Abstract
The new β2 Adrenoceptor (β2AR) crystal structures provide a high-resolution snapshot of receptor interactions with two particular partial inverse agonists, (−)-carazolol and timolol. However, both experimental and computational studies of GPCR structure are significantly complicated by the existence of multiple conformational states coupled to ligand type and receptor activity. Agonists and antagonists induce or stabilize distinct changes in receptor structure that mediate a range of pharmacological activities. In this work, we (1) established that the existing β2AR crystallographic conformers can be extended to describe ligand/receptor interactions for additional antagonist types, (2) generated agonist-bound receptor conformations, and (3) validated these models for agonist and antagonist virtual ligand screening (VLS). Using a ligand directed refinement protocol, we derived a single agonist-bound receptor conformation that selectively retrieved a diverse set of full and partial β2AR agonists in VLS trials. Additionally, the impact of extracellular loop 2 conformation on VLS was assessed by docking studies with rhodopsin-based β2AR homology models, and loop-deleted receptor models. A general strategy for constructing and selecting agonist-bound receptor pocket conformations is presented, which may prove broadly useful in creating agonist and antagonist bound models for other GPCRs.
Keywords: β2 Adrenoceptor, homology model, G-protein coupled receptor, agonist, virtual ligand screening, GPCR, MMFF
Introduction
G-protein coupled receptors (GPCRs) are critical targets for drug discovery, yet remain technically challenging to study by X-ray crystallography. Only a handful of GPCR structures have been solved, including those for bovine rhodopsin covalently linked to retinal [1], unliganded bovine opsin [2], turkey β1AR in complex with the inverse agonist cyanopindolol [3], and β2AR with the partial inverse agonists (−)-carazolol and timolol [4; 5]. The β2AR structures elucidate interactions between the receptor and two diffusible inverse agonists in atomic detail, and identify several key distinctions between the β2AR and bRho structures. Nonetheless, experimental evidence demonstrates that GPCRs exhibit ligand-selective conformational states corresponding to differences in ligand potency and functionality [6; 7; 8; 9]. In isolation, the structure of a single GPCR/antagonist complex is unable to address the ligand/receptor interactions and binding pocket conformational changes necessary for recognition of multiple structurally diverse antagonists and agonists. The conformational flexibility and heterogeneity intrinsic to GPCRs makes crystallography of additional ligand/receptor complexes a daunting task. In the absence of comprehensive experimental structural data, computational modeling can extend and expand existing crystallographic information to provide insight into other ligand-bound receptor conformations, and ultimately even to other receptor types. Here, we present a method to generate ligand-selective receptor models using the β2AR crystal structures as a starting point. We evaluated the capacity of the new receptor models to describe ligand/receptor interactions and identify new antagonists and agonists by VLS. Additionally, we investigated the deletion of extracellular loop 2 as a strategy for limiting conformational uncertainty in GPCR homology models.
Importantly, in docking and VLS a small number (usually n=1) of static receptor conformations is used to represent the ligand binding pocket while each small molecule ligand is treated as fully flexible. This is in stark contrast to the experimental picture of GPCR structure as a dynamic and responsive conformational ensemble. Previous estimates based upon a general protein/ligand benchmark set have found that at most 50% of ligands cross-dock correctly to a single receptor conformation [10]. The structural features of the ligands selected and the overall VLS yield then depend on the nature of the receptor conformation employed for screening. Changes in binding pocket conformation can then manifest themselves as biases towards ligands with a particular pharmacological activity, providing selective enrichment for agonist, inverse agonist, or antagonist compounds. For example, bRho-based homology models of the dopamine D3, muscarinic M1 and vasopressin V1a receptors were found to be antagonist selective in a VLS application, following optimization of the binding pocket conformation with known antagonist compounds [11]. Additionally, the VLS performance of bRho-based GPCR homology models is typically somewhat limited, particularly if empirical information regarding ligand/receptor interactions is not included as restraints. Lower VLS enrichment factors and poor yields may originate from multiple sources of error, including overall distortions in the three dimensional structure of the homology models and/or the failure of a single receptor conformation to act as a suitable model for all known antagonists. The β2AR crystal structures provide an opportunity to parse apart these two sources of error. As a high-resolution crystal structure, the β2AR coordinates should provide a more accurate representation of binding pocket geometry, but are nonetheless restricted to a static conformation. β2AR was crystallized with (−)-carazolol and timolol, both partial inverse agonists that decrease the basal activity of the receptor. However, in contrast to the bRho structure, which was crystallized with the full inverse agonist 11-cis-retinal, several structural signatures of receptor inactivation are not observed. These include the absence of an ionic lock between β2AR residues Glu2686.30 and Arg1313.50, as well as lack of a direct interaction between carazolol and the proposed rotamer toggle residue Trp2656.48 (residue indices are specified for the β2AR structure, followed by Ballesteros and Weinstein numbering [12] in superscript) [4; 13; 14]. In this work, we conducted VLS with full atom β2AR models derived from the crystal structure of either β2AR with carazolol or bRho to compare the ligand selectivity and yield for these conformations. With the β2AR-based model as template, high yields and enrichment factors were obtained for a set of structurally diverse antagonists, indicating that a single receptor conformation can provide a reasonable representation of the antagonist bound state. In comparison, the bRho-based homology model failed to retrieve antagonists, due to occlusion of the ligand binding pocket by extracellular loop 2 (EL2). To further energetically minimize the conformation of the β2AR-based model, Monte Carlo optimization of the β2AR ligand binding pocket sidechains was performed in the presence of a flexible (−)-carazolol molecule. This gave rise to a new model that includes five additional hydrogen bonds in the ligand binding pocket that are missing or suboptimal in the PDB-deposited coordinates.
While the protonated β2AR crystallographic coordinates provide a reasonable model and excellent enrichment for antagonists, agonist/receptor interactions are not well described using any of the four currently solved GPCRs. For many drug development efforts agonist compounds exhibit the desired pharmaceutical effect, and a suitable representation of the agonist-bound receptor is required for VLS. Several previous computational studies have developed bRho-based or de novo agonist bound β2AR receptor models, each following a distinct methodology[11; 15; 16]. In one approach, experimental evidence from electron paramagenetic resonance and fluorescence spectroscopy studies was used to guide manual rotations of TM6 in the bRho structure. This was performed to generate bRho based homology models of the activated dopamine D3, δ-opioid, and β2 adrenergic receptors[11]. These starting structures were refined with a set of superimposed agonists and tested by VLS. The models were successful in retrieving full agonists from a decoy set when combined with a scoring scheme that produced consensus hit lists from the results of multiple docking and scoring functions. However, the combination of docking and scoring functions used to generate the hit list was optimized separately for each receptor to produce the best hit rates, and a single, general scoring scheme independent of receptor type was not identified. In a separate study, Gouldson et al. used distance constraints compiled from zinc binding and site directed spin labeling experiments to generate agonist and antagonist bound β2AR models derived from bRho[16]. The experimental data were separated into constraint sets consistent with agonist or antagonist binding, and used in restrained molecular dynamics simulations. The resulting agonist and antagonist models performed well in VLS trials, demonstrating that appropriate binding pocket conformations can provide agonist and antagonist selectivity. Nonetheless, the computational expense required to generate a convergent molecular dynamics trajectory restricts the number of possible starting receptor conformations. Techniques that allow rapid generation of low energy yet structurally diverse receptor conformations would allow more thorough exploration of receptor conformational space. In other work, de novo methods that do not consider a template structure were used to prepare bound conformations of β2AR with a set of full, partial, and inverse agonists[15]. These models describe distinct minimum energy arrangements of the TM domains for each compound. Impressively, a strong agreement was found between experimentally observed conformational switches, such as the rotamer toggle and ionic lock, with those predicted computationally. Virtual screening results were reported for two of the de novo models, though selectivity for agonist or antagonist compounds was not discussed.
In the present work, we employ a ligand directed modeling approach to adapt the existing β2AR crystal structure to capture critical agonist/receptor interactions and conduct selective VLS for agonist compounds [17; 18]. In this procedure, the receptor ligand binding pocket sidechains and ligand conformation are flexibly sampled under an empirical energy potential to optimize ligand/receptor interactions and provide a more thorough description of ligand recognition. Here, we additionally include rigid body movements of TM5. A large set of low energy structures is generated, and a representative set of conformationally diverse models is selected for evaluation. As no agonist-bound crystal structure is available for comparison to the calculated structures, evaluation of the resulting conformations must be accomplished by other means. In a ligand directed strategy, VLS on a small set of known agonists and decoy compounds is instead employed as the criteria for selecting a receptor binding conformation. This has the advantages that: 1) it does not rely on the existence of an experimental structure 2) it does not depend on experimental mutagenesis data that may not provide an unambiguous description of ligand/receptor contacts. Through this process, a single receptor conformation was identified that reliably discriminates full and partial β2AR agonists from a set of decoy compounds composed of β2AR antagonists as well as biogenic amine receptor and other GPCR ligands.
Ultimately the structural information regarding antagonist and agonist binding modes in β2AR should be applied to generate homology models of other receptor types. Notably, β2AR is a member of the biogenic amine receptor family, a receptor class that includes drug targets for cardiovascular disease, asthma, depression, and psychosis. Relative to bRho, β2AR exhibits greater sequence identity to the other biogenic amine receptors and may improve modeling and VLS results for this important receptor class. However, it was observed here that bRho-based homology models of β2AR perform poorly in VLS relative to the actual crystal structure. How can one ensure that future β2AR-based or bRho-based homology models do not suffer from the same structural inaccuracies? The modeling of EL2 was a major source of error in the bRho-based homology model of β2AR. This loop assumes distinct conformations among GPCR types: in the β1AR and β2AR structures, EL2 forms a short helical structure and is removed from the binding pocket, while in both bRho and unliganded opsin EL2 exhibits a β hairpin conformation that blocks the transmembrane ligand binding pocket [1; 2; 3; 4; 5]. Additionally, loop regions are inherently flexible, and in many cases are best approximated by an ensemble of conformations, rather than a single rigid structure. For membrane proteins, further inaccuracies arise, as interactions amongst the receptor loops, receptor termini, transmembrane helical bundle, and phospholipid membrane may affect loop conformation and are frequently not considered during the modeling procedure. These errors may be worsened for GPCR homology models, as the end-to-end distances of the helices are uncertain, and may even be inappropriate for the target loop length. Recently, Mehler et al. developed a de novo loop prediction algorithm for membrane proteins that explicitly considers a loop in the context of the full protein and other predicted loop conformations. This procedure also attempts to minimize error by predicting an ensemble of compatible loop conformations rather than a single low energy structure [19]. For shorter loops, such as intracellular loop 1 and extracellular loop 1, this method was found to be quite effective, however the performance on longer loops has yet to be extensively verified [20]. Another possibility for minimizing conformational uncertainty in EL2 is to simply delete the loop. This tactic of eliminating regions of conformational uncertainty follows from a similar strategy implemented in the recently validated SCARE algorithm, wherein pairs of sidechains are systematically deleted to allow for induced fit docking [10]. Accordingly, VLS was conducted for loop-deleted variants of the agonist and antagonist structures to evaluate the impact of EL2 on selective ligand retrieval, and to suggest possible strategies for future homology modeling efforts.
Materials and Methods
All-atom molecular models, virtual ligand screening results and images in this work were generated with the ICM-Pro software package, version 3.5-1[21]. Molecular conformation was specified by internal coordinate variables, rather than cartesian coordinates, and a modified ECEPP/3 potential was used, as implemented in the ICM program [22]. Charges for ligands were taken from the MMFF94 description [23].
Ligand directed refinement of the β2AR crystal structure
An initial all-atom β2AR model was constructed from the PDB-deposited crystallographic coordinates of β2AR bound to carazolol (PDB ID: 2RH1) by the addition and systematic search/local minimization of hydrogen atoms. This model was then sampled by biased probability Monte Carlo (BPMC) in internal coordinates [21]. During the BPMC sampling, both selected receptor sidechains and the (−)-carazolol ligand were treated as fully flexible. Sidechain conformations were globally minimized in an 8 Å radius from the starting carazolol position, and a harmonic distance restraint was imposed between the od1 oxygen of Asp1133.32 and one of the carazolol amine hydrogens to ensure that the receptor/amine hydrogen bond was maintained. All backbone coordinates were held fixed. Two rounds of BPMC were performed: an initial refinement under a softened van der Waals (vdW) potential, and a second refinement with the full vdW potential. A final stack of 50 distinct conformations was generated; conformations displaying a root mean squared deviation (RMSD) less than 0.2 Å in the pocket sidechains and ligand conformation were clustered and compressed. Each of the 50 conformations was scored for binding affinity to the ligand, and the best scoring conformation was selected for VLS. Ligand receptor binding affinity was estimated as the energy of the bound ligand/receptor complex, and included the ICM energy terms for van der Waals, generalized Born electrostatics, torsion, hydrogen bonding, and hydrophobic desolvation with constant surface tension, as described in [24].
Ligand directed agonist bound β2AR model construction
Ten agonist bound models were constructed from the β2AR structure: one model retaining the crystallographic backbone conformation, and nine models with a systematically shifted and/or rotated TM5. Residues 196 and 197 of extracellular loop 2 (EL2) were deleted in all of the TM-5 modified models to eliminate possible clashes between EL2 and TM5. All shifts were directed along a vector extending from the N-terminus of TM5 to the N-terminus of TM1, effectively translating TM5 towards TM3 and 6. TM5 was shifted by four increments: 0.5 Å, 1.0 Å, 1.5 Å, and 2 Å. A rotation of 5° was then applied to each of the four shifted models plus the original crystallographic coordinates, to generate a total of ten TM5 conformations. Rotations of TM5 were counter-clockwise in direction when viewed from the extracellular side of the receptor, about a longitudinal axis parallel to the helix. The rotational axis was described by a vector from the N-terminus of TM5 to the center of mass of TM5. These rotations were introduced to position Ser2045.43, a residue expected to interact with agonist ligands, closer to the binding pocket interior. The 10 models underwent flexible sidechain/ligand BPMC refinement as described for the antagonist bound models with (−)-isoproterenol, a strong agonist. The starting conformation of isoproterenol was generated by flexibly aligning it to the crystallographic conformation of carazolol using atomic property fields, and this starting orientation of isoproterenol was held identical for all agonist models [25]. Alternatively, in the absence of a crystal structure to guide ligand placement, a known ligand could be positioned using experimentally determined distance restraints, or several positions of the ligand (generated through docking simulations) could be selected as starting points for model refinement (as in Cavasotto et al. [18]). For each of the 10 starting models, a conformational stack of 50 low energy binding pocket conformations resulted from the BPMC simulation. For several models, the predicted binding mode was not consistent with known experimental mutagenesis data. This led to the elimination of the 1.5 Å shift, 1.5 Å shift + 5° rotated, 2.0 Å shift + 5° rotated and no shift + 5° rotated TM5 structures. The conformation with the best ligand binding score was retained for the remaining models, with the exception of the 1.0 Å shift plus 5° rotated TM5 conformation: in this case two distinct low energy ligand binding conformations were found. This resulted in seven final agonist bound models, all of which were evaluated by VLS.
bRho-based antagonist bound β2AR homology model construction
A homology model of β2AR was built from the bRho crystallographic coordinates (PDB ID: 1F88) for comparison with the β2AR structures. The sequence alignment was constructed using ZEGA alignment with gap opening and extension penalties of 2.4 and 0.15 respectively, and a position-specific matrix of alignment weights was used alongside the default substitution matrix to enforce alignment of conserved class A GPCR sequence motifs (Fig. 1) [26]. Some manual adjustment was necessary to eliminate gaps in the TM regions. Following sequence alignment, the initial homology model was generated and energetically minimized under distance restraints to the starting template with the goal of eliminating conformational strain in bond angles and lengths. Intracellular loop three was not included in the model, while all other loops were modeled by the bRho receptor conformation. Carazolol was positioned in the ligand binding pocket of the homology model by analogy to the β2AR crystal structure, and flexible receptor sidechain/ligand refinement was performed by the Monte Carlo procedure described above. The ligand/receptor interaction energy was scored for a stack of 38 binding pocket conformations, and the model with the best binding score was selected for VLS.
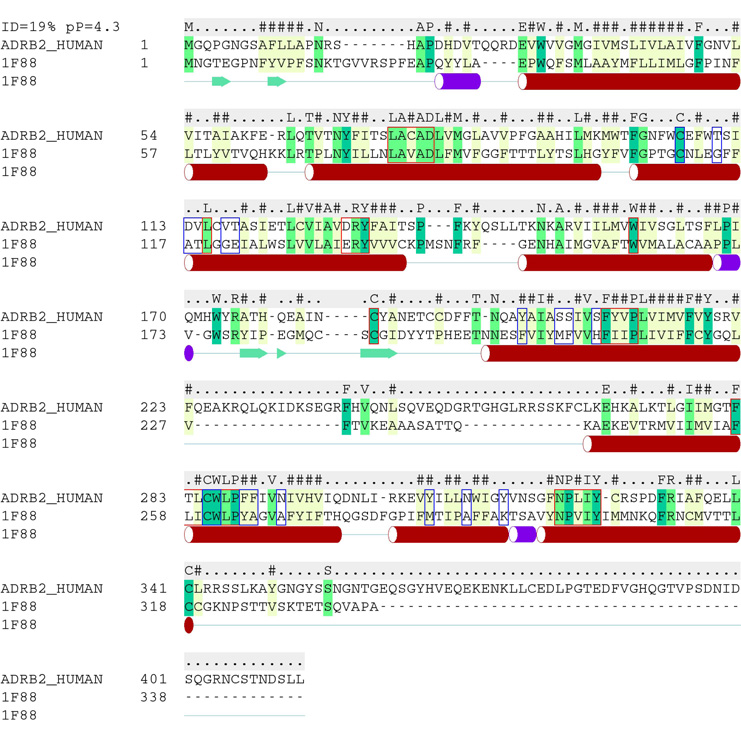
Fig. 1.
Pairwise sequence alignment of bRho and β2AR used to construct the β2AR homology model. The sequence for β2AR was obtained from Swiss-Prot (ID #: P07550), while that of bRho was extracted from PDB file 1F88. The secondary structure of bRho is shown beneath the alignment, and the consensus sequence is given above. The consensus sequence symbols are as follows: letters indicated conserved positions, number signs (‘#”) are used to denote hydrophobic positions, and dots (‘.’) indicate non-conserved positions. Comparison of the two full-length sequences yields a sequence identity of 19%. Conserved GPCR sequence motifs are outlined with red boxes, and binding pocket residues (defined as those within 4 Å of carazolol) are outlined in blue. Sequence identity within the binding pocket is 16.7%
Docking and virtual ligand screening (VLS)
For all VLS results, fully flexible ligand docking was performed by ICM BPMC to a rigid receptor model under softened van der Waals conditions[27]. The receptor binding pocket was represented by six grid potentials of 0.5 Å spacing, including three van der Waals grid potentials for a carbon probe, large atom probe or hydrogen probe, a hydrogen bonding grid potential, an electrostatic grid potential, and a hydrophobic grid potential. For initial VLS evaluations of the antagonist and agonist bound models, the docking grids encompassed a ligand binding pocket described by all receptor residues within 4.5 Å of the ligand in the β2AR crystal structure (Table 2 and Table 4). However, it was observed that the grid could be extended by 8 Å towards TMs 1 and 2 without compromising the screening results. This expanded docking box allowed several of the longer ligands to occupy a sub-pocket between TM2, 3 and 7. Thus, for all other docking runs the extended grid box was used (Table 3 and Table 5). Each ligand was docked twice and the best scoring conformation retained to account for the stochastic nature of the docking process.
Table 2.
Comparison of VLS performance for several antagonist-bound β2AR binding pocket conformations, evaluated upon the 1K test set
| bRho homology Model |
PDB-based β2AR model |
carazolol-refined β2AR model |
Maximum (1K Test Set) |
|
|---|---|---|---|---|
| Antagonists | ||||
| EF / Hit Rate (1 %) | 6.4 / 10% | 50.9 / 80% | 50.9 / 80% | 63.6 / 100% |
| EF / Hit Rate (5 %) | 2.7 / 4.2% | 14.6 / 22.9% | 15.9 / 25% | 19.9 / 31.2% |
| EF / Hit Rate (10 %) | 3.3 / 5.3% | 8.0 / 12.6% | 8.0 / 12.6% | 10.0 / 15.8% |
| Yield (10 %) | 33.3% | 80% | 80% | 100% |
| Agonists | ||||
| EF / Hit Rate (1 %) | 0 / 0% | 0 / 0% | 6.4 / 10% | 63.6 / 100% |
| EF / Hit Rate (5 %) | 5.3 / 8.3% | 0 / 0% | 6.6 / 10.4% | 19.9 / 31.2% |
| EF / Hit Rate (10 %) | 4.0 / 6.3% | 2.7 / 4.2% | 5.4 / 8.4% | 10.0 / 15.8% |
| Yield (10 %) | 40% | 26.7% | 53.3% | 100% |
Table 4.
Evaluation of hypothetical agonist bound β2AR conformations by VLS, using the 1K test set. The labels “conf 1” and “conf 2” refer to two distinct low energy ligand binding conformations observed the 1.0 Å shift plus 5° rotated TM5 model.
| 1.0 Å, | 1.0 Å, | |||||||
|---|---|---|---|---|---|---|---|---|
| no | 0.5 Å, | 5 ° rot, | 5 °rot, | |||||
| shift | 0.5 Å | 1.0 Å | 2.0 Å | 5 °rot | conf 1 | conf 2 | Max | |
| Antagonists | ||||||||
| EF / Hit Rate (1 %) | 6.4 / 10% | 12.7 / 20% | 6.4 / 10% | 6.4 / 10% | 12.7 / 20% | 31.8 / 50% | 0 / 0% | 63.6 / 100% |
| EF / Hit Rate (5 %) | 1.3 / 2.1% | 2.7 / 4.2% | 1.3 / 2.1% | 4.0 / 6.3% | 8.0 / 12.5% | 13.3 / 20.8% | 0 / 0% | 19.9 / 31.2% |
| EF / Hit Rate (10 %) | 2.7 / 4.2% | 4.0 / 6.3% | 2.0 / 3.2% | 3.3 / 5.2% | 5.4 / 8.4% | 7.4 / 11.6% | 1.3 / 2.1% | 10.0 / 15.8% |
| Yield (10 %) | ||||||||
| 26.7% | 40.0% | 20.0% | 33.3% | 53.3% | 73.3% | 13.3% | 100% | |
| Agonists | ||||||||
| EF / Hit Rate (1 %) | 19.1 / 30% | 31.8 / 50% | 50.9 / 80% | 19.1 / 30% | 0 / 0% | 31.8 / 50% | 0 / 0% | 63.6 / 100% |
| EF / Hit Rate (5 %) | 11.9 / 18.8% | 13.3 / 20.8% | 18.6 / 29.2% | 11.9 / 18.8% | 8.0 / 12.5% | 11.9 / 18.8% | 4.0 / 6.3% | 19.9 / 31.2% |
| EF / Hit Rate (10 %) | 6.7 / 10.5% | 8.0 / 12.6% | 9.4 / 14.7% | 6.7/ 10.5% | 6.7/ 10.5% | 8.0 / 12.6% | 4.7 / 7.4% | 10.0 / 15.8% |
| Yield (10 %) | 66.7% | 80.0% | 93.3% | 66.7% | 66.7% | 80.0% | 46.7% | 100% |
Table 3.
Comparison of VLS performance for several agonist and antagonist bound β2AR models, evaluated upon the 14K test set
| PDB-based β2AR model |
carazolol-refined β2AR model |
β2AR Agonist Model - 1Å shift |
Maximum (14K test set) |
|
|---|---|---|---|---|
| Antagonists | ||||
| EF / Hit Rate (1 %) | 36.5 / 38.6% | 33.1 / 35% | 1.4 / 1.4% | 94.6 / 100% |
| EF / Hit Rate (5 %) | 10.7 / 11.3% | 10.8 / 11.4% | 1.5 / 1.6% | 20.0 / 21.1% |
| EF / Hit Rate (10 %) | 6.4 / 6.8% | 6.0 / 6.4% | 1.4 / 1.4% | 10.0 / 10.6% |
| Yield (10 %) | 64.2% | 60.14% | 13.5% | 100% |
| Agonists | ||||
| EF / Hit Rate (1 %) | 1.6 / 2.1% | 13.0 / 17.9% | 38.4 / 52.9% | 72.6 / 100% |
| EF / Hit Rate (5 %) | 3.5 / 4.9% | 6.6 / 9.1% | 12.7 / 17.4% | 20.0 / 27.6% |
| EF / Hit Rate (10 %) | 3.3 / 4.5% | 4.9 / 6.7% | 7.4 / 10.2% | 10.0 / 13.8% |
| Yield (10 %) | 32.6% | 48.7% | 74.1% | 100% |
Table 5.
Comparison of VLS performance for the full length and EL2-deleted β2AR structures, using the 1K test set
| PDB-based | β2AR agonist | |||
|---|---|---|---|---|
| PDB-based β2AR | β2AR model – | β2AR agonist | model – no EL2 | |
| model | no EL2 loop | model | loop | |
| Antagonists | ||||
| EF / Hit Rate (1 %) | 50.9 / 80% | 50.9 / 80% | 6.4 / 10% | 6.4 / 10% |
| EF / Hit Rate (5 %) | 16.0 / 25% | 13.3 / 20.8% | 1.3 / 2.1% | 1.3 / 2.1% |
| EF / Hit Rate (10 %) | 8.7 / 13.7% | 6.7 / 10.5% | 2.0 / 3.2% | 1.3 / 2.1% |
| Yield (10 %) | 86.70% | 66.7% | 20% | 13.3% |
| Agonists | ||||
| EF / Hit Rate (1 %) | 0 / 0% | 0 / 0% | 50.9 / 80% | 31.8 / 50% |
| EF / Hit Rate (5 %) | 2.7 / 4.2% | 0 / 0% | 17.2 / 27.1% | 14.6 / 23% |
| EF / Hit Rate (10 %) | 2.0 / 3.2% | 2.0 / 3.2% | 9.4 / 14.7% | 7.4 / 11.6% |
| Yield (10 %) | 20% | 20% | 93.3% | 73.3% |
Ligand screening test set construction
Two datasets were compiled for VLS, both derived from the GPCR ligand database (GLIDA) (Table 1)[28]. Both test sets exclusively contained small molecule GPCR ligands, and should therefore rigorously test the discrimination of decoy and non-decoy ligands that share physical properties consistent with binding the GPCR TM domain. All ligands were limited in size to a molecular weight between 100 and 500 daltons, and the annotations for receptor agonist and antagonist activity were taken from GLIDA. The full dataset included 14006 ligands, 347 of them annotated as β2AR binders (referred to as the 14K test set). A smaller test set of 954 compounds was constructed by choosing 15 well-known β2AR antagonist and agonist compounds from GLIDA, and adding 924 randomly selected decoy compounds (referred to as the 1K test set). Isoproterenol and carazolol were omitted from this test set, since these ligands were used in the receptor refinement step. Formal charges were assigned to both test sets using the pH-model functionality of openBabel version 2.0.1[29]. As the most active stereoisomer of each compound was not always known or annotated, all stereoisomers were generated for both test sets to yield a total of 55711 (14K test set) or 3757 (1K test set) compounds. Following docking, all stereoisomers of a given compound were compared, and the best-scoring stereoisomer was retained.
Table 1.
Compound test sets for VLS trials
| Dataset | Total Number of Compounds |
Number of β2AR ligands |
Number of β2AR antagonists |
Number of β2AR agonists |
|---|---|---|---|---|
| 1K Test Set | 954 | 30 (3.1 %) | 15 (1.6 %) | 15 (1.6%) |
| 14K Test Set | 14006 | 347 (2.5%) | 148 (1.1%) | 193 (1.4%) |
Evaluation of VLS results
The VLS results were quantitatively assessed by calculating enrichment factors, hit rates, and yields for the top scoring 1%, 5%, and 10% of all compounds. Receiver operator characteristic (ROC) plots were also generated, which depict the true positive rate on the y axis, and false positive rate on the x axis. The equations used for assessing the VLS results are shown below:
| (1) |
In the above equations, Lt and Dt are the total number of known ligands and decoy compounds respectively, where Lt + Dt equals the total number of compounds in the test set. Lf and Df are the number of known ligands and decoy compounds in some fractional subset of the database (f).
Results
Unrestrained VLS with the unmodified β2AR crystallographic coordinates achieves high antagonist enrichment
The crystal structure coordinates of β2AR in complex with carazolol were prepared for VLS by the addition and optimization of hydrogen atoms in internal coordinates space to yield a full atom PDB-based β2AR model. Coordinates of β2AR heavy atoms were not modified from those in the crystal structure. To evaluate docking performance, VLS was conducted upon challenging 1K and 14K ligand test sets consisting of only GPCR ligands (Table 1). For the 1K test set, the PDB-based β2AR model provided an enrichment factor of 50.9 for antagonist compounds, and a hit rate of 80% for the top scoring 1% of the database (10 compounds) (Table 2). Similarly, an enrichment factor of 36.5 and hit rate of 38.6% were obtained for the top scoring 1% (140 compounds) of the 14K test set (Table 3). High yields of known antagonist compounds in the hit lists indicated that the PDB-based model is able to retrieve a number of structurally diverse small molecules − 80% of the known antagonist compounds contained in the 1K test set were retrieved in the top-scoring 10%. The yield was slightly lower for the 14K test set, though this may be due in part to inaccuracies in database annotation of antagonist/agonist ligand function. Recently Topiol et al. also reported qualitatively successful antagonist VLS results for the β2AR crystal structure using a proprietary compound database, though values for enrichment factors or yields were not provided [30]. Our quantitative results are consistent with their findings, and are extended to a stringent test set that is restricted to only known GPCR ligands. Notably, the high enrichment factors for antagonist compounds were obtained without the use of distance restraints or a template ligand during docking. Several previous VLS studies with bRho derived homology models have included ligand/receptor distance restraints or pharmacophoric information to guide docking[31; 32; 33; 34]. While the inclusion of experimental information during docking is an important strategy for improving pose prediction and ligand retrieval, such information is not available for all receptor types. The encouraging results here suggest that successful VLS results may be obtained for GPCRs given an accurate structural model, even in the absence of experimental restraints.
VLS with a bRho-based β2AR homology model fails due to inaccuracies in EL2 loop modeling
For comparison to the crystal structure, a β2AR homology model was constructed from bovine rhodopsin. The homology model was generated using standard sequence alignment and homology modeling procedures within ICM (Fig. 1). The backbone conformation of bRho was kept fixed, including all loop regions, and ligand-directed refinement of the binding pocket sidechains with fully flexible (−)-carazolol was performed to optimize the conformation of the homology model ligand binding pocket. A number of bRho homology models presented in the literature have been constructed following a similar procedure of retaining the conformation of EL2 from bRho, and refining the binding pocket conformation in the presence of a manually placed ligand[11; 31]. In other studies, more sophisticated homology models of β2AR have been constructed, which made use of balloon potentials, flexible loop modeling, and/or molecular dynamics to refine the receptor binding pocket conformation[16; 35; 36; 37; 38]. Such models may well perform better in VLS than the model presented here. However, the goal of this study was to compare VLS results for the β2AR crystal structure with a reference homology model generated by a relatively straightforward process of ligand-directed refinement.
Structural examination of the homology model and the carazolol-bound β2AR crystallographic coordinates identifies substantial differences in binding pocket shape and ligand placement (Fig. 2a,b). The RMSD between heavy atoms of carazolol in the homology model and crystal structure is 3.72 Å. The orientation of EL2 in bRho generates a clash between Tyr185 and the crystallographic conformation of carazolol, thus in the homology model carazolol is shifted away from the extracellular part of the GPCR and deeper into the TM pocket. This docked conformation of carazolol is similar to that observed by Costanzi in his description of a bRho derived β2AR homology model wherein EL2 was constructed by analogy to that in β2AR. However, we note that Costanzi et al. achieved an improved accuracy in the positioning of the docked carazolol in a model where EL2 was built de novo. In their de novo EL2 model, the loop is significantly more solvent exposed and allows carazolol to dock with an RMSD of 2.90 Å to the crystallographic pose, in comparison to a 3.69 Å RMSD as observed for the bRho-like EL2 conformation. A further improvement to an RMSD of 1.70 Å was made when experimental evidence was used to adjust the rotameric conformation of Phe2906.52, though this step was in part rationalized using information from the β2AR crystal structure [39]. The results of Costanzi et al. highlight the importance of EL2 conformation in determining the shape of the ligand binding pocket, and suggest that for some GPCR homology models, an accurate docked position may be obtained when sufficient experimental data are available to guide modeling.
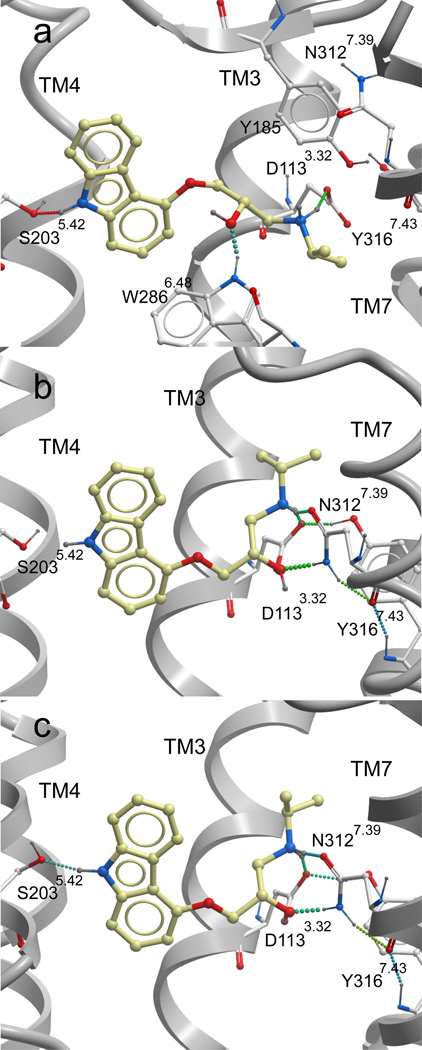
Fig. 2.
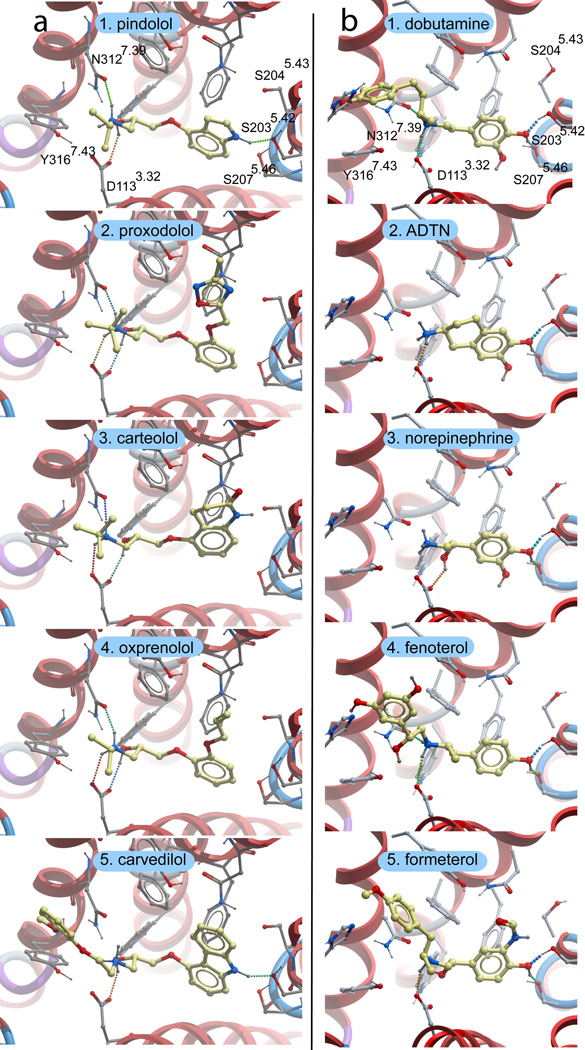
Comparison of the β2AR ligand binding pocket for several antagonist-bound receptor models. Panel (a) depicts the bRho-based β2AR homology model, panel (b) the PDB-based β2AR model and panel (c) the ligand-refined β2AR binding pocket. For all structures, the GPCR backbone is displayed in grey ribbons, and hydrogen bonds depicted with dotted lines. Key binding pocket sidechains are shown in white sticks and the partial inverse agonist carazolol is displayed in yellow. For clarity, the TM6 backbone is not shown, though the W286 sidechain is part of this helix. Ballesteros and Weinstein numbering (in superscript) is not provide for Y185, as this residue is part of the EL2 loop.
Additional inaccuracies in the bRho-based β2AR homology model arise due to the fact that the extracellular portions of TM3 and TM5 are slightly further apart in bRho than the β2AR crystal structure. For comparison, the distance between the cα atoms of Asp1130 (in TM3) and Ser2035.42 (in TM5) is 14.5 Å in the β2AR homology model and only 13.3 Å in the β2AR crystal structure. As a result, in the homology model the ligand is placed closer to TM5 and further from TM3 to allow hydrogen bond formation with both Ser2035.42 and Asp1133.32. As a result of this change in ligand position, the beta hydroxyl of carazolol interacts with the sidechain nitrogen of Trp2866.48 rather than that of Asn3127.39 as observed in the β2AR crystal structure. The total binding pocket RMSD between the crystal structure and homology model is 5.26 Å, when all ligand and receptor sidechain heavy atoms within 4 Å of the crystallographic conformation of carazolol are included.
The VLS performance of the β2AR homology model was evaluated with the 1K ligand test set (Table 2). The homology model achieves far lower enrichment factors and hit rates than those obtained with the crystal structure. A hit rate of 10% was obtained for the top scoring 1% of the database when the homology model was used, in comparison to a hit rate of 80% for the crystal structure. Additionally, the homology model exhibits rather poor agonist/antagonist selectivity. Enrichment factors of 6.4, 2.7, and 3.3 were obtained for antagonist compounds in the top scoring 1%, 5%, and 10% of the test set. The corresponding enrichment factors for agonist compounds were 0, 5.3, and 4.0. These results highlight the inadequacies of the bRho backbone in creating GPCR homology models, particularly in the region of EL2, which exerts a profound effect upon docked ligand conformation. Other GPCR homology models, whether based on bRho or β2AR, may suffer from similar discrepancies in correct conformational prediction of the highly flexible and sequence variable EL2. Removal of this loop may be the most effective means to reduce conformational uncertainty in this region of the receptor structure, and this approach is further evaluated below.
Ligand-directed refinement of the carazolol bound β2AR crystal structure identifies new hydrogen bonding interactions and moderately enhances agonist recognition
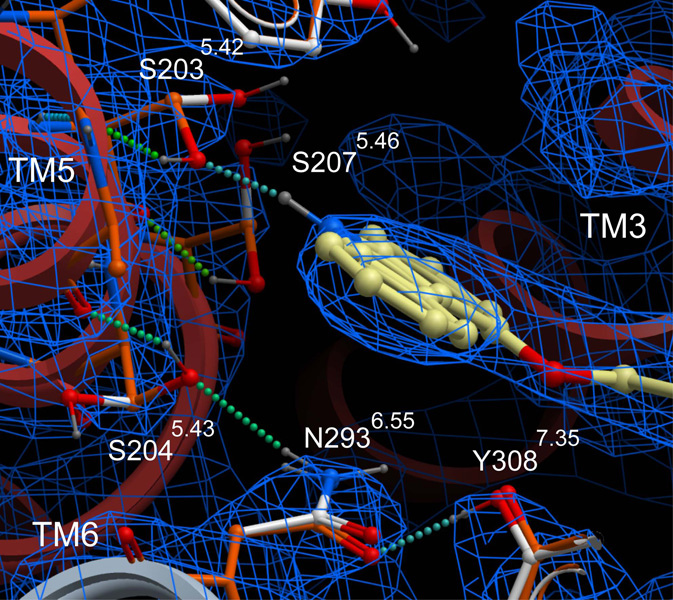
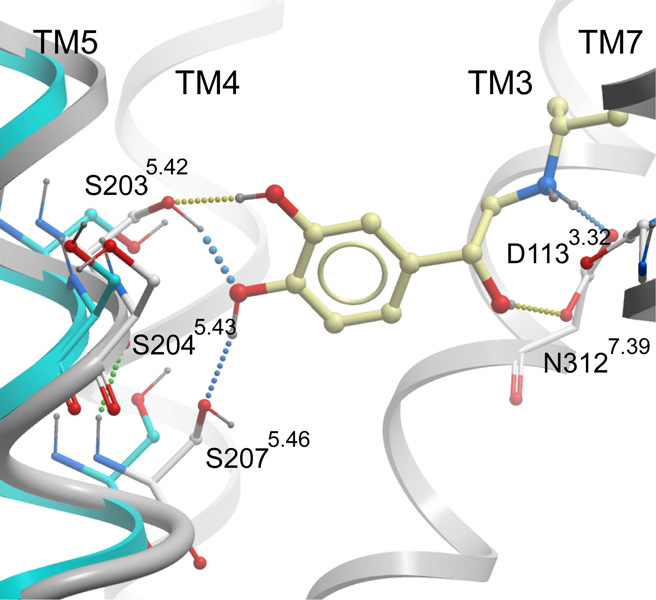
While the protonated crystallographic coordinates performed well as an initial template for VLS, inaccuracies in conformation arising from the uncertainties of electron density fitting can lead to diminished docking and VLS results. Such errors may include incorrect (flipped) orientations of asparagine and glutamine, suboptimal hydrogen bonding networks, and misplaced sidechains. Flexible ligand directed refinement of the receptor sidechains and (−)-carazolol conformation was performed to optimize receptor/ligand interactions and further explore low-energy ligand bound conformations. This refinement introduced only subtle changes in binding pocket conformation, and the heavy atom RMSD for pocket sidechains is 0.62 Å. While the conformation of most sidechains did not change, Ser2035.42, Ser2045.43 and Ser2075.46 were repositioned to form a more optimal hydrogen bonding network (Fig. 3). The new conformation of Ser2045.43 permits a hydrogen bonding interaction with Asn2936.55, −though with a somewhat suboptimal hydrogen bonding distance (3.3 Å). A similar interaction is observed in the structure of β2AR with timolol [5]. Notably, the new conformation of Ser2035.42 improves the geometry of the hydrogen bonding interaction with the carbazole nitrogen of carazolol, and allows formation of a hydrogen bond between the Ser2035.42 hydroxyl and the backbone oxygen of Tyr1995.38.
Fig. 3.
Overlay of the PDB-based and carazolol-refined β2AR models. The β2AR backbone is displayed in grey ribbons. The sidechains of the PDB-based and carazolol-refined β2AR models are displayed as white sticks and green sticks respectively. For clarity, carazolol is shown for the refined β2AR model only, and is depicted in yellow sticks. The electron density is contoured at 1.2 σ and is displayed in blue mesh.
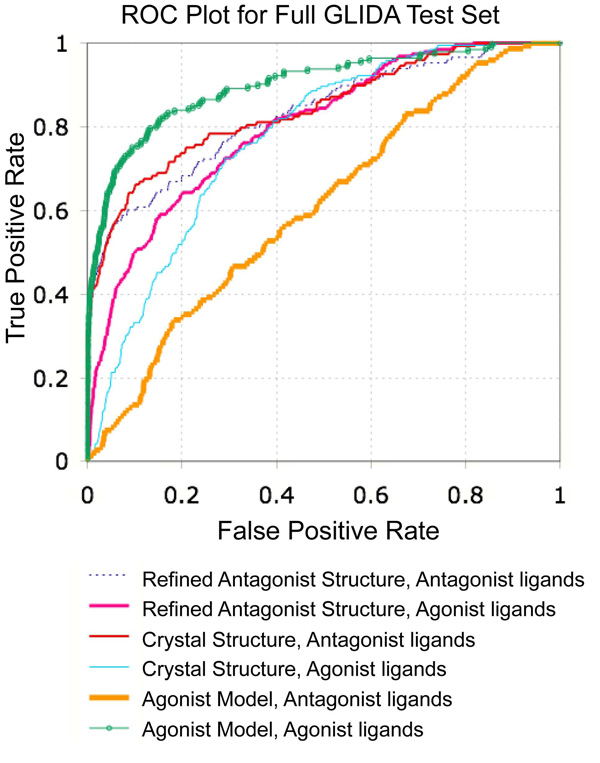
The carazolol-refined binding pocket was evaluated by VLS, using both the 1K and 14K test sets (Table 2 and Table 3). The small changes in pocket conformation had little impact upon antagonist VLS performance; a high enrichment factor of 50.9 is obtained for the top scoring 1% of the 1K database with either the carazolol-refined or PDB-based coordinates. For the top scoring 1% of the 14K data set, enrichment factors of 36.5 and 33.1 were achieved with the PDB-based and carazolol-refined coordinates respectively. Overall, the areas under the ROC curves are similar for both conformations, with close to perfect values of 0.84 and 0.83 for the PDB-based and carazolol-refined conformations (Fig. 4). Importantly, the compounds retrieved by the carazolol-refined coordinates vary in both size and overall structural similarity (Fig. 5a), indicating that the binding pocket conformation is suitable for recognition of other antagonist chemotypes despite refinement with a single antagonist. Though the VLS enrichment factors and hit rates are quantitatively very similar, visual inspection of the docked ligand/receptor conformations finds that repositioning of Ser2035.42 allows several of the top ranked hits to include a hydrogen bonding interaction between the ligand and Ser2035.42 which was overlooked or underestimated by the PDB-based model (Fig. 6a). As such, the carazolol-refined binding pocket conformation provides a more complete description of ligand/receptor interactions. Additionally, for both the 1K and 14K test sets, the yield of agonists retrieved during VLS was moderately increased (Table 2 and Table 3). The carazolol-refined orientations of Ser2035.42 and Ser2075.46 thus provide for somewhat improved agonist recognition, though this decreases the overall antagonist selectivity of the model (Fig. 4).
Fig. 4.
ROC plot comparing VLS results on the 14K test set for the PDB-based β2AR model, carazolol-refined antagonist bound model, and 1 Å shifted agonist bound receptor model. For each of the three receptor conformations, results are indicated separately for the retrieval of agonist and antagonist ligands
Fig. 5.
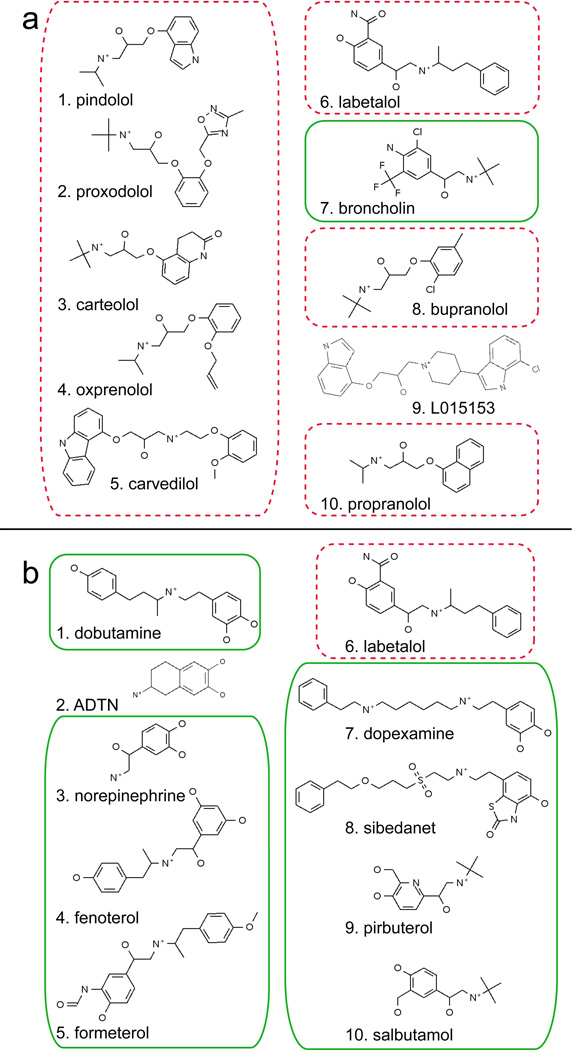
Compound hit lists resulting from VLS with (a) the carazolol-refined antagonist bound model and (b) the 1 Å shifted agonist bound model. The top scoring 1% of compounds from the 1K test set is shown (10 out of 954 in total). Known agonists and antagonists are boxed, the former with a solid green line and the latter by a red dashed line
Fig. 6.
Docked conformations of top-scoring ligands for the (a) carazolol-refined antagonist bound model and (b) 1 Å shifted agonist-bound receptor model. The compounds shown correspond to the five top scoring ligands from panels (a) and (b) of Fig. 5. For all models, the receptor backbone is drawn in ribbons. Ligand is in shown in yellow sticks, receptor sidechains in grey sticks, and hydrogen bonds are depicted with dotted lines. Residue numbering is shown in the two top panels as a reference for the sidechains in the panels below
The antagonist bound β2AR models fail in agonist VLS and do not capture key agonist/receptor interactions
Though the carazolol-refined model of β2AR displays an improved yield for agonist compounds, the agonist enrichment factors for both the PDB-based β2AR model and carazolol-refined β2AR model remain very low. For the PDB-based β2AR model, agonist enrichment factors of 0 are obtained for both the top scoring 1% and 5% of the 1K test set, while the carazolol-refined structure exhibits enrichment factors of 6.4 and 6.6 (Table 2). Docking of isoproterenol, a known full agonist, to the rigid crystal structure finds that the distance between TM3 and TM5 is too great for the simultaneous formation of hydrogen bonding interactions between the ligand amine with Aspartate 1133.32 in TM3 and the ligand hydroxyls with Serines 2035.42,2045.43 and 2075.46 in TM5. However, mutagenesis studies have demonstrated that both interaction points are critical for agonist binding affinity [40]. Thus, in the present work small shifts (0.5 Å) and rotations (5°) are introduced in TM5 to decrease the distance between the TM5 Serines and D1133.32.
A single binding pocket conformation retrieves both full and partial agonist ligands
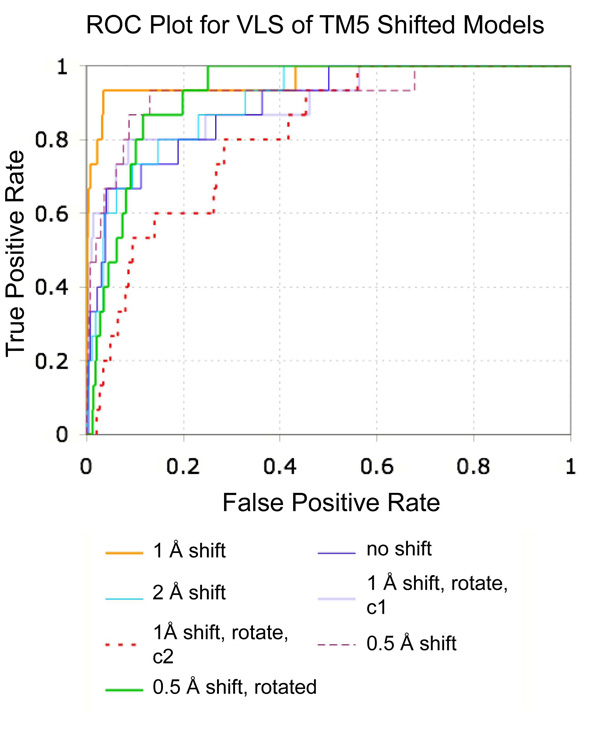
A series of seven hypothetical agonist bound β2AR models was constructed by rigid body translation and rotation of TM5, followed by ligand directed receptor refinement with (−)-isoproterenol, a known full agonist. All seven models were evaluated in VLS trials using the 1K test set (Table 4 and Fig. 7). The 1 Å shifted conformation of TM5 provided the best enrichment factors and hit rates for known agonists. Examination of the binding pocket conformation for this model finds that the 1 Å shift of TM5 allows formation of two hydrogen bonds between the para hydroxyl of the catecholamine ring and Ser2035.42 and 2075.46 while maintaining the hydrogen bond between the ligand amine and D1133.32 (Fig. 8). The meta hydroxyl of isoproterenol additionally hydrogen bonds to Ser2035.42, though no interaction was observed with Ser2045.43, in contrast to the conformation proposed by Strader et al. in keeping with their mutagenesis experiments [40]. Ser2045.43 is instead predicted to form a hydrogen bond to the backbone of Ala2005.39, and is in a conformation similar to that observed in the carazolol bound crystal structure. During conformational optimization of the 1 Å shifted model, an additional binding pocket conformation was obtained that includes a hydrogen bond between isoproterenol and Ser2045.43 hydrogen bond. However, the calculated energy of the conformation was lower than that obtained for the Ser2045.43-Ala2005.39 structure, and this binding pocket conformation displayed significantly poorer VLS enrichment. Interestingly, previous work by Vaidehi and co-workers yielded a norepinephrine bound model of β2AR wherein the para hydroxyl of norepinephrine forms a hydrogen bond with Ser2035.42 and the meta hydroxyl of norepinephrine interacts with both Ser2045.43 and Ser2075.46. These models were generated by a de novo modeling procedure that included systematic rotations of TM5. However, the agonist vs. antagonist selectivity of these models was not tested in VLS. We note that our model may not reflect the optimal geometry of TM5, as only a discrete set of TM5 positions was sampled. Additional refinement of the TM5 position that includes smaller translational steps, rotations and helical tilts could possibly improve the accuracy of the binding pocket conformation, though we find that the present model performs quite well in VLS.
Fig. 7.
ROC plot comparing VLS results for seven hypothetical agonist bound models of β2AR. The shift and/or rotation of TM5 for each model is noted in the legend. The labels “conf 1” and “conf 2” refer to two distinct low energy ligand binding conformations observed for the 1.0 Å shift plus 5° rotated TM5 model. Known agonist compounds were considered as true positives, while antagonists and ligands for other receptors were considered false positives. All seven models were assessed using the 1K test set consisting of 954 compounds
Fig. 8.
Binding pocket conformation for the agonist bound receptor model that performs best in VLS trials. The crystallographic conformation of TM5 is shown in cyan, while the 1 Å shifted (agonist-bound) conformation of TM5 is shown in grey. Isoproterenol is shown in yellow sticks, and hydrogen bonds are depicted with dotted lines
Notably, in the agonist bound model the position of (−)-isoproterenol is shifted closer to TM3 (and away from TM7) relative to the conformation of (−)-carazolol, disrupting the hydrogen bond network between ligand, Tyr3167.43, and Asn3127.39. This change in hydrogen bonding pattern could again arise from insufficient sampling of TM5 position and orientation. Alternatively, the 1 Å shift identified here may be a suboptimal distance for the docking of isoproterenol, while allowing broader recognition of multiple ligand types during VLS. Screening with this single pocket conformation achieved agonist enrichment factors of 50.9, 18.6, and 9.4 for cutoffs at the top scoring 1%, 5%, and 10% of database molecules, respectively. Inspection of the hit list found that the top scoring 1% of the database contained both catecholamine (norepinephrine, dobutamine, dopexamine) and non-catecholamine (salmeterol, pirbuterol, sibenadet, formoterol, and fenoterol) ligands, as well as full (norepinephrine) and partial (salbutamol) agonists (Fig. 5b). Importantly, the top scoring agonist VLS hits all formed hydrogen bonding interactions between the ligand para hydroxyl and Ser2035.42 and 2075.46, demonstrating the importance of this feature for agonist/antagonist selectivity (Fig. 6b). For several top-ranking VLS hits, the meta hydroxyl of the catecholamine ring was oriented “downwards” in the pocket, and away from Ser2035.42 and Ser2045.43. This conflicts with previous experimental mutagenesis data indicating a hydrogen bonding interaction between the catecholamine meta hydroxyl, Ser2035.42, and possibly Ser2045.43 [40; 41]. We note that during VLS the receptor pocket is represented as a softened 0.5 Å grid. As such, the grid-docked ligand conformations may not be as accurate as calculations performed in the context of a full atom model. However, it is interesting to note that when superimposed to the recent 2.8 Å structure of β2AR with timolol, the docked position of the agonist meta-hydroxyl (for both dobutamine and norepinephrine) aligns exactly to the sulfur of the timolol thiadiazole ring [5]. In the timolol-bound β2AR structure, this results in formation of a hydrogen bond between the thiadiazole ring and Thr118 in TM3. The docked poses here suggest the possibility of a similar interaction between Thr118 and the agonist meta hydroxyl as an alternative agonist binding conformation.
Following selection of the 1Å shift structure using the 1K VLS test set, the performance of this model was further assessed with the full 14K test set (Table 3). With the expanded test set, enrichment factors of 38.4, 12.7 and 7.4 were calculated for 1%, 5% and 10% scoring cutoffs. ROC curves comparing the agonist and antagonist selectivity of the 1 Å shift agonist bound model with that of the PDB-based β2AR model and carazolol-refined antagonist binding pocket are shown in Figure 4. Interestingly, analysis of the hit list identified several low-ranking agonist compounds (including deoxyepinephrine and ritodrine) that were not annotated as agonists within GLIDA and therefore counted as false positives within the 14K test set. Several known β2AR antagonists were also mis-labeled within the 14K test set as agonists or inactives, including carazolol, fluorocarazolol, isamoltan, and cyanopindolol. While the assignment of agonist or antagonist was confirmed manually for the β2AR ligands of the 1K test set, the annotations for the 14K test set were taken directly from GLIDA. These incorrect assignments of agonist and antagonist in GLIDA, combined with the possible presence of test set ligands with uncharacterized β2AR activity, may have artificially lowered hit rates and enrichment factors in the evaluation of both agonist and antagonist models.
EL2 deletion does not alter docked ligand pose, and only moderately affects VLS enrichment
The second extracellular loop (EL2) connects TM4 and TM5, and helps to define the overall size and shape of the transmembrane ligand binding pocket. While atomic contacts between EL2 and carazolol are limited in the β2AR crystal structure, this loop may form specific contacts with other agonists and antagonists critical for ligand binding and subtype specificity [42; 43]. For several reasons, the EL2 loop poses a special challenge in GPCR homology modeling: it is conformationally flexible, varies widely in length and amino acid composition between receptor type, and has been crystallized in two distinct conformations for the rhodopsin and β1AR/β2AR receptors. One strategy for coping with the conformational uncertainty of this loop is to omit it from the structure [10]. In recent work by de Graaf et al., VLS performance was evaluated for bRho based models of the dopamine D2, adenosine A3 and thromboxane A2 receptors, where EL2 was either deleted or modeled by homology to bovine rhodopsin[44]. In two of the three cases, EL2-deleted models performed better or as well as the rhodopsin-modeled loop structures.
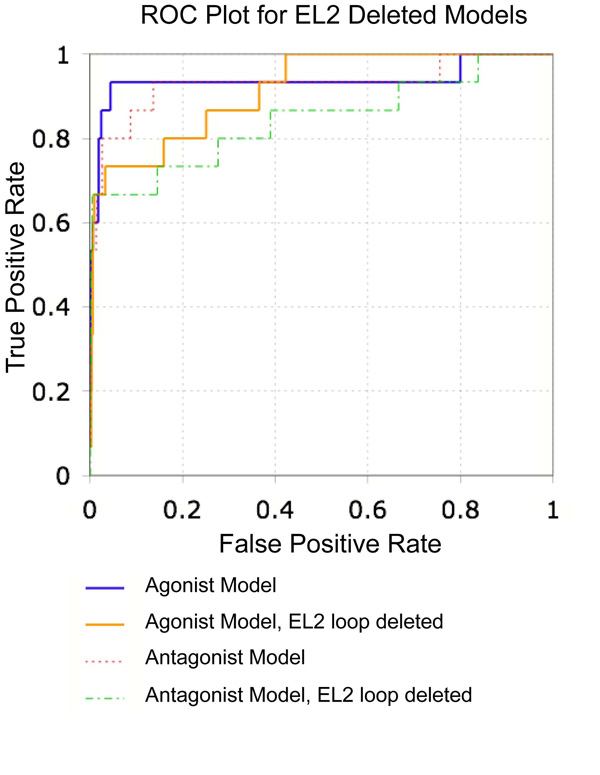
The impact of this loop on β2AR docking was assessed by deleting EL2 (residues 170–197) from both the PDB-based β2AR model and best agonist model and conducting VLS trials. The absence of EL2 did not affect the docked conformation of either (−)- carazolol with the crystal structure, or (−)-isoproterenol with the agonist-bound model. Carazolol docked to the EL2 truncated version of the receptor with an RMSD of 0.23 Å to the crystallized orientation, while the RMSD of isoproterenol for the loop deleted and intact agonist bound models was 0.16 Å. Omission of EL2 had a relatively small impact on VLS performance. Antagonist enrichment factors for the top scoring 1% and 5% of the 1K test set is comparable in the presence and absence of EL2, though the total yield of antagonists in the top scoring 10% was somewhat decreased, from 86.7% to 66.7%. Enrichment factors were also decreased for the agonist bound model in VLS (Table 5, Fig. 9). Inspection of the hit lists indicated that the overall selectivity of the agonist bound model was reduced. For example, when the full length agonist bound model was used for VLS, the top scoring 1% of the 1K test set (10 compounds) was composed of eight known β2AR agonists, one β2AR antagonist, and a serotonin 5HT2c antagonist. When the loop was deleted, the hit list consisted of five known β2AR agonists, one β2AR antagonist, one β3AR antagonist, an α-adrenergic antagonist, and two D1 dopamine receptor agonists. Notably, both the serotonin and dopamine receptors share approximately 70% sequence identity to β2AR within the ligand binding pocket. Nonetheless, VLS in the absence of EL2 provides high enrichment factors and hit rates, with enrichment factors of 50.9 (antagonists) and 31.8 (agonists) for the top scoring 1% of the 1K test set. This indicates that omission of EL2 from receptor homology models is a suitable alternative when experimental structural data or modeling techniques are unable to provide accurate conformational information.
Fig. 9.
ROC plot comparing VLS results for the β2AR crystal structure and best agonist bound model with and without the EL2 loop. The 1K test set was used for evaluation. For each conformation, results are shown separately for the retrieval of agonist and antagonist compounds
Discussion
The β2AR crystal structures are a major step forward in conducting structure based drug discovery for adrenergic receptors. Enrichment factors and hit rates for antagonist compounds were greatly improved when the carazolol bound PDB-based β2AR model was used for screening relative to those obtained with a bRho-based homology model. Additionally, while the PDB-based β2AR model is antagonist selective, a 1 Å shift of TM5 yielded a receptor conformation that selectively retrieves structurally diverse agonist compounds. Importantly, it is not the case that shifting of TM5 merely positions the receptor to recognize a trivial catecholamine pharmacophore, composed of an appropriately positioned positively charged amine group and two hydrogen bond acceptors. If the only recognition provided by the full binding pocket structure was a recapitulation of these two features, one would anticipate vastly deteriorated β2AR selectivity. However, it is observed that the agonist bound receptor conformation is specific for β2AR agonists, regardless of the high structural similarity of many dopamine and serotonin agonists contained in both GPCR ligand test sets. Modeling of the agonist-bound receptor pocket is especially important, as this conformational state may prove particularly difficult to crystallize. Fluorescence lifetime spectroscopy experiments have shown that even under saturating concentrations of agonist ligand, at least two β2AR conformational states exist - a less populated fully activated conformation, and a more abundant partially activated intermediate[45]. This conformational heterogeneity is likely to effect the formation of well-organized agonist-bound receptor crystals. Here we find that in the absence of crystallographic information, construction of agonist models that attain high VLS enrichment factors is possible.
The 1Å shifted model provides excellent enrichment factors and yields for a large set of structurally diverse agonist compounds including both full and partial agonists. However, as noted in the results, in some cases the ligand/receptor interactions anticipated from experimental mutagenesis data are not always achieved during docking. For example, a hydrogen bond is not observed between the meta-hydroxyl of (−)-isoproterenol and Ser2045.43, and for several agonists the meta-hydroxyl is observed to point deeper into the ligand binding pocket away from Ser2035.42/Ser2045.43. This highlights a potential weakness in our method: because VLS is used as the criteria for model selection, the “best” model may be a binding pocket conformation that approximates for recognition of many ligand types, but does not capture the optimum conformation for any particular compound. As such, the 1Å shift of TM5 may represent an intermediate distance suitable for retrieval of both full and partial agonists. Further optimization of TM5 position may yield receptor conformations exquisitely tuned for the recognition of full or partial agonist compounds. In ongoing work in our lab, optimization of β2AR with various agonists has been found to introduce different shifts in TM5 that roughly correlate with agonist strength (Katritch V., Cherezov V., Hanson M.A., Roth C.B., Reynolds K.A. and Abagyan R., manuscript submitted). Interestingly, recent work from de Graaf and Rognan has also used a structure based approach to screen for β2AR agonists[46]. In their model, the TM5 serine sidechains are repositioned to allow interaction with isoproterenol, but the backbone position of TM5 is unaltered. In VLS trials, this conformation retrieves both agonists and antagonists. Selectivity for full and partial agonists is only achieved when molecular interaction fingerprints are included as an additional constraint. Here we find that structure based screening can be made fully selective for agonists without additional constraints by a simple modification of the TM5 helix position.
The protocol employed here for agonist bound model generation consists of systematically introducing TM5 shifts in 0.5 Å increments, performing ligand-directed binding pocket sidechain optimization, and conducting receptor conformational selection by VLS. Future work could expand upon this methodology to incorporate additional rigid body motions of TM5, and/or TM6, including shifts, tilts, rotations, and vertical translations, in either isolation or combination. This ligand directed procedure is fairly simple and should be straightforward to apply to other receptor types, for which TM5 shifts can play a role in receptor recognition. However, homology modeling is complicated by EL2, a region of low sequence identity and high conformational flexibility. Importantly, ligand docking to the β2AR models in the absence of EL2 yielded a ligand conformation identical to the full-length receptor models, indicating that omission of EL2 still allows for correct ligand docking and evaluation of receptor/ligand interactions. The VLS results were somewhat worsened by removing EL2, but nonetheless provided significant enrichment for known agonist and antagonist compounds. One possible modeling strategy could take advantage of the ability for ligands to dock correctly in the absence of EL2. First, the receptor could be modeled without EL2, then a known ligand then docked into the binding pocket, and conformation of the loop subsequently refined in the presence of docked ligand. A similar strategy was followed by Evers and colleagues in the construction of an α1A Adrenergic receptor model, and our results here demonstrate that this is a highly valid approach [34].
Conclusions
In summary, the β2AR crystallographic coordinates provide an excellent template for conducting antagonist VLS, while a bRho-based homology model fails due to inaccuracies in backbone conformation, particularly in EL2. Through systematic variation of the antagonist bound conformation, agonist bound models can be generated that selectively retrieve agonist compounds. Exceptionally high hit rates and enrichment factors were obtained using these structures, even when the screening test set was restricted to GPCR ligands, a significant fraction of which bind to other adrenergic subtypes and aminergic receptors. These results were accomplished using a standard docking and scoring procedure without need for a customized receptor specific scoring potential, incorporation of pharmacophoric information, or the inclusion of ligand/receptor distance restraints. Though further studies with other receptor types are necessary to evaluate the general application of this procedure, we have demonstrated that deletion of EL2 may be a suitable strategy for limiting structural uncertainty in future GPCR homology models. Full atom structures of both the agonist bound and carazolol-refined β2AR models are provided as supplementary information.
Supplementary Material
The 1 Å shifted agonist bound and carazolol-refined antagonist bound β2AR models are provided in PDB format as electronic supplementary material.
Acknowledgements
K.A.R. and R.A. gratefully acknowledge support from the National Institutes of Health (GM074832).
References
- 1.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 2.Park JH, Scheerer P, Hofmann KP, Choe H-W, Ernst OP. Nature. 2008 doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 3.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Nature. 2008 doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Kuhn P, Weis WI, Kobilka BK, Stevens RC. Science. 2007;318:1258. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola V-P, Chien EYT, Velasquez J, Kuhn P, Stevens RC. Structure. 2008;16:897. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenakin T. Trends in Pharmacological Sciences. 2003;24:346. doi: 10.1016/S0165-6147(03)00167-6. [DOI] [PubMed] [Google Scholar]
- 7.Kobilka BK, Deupi X. Trends in Pharmacological Sciences. 2007;28:397. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Urban JD, Clarke WP, von Zastrow M, Nichols DE, Kobilka B, Weinstein H, Javitch JA, Roth BL, Christopoulos A, Sexton PM, Miller KJ, Spedding M, Mailman RB. J Pharmacol Exp Ther. 2007;320:1. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. Annual Review of Pharmacology and Toxicology. 2006;46:481. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 10.Bottegoni G, Kufareva I, Totrov M, Abagyan R. Journal of Computer-Aided Molecular Design. 2008;22:311. doi: 10.1007/s10822-008-9188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bissantz C, Bernard P, Hibert M, Rognan D. Proteins: Structure, Function, and Genetics. 2003;50:5. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 12.Ballesteros JA, Weinstein H. Methods Neurosci. 1995;25:366. [Google Scholar]
- 13.Rasmussen SGF, Choi H-J, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK. Nature. 2007;450:383. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 14.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Yao X-J, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharya S, Hall SE, Li H, Vaidehi N. Biophys. J. 2008;94:2027. doi: 10.1529/biophysj.107.117648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouldson PR, Kidley NJ, Bywater RP, Psaroudakis G, Brooks HD, Diaz C, Shire D, Reynolds CA. Proteins. 2004;56:67. doi: 10.1002/prot.20108. [DOI] [PubMed] [Google Scholar]
- 17.Bisson WH, Cheltsov AV, Bruey-Sedano N, Lin B, Chen J, Goldberger N, May LT, Christopoulos A, Dalton JT, Sexton PM, Zhang X-K, Abagyan R. PNAS. 2007;104:11927. doi: 10.1073/pnas.0609752104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavasotto CN, Orry AJW, Murgolo NJ, Czarniecki MF, Kocsi SA, Hawes BE, x , Neill KA, Hine H, Burton MS, Voigt JH, Abagyan RA, Bayne ML, Monsma FJ. J. Med. Chem. 2008;51:581. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 19.Mehler EL, Hassan SA, Kortagere S, Weinstein H. Proteins: Structure, Function, and Bioinformatics. 2006;64:673. doi: 10.1002/prot.21022. [DOI] [PubMed] [Google Scholar]
- 20.Kortagere S, Roy A, Mehler E. Journal of Computer-Aided Molecular Design. 2006;20:427. doi: 10.1007/s10822-006-9056-0. [DOI] [PubMed] [Google Scholar]
- 21.Abagyan R, Totrov M. Journal of Molecular Biology. 1994;235:983. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 22.Nemethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA. J.Phys. Chem. 1992;96:6472. [Google Scholar]
- 23.Halgren TA. J Comp Chem. 1996;17:490. [Google Scholar]
- 24.Raffa RB, editor. Drug-Receptor Thermodynamics Introduction and Applications. Wiley: VCH; 2001. [Google Scholar]
- 25.Totrov M. Chem Biol Drug Des. 2008;71:15. doi: 10.1111/j.1747-0285.2007.00605.x. [DOI] [PubMed] [Google Scholar]
- 26.Abagyan RA, Batalov S. Journal of Molecular Biology. 1997;273:355. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- 27.Totrov M, Abagyan R. Proteins. 1997 Suppl 1:215. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 28.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, Kunimoto R, Feng C. Nucl. Acids Res. 2008;36:D907. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. J. Chem. Inf. Model. 2006;46:991. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topiol S, Sabio M. Bioorganic & Medicinal Chemistry Letters. 2008;18:1598. doi: 10.1016/j.bmcl.2008.01.063. [DOI] [PubMed] [Google Scholar]
- 31.Bissantz C, Wolfgang CS, Stahl GM. Proteins: Structure, Function, and Bioinformatics. 2005;61:938. doi: 10.1002/prot.20651. [DOI] [PubMed] [Google Scholar]
- 32.Chen JZ, Wang J, Xie XQ. J. Chem. Inf. Model. 2007;47:1626. doi: 10.1021/ci7000814. [DOI] [PubMed] [Google Scholar]
- 33.Evers A, Hessler G, Matter H, Klabunde T. J. Med. Chem. 2005;48:5448. doi: 10.1021/jm050090o. [DOI] [PubMed] [Google Scholar]
- 34.Evers A, Klabunde T. J. Med. Chem. 2005;48:1088. doi: 10.1021/jm0491804. [DOI] [PubMed] [Google Scholar]
- 35.Krystek SR, Kimura SR, Tebben AJ, Langley DR. Journal of Computer-Aided Molecular Design. 2006;20:463. doi: 10.1007/s10822-006-9065-z. [DOI] [PubMed] [Google Scholar]
- 36.Kimura SR, Tebben AJ, Langley DR. Proteins: Structure, Function, and Bioinformatics. 2007 Epub ahead of print. NA. [Google Scholar]
- 37.Patny A, Desai PV, Avery MA. Proteins: Structure, Function, and Bioinformatics. 2006;65:824. doi: 10.1002/prot.21196. [DOI] [PubMed] [Google Scholar]
- 38.Varady J, Wu X, Fang X, Min J, Hu Z, Levant B, Wang S. J. Med. Chem. 2003;46:4377. doi: 10.1021/jm030085p. [DOI] [PubMed] [Google Scholar]
- 39.Costanzi S. J. Med. Chem. 2008 doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strader CD, Candelore MR, Hill WS, Sigal IS, Dixon RA. J. Biol. Chem. 1989;264:13572. [PubMed] [Google Scholar]
- 41.Liapakis G, Ballesteros JA, Papachristou S, Chan WC, Chen X, Javitch JA. J. Biol. Chem. 2000;275:37779. doi: 10.1074/jbc.M002092200. [DOI] [PubMed] [Google Scholar]
- 42.Avlani VA, Gregory KJ, Morton CJ, Parker MW, Sexton PM, Christopoulos A. J. Biol. Chem. 2007;282:25677. doi: 10.1074/jbc.M702311200. [DOI] [PubMed] [Google Scholar]
- 43.Shi L, Javitch JA. Proceedings of the National Academy of Sciences. 2004;101:440. doi: 10.1073/pnas.2237265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Graaf C, Foata N, Engkvist O, Rognan D. Proteins: Structure, Function, and Bioinformatics. 2008;71:599. doi: 10.1002/prot.21724. [DOI] [PubMed] [Google Scholar]
- 45.Ghanouni P, Gryczynski Z, Steenhuis JJ, Lee TW, Farrens DL, Lakowicz JR, Kobilka BK. J. Biol.Chem. 2001;276:24433. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 46.de Graaf C, Rognan D. J Med Chem. 2008;51:4978. doi: 10.1021/jm800710x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 1 Å shifted agonist bound and carazolol-refined antagonist bound β2AR models are provided in PDB format as electronic supplementary material.