Abstract
The proper segregation of chromosomes during meiosis or mitosis requires the assembly of well organized spindles. In many organisms, meiotic spindles lack centrosomes. The formation of such acentrosomal spindles seems to involve first assembly or capture of microtubules (MTs) in a random pattern around the meiotic chromosomes and then parallel bundling and bipolar organization by the action of MT motors and other proteins. Here, we describe the structure, distribution, and function of KLP-18, a Caenorhabditis elegans Klp2 kinesin. Previous reports of Klp2 kinesins agree that it concentrates in spindles, but do not provide a clear view of its function. During prometaphase, metaphase, and anaphase, KLP-18 concentrates toward the poles in both meiotic and mitotic spindles. Depletion of KLP-18 by RNA-mediated interference prevents parallel bundling/bipolar organization of the MTs that accumulate around female meiotic chromosomes. Hence, meiotic chromosome segregation fails, leading to haploid or aneuploid embryos. Subsequent assembly and function of centrosomal mitotic spindles is normal except when aberrant maternal chromatin is present. This suggests that although KLP-18 is critical for organizing chromosome-derived MTs into a parallel bipolar spindle, the order inherent in centrosome-derived astral MT arrays greatly reduces or eliminates the need for KLP-18 organizing activity in mitotic spindles.
INTRODUCTION
In eukaryotes meiosis allows the exchange of genetic material between parental chromosomes and leads to the formation of haploid gametes. Reliable segregation of meiotic chromosomes depends on the correct assembly of microtubules (MTs) into a bipolar spindle. In most animal systems, female meiotic spindles lack centrosomes and their MT nucleating activity (Sawada and Schatten, 1988; Gard, 1992; Theurkauf and Hawley, 1992; Albertson and Thomson, 1993). Interestingly, vertebrate cultured cells in which centrosomes have been destroyed can use a centrosome-independent pathway to build a functional bipolar spindle (Khodjakov et al., 2000). These and other results suggest that although the diastral mode of spindle assembly dominates when centrosomes are present, chromatin-based spindle assembly can be used when centrosomes are absent. The question of how meiotic MTs become organized into a bipolar structure remains largely unanswered.
Studies in Xenopus egg extracts have provided some important insights into acentrosomal spindle assembly. The observation that bipolar spindles can form around DNA-coated beads confirmed that chromatin itself can provide a platform for the nucleation, stabilization, and/or capture of MTs (Heald et al., 1996). Analysis of the effects of inactivating or depleting specific MT motor proteins from Xenopus extracts along with analysis of mutations that affect assembly of acentrosomal meiotic spindles in Drosophila (Merdes and Cleveland, 1997; Walczak et al., 1998; Walczak, 2000) have led to the following model for chromatin-based assembly of bipolar spindles. Chromatin-associated kinesins, e.g., Xenopus klp1, participate in the interaction of MTs with chromatin and may help push the minus ends of captured MTs away from the chromatin by walking toward plus ends (McKim and Hawley, 1995; Vernos et al., 1995; Matthies et al., 1996; Walczak et al., 1998; Antonio et al., 2000; Funabiki and Murray, 2000). Plus-end–directed BimC kinesins, e.g., Xenopus Eg5 and Drosophila KLP61F, participate in parallel bundling by lateral cross-linking of MTs. These motors also sort the bundled MTs by sliding antiparallel MTs such that plus ends move toward one another at the spindle equator, whereas minus ends move away from one another toward the poles (Sawin and Mitchison, 1995; Kashina et al., 1996; Sharp et al., 1999). The minus-end–directed motor cytoplasmic dynein then helps focus the poles, perhaps by cross-linking MT minus ends and by moving other motors and/or MT cross-linking proteins toward minus ends (Heald et al., 1997; Walczak et al., 1998; Wittmann et al., 1998, 2000; Merdes et al., 2000). Other minus-end–directed motors, e.g., the Xenopus XCTK2 and Drosophila NCD kinesins, also seem to be required for focusing of poles (Hatsumi and Endow, 1992; Matthies et al., 1996; Walczak et al., 1998).
The function of another kinesin, Xenopus klp2 (Xklp2), in spindle assembly has been controversial. Founder of the Klp2 kinesin subfamily, Xklp2 is a slow plus-end–directed motor that can associate with MTs when TPX2, an MT-associated protein, is active. Early reports suggested that Xklp2 was a core centrosomal protein essential for the assembly of bipolar spindles in mitotic egg extracts (Boleti et al., 1996; Robb et al., 1996). However, subsequent work indicated that XKlp2 is not centrosomal and that its activity is not needed in extracts for the assembly of centrosomal spindles (Wittmann et al., 2000). Xklp2 was observed to play a minor role in the assembly of chromatin-based acentrosomal spindles, functioning redundantly with cytoplasmic dynein and XCTK2 in focusing MTs at poles (Walczak et al., 1998). The first in vivo disruption tests of a Klp2 kinesin, sea urchin KRP180, used antibody injection to examine its function in mitosis. A partial inhibition of centrosome separation during early prometaphase suggests a role in spindle assembly that could depend on MT-MT cross-linking and antiparallel sliding, similar to Eg5 and other BimC kinesins (Rogers et al., 2000).
This article focuses on identification and functional tests of KLP-18, a Caenorhabditis elegans Klp2 kinesin. In the female germ line of C. elegans, mature oocytes arrest at diakinesis of prophase I. The oocyte nuclear envelope breaks down before ovulation, and then shortly after fertilization, a disordered array of MTs occurs around the bivalent chromosomes. During meiotic prometaphase and metaphase, spindle MTs become parallel and organize into a bipolar spindle with chromosomes at the equator (Albertson and Thomson, 1993). In C. elegans, relatively few gene products are known to affect meiotic spindle formation. MEI-1 and MEI-2 form a katanin-like heterodimer concentrated at poles, which is thought to limit the length of spindle MTs (Mains et al., 1990; Clark-Maguire and Mains, 1994a,b; Srayko et al., 2000). The MT-associated protein ZYG-9 (Msps) also is required for meiotic spindle assembly (Kemphues et al., 1986). ZYG-9 may function to increase MT stability and length, thus antagonizing destabilization by the MEI-1/MEI-2 complex, as postulated for mitotic spindles (Matthews et al., 1998). Our studies demonstrate that KLP-18 is critical for assembly of acentrosomal meiotic spindles. It participates in converting the disordered MTs that collect around prometaphase meiotic chromosomes into an ordered, parallel, bipolar array. Subsequent formation of a centrosomal mitotic spindle does not seem to depend on KLP-18 function, suggesting that KLP-18's MT organizing role is dispensable when centrosomes are present.
MATERIALS AND METHODS
C. elegans Strains and Alleles
Maintenance and handling of C. elegans were carried out as described previously (Brenner, 1974). Bristol N2 was used as the wild-type strain. The following mutations were used: LGI, glp-4(bn2ts); LGII, emb-27(g48ts); LGIII, fem-2(b245ts); LGIV, fem-3(q20ts), klp-10(ok704). Temperature-sensitive strains were maintained at permissive temperature (15°C) and shifted to restrictive temperature [25°C or 25.5°C in the case of emb-27(g48ts)]. AZ212(ruIs32) contains an integrated GFP::H2B histone transgene on LGIII (Praitis et al., 2001). Wild-type or mutant worms were synchronized as described previously (Lewis and Fleming, 1995).
Molecular Analysis of klp-18
KLP-18 corresponds to the predicted open reading frame (ORF) C06G3.2, which is located on the right arm of chromosome IV. Sequenced parts of overlapping cDNA clones (see http://www.wormbase.org for further information) cover most of the coding region; remaining gaps were filled by sequencing polymerase chain reaction (PCR) products from cDNA clone yk167g6 (2.67 kb; kindly provided by Y. Kohara, Gene Network Lab, National Institute of Genetics, Mishima 411, Japan), which also contains the entire 3′ untranslated region. Additional 5′ cDNA sequence, including the SL1 transsplice leader sequence, was amplified by 5′-rapid amplification of cDNA ends (RACE) from mixed stage polyA+ RNA by using the 5′/3′-RACE kit (Roche Diagnostics, Indianapolis, IN) and the following primer: 5′-CAACAGAAACGGAGAACGTC-3′.
To determine whether the predicted ORF C33H5.4 is also transcribed, we performed reverse transcription (RT)-PCR, by using primers that exactly match both klp-18 and C33H5.4: forward, 5′-CCACCTTGATGTTCGCTCAGTCGTG-3′ and reverse, 5′-TCCAGTTGACGCTTAGTAGAGCTC-3′; and a second nested set: forward, 5′-TATGACCGGCGACCAGGAGAGCAG-3′ and reverse, 5′-TCCAGCCGAGTGATCATCTGAGACT-3′. The predicted RTPCR products were 1.1 kb for klp-18 and 0.8 kb for C33H5.4 (Figure 1C). RT-PCR was performed on mixed stage polyA+ RNA by using Omniscript RT kit (QIAGEN, Valencia, CA).
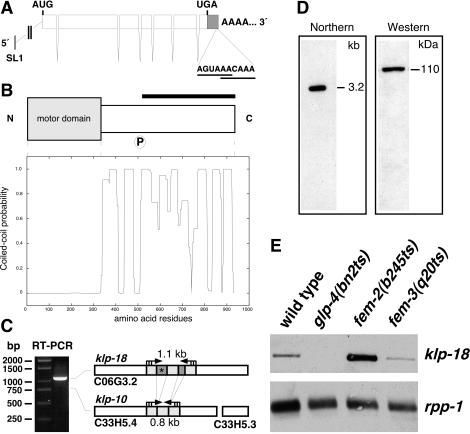
Figure 1.
Molecular characterization of klp-18, which encodes a C. elegans kinesin-like protein. (A) Schematic illustration of the klp-18 transcript (3059 nt; GenBank accession no. AY211948). The klp-18 gene contains seven exons and is trans-spliced to SL1 at the 5′ end. AUG and UGA mark the translation initiation and termination codons, respectively. The 3′-UTR (≥273 nt) contains two putative polyadenylation signals (AGUAAA and AACAAA) upstream (19 and 15 nt, respectively) of the poly-A tail. (B) klp-18 encodes a protein of 932 aa. The N terminus contains the predicted motor domain (aa 2–338). The C terminus is predicted to form an extensive α-helical coiled-coil (Lupas plot; Lupas et al., 1991). P, a putative cdc2 kinase phosphorylation site (SPAR). The bar marks the protein fragment used for generation of anti-KLP-18 antibodies. (C) RTPCR amplified a 1.1-kb fragment of klp-18 but not a 0.8-kb fragment of a related predicted gene, C33H5.4/klp-10. The darker shaded regions of klp-18 are absent from C33H5.4. The region with an asterisk was used to produce klp-18-specific dsRNA for RNAi. (D) Northern blot and Western blot analyses both detected only a single band, corresponding to the klp-18 transcript (∼3.2 kb) and protein (∼110 kDa), respectively. (E) klp-18 transcript levels in wild-type adult hermaphrodites and temperature-sensitive mutants raised at restrictive temperature: glp-4(bn2ts) hermaphrodites contain a severely underproliferated germ line; fem-2(b245ts) hermaphrodites contain only oocytes; and fem-3(q20ts) hermaphrodites contain only sperm. For comparison of relative transcript levels, the ribosomal protein mRNA (rpp-1) served as a loading control.
RNA-mediated Interference (RNAi)
To isolate a yk167g6 cDNA clone, phagemid DNA was prepared as described in the Stratagene ExAssist protocol. For in vitro transcription (RiboMAX; Promega, Madison, WI), the template was linearized with EcoRI or XhoI. To generate double-stranded RNA (dsRNA), equal amounts of sense and antisense RNA were mixed in nuclease-free water, incubated at 70°C for 15 min, and annealed by cooling down to room temperature. dsRNA specific for klp-18 was prepared from a 194-base pair PCR product derived from the middle of klp-18, a region absent from klp-10 (Figure 1C). For RNAi tests, dsRNA diluted to a final concentration of 0.5 μg/μl was injected into the gonads of either young adult wild-type hermaphrodites or young adult emb-27(g48) animals (maintained at 15°C and shifted to 25.5°C after injection). After 24–48 h, the progeny of injected worms were prepared for either antibody staining or videomicroscopy. RNAi by feeding was also done, performed as described by others (Kamath et al., 2001). A 1.2-kb XhoI/PstI fragment of yk167g6 was cloned between the T7 promoters of the pPD129.36 (L4440) feeding vector. The recombinant plasmid was transformed into the RNase III-deficient Escherichia coli strain HT115 (DE3), carrying IPTG-inducible T7 polymerase. After amplification of a single colony overnight (37°C, LBamp tet medium), bacteria were seeded on NGMamp tet plates, containing IPTG (1 mM) and further incubated overnight at room temperature to allow the expression of dsRNA. Ten to 20 wild-type embryos were transferred to a single plate and allowed to grow to adulthood (15°C), and their offspring were analyzed.
KLP-18 Antibody Production
PCR was used to amplify a 1.2-kb fragment of yk167g6, encoding the C-terminal 425 aa of KLP-18 (PARMA.... TTKP): forward, 5′-GAGGATCCCCTGCACGAATGGCTTTGTT-3′ and reverse, 5′-ATGCGGTACCTGGCTTTGTGGTCTGACT-3′ (synthetic restriction sites for BamH1 and Asp718 are underlined). The fragment was cloned in frame into the 6 × His-tag expression vector pQE-30 (QIAGEN), transformed, and induced in E. coli strain M15[pREP4] (QIAGEN). Recombinant protein was purified on Ni2+-NTA matrix (QIAGEN) and sent to Eurogentec (Seraing, Belgium) for immunization. Final bleeds of one rabbit and one rat were purified using the 6 × His-KLP-18 fragment conjugated to AminoLink Plus (Pierce Chemical, Rockford, IL). For elution of anti-KLP-18 antibodies, columns were first rinsed with 4.5 M MgCl2, 50 mM sodium HEPES, pH 7.5, followed by 100 mM glycine, pH 2.5. Both fractions were dialyzed against phosphate-buffered saline, collected, and concentrated using a Centricon YM-100 column (Millipore, Bedford, MA).
Northern Blot Analysis
PolyA+ RNA was prepared from mixed or synchronous worm populations by using the μMACS mRNA isolation kit (Miltenyi Biotec, Bergisch Gladbach, Germany). RNA was fractionated in formaldehyde-containing (1.8%) agarose gels, transferred to a Hybond-N+ membrane (Amersham Biosciences, Piscataway, NJ) by using 20× SSC, and cross-linked with UV light. Digoxigenin (Roche Diagnostics)-labeled RNAs, generated from yk167g6 or rpp-1 cDNAs (Evans et al., 1997), were used as probes at a final concentration of 100 and 10 ng/ml, respectively. The membrane was prehybridized (30 min, 68°C) and hybridized (overnight, 68°C) in hybridization solution [5× SSC; 50% deionized formamide, 0.1% (wt/vol) sodium-laurylsarcosine, 0.02% (wt/vol) SDS, 2% (vol/vol) blocking reagent (Roche Diagnostics)] and washed (2–5 min in 2× SSC/0.1% SDS at room temperature and 2–15 min in 0.1× SSC/0.1% SDS at 68°C). Detection was performed with horseradish peroxidase (HRP)-conjugated anti-digoxigenin antibodies (dilution 1:10,000) by using BM chemiluminescence substrate (Roche Diagnostics). Sizes of transcripts were determined by comparison with RNA Mr standards (Promega).
Western Blot Analysis
Young adult hermaphrodites (100 or 20 worms per lane; Figures 1D and 4A, respectively) were transferred to 10 μl of M9 buffer and frozen in liquid nitrogen. Then 10 μl of 2× SDS sample buffer was added, and samples were immediately boiled (10 min), chilled on ice (5 min), and loaded on a 10% SDS polyacrylamide gel. Separated proteins were transferred to nitrocellulose membrane, incubated with blocking buffer containing TBT [Tris-buffered saline (25 mM Tris), plus 0.1% Tween 20 or Triton X-100] plus 1% bovine serum albumin, 3% nonfat dry milk powder, and 0.02% sodium azide for 1 h at room temperature, further incubated at 4°C overnight with rabbit anti-KLP-18 antibody (1:500) in blocking buffer, washed three times for 5 min each with TBT at room temperature, and finally incubated at room temperature for 1.5 h with anti-rabbit secondary antibody (HRP-conjugated, 1:10,000) in blocking buffer (without sodium azide). After washing three times for 5 min each in TBT, detection was performed using BM chemiluminescence substrate. The membrane was incubated with TBT blocking buffer again (sodium azide irreversibly inactivates HRP) and reprobed with anti-α-tubulin monoclonal (mouse, 1:500; Sigma-Aldrich) and an anti-mouse secondary antibody, as a loading control.
Figure 4.
Depletion of KLP-18 by RNA interference. (A) Equal amounts of protein from wild-type and klp-18(RNAi) young adults were immunoblotted after depletion of KLP-18 via injection or feeding of dsRNA. In both cases, KLP-18 was almost undetectable. The blot was reprobed with anti-α-tubulin antibodies as a loading control. (B–D) In comparison to wild-type, klp-18(RNAi) meiotic spindles at equivalent stages showed either greatly reduced levels of KLP-18 staining (C; n = 9) or no detectable staining (D; n = 9). Anterior is to the left. Bar, 5 μm.
Immunostaining of Embryos
Gravid adults were transferred with a drawn-out pipette to a microscope slide coated with a thin layer of polylysine in a drop of sterile M9 buffer and cut with a scalpel. Embryos were immediately permeabilized by the freeze-crack method (Strome and Wood, 1983) and fixed in 100% methanol (10 min), 100% acetone (20 min), 90% ethanol (10 min), 60% ethanol (10 min), and 30% ethanol (10 min). Slides were washed twice for 10 min each with TBT (see above), incubated at 4°C overnight with primary antibodies (see below) in blocking buffer (TBT plus 1% bovine serum albumin and 1% nonfat dry milk powder), washed three times for 10 min each with TBT at room temperature, and incubated at room temperature for 1–3 h, with secondary antibodies (see below) in blocking buffer. Finally, slides were washed three times for 10 min each in TBT and mounted in Mowiol containing 1,4-diazabicyclo(2.2.2)octane (Sigma-Aldrich) as an antifade reagent.
The following primary and secondary antibodies were used at the dilutions (in blocking buffer) indicated: anti-KLP-18 (rabbit, 1:3,000–1:40,000), anti-KLP-18 (rat, 1:200), anti-α-tubulin (mouse, 1:40, 4A1; Piperno and Fuller, 1985), anti-PGL-1 (rabbit, 1:5,000, Kawasaki et al., 1998), anti-MEI-1 (rabbit, 1:40; Clark-Maguire and Mains, 1994a), and anti-ZYG-9 (rabbit, 1:25; Matthews et al., 1998). Secondary antibodies were Cy2, Cy-3 (1:200; Jackson Immunoresearch Laboratories, West Grove, PA), and Alexa 647 (1:200; Molecular Probes) conjugated. Yoyo-1 (1:20,000; Molecular Probes) and RNase A (1:200; 10 μg/μl) were usually added with secondary antibodies for DNA staining. To test specificity, anti-KLP-18 antibodies were incubated with an excess of 6 × His-KLP-18 protein (0.5–1 μg/μl) in blocking solution and incubated for 30 min at room temperature.
Microscopy, Live Imaging, and Image Processing
Development of embryos and microinjections into the gonad of hermaphrodites were observed with Nomarski optics by using an Axiophot microscope (Carl Zeiss, Jena, Germany) equipped with a 100× PL Fluotar oil-immersion objective and a DM IRBE inverted microscope (Leica, Wetzlar, Germany) equipped with a 40× PL Fluotar objective, respectively. Immunofluorescence analyses of embryos were performed on a confocal laser scanning microscope (TCS-NT; Leica) equipped with a 100× PL Fluotar oil-immersion objective. Each recorded image represents a projection of a Z-series of 0.5-μm optical sections. For live imaging of green fluorescent protein (GFP)-tagged chromosomes during meiosis and mitosis, GFP::histone H2B-expressing embryos were grown to adulthood at room temperature on plates containing either nontransformed OP50 or HT115 bacteria expressing klp-18 dsRNA (see above). These “RNAi-fed” worms were placed in a solution of 0.01% tricane/0.01% tetramisole, an anesthetizing agent, for 15–20 min, and then on 2% agar pads on slides. Embryos inside the anesthetized worms were imaged using an MRC600 laser scanning confocal system (Bio-Rad, Hercules, CA) with an argon/krypton laser. Images were collected every 5 s on slow scan, by using a 60× objective. Temperature ranged from 21°C to 24°C. Images were manipulated to generate movies and figures in NIH Image (version 1.62f, developed by Wayne Rasband at the National Institutes of Health and available at http://rsb.info.nih.gov/nih-image/), Adobe Photoshop (Adobe Systems, Mountain View, CA) and Canvas (ACD Systems, Miami, FL).
RESULTS
Molecular Characterization of klp-18
The klp-18 gene (ORF C06G3.2; Figure 1A) is predicted to encode a kinesin-like protein (The C. elegans Sequencing Consortium, 1998; Siddiqui, 2002). To determine the sequence of full-length klp-18, we used 5′-RACE, PCR, and overlapping cDNAs. The klp-18 gene encodes a protein of 932 aa with a motor domain at its N terminus (Figure 1B). Based on sequence alignment of its motor domain, KLP-18 is most similar to Xenopus Xklp2 (43% identity/60% similarity; Boleti et al., 1996), Homo sapiens HKlp2 (45%/61%; Sueishi et al., 2000), and sea urchin KRP180 (43%/61%; Rogers et al., 2000). Furthermore, in a recent evolutionary analysis of kinesin-related proteins maximum likelihood methods indicate that KLP-18 is a member of the Klp2 clade (Lawrence et al., 2002). Between its motor domain and C terminus, KLP-18 does not show significant sequence similarity to other proteins in the database. However, that region has a high probability of forming an extended α-helical domain with the potential to form coiled-coil interactions (Figure 1B). Also, amino acids 507–510 (SPAR) in this “stalk” region provide a potential cdc2 kinase phosphorylation site ([T/S]PX[K/R]; Nigg, 1993). These features are similar to those of the stalk regions of other members of the Klp2 subfamily (Boleti et al., 1996; Wittmann et al., 1998; Rogers et al., 2000).
Database searches revealed that ∼750 kb away from klp-18 in the C. elegans genome is a pair of predicted ORFs (C33H5.4 and C33H5.3) with high sequence similarity to the KLP-18 amino acid sequence. The C33H5.4 ORF, previously designated as klp-10 (Siddiqui, 2002), is separated from C33H5.3 by only 19 base pairs of genomic DNA. The two are colinear with KLP-18 (Figure 1C), suggesting that C33H5.3 actually encodes the C terminus of KLP-10. The high degree of sequence identity between klp-18 and the composite klp-10 ORF (85% identity at the DNA level and 90% identity/95% similarity at the amino acid level) suggests that they arose from a recent gene duplication event and that KLP-18 and KLP-10 could be redundant. However, the results of RTPCR, Northern blots, and Western blots suggest that klp-10 is not expressed at appreciable levels (Figure 1, C and D). Furthermore, a KLP-10 protein would not be likely to engage in kinesin-like motor activity, because it lacks the highly conserved “switch I” amino acids “SSR,” which are thought to be critical for interaction with ATP (Kull et al., 1996; Sablin et al., 1996).
Despite these indications that klp-10 may not produce a MT motor, two recent studies reported that RNAi intended to deplete KLP-10 caused embryonic lethality (Piano et al., 2000; Kamath et al., 2003). Sequence comparison suggests that the dsRNAs used in those studies should be effective in eliminating KLP-18 as well as KLP-10. This raised the question of whether the embryonic lethality was due to depletion of KLP-10, KLP-18, or both. To address this issue, we obtained a deletion allele of klp-10 [klp-10(ok704)] from the C. elegans Gene Knock-Out Project at Oklahoma Medical Research Foundation (http://www.mutantfactory.ouhsc.edu/). The deletion removes codons for the predicted motor domain amino acids 178–662 of KLP-10. Homozygous klp-10(ok704) worms are viable and fertile, and their embryos show no obvious defects. We also designed a dsRNA that is specific for klp-18 (see MATERIALS AND METHODS). That specific probe, when tested by RNAi, caused 100% embryonic lethality. Consequently, we conclude that the embryonic lethality previously attributed to KLP-10 depletion was actually due to KLP-18 depletion.
Germline Expression of klp-18
In adult C. elegans, klp-18 mRNA expression is strongest in the female germ line. In glp-4 hermaphrodites, which have a severely underproliferated germ line (Beanan and Strome, 1992), no klp-18 transcript is detectable (Figure 1E). In fem-2 hermaphrodites, which produce only oocytes (Kimble et al., 1984), klp-18 transcript is present at high levels, and in fem-3 gain-of-function hermaphrodites, which produce only sperm (Barton et al., 1987), it is present at approximately wild-type levels. Additional Northern blot analysis revealed increasing klp-18 transcript levels at progressively later stages of wild-type larval development (our unpublished data). This increase is likely due to a growing population of germ cells (also see Figure 2).
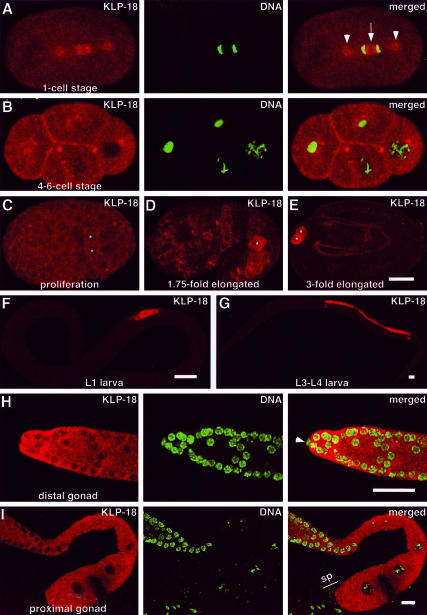
Figure 2.
KLP-18 localization during development. Embryonic (A–E) and postembryonic stages (F–I) were stained for KLP-18 (red) and DNA (Yoyo-1, green; in A, B, H, and I). Anterior is to the left. (A) During mitotic anaphase KLP-18 is concentrated on the centrosomes (arrowheads) and on the MTs between the separating masses of chromatin, with a dim zone at the spindle equator (arrow). (B) KLP-18 becomes concentrated near plasma membranes at sites of cell-cell contact. (C–E) During later embryo development, KLP-18 signal begins to increase in the germ line precursor cells, Z2 and Z3, which were identified by double staining with anti-PGL-1 antibodies (Kawasaki et al., 1998; our unpublished data) and are marked with asterisks. (F and G) KLP-18 accumulates in the germ line throughout larval development. (H and I) KLP-18 is present in the adult germ line. The gonad distal tip cell, which is somatic (arrow-head in H), and mature sperm (“sp” in I) do not contain detectable KLP-18. Bars, 10 μm.
KLP-18 Localization in Mitotic Spindles
To investigate where KLP-18 is localized and may function, two antisera against the C terminus of the protein were generated, affinity purified, and used for immunofluorescence staining. Both antisera produced the same staining pattern (Figures 2 and 3). Both are specific for KLP-18, as demonstrated by a dramatic reduction of staining in embryos after preincubation of the antibody with an excess of KLP-18 fusion protein (our unpublished data). Staining was also greatly reduced or eliminated in embryos subjected to RNAi depletion of KLP-18 (Figure 4). KLP-18 staining was most concentrated in mitotic spindles toward the poles. During anaphase, KLP-18 staining also concentrated between the separating groups of chromosomes, with a dim zone at the spindle equator (Figure 2A). These patterns are similar to those seen for Xklp2 in mitotic Xenopus cultured cells and KRP180 in mitotic sea urchin embryos (Boleti et al., 1996; Rogers et al., 2000), although a dim zone at the anaphase equator has not been reported previously.
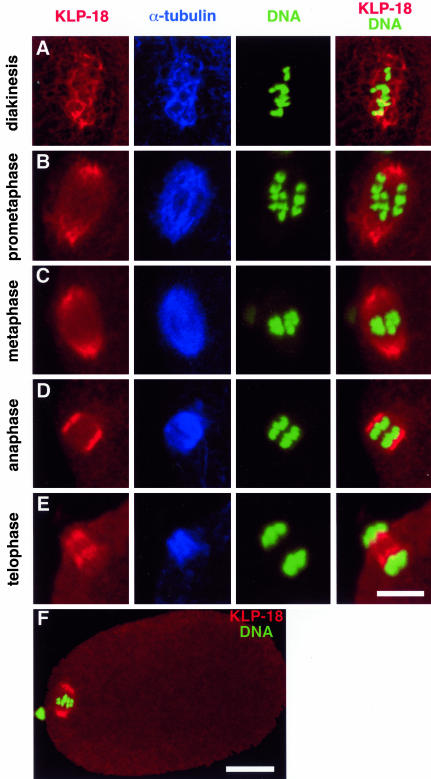
Figure 3.
Localization of KLP-18 during female meiosis. Different stages of wild-type female meiosis I (A, B, D, and E) or meiosis II (C and F) stained with anti-KLP-18 (red), anti-α-tubulin (blue), and Yoyo-1 (green) to visualize KLP-18, MTs, and DNA, respectively. Anterior is to the left. (A) After nuclear envelope breakdown, KLP-18 colocalizes with MTs around the condensed chromosomes. (B–D) During prometaphase, metaphase, and anaphase KLP-18 is highly enriched at the spindle poles. (E) During telophase, KLP-18 and MTs are concentrated between separating chromosomes. Bar, 2.5 μm (A–E). (F) For comparison of meiotic spindle brightness relative to cytoplasmic levels an entire one-cell embryo (metaphase II) is shown. Bar, 10 μm.
After cytokinesis, KLP-18 accumulated somewhat around the nucleus and at the cortex near sites of cell-cell contact (Figure 2B). During later stages of embryogenesis, KLP-18 levels gradually diminished in somatic cells but increased in the primordial germ cells Z2 and Z3, presumably due to expression in the embryonic germ line (Figure 2, C–E). The germ line stained brightly for KLP-18 at all stages of larval development (Figure 2, F and G). Adult hermaphrodites contained high levels of cytoplasmic stain in the distal gonad arm, where germ cells proliferate and begin meiosis (Figure 2H). Adults also showed high levels of staining in the proximal gonad arm, where oocytes mature during the late stages of meiotic prophase I (Figure 2I).
KLP-18 Localization in Meiotic Spindles
In C. elegans, mature oocytes, and thus female meiotic spindles, lack centrosomes. After the oocyte is fertilized in the spermatheca (Figure 2I, sp), disorganized MTs accumulate around the chromosomes and subsequently become ordered into a bipolar metaphase spindle that lacks astral MTs (Figure 3). Immunostaining showed KLP-18 colocalized with the disorganized MTs during prometaphase (Figure 3A). As the MTs became ordered into a parallel bipolar array, KLP-18 concentrated most at the poles but was also present between the poles and chromosomes (Figure 3B). The concentration at poles persisted during metaphase and early anaphase (Figure 3, C and D), but then shifted to the interzone between the separating chromosomes during late anaphase/telophase (Figure 3E). Similar KLP-18 staining patterns were observed during meiosis II (Figure 3, C and F; our unpublished data). The concentration of KLP-18 at the spindle poles of female meiotic spindles, which lack centrosomes, supports the view that Klp2 kinesins are not core centrosomal proteins but instead concentrate toward MT minus ends (Rogers et al., 2000; Wittmann et al., 2000).
During the meiotic divisions that occur in the male germ line, anti-KLP-18 staining patterns were similar to those seen in female meiosis and mitosis. KLP-18 was concentrated at the spindle poles, which have centrosomes, and in the anaphase interzone (our unpublished data). There was also a significant amount of KLP-18 staining on male meiotic chromosomes. Attempts to deplete KLP-18 from the male germ line by RNAi failed, as has been observed for other proteins expressed during spermatogenesis (S. L'Hernault, personal communication). Consequently, the specificity of KLP-18 antisera in staining the male meiotic chromosomes remains uncertain.
Depletion of KLP-18 Impairs Formation of the Female Pronucleus and Causes Cortical Instability
To test the function of KLP-18, we studied the consequences of depleting it by RNAi. Double-stranded RNA, transcribed from a klp-18 cDNA clone or from a smaller region of the klp-18 gene that is not present in klp-10, was introduced into the syncytial gonad of young adult hermaphrodites either by injection or by feeding. Western blots of adults and immunostaining of embryos demonstrated that 24–48 h after dsRNA treatment, KLP-18 levels were dramatically reduced (Figure 4). This confirmed the specificity of the antibodies and showed that the RNAi depletion was effective. Most experiments used dsRNA derived from cDNA, which could deplete klp-10 transcripts if they were present. Two results suggest that dsRNA-induced embryonic defects and lethality were due solely to KLP-18 depletion. First, cDNA-derived RNAi and klp-18 gene-specific RNAi caused identical defects. Second, RNAi depletion of KLP-18 from wild-type and from klp-10(ok704) embryos caused identical defects (our unpublished data).
We first recorded the consequences of KLP-18 depletion in live klp-18(RNAi) embryos by using Nomarski video differential interference contrast microscopy. In wild-type, completion of female meiosis results in the formation of two polar bodies and a female pronucleus at the anterior end of the newly fertilized embryo. The female pronucleus migrates to meet the posteriorly located male pronucleus, and the joined pronuclei move to the center of the embryo where they begin mitosis (Figure 5, A–D). In klp-18(RNAi) embryos (n = 12 movies), severe but variable defects were seen in meiosis. The defects ranged from the complete absence of a female pronucleus (Figure 5, E and F; 3 of 12) to the formation of multiple small female pronuclei (Figure 5, I and J; 9 of 12), only some of which fused with the male pronucleus. The defects in formation of the female pronucleus suggested a failure in meiotic chromosome segregation. However, in most cases, subsequent formation of a bipolar mitotic spindle seemed normal, even when klp-18(RNAi) was done in the klp-10 deletion background. Occasionally, extra nuclei became visible after mitosis in the anterior daughter cell AB (Figure 5L). Extra nuclei may have been remnants of extra female pronuclei. Later in development, klp-18(RNAi) embryos displayed various signs of tissue differentiation but did not undergo morphogenesis (Figure 5N).
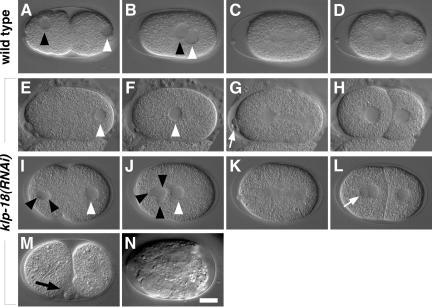
Figure 5.
klp-18(RNAi) embryos show defects in formation of a female pronucleus. Nomarski images of wild-type embryos (A–D) and two klp-18(RNAi) embryos (E–L). Anterior is to the left. Black and white arrowheads mark the female and male pronuclei, respectively. (A–D) The female and male pronuclei migrate to meet posteriorly (B), the first mitotic spindle forms (C), and the embryo undergoes cytokinesis to form a 2-cell (D). (E–H) In this klp-18(RNAi) embryo, a female pronucleus did not form (E and F), but mitosis and cytokinesis occurred apparently normally (G and H). Note the abnormal number of polar bodies (arrow in G). (I–L) In another klp-18(RNAi) embryo, three nuclear structures became visible in the anterior cytoplasm (J), and an additional nuclear structure formed in the anterior cell after cytokinesis (arrow in L). (M) Shortly after cytokinesis (e.g., two-cell stage) klp-18(RNAi) embryos often have transient membrane ruffling and blebbing (arrow). (N) They do not undergo morphogenesis. Bar, 10 μm.
It was interesting that after cytokinesis klp-18(RNAi) embryos usually displayed aberrant ruffling and blebbing of the plasma membrane, especially at sites of cell-cell contact (Figure 5M; n = 14 of 19). These defects and the accumulation of KLP-18 seen near the plasma membrane (Figure 2B) suggest a role for KLP-18 in cortex dynamics during postmeiotic development. It has been demonstrated that proper interactions of astral MTs with the plasma membrane are necessary for stabilization of the cell cortex in early C. elegans embryos (Hird and White, 1993).
KLP-18 Is Required for Assembly of Acentrosomal Meiotic Spindles
To analyze the formation and organization of female meiotic spindles in klp-18(RNAi) embryos, we imaged microtubules and DNA at the anterior end of fixed early embryos by confocal fluorescence microscopy. In klp-18(RNAi) embryos (n >40), MTs were clustered around female meiotic chromosomes, but those MTs were never seen in a bipolar array (compare Figure 6, A and B, with Figure 3), and meiotic chromosomes never showed signs of normal anaphase segregation.
Figure 6.
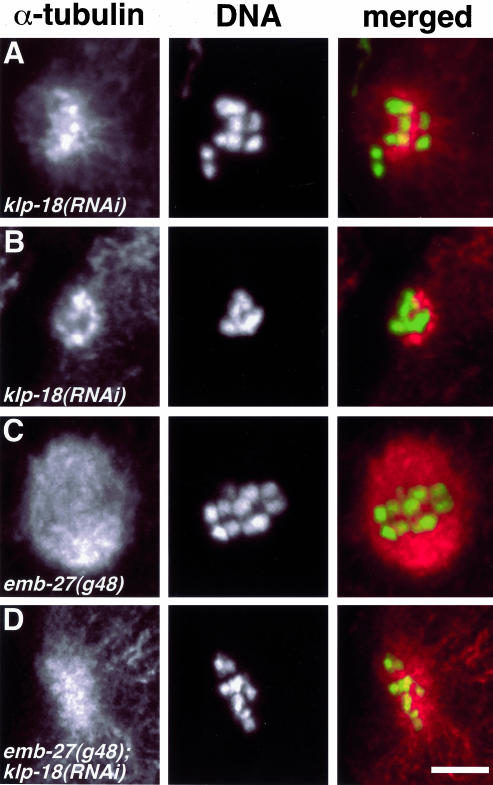
Immunofluorescence analysis of female meiotic spindle formation after depletion of KLP-18. klp-18(RNAi) hermaphrodites were cut open to release embryos, fixed, and stained for α-tubulin (red) and DNA (Yoyo-1, green). Anterior is to the left. (A and B) After depletion of KLP-18, chromosomes fail to align properly. MTs are concentrated in the vicinity of the chromosomes, arranged in a disorganized astral-like array (A), and often are reduced in abundance (B). (C) emb-27 mutant embryos arrest at metaphase I. (D) Depletion of KLP-18 in emb-27 mutant embryos produces a klp-18(RNAi) phenotype. Bar, 5 μm.
The disorder of meiotic microtubules and chromosomes in klp-18(RNAi) embryos suggested an effect on initial acentrosomal spindle assembly. However, judging a temporal sequence of events from fixed samples is uncertain. For example, a period of spindle assembly could have occurred followed quickly by a loss of order during anaphase. To test this possibility, we studied the effects of klp-18(RNAi) in emb-27 mutants, which arrest at meiotic metaphase I. The emb-27(g48ts) allele encodes a temperature-sensitive CDC-16/APC-6 that at restrictive temperature is defective in promoting the metaphase-to-anaphase transition (Golden et al., 2000). Examination of meiotic figures in klp-18(RNAi);emb-27(g48ts) embryos (n = 14) revealed disordered spindles (Figure 6D) that were indistinguishable from those seen in klp-18(RNAi) embryos. These results confirm that KLP-18 is required during prometaphase/metaphase to organize chromatin-associated MTs into a bipolar spindle.
klp-18(RNAi) embryos often had a reduced accumulation of MTs around meiotic chromosomes (Figure 6B). This may reveal an additional role for KLP-18 in the initial nucleation, stabilization, or capture of MTs by chromatin. Alternatively, failure to organize a bipolar meiotic spindle may lead to subsequent loss of MTs. Future time-lapse analysis of MT behavior will discriminate between these possibilities.
Depletion of KLP-18 Prevents Proper Meiotic Chromosome Alignment and Segregation
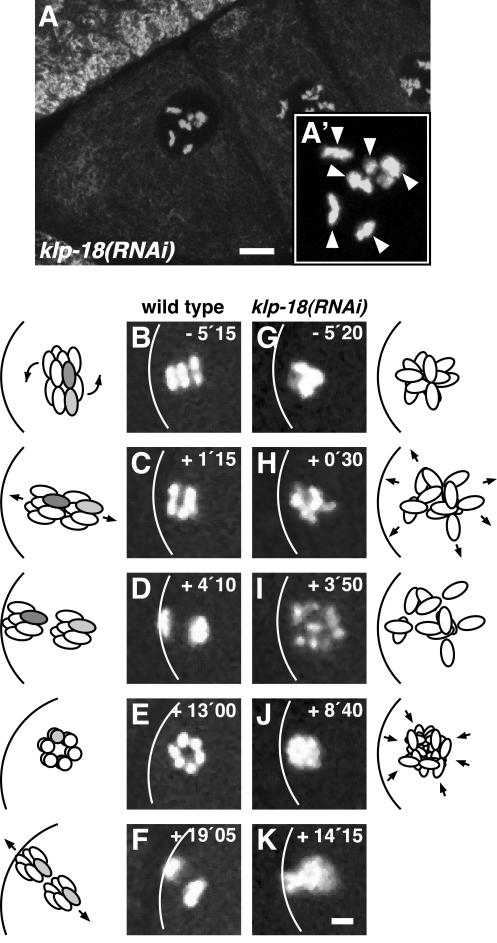
To gain a clearer understanding of female meiosis and how KLP-18 depletion affects it, we studied chromosome dynamics in oocytes and early embryos. In fixed and stained klp-18(RNAi) hermaphrodites, diakinesis-arrested oocytes were indistinguishable from their wild-type counterparts; they contained six properly condensed bivalents, suggesting normal execution of early meiotic events (Figure 7A). In live hermaphrodites expressing a GFP::histone H2B fusion protein, meiotic chromosome behavior was studied by timelapse confocal fluorescence microscopy (see on-line video supplement Movies 1–4). In wild-type embryos (Movie 1) the six bivalents initially congressed to a metaphase I plate perpendicular to the anterior cortex (Figure 7B). The metaphase plate then rotated 90° to orient parallel to the cell surface during the transition to anaphase I (Figure 7C). After separation (Figure 7D), the set of chromosomes near the cortex was expelled as a polar body, whereas the remaining set of chromosomes engaged in meiosis II. As in meiosis I, the meiosis II metaphase plate formed perpendicular to the anterior cortex (Figure 7E) and then rotated to a parallel orientation at the beginning of anaphase II (Figure 7F). During telophase II, the set of sister chromatids near the cortex was expelled as a second polar body and the remaining set decondensed to form the female pronucleus.
Figure 7.
Time-lapse images of GFP::histone H2B in wild-type and klp-18(RNAi) embryos during female meiosis. (A) The most proximal oocytes of a klp-18(RNAi) gonad contain the normal number of six bivalents, as indicated by arrowheads in the higher magnification view (A′). (B–K) Time-lapse confocal fluorescence series of embryos containing GFP::histone H2B to mark chromosomes. Anterior is to the left. (B–F) Meiotic chromosome behavior in a wild-type embryo. (B) Metaphase I: the metaphase plate is aligned perpendicular to the anterior cortex, shown as a white curve. (C) Anaphase I: after a 90° rotation, the two sets of homologs are aligned parallel to the cortex. (D) Telophase I. (E) Metaphase II: the metaphase plate is aligned parallel to the image plane and perpendicular to the cortex. (F) Telophase II: after a 90° rotation, the two sets of sister chromatids are aligned parallel to the cortex. Note that the first polar body is out of the focal plane. In the accompanying cartoons, during wild-type meiosis I a bivalent is represented as a pair of rods (the two homologs of one bivalent are shown as light gray and dark gray). During wild-type meiosis II a pair of sister chromatids is represented as a pair of light gray rods. (G–K) Meiotic chromosome behavior in a klp-18(RNAi) embryo. Bivalents failed to align correctly and were not segregated into discrete masses of chromatin. In klp-18(RNAi) embryos, it was not possible to identify bivalents and sister chromatids, so they are not shaded in the accompanying cartoon. Elapsed time is in minutes seconds. t = 0 indicates onset of anaphase I in wild type or the initial spreading of chromosomes in klp-18(RNAi) embryos (see video in supplementary material). Bars, 5 μm.
In klp-18(RNAi) embryos containing GFP::histone H2B (Movies 2–4), bivalent chromosomes looked morphologically normal immediately after fertilization, but they did not align on a metaphase plate. Instead, the bivalents formed a disorganized cluster near the anterior cortex (Figure 7G; Movie 2). At a time when one would expect anaphase I, chromosomes moved slightly outward, making the cluster less compact, but did not separate into two sets (Figure 7, H and I). In most klp-18(RNAi) embryos, we observed more than six chromatin masses at this stage (Figure 7I), suggesting that homologous chromosomes, or perhaps sister chromatids, did disjoin. Coincident with the period expected for meiosis II, the chromosomes clustered together again (Figure 7J). Part or all of the chromatin cluster then usually moved toward the cell surface in an attempt at polar body extrusion (Figure 7K; Movie 2). In 4 of the 18 embryos observed, the entire maternal complement of DNA was extruded (Figure 8, G–J; Movie 3). In 3 of the remaining embryos, the entire maternal complement of DNA remained in the embryo, forming one or more pronuclei. In the other 11 embryos, some maternal DNA was extruded as a polar body and some was packaged into one or more pronuclei (Figure 8, K–N; Movie 4). These results and those described above show that KLP-18 is necessary during prometaphase I for the correct alignment of meiotic chromosomes and for their subsequent segregation. Prior tests of Xklp2 function in Xenopus extract-driven assembly of acentrosomal spindles around chromatin beads revealed a minor contribution that overlaps with those of pole-focusing motors (Walczak et al., 1998). In contrast, our in vivo tests of KLP-18, a C. elegans Xklp2 homolog, indicate a major contribution to early stages of acentrosomal spindle assembly.
Figure 8.
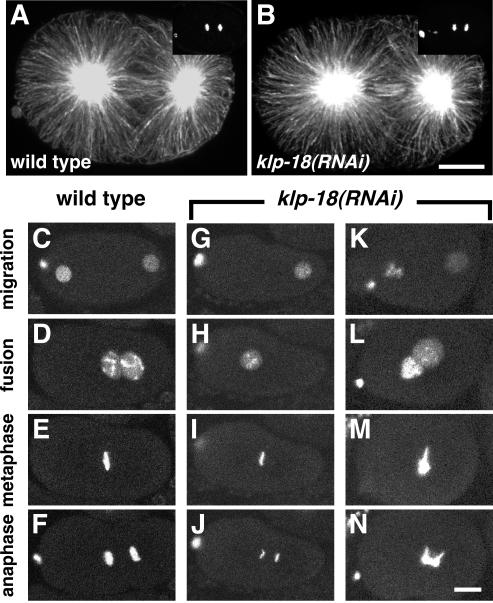
Spindle organization and chromosome behavior during first mitosis in klp-18(RNAi) embryos. (A and B) Staining of the first mitotic spindle with α-tubulin antibodies reveals a similar pattern in wild-type and klp-18(RNAi) embryos. Yoyo-1 staining of DNA, shown in the top right corner, reveals that both embryos were in late anaphase. Anterior is to the left. (C–N) Time-lapse image series of GFP::histone H2B in live wild-type and klp-18(RNAi) embryos during pronuclear migration and first mitosis. (C–F) In wild-type, the female pronucleus migrates to meet the male pronucleus in the posterior hemisphere (D), the pronuclei move to the center of the embryo, and the chromosomes condense into a tight metaphase plate (E) and cleanly separate into two distinct masses during anaphase (F). (G–J) In this klp-18(RNAi) embryo an oocyte pronucleus did not form. The sperm-derived haploid complement of chromosomes underwent apparently normal mitosis (I and J). (K–N) In this klp-18(RNAi) embryo, the oocyte pronucleus contained an abnormally large amount of DNA. The oocyte-derived chromatin complement did not condense or congress normally at metaphase (M) and displayed bridges during anaphase (N). The sperm-derived chromosomes seemed to behave relatively normally. Bars, 10 μm.
KLP-18 Is Not Required for Mitosis
To investigate the possibility that KLP-18 is also important for the assembly and function of centrosomal spindles, we examined the effects of KLP-18 depletion on mitotic spindle organization and chromosome behavior in embryos. The only prior report of in vivo function disruption tests of a Klp2 kinesin, KRP180 in sea urchin embryos, suggested that although it is not essential for mitosis, it helps maintain spindle pole separation in prometaphase and metaphase, perhaps by cross-linking and sliding overlapped antiparallel MTs (Rogers et al., 2000). To investigate this possibility in C. elegans, pole-to-pole distances were compared in Nomarski recordings of klp-18(RNAi) and wild-type one-cell embryos. The mean distance at metaphase in klp-18(RNAi) embryos (14.3 ± 0.7 μm; n = 11) was not significantly different from wild type (14.6 ± 0.8 μm; n = 11). Immunostaining of fixed embryos concurred that mitotic MT organization was normal in klp-18(RNAi) embryos (Figure 8, A and B) and in klp-10(ok704) klp-18(RNAi) embryos (our unpublished data). Even small mitotic spindles in older embryos, similar in size to female meiotic spindles, seemed normal, arguing against the possibility that it is small spindle size that dictates a need for KLP-18. Furthermore, comparison of wild-type (n = 12) and klp-18(RNAi) (n = 5) embryos (2- to 26-cell stage) revealed no significant differences in cell cycle timing, spindle orientation/position, or cytokinesis (our unpublished data). However, the observation of some binucleate cells highlighted the possibility of defective chromosome segregation.
To study mitotic chromosome behavior in more detail, the first embryonic cell cycle was studied by time-lapse imaging of GFP::histone H2B. In wild-type embryos, the female and male pronuclei migrate toward each other, meet in the posterior hemisphere, and move together to the center of the embryo (Figure 8, C and D; Movie 1). The nuclear envelopes then break down and the maternal and paternal chromosomes congress to a tight metaphase plate (Figure 8E). Sister chromatids separate cleanly into two distinct groups during anaphase (Figure 8F). As noted above, klp-18(RNAi) embryos displayed variable defects in formation of the female pronucleus. In embryos with a larger than normal complement of maternal chromatin in one or more female pronuclei (12 of 18 embryos), chromosome behavior during mitosis was defective: the maternal complement of chromatin failed to condense tightly during metaphase (Figure 8M) and failed to segregate normally, forming massive chromosome bridges during anaphase (Figure 8N; Movie 4). In embryos with no female pronucleus (4 of 18 embryos), chromosomes derived solely from the male pronucleus, although haploid, underwent a normal mitotic division (Figure 8, G–J; Movie 3). Thus, although depletion of KLP-18 resulted in severe meiotic defects in all cases analyzed, it resulted in mitotic defects only in those embryos that delivered an aberrant complement of maternal chromatin to the mitotic spindle. These results suggest that mitotic chromosome congression and segregation problems caused by KLP-18 depletion do not reflect a direct function for KLP-18 in the mitotic spindle. Rather, they are a consequence of the presence of maternal chromosomes that have suffered aberrant meiotic segregation. In summary, the assembly of centrosomal mitotic spindles is significantly less dependent on KLP-18 than is the assembly of acentrosomal female meiotic spindles. KLP-18 may function in mitotic spindles, but if so, its function is redundant.
DISCUSSION
KLP-18 has an N-terminal kinesin-related motor domain that is most similar to members of the Klp2 kinesins (Lawrence et al., 2002). Like Klp2 homologs in other organisms, C. elegans KLP-18 concentrates in spindles, especially at spindle poles during prometaphase, metaphase, and early anaphase, and then in the interzone during late anaphase/telophase. Although this localization pattern is similar in both acentrosomal (female meiotic) and centrosomal (mitotic) spindles, KLP-18 depletion blocks assembly only of acentrosomal meiotic spindles. That assembly failure prevents normal segregation of maternal chromosomes, leading to embryos with either no maternal DNA or an abnormal complement of maternal DNA. In the division cycle that follows meiosis, mitotic spindle assembly seems normal, but there are usually defects in congression and anaphase separation for some chromosomes. Time-lapse imaging suggests that the misbehaving chromosomes are maternal. Why chromatin from a defective female meiosis behaves aberrantly in the subsequent mitotic division remains an interesting unanswered question.
As described in the INTRODUCTION, it is thought that acentrosomal spindle assembly requires 1) chromatin-stimulated tubulin polymerization that leads to the accumulation of disordered MTs around chromosomes, 2) MT cross-linking that generates parallel bundles, 3) sorting of polarity by MT-chromatin and MT-MT force generation that moves minus ends away from chromosomes, and 4) focusing of minus ends to form more compact spindle poles (Theurkauf and Hawley, 1992; Albertson and Thomson, 1993; Heald et al., 1996). Excellent progress has been made in understanding the initial chromatin-stimulated polymerization, but subsequent steps are not as well understood. TPX2, a MT-binding protein, which is activated by high levels of RanGTP near chromatin, stimulates MT polymerization (Wittmann et al., 2000; Gruss et al., 2001, 2002). TPX2 also mediates attachment of the nonmotor C-terminal tail of Xklp2 to MTs (Wittmann et al., 1998, 2000). It is reasonable to think that the N-terminal motor domain of Xklp2 then binds to neighboring MTs, forming force-producing cross-links that could contribute to the intermediate steps of acentrosomal spindle assembly, i.e., parallel bundling and MT-MT sliding. However, Xklp2 is dispensable for extract spindle assembly, making minor or redundant contributions (Walczak et al., 1998). Instead, a different MT cross-linking kinesin, the BimC homolog Eg5, is required for MT bundling/sorting. A chromosome-associated kinesin, Xklp1, also contributes to those intermediate assembly steps (Walczak et al., 1998).
Our results suggest a switch in the relative importance of kinesins in C. elegans meiotic spindle assembly. KLP-18 depletion leads to a failure in MT bundling and organization around female meiotic chromosomes. Our depletion/disruption tests of the worm BimC homolog (BMK-1) and of chromokinesin homologs (KLP-12 and KLP-19) suggest that they are dispensable in female meiosis (Saunders, Rose, Powers, Strome, and Saxton, unpublished data). The contrast between the Xenopus extract and worm results could be due to differences between the two organisms or between the extract versus in vivo approaches. The extract system can provide powerful new insights through the analysis of processes under defined conditions, but it cannot be expected to reveal all aspects of in vivo mechanisms. Thus, it is possible that in vivo acentrosomal meiotic spindle assembly in Xenopus oocytes does require Xklp2. Alternatively, Xenopus and C. elegans may have evolved different strategies for acentrosomal spindle assembly, emphasizing the use of different motors to accomplish the same MT accumulation, bundling, and sorting objectives.
Our results suggest that although KLP-18 concentrates in the mitotic spindles of early C. elegans embryos, it is dispensable for their assembly and function. MTs nucleated by mitotic centrosomes are well ordered relative to those organized by chromatin; their radial arrangement makes neighboring MTs nearly parallel, with most minus ends oriented toward the nearest centrosome. Hence, a KLP-18 MT bundling/sorting activity that is essential in acentrosomal meiotic spindles may be superfluous when centrosomes are present. Similarly, the human kinesin-related protein HSET localizes to both meiotic and mitotic spindles, perhaps cross-linking neighboring MTs, but HSET inhibition seems to cause significant assembly defects only in acentrosomal meiotic spindles. Additional evidence that HSET is needed for taxol-induced, acentrosomal aster formation argues that it is the presence of centrosomes that eliminates the need for HSET during mitosis (Mountain et al., 1999). The Drosophila kinesins NCD and SUB also seem to play important roles in acentrosomal spindle assembly and lesser roles in embryonic mitotic spindle assembly (Matthies et al., 1996; Giunta et al., 2002).
The need for TPX2 activity in localizing Xklp2 to Xenopus spindle MTs and the role of TPX2 in chromatin-stimulated spindle assembly suggest that a TPX2 homolog and associated proteins might contribute to KLP-18 localization/function in C. elegans. An obvious homolog of TPX2 has not been found in searches of the C. elegans genome sequence (our unpublished data; Askjaer et al., 2002). Furthermore, it was recently noted that RNAi of the worm homologs of Ran, RanGAP, and RanBP2, although sufficient to prevent organization of kinetochore and interpolar MTs in mitotic spindles, had no apparent effects on meiotic chromosome segregation (Askjaer et al., 2002; Bamba et al., 2002). These results raise the possibility that acentrosomal meiotic spindles use an assembly pathway that is independent of Ran and TPX2. In this context, the decreased accumulation of MTs around meiotic chromosomes often seen in klp-18(RNAi) embryos is interesting; KLP-18 may contribute to the MT stabilization that is done by Ran and TPX2 in vertebrates. Our results do not address whether KLP-18 contributes to the last stage of acentrosomal assembly, pole-focusing. A fast-acting temperature-sensitive KLP-18 mutation would be ideal for studying this and other questions that require time-resolved loss of function. Time-lapse analysis of MTs and studies of meiotic proteins that bind KLP-18 should also be informative and may provide key insights into the principles underlying chromosome-mediated acentrosomal spindle assembly.
Supplementary Material
Acknowledgments
We thank Y. Kohara for the generous gift of cDNAs; M. Fuller, I. Kawasaki, K. Kemphues, and P. Mains for antibodies; and K. Johnson, A. Bachmann, E. Knust, and C. Walczak for discussions and critical reading of the manuscript. Some strains were obtained from the C. elegans Genetic Stock Center, which is funded by a grant from the National Institutes of Health National Center for Research Support. C.S. was a Boehringer Ingelheim Fonds fellow. This work was supported by National Institutes of Health grant GM-58811 to W.M.S. and S.S. Work in the laboratory of O.B. is supported by grants from the Deutsche Forschungsgemeinschaft.
Abbreviations used: aa, amino acid, dsRNA, double-stranded RNA, KLP, kinesin-like protein, MT, microtubule, RNAi, RNA-mediated interference.
This article is dedicated to the memory of Christoph Segbert, an outstanding young scientist.
Online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org
References
- Albertson, D.G., and Thomson, J.N. (1993). Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1, 15-26. [DOI] [PubMed] [Google Scholar]
- Antonio, C., Ferby, I., Wilhelm, H., Jones, M., Karsenti, E., Nebreda, A.R., and Vernos, I. (2000). Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102, 425-435. [DOI] [PubMed] [Google Scholar]
- Askjaer, P., Galy, V., Hannak, E., and Mattaj, I.W. (2002). Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol. Cell 13, 4355-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamba, C., Bobinnec, Y., Fukuda, M., and Nishida, E. (2002). The GTPase Ran regulates chromosome positioning and nuclear envelope assembly in vivo. Curr. Biol. 12, 503-507. [DOI] [PubMed] [Google Scholar]
- Barton, M.K., Schedl, T.B., and Kimble, J. (1987). Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanan, M.J., and Strome, S. (1992). Characterization of a germ-line proliferation mutation in C. elegans. Development 116, 755-766. [DOI] [PubMed] [Google Scholar]
- Boleti, H., Karsenti, E., and Vernos, I. (1996). Xklp2, a novel Xenopus centrosomal kinesin-like protein required for centrosome separation during mitosis. Cell 84, 49-59. [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire, S., and Mains, P.E. (1994a). Localization of the mei-1 gene product of Caenorhaditis elegans, a meiotic-specific spindle component. J. Cell Biol. 126, 199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Maguire, S., and Mains, P.E. (1994b). mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics 136, 533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, D., Zorio, D., MacMorris, M., Winter, C.E., Lea, K., and Blumenthal, T. (1997). Operons and SL2 trans-splicing exist in nematodes outside the genus Caenorhabditis. Proc. Natl. Acad. Sci. USA 94, 9751-9756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., and Murray, A.W. (2000). The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102, 411-424. [DOI] [PubMed] [Google Scholar]
- Gard, D.L. (1992). Microtubule organization during maturation of Xenopus oocytes: assembly and rotation of the meiotic spindles. Dev. Biol. 151, 516-530. [DOI] [PubMed] [Google Scholar]
- Giunta, K.L., Jang, J.K., Manheim, E.A., Subramanian, G., and McKim, K.S. (2002). subito encodes a kinesin-like protein required for meiotic spindle pole formation in Drosophila melanogaster. Genetics 160, 1489-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, A., Sadler, P.L., Wallenfang, M.R., Schumacher, J.M., Hamill, D.R., Bates, G., Bowerman, B., Seydoux, G., and Shakes, D.C. (2000). Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151, 1469-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss, O.J., Carazo-Salas, R.E., Schatz, C.A., Guarguaglini, G., Kast, J., Wilm, M., Le Bot, N., Vernos, I., Karsenti, E., and Mattaj, I.W. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83-93. [DOI] [PubMed] [Google Scholar]
- Gruss, O.J., Wittmann, M., Yokoyama, H., Pepperkok, R., Kufer, T., Sillje, H., Karsenti, E., Mattaj, I.W., and Vernos, I. (2002). Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4, 871-879. [DOI] [PubMed] [Google Scholar]
- Hatsumi, M., and Endow, S.A. (1992). The Drosophila NCD microtubule motor protein is spindle-associated in meiotic and mitotic cells. J. Cell Sci. 103, 1013-1020. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Blank, T., Sandaltzopoulos, R., Becker, P., Hyman, A., and Karsenti, E. (1996). Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382, 420-425. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Habermann, A., Karsenti, E., and Hyman, A. (1997). Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138, 615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird, S.N., and White, J.G. (1993). Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J. Cell Biol. 121, 1343-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231-237. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Martinez-Campos, M., Zipperlen, P., Fraser, A.G., and Ahringer, J. (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, research 0002.1-0002.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina, A.S., Baskin, R.J., Cole, D.G., Wedaman, K.P., Saxton, W.M., and Scholey, J.M. (1996). A bipolar kinesin. Nature 379, 270-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, I., Shim, Y.H., Kirchner, J., Kaminker, J., Wood, W.B., and Strome, S. (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635-645. [DOI] [PubMed] [Google Scholar]
- Kemphues, K.J., Wolf, N., Wood, W.B., and Hirsh, D. (1986). Two loci required for cytoplasmic organization in early embryos of Caenorhabditis elegans. Dev. Biol. 113, 449-460. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., Cole, R.W., Oakley, B.R., and Rieder, C.L. (2000). Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10, 59-67. [DOI] [PubMed] [Google Scholar]
- Kimble, J., Edgar, L., and Hirsh, D. (1984). Specification of male development in Caenorhabditis elegans: the fem genes. Dev. Biol. 105, 234-239. [DOI] [PubMed] [Google Scholar]
- Kull, F.J., Sablin, E.P., Lau, R., Fletterick, R.J., and Vale, R.D. (1996). Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380, 550-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, C.J., Malmberg, R.L., Muszynski, M.G., and Dawe, R.K. (2002). Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 54, 42-53. [DOI] [PubMed] [Google Scholar]
- Lewis, J.A., and Fleming, J.T. (1995). Genetic and culture methods. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism, vol. 48, ed. F.H. Epstein and D.C. Shakes, San Diego: Academic Press, 3-29. [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- Mains, P.E., Kemphues, K.J., Sprunger, S.A., Sulston, I.A., and Wood, W.B. (1990). Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics 126, 593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, L.R., Carter, P., Thierry-Mieg, D., and Kemphues, K. (1998). ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 141, 1159-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies, H.J., McDonald, H.B., Goldstein, L.S., and Theurkauf, W.E. (1996). Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134, 455-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, K.S., and Hawley, R.S. (1995). Chromosomal control of meiotic cell division. Science 270, 1595-1601. [DOI] [PubMed] [Google Scholar]
- Merdes, A., and Cleveland, D.W. (1997). Pathways of spindle pole formation: different mechanisms; conserved components. J. Cell Biol. 138, 953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes, A., Heald, R., Samejima, K., Earnshaw, W.C., and Cleveland, D.W. (2000). Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149, 851-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain, V., Simerly, C., Howard, L., Ando, A., Schatten, G., and Compton, D.A. (1999). The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147, 351-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A. (1993). Targets of cyclin-dependent protein kinases. Curr. Opin. Cell Biol. 5, 187-193. [DOI] [PubMed] [Google Scholar]
- Piano, F., Schetterdagger, A.J., Mangone, M., Stein, L., and Kemphues, K.J. (2000). RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10, 1619-1622. [DOI] [PubMed] [Google Scholar]
- Piperno, G., and Fuller, M.T. (1985). Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101, 2085-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis, V., Casey, E., Collar, D., and Austin, J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb, D.L., Heasman, J., Raats, J., and Wylie, C. (1996). A kinesin-like protein is required for germ plasm aggregation in Xenopus. Cell 87, 823-831. [DOI] [PubMed] [Google Scholar]
- Rogers, G.C., Chui, K.K., Lee, E.W., Wedaman, K.P., Sharp, D.J., Holland, G., Morris, R.L., and Scholey, J.M. (2000). A kinesin-related protein, KRP(180), positions prometaphase spindle poles during early sea urchin embryonic cell division. J. Cell Biol. 150, 499-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin, E.P., Kull, F.J., Cooke, R., Vale, R.D., and Fletterick, R.J. (1996). Crystal structure of the motor domain of the kinesin-related motor NCD. Nature 380, 555-559. [DOI] [PubMed] [Google Scholar]
- Sawada, T., and Schatten, G. (1988). Microtubules in ascidian eggs during meiosis, fertilization, and mitosis. Cell Motil. Cytoskeleton 9, 219-230. [DOI] [PubMed] [Google Scholar]
- Sawin, K.E., and Mitchison, T.J. (1995). Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA 92, 4289-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., McDonald, K.L., Brown, H.M., Matthies, H.J., Walczak, C., Vale, R.D., Mitchison, T.J., and Scholey, J.M. (1999). The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, S.S. (2002). Metazoan motor models: kinesin superfamily in C. elegans. Traffic 3, 20-28. [DOI] [PubMed] [Google Scholar]
- Srayko, M., Buster, D.W., Bazirgan, O.A., McNally, F.J., and Mains, P.E. (2000). MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 14, 1072-1084. [PMC free article] [PubMed] [Google Scholar]
- Strome, S., and Wood, W.B. (1983). Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35, 15-25. [DOI] [PubMed] [Google Scholar]
- Sueishi, M., Takagi, M., and Yoneda, Y. (2000). The forkhead-associated domain of Ki-67 antigen interacts with the novel kinesin-like protein Hklp2. J. Biol. Chem. 275, 28888-28892. [DOI] [PubMed] [Google Scholar]
- Theurkauf, W.E., and Hawley, R.S. (1992). Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J. Cell Biol. 116, 1167-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012-2018. [DOI] [PubMed] [Google Scholar]
- Vernos, I., Raats, J., Hirano, T., Heasman, J., Karsenti, E., and Wylie, C. (1995). Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell 81, 117-127. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E. (2000). Molecular mechanisms of spindle function. Genome Biol 1, reviews 101.101-101.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C.E., Vernos, I., Mitchison, T.J., Karsenti, E., and Heald, R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8, 903-913. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., Boleti, H., Antony, C., Karsenti, E., and Vernos, I. (1998). Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J. Cell Biol. 143, 673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, T., Wilm, M., Karsenti, E., and Vernos, I. (2000). TPX2, A novel xenopus MAP involved in spindle pole organization. J. Cell Biol. 149, 1405-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.