassa genome sequence with a large-scale genetic approach. By generating a representative mutant collection and identifying proteins involved in polar and directed growth, we have established a solid foundation for studies of cellular morphogenesis in a filamentous fungus.
For the generation of a normal-shaped fungal hypha, two aspects of polarity are important: 1) the establishment of polarity during germination or branching and 2) the maintenance of polar growth during tip elongation. N. crassa hyphae are capable of very fast extension rates (>1 μm/s), and it has been estimated that a growing hyphal tip requires the fusion of ≈40,000 secretory vesicles per minute to maintain this extension rate (Collinge and Trinci, 1974). Thus, the finding that many of the hyphal morphology mutations affect the secretory pathway may reflect the high demand placed on this pathway during hyphal elongation. The lysis phenotype of these mutants was unexpected and contrasts sharply with the phenotype of the corresponding mutants in S. cerevisiae, where defects in secretion do not lead to lysis but rather to the generation of denser cells due to the accumulation of various endomembranes (Novick et al., 1980). Genetic analysis of hyphal growth in Aspergillus nidulans has also shown that mutations blocking the secretory pathway lead to lysis (Whittaker et al., 1999), suggesting that this may be a typical phenotype for secretion-defective mutants of filamentous fungi. One possible explanation is that because of the extremely fast growth rate in filamentous fungi, their cell wall synthesis is more dynamic than in S. cerevisiae, and a continuous flow of wall material and enzymes is needed to maintain a delicate balance between rapid cell wall degradative and biosynthetic activities (Bartnicki-Garcia, 1973). Continuing degradation of the wall at sites of growth would then result in the characteristic rapid lysis phenotype of secretory mutants. As a caveat, it should be noted that although cell wall degradative activities have been proposed for many years to act at hyphal tips, experimental evidence is still lacking (Wessels, 1986; Bartnicki-Garcia, 1999).
Given the lysis phenotypes of known secretory-pathway mutants, the similar lysis phenotypes of the pod-1, pod-2, and pod-3 mutants (Table 1) suggest that the products of these genes also function in the secretory pathway. The suppression of the pod-3 phenotype by the addition of 1 M sorbitol to the growth medium suggests that POD3 may be involved in cell wall stability. However, it should be noted that a large fraction of S. cerevisiae temperature-sensitive mutants not defective in cell wall synthesis were also rescued by high osmolarity (Hawthorne and Friis, 1964). It is worth noting that the essential nature of these novel proteins and their apparent restriction to the fungal kingdom (Table 1) may make them attractive targets for the development of antifungal drugs.
Given the many secretory-pathway mutants isolated in our screen, it is surprising that unequivocal endocytotic mutants were missing. The only real candidate is cytosolic heat shock protein 70 (HSP70), a protein with a variety of cellular roles, including the uncoating of clathin-coated vesicles during endocytosis (Newmyer and Schmid 2001; Chang et al., 2002). Interestingly, the mutations in this gene resulted in a swollen-tip phenotype with delayed lysis at the tip, as in many secretory mutants.
Many studies have indicated that polarization of the actin cytoskeleton is a prerequisite for the induction of cell polarization, whereas cytoplasmic microtubules act as a structural scaffold to reinforce polarized morphology and contribute to the functional compartmentalization of the cell (Mata and Nurse, 1998; Schmidt and Hall, 1998; Shulman and St. Johnston, 1999; Geitmann and Emons, 2000; Pruyne and Bretscher, 2000). Similarly, microtubules have been shown to be an important cytoskeletal element regulating proper and directed growth in filamentous fungi (That et al., 1988; Plamann et al., 1994; Seiler et al., 1997, 1999; Riquelme et al., 1998; Heath et al., 2000). In line with this view, we found that mutants affected in the microtubule cytoskeleton displayed defects in growth directionality (alf-1, ro-1, ro-3, and ro-10), whereas mutants presumably defective in actin organization (cdc-24, bem-1, stt-4, and mss-4) showed general polarity defects. As expected, actively growing hyphal tips appear to be especially sensitive to changes in actin organization, as we observed tip-branched cells in some cdc-24 mutants and in the one actin mutant isolated. The needle phenotype of lrg-1 mutants (encoding a regulatory factor for Rho-type GTPases) may also be due to actin misorganization, resulting in the cessation of tip growth at a very early stage. Some cdc-24 mutants and all bem-1 mutants showed a more extensive loss of polarity throughout the length of the hyphae upon shift to restrictive temperature. The chains of spherically growing cells observed in these mutants were apparently the result of increased septation in combination with delocalized growth all over the hypha. This phenotype might be the consequence of failed attempts to branch due to an inability to localize new growth. Thus, coordinated actin organization appears to be essential both to establish polarity during germination and to maintain polar growth during tip elongation. Surprisingly, the strikingly different phenotypes of the cdc-42 alleles isolated suggest an additional influence of actin in the maintenance of growth directionality. In mammalian cells, there is evidence that Rho GTPases can directly control microtubule dynamics (Palazzo et al., 2001a, 2001b), and this mechanism would help to coordinate both cytoskeletal elements during axis formation and directed growth in filamentous fungi. The lack of cdc-42 mutants showing a complete loss of polarization may reflect the difficulty of obtaining complete loss-of-function temperature-sensitive alleles of this gene (Johnson, 1999).
The pod-4, pod-8, and other bunches of grapes mutants share the multipolar germination phenotype that is characteristic of microtubule-defective mutants, but they are also defective in the generation of mature tips on new branches. Functional tips in these mutants grow in an apparently wild-type manner and can be generated by apical branching. Subapical branches also form; however, >95% of such branches grow in a contorted way after they have produced a normal looking stalk of several micrometers in length. This defect suggests that microtubules might be involved in the stable establishment of polarity at new sites in addition to their function in regulating growth directionality. For example, they might locally modify actin dynamics, thereby influencing the growth mode at the hyphal apex and/or the future branch points. The fact that budding yeast apparently lacks such microtubule-dependent regulation of growth polarity (Winsor and Schiebel, 1997), whereas the more elongated fission yeast is highly reliant on this system (Hayles and Nurse, 2001), raises the possibility that this is a common characteristic of elongated cell types.
Important roles during hyphal morphogenesis were recently established for both the cAMP and MAP kinase pathways in a wide variety of fungi (Madhani and Fink, 1998; Borges-Walmsley and Walmsley, 2000). Consistent with these previous studies, several components of these signaling cascades were identified in our screen. The phenotypes of mutants defective in either the Ras GDP-GTP exchange factor CDC25 or the regulatory subunit of protein kinase A (MCB) suggest that neither pathway is necessary for polarized growth per se, although they are important for localizing growth to specific sites during germination, hyphal elongation, and the initiation of new branches. The phenotypes are reminiscent of cdc-24 mutants, suggesting that both pathways may act by modulating the actin cytoskeleton, as described for budding yeast (Ho and Bretscher, 2001). Interestingly, the cellular response is quite different between yeast and N. crassa. Although activation of PKA in budding yeast leads to increased polarization and pseudohyphal differentiation, activation of PKA in N. crassa results in apolar growth (Bruno et al., 1996b; this study).
Mutations affecting mevalonate biosynthesis (erg-9, erg-13, and hmg-1) result in highly bulbous, branched hyphae very similar to those formed by mutants defective in phosphatidylinositol signaling (stt-4, mss-4). Interestingly, these phenotypes can be mimicked through the growth of wild-type cultures on the detergent SDS (our unpublished observations). It is possible that the effects of these mutations on hyphal growth might be linked to the proper function of the Rho-type GTPases that regulate actin dynamics. Proper membrane association through isoprenylation is necessary for normal function of these GTPases, and blocking mevalonate biosynthesis depletes the isoprenoid pool of the cell and thus alters the expression and localization of isoprenylated proteins (Holstein et al., 2002). In addition, lipid rafts have recently been implicated in polar growth in S. cerevisiae (Bagnat and Simons, 2002), and sphingolipid and ergosterol biosynthetic mutants fail to localize proteins toward the polarized growth site that forms in response to mating pheromone. Also consistent with this finding is the observation that lipid biosynthesis plays a central role in cellular polarity in A. nidulans (Cheng et al., 2001).
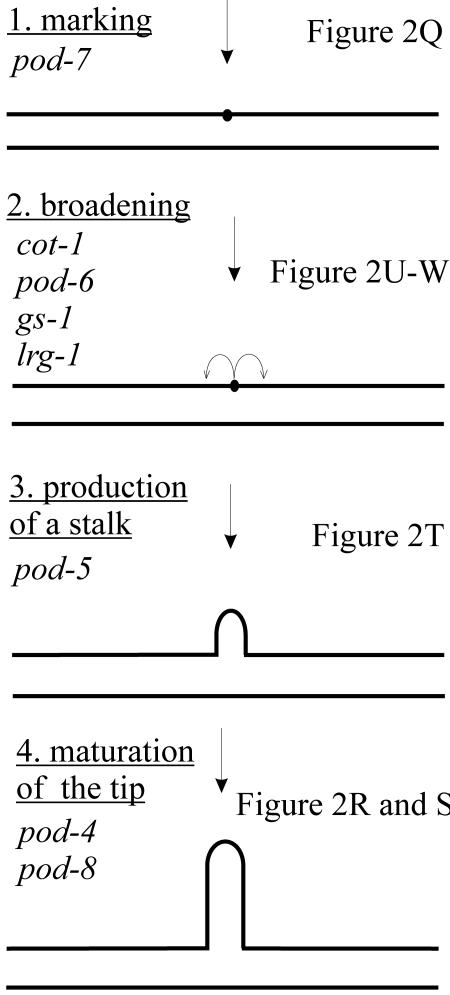
An important aspect of hyphal growth is the generation of new tips during branch formation. The first sign of the formation of a new branch in wild-type hyphae is a small bud-like structure, whereas the next visible step is a fully mature branch of several micrometers in length. These observations suggest a series of successive steps during branching, similar to the establishment of the initial germ tube during germination (Momany et al., 1999). The isolation of mutants that appear to be blocked in discrete steps supports this idea and might provide insights into a morphogenetic pathway for the formation of new tips (Figure 3). We propose the existence of at least four discrete and genetically separable steps: 1) the selection of a new branch site; 2) the broadening of the marked spot into a zone of growth; 3) the production of a small stalk-like branch; and 4) the maturation of the new tip. Theoretically, the selection of a new branch site could be a statistical process, dependent only on the distance between the tip of the major hypha and the nearest existing branch. Exceeding a certain threshold (cytoplasmic volume vs. number of hyphal tips) would signal the initiation of a new branch. On the other hand, the isolation of mutants that branch only on one side of the hypha is a strong indication that determination of the sites of branch formation is a highly regulated process. Conversion of the initial signal into a zone of active growth in a previously silent subapical area requires the transport of components necessary for growth toward the nascent bud and the remodeling of the cell wall at this point. Candidates upon which these signals might act to regulate actin organization and to initiate growth are the products of genes whose mutants display needle-like branches. The transition from growth only at the marked spot to a broader zone of active growth appears to require LRG1, a putative Rho-GAP, suggesting that excessive or prolonged activity of a small GTPase may restrict the zone of growth. Interestingly, we observed intergenic noncomplementation between lrg-1, cot-1, and pod-6 mutations, suggesting that these three gene products may interact. However, in contrast to lrg-1, the cot-1 and pod-6 phenotype appears to reflect arrested tip elongation at a later stage, and these mutants produce hyperbranched hyphae with branches up to 30 μm in length. An attractive hypothesis is that these proteins link signal transduction with the cytoskeleton by acting together in a stepwise manner. Finally, the maturation into a fully functional tip is dependent on microtubules and the proteins defined by the bunches of grapes mutants.
Figure 3.
Model for hyphal branching. The phenotypes of several of the isolated mutants suggest that branching and the generation of new hyphal tips is a genetically separable process consisting of at least four discrete steps: (1) the selection of a new branch site; (2) the broadening of the marked spot into a zone of growth; (3) the production of a small stalk-like branch; and (4) a microtubule dependent maturation step to form a functional tip. On the left, the proposed steps and the gene(s) involved are indicated; on the right are references to the corresponding images of mutant phenotypes in Figure 2. See text and Table 1 for detailed descriptions of the mutants. Bars, 20 μm.
The mutant collection described here should be a valuable resource for elucidation of the molecular network that coordinates morphogenesis in the filamentous fungus N. crassa. The large number of complementation groups clearly illustrates the complexity of hyphal growth and the necessity for further large-scale genetic approaches. Although our screen was quite extensive, a look at the number of alleles per complementation group (Table 1) strongly suggests that we have defined only a relatively small subset of the genes required for hyphal morphogenesis. In addition, our screening procedure focused on the isolation of mutants with strong, distinctive growth defects and did not allow the isolation of mutants with subtle phenotypes. Additional screens using different approaches and the generation of genome-wide knock-out libraries will be necessary to fully exploit the potential of filamentous fungi.
Acknowledgments
We acknowledge the help of In Hyung Lee in the initial phase of this work and the helpful suggestions of John Pringle in the preparation of the manuscript. A University of Missouri Research Grant to M.P. (340975) and an EMBO two-year postdoctoral fellowship to S.S. (ALTF 663-1999) supported this work.
References
- Adams, M.D. et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185-2195. [DOI] [PubMed] [Google Scholar]
- Bagnat, M., and Simons, K. (2002). Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA 99, 14183-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnicki-Garcia, S. (1973). Fundamental aspects of hyphal morphogenesis. In: Microbial Differentiation, ed. J.M. Ashworth and J.E. Smith, Cambridge University Press, 245-267.
- Bartnicki-Garcia, S. (1999). Glucans, walls, and morphogenesis: on the contributions of J. G. H. Wessels to the golden decades of fungal physiology and beyond. Fungal Genet. Biol. 27, 119-127. [DOI] [PubMed] [Google Scholar]
- Bartnicki-Garcia, S., Hergert, F., and Gierz, G. (1989). Computer simulation of fungal morphogenesis and the mathematical basis for hyphal (tip) growth. Protoplasma 153, 46-57. [Google Scholar]
- Bartnicki-Garcia, S., Bartnicki, D.D. Gierz, G., Lopezfranco, R., and Bracker, C.E. (1995). Evidence that the Spitzenkörper behavior determines the shape of a fungal hypha—a test of the hyphoid model. Exp. Mycol. 19, 153-159. [DOI] [PubMed] [Google Scholar]
- Borges-Walmsley, M.I., and Walmsley, A.R. (2000). cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8, 133-141. [DOI] [PubMed] [Google Scholar]
- Bruno, K.S., Tinsley, J.H., Minke, P.F., and Plamann, M. (1996a). Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc. Natl. Acad. Sci. USA 93, 4775-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, K.S., Aramayo, R., Minke, P.F., Metzenberg, R.L., and Plamann, M. (1996b). Loss of growth polarity and mislocalization of septa in a Neurospora mutant altered in the regulatory subunit of cAMP-dependent protein kinase. EMBO J. 15, 5772-5782. [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012-2018. [DOI] [PubMed] [Google Scholar]
- Chang, H.C., Newmyer, S.L., Hull, M.J., Ebersold, M., Schmid, S.L., and Mellman, I. (2002). Hsc70 is required for endocytosis and clathrin function in Drosophila. J. Cell Biol. 159, 477-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., Park, T.S., Fischl, A.S., and Ye, X.S. (2001). Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol. Cell Biol. 21, 6198-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge, A.J., and Trinci, A.J.P. (1974). Hyphal tips of wild-type and spreading colonial mutants of Neurospora crassa. Arch. Microbiol. 99, 353-368. [DOI] [PubMed] [Google Scholar]
- Davis, R.H. (2000). Neurospora—Contributions of a Model Organism. Oxford University Press.
- Davis, R.H., and de Serres, F.J. (1970). Genetic and microbial research techniques for Neurospora crassa. Methods Enzymol. 17A, 79-143. [Google Scholar]
- Davis, R.H., and Perkins, D.D. (2002). Neurospora: a model of model microbes. Nature Rev. Genet. 3, 7-13. [DOI] [PubMed] [Google Scholar]
- De Backer, M.D. et al. (2001). An antisense-based functional genomics approach for identification of genes critical for growth of Candida albicans. Nat. Biotechnol. 19, 235-241. [DOI] [PubMed] [Google Scholar]
- Enderlin, C.S., and Selitrennikoff, C.P. (1994). Cloning and characterization of a Neurospora crassa gene required for (1, 3) beta-glucan synthase activity and cell wall formation. Proc. Natl. Acad. Sci. USA 91, 9500-9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan, J.E. et al. (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859-868. [DOI] [PubMed] [Google Scholar]
- Geitmann, A., and Emons, A.M. (2000). The cytoskeleton in plant and fungal cell tip growth. J. Microsc. 198, 218-245. [DOI] [PubMed] [Google Scholar]
- Girbardt, M. (1969). Die Ultrastruktur der Apikalregion von Pilzhyphen. Protoplasma 67, 413-441. [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509-514. [DOI] [PubMed] [Google Scholar]
- Hall, A., and Nobes, C.D. (2000). Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer, L. et al. (2001). Gene discovery and gene function assignment in filamentous fungi. Proc. Natl. Acad. Sci. USA 98, 5110-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L.H., Culotti, J., Pringle, J.R., and Reid, B.J. (1974). Genetic control of the cell division cycle in yeast. Science 183, 46-51. [DOI] [PubMed] [Google Scholar]
- Hawthorne, D.C., and Friis, J. (1964). Osmotic-remedial mutants. A new classification for nutritional mutants in yeast. Genetics 50, 829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles, J., and Nurse, P. (2001). A journey into space. Nat. Rev. Mol. Cell. Biol. 2, 647-656. [DOI] [PubMed] [Google Scholar]
- Heath, I.B., Gupta, G., and Bai, S. (2000). Plasma membrane-adjacent actin filaments, but not microtubules, are essential for both polarization and hyphal tip morphogenesis in Saprolegnia ferax and Neurospora crassa. Fungal Genet. Biol. 30, 45-62. [DOI] [PubMed] [Google Scholar]
- Ho, J., and Bretscher, A. (2001). Ras regulates the polarity of the yeast actin cytoskeleton through the stress response pathway. Mol. Biol. Cell 12, 1541-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein, S.A., Wohlford-Lenane, C.L., and Hohl, R.J. (2002). Consequences of mevalonate depletion: differential transcriptional, translational and post-translational up-regulation of Ras, Rap1a, RhoA, and RhoB. J. Biol. Chem. 277, 10678-10682. [DOI] [PubMed] [Google Scholar]
- Johnson, D.I. (1999). Cdc 42, An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63, 54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani, H.D., and Fink, G.R. (1998). The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 8, 348-353. [DOI] [PubMed] [Google Scholar]
- Mata, J., and Nurse, P. (1998). Discovering the poles in yeast. Trends Cell Biol. 8, 163-167. [DOI] [PubMed] [Google Scholar]
- McGoldrick, C.A., Gruver, C., and May, G.S. (1995). MyoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J. Cell Biol. 128, 577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke, P.F., Lee, I.H., Tinsley, J.H., Bruno, K.S., and Plamann, M. (1999). Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 32, 1065-1076. [DOI] [PubMed] [Google Scholar]
- Momany, M., Westfall, P.J., and Abramowsky, G. (1999). Aspergillus nidulans swo mutants show defects in polarity establishment, polarity maintenance and hyphal morphogenesis. Genetics 151, 557-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmyer, S.L., and Schmid, S.L. (2001). Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J. Cell Biol. 152, 607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick, P., Field, C., and Schekman, R. (1980). Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21, 205-215. [DOI] [PubMed] [Google Scholar]
- Orbach, M.J. (1994). A cosmid with a HyR marker for fungal library construction and screening. Gene 150, 159-162. [DOI] [PubMed] [Google Scholar]
- Osherov, N., and May, G. (2000a). Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155, 647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov, N., and May, G. (2000b). Polarity-defective mutants of Aspergillus nidulans. Fungal Genet. Biol. 31, 181-188. [DOI] [PubMed] [Google Scholar]
- Palazzo, A.F., Joseph, H.L., Chen, Y.J., Dujardin, D.L., Alberts, A.S., Pfister, K.K., Vallee, R.B., and Gundersen, G.G. (2001a). Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr. Biol. 11, 1536-1541. [DOI] [PubMed] [Google Scholar]
- Palazzo, A.F., Cook, T.A., Alberts, A.S., and Gundersen, G.G. (2001b). mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 3, 723-729. [DOI] [PubMed] [Google Scholar]
- Paluh, J.L., Orbach, M.J., Legerton, T.L., and Yanofsky, C. (1988). The crosspathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc. Natl. Acad. Sci. USA 85, 3728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D.D., Radford, A., and Sachs, M.S. (2001). The Neurospora Compendium. Academic Press.
- Plamann, M., Minke, P.E., Tinsley, J.H., and Bruno, K.S. (1994). Cytoplasmic dynein and actin-related protein arp1 are required for normal nuclear distribution in filamentous fungi. J. Cell Biol. 127, 139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000). Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113, 365-375. [DOI] [PubMed] [Google Scholar]
- Riquelme, M., Reynagapena, C.G., Gierz, G., and Bartnicki-Garcia, S. (1998). What determines growth direction in fungal hyphae? Fungal Genet. Biol. 24, 101-109. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., and Hall, M.N. (1998). Signaling to the actin cytoskeleton. Annu. Rev. Cell Biol. 14, 305-338. [DOI] [PubMed] [Google Scholar]
- Schulte, U., Becker, I., Mewes, H.W., and Mannhaupt, G. (2002). Large scale analysis of sequences from Neurospora crassa. J. Biotechnol. 94, 3-13. [DOI] [PubMed] [Google Scholar]
- Seiler, S., Nargang, F.E., Steinberg, G., and Schliwa, M. (1997). Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16, 3025-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, S., Plamann, M., and Schliwa, M. (1999). Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr. Biol. 9, 779-785. [DOI] [PubMed] [Google Scholar]
- Shulman, J.M., and St. Johnston, D. (1999). Pattern formation in single cells. Trends Cell Biol. 9, M60-M64. [PubMed] [Google Scholar]
- Springer, M.L., and Yanofsky, C. (1989). A morphological and genetic analysis of conidiophore development in Neurospora crassa. Genes Dev. 3, 559-571. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., and Schliwa, M. (1993). Organelle movements in the wild-type and wall-less fz;sg;os-1 mutants of Neurospora crassa are mediated by cytoplasmic microtubules. J. Cell Sci. 106, 555-564. [DOI] [PubMed] [Google Scholar]
- Steinberg, G., Wedlich-Soldner, R., Brill, M., and Schulz, I. (2001). Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 114, 609-622. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Park, H., Hollingsworth, N.M., Sternglanz, R., and Lennarz, W.J. (2000). PNG1, a yeast gene encoding a highly conserved peptide:N-glycanase. J. Cell Biol. 149, 1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft, C.S., Zugel, M., and Selitrennikoff, C.P. (1991). In vitro inhibition of stable 1, 3-beta-D-glucan synthase activity from Neurospora crassa. J. Enzyme Inhibition 5, 41-49. [DOI] [PubMed] [Google Scholar]
- Takaku, H., Horiuchi, H., Takagi, M., and Ohta, A. (2002). Pseudohyphal growth in a dimorphic yeast, Candida maltosa, after disruption of the C-GCN4 gene, a homolog of Saccharomyces cerevisiae GCN4. Biosci. Biotech. Biochem. 66, 1936-1939. [DOI] [PubMed] [Google Scholar]
- Tripathi, G., Wiltshire, C., Macaskill, S., Tournu, H., Budge, S., and Brown, A.J. (2002). Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21, 5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- That, C.T.C., Rossier, C., Barja, F., Turian, G., and Ross, U.-P. (1988). Induction of multiple germ tubes in Neurospora crassa by antitubulin agents. Eur. J. Cell Biol. 46, 68-79. [PubMed] [Google Scholar]
- Wendland, J. (2001). Comparison of morphogenetic networks of filamentous fungi and yeast. Fungal Genet. Biol. 34, 63-82. [DOI] [PubMed] [Google Scholar]
- Wendland, J., and Philippsen, P. (2001). Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics 157, 601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, J.G.H. (1986). Cell wall synthesis in apical hyphal growth. Int. Rev. Cytol. 104, 37-79. [Google Scholar]
- Whittaker, S.L., Lunness, P., Milward, K.J., Doonan, J.H., and Assinder, S.J. (1999). sod(VI)C is an alpha-COP-related gene, which is essential for establishing and maintaining polarized growth in Aspergillus nidulans. Fungal Genet. Biol. 26, 236-252. [DOI] [PubMed] [Google Scholar]
- Winsor, B., and Schiebel, E. (1997). An overview of the Saccharomyces cerevisiae microtubule and microfilament cytoskeleton. Yeast 13, 399-434. [DOI] [PubMed] [Google Scholar]
- Wood, V., Rutherford, K.M., Ivens, A., Rajandream, M.-A., and Barrell, B. (2001). A re-annotation of the Saccharomyces cerevisiae genome. Comp. Funct. Genom. 2, 143-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, V. et al. (2002). The genome sequence of Schizosaccharomyces pombe. Nature 415, 871-880. [DOI] [PubMed] [Google Scholar]
- Verde, F., Mata, J., and Nurse, P. (1995). Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 131, 1529-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer, S.J., and Yanofsky, C. (1986). Efficient cloning of genes of Neurospora crassa. Proc. Natl. Acad. Sci. USA 83, 4869-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden, O., Plamann, M., Ebbole, D.J., and Yanofsky, C. (1992). Cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 11, 2159-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]