Abstract
In budding yeast, the protein phosphatase Cdc14 controls exit from mitosis. Its activity is regulated by a competitive inhibitor Cfi1/Net1, which binds to and sequesters Cdc14 in the nucleolus. During anaphase, Cdc14 is released from its inhibitor by the action of two regulatory networks. The Cdc Fourteen Early Anaphase Release (FEAR) network initiates Cdc14 release from Cfi1/Net1 during early anaphase, and the Mitotic Exit Network (MEN) promotes Cdc14 release during late anaphase. Here, we investigate the relationship among FEAR network components and propose an order in which they function to promote Cdc14 release from the nucleolus. Furthermore, we examine the role of the protein kinase Cdc5, which is a component of both the FEAR network and the MEN, in Cdc14 release from the nucleolus. We find that overexpression of CDC5 led to Cdc14 release from the nucleolus in S phase-arrested cells, which correlated with the appearance of phosphorylated forms of Cdc14 and Cfi1/Net1. Cdc5 promotes Cdc14 phosphorylation and, by stimulating the MEN, Cfi1/Net1 phosphorylation. Furthermore, we suggest that Cdc14 release from the nucleolus only occurs when Cdc14 and Cfi1/Net1 are both phosphorylated.
INTRODUCTION
During exit from mitosis, cells eliminate mitotic determinants and establish conditions permissive for budding, DNA replication, and spindle pole body duplication. In the budding yeast Saccharomyces cerevisiae, the protein phosphatase Cdc14 triggers this transition by reversing cyclin-dependent kinase (CDK) phosphorylation (reviewed in (Morgan, 1999; Bardin and Amon, 2001; McCollum and Gould, 2001). The activity of Cdc14 is controlled by Cfi1/Net1, which functions as a competitive inhibitor of this protein phosphatase (Shou et al., 1999; Visintin et al., 1999; Traverso et al., 2001). The association of Cdc14 with its inhibitor is cell cycle regulated. During G1, S phase, G2, and metaphase Cfi1/Net1 sequesters and inhibits Cdc14 in the nucleolus (Shou et al., 1999; Visintin et al., 1999). During anaphase the phosphatase dissociates from its inhibitor in the nucleolus and spreads throughout the cell, where it is presumably active and able to dephosphorylate its targets.
Two regulatory networks have been identified that control the localization of Cdc14. The Cdc Fourteen Early An-aphase Release (FEAR) network promotes release of Cdc14 from the nucleolus during early anaphase and the Mitotic Exit Network (MEN) maintains Cdc14 in this released state during late stages of anaphase and telophase (Shou et al., 1999; Visintin et al., 1999; Pereira et al., 2002; Stegmeier et al., 2002; Yoshida et al., 2002). Inactivation of FEAR network components prevents Cdc14 release from the nucleolus during early anaphase, when the mitotic spindle is between 3 and 7 μm in length. In contrast, inactivation of the MEN abolishes Cdc14 release from the nucleolus during late stages of anaphase, when the mitotic spindle measures 7-10 μm in length (Pereira et al., 2002; Stegmeier et al., 2002; Yoshida et al., 2002). MEN-dependent release of Cdc14 from the nucleolus is essential for exit from mitosis, because temperature-sensitive mutants in MEN components fail to exit from mitosis but arrest in telophase. The FEAR network-dependent release of Cdc14 from the nucleolus during early anaphase is required for the timely exit from mitosis but is not essential, because mutations in FEAR network components delay, but do not prevent, exit from mitosis.
The MEN resembles a Ras-like signaling cascade with the GTPase Tem1 functioning at or near the top of the MEN. Tem1 activity is thought to be regulated both by the putative GTP exchange factor (GEF) Lte1 and a two-component GT-Pase-activating protein (GAP) complex composed of Bub2 and Bfa1 (reviewed in Bardin and Amon, 2001; McCollum and Gould, 2001). Tem1-GTP is believed to activate the protein kinase Cdc15, which then activates the protein kinase Dbf2 in a manner dependent on the Dbf2-associated factor Mob1 (Lee et al., 2001; Mah et al., 2001; Visintin and Amon, 2001). The spindle pole body (SPB) component Nud1 functions as a scaffold for MEN components at the SPB (Gruneberg et al., 2000). The polo kinase Cdc5 has also been implicated in MEN signaling, impinging on the pathway in at least two ways. Cdc5 phosphorylates Bfa1 and Bub2, thereby inactivating Tem1's GAP (Hu et al., 2001; Hu and Elledge, 2002; Geymonat et al., 2003) and functions to promote Dbf2 activity by an unknown mechanism (Lee et al., 2001).
CDC5 is also a component of the FEAR network, because the gene is not only required for the release of Cdc14 from the nucleolus during late stages of anaphase but also during early anaphase (Pereira et al., 2002; Stegmeier et al., 2002). Other components of the FEAR network include ESP1, SLK19, and SPO12 (Stegmeier et al., 2002). Esp1, which is also known as Separase, encodes a protease that is responsible for triggering sister chromatid separation at the onset of anaphase (reviewed in Nasmyth, 2001; Uhlmann, 2001). ESP1 is also required for exit from mitosis functioning upstream of SLK19 and CDC5 (Tinker-Kulberg and Morgan, 1999; Stegmeier et al., 2002; Sullivan and Uhlmann, 2003). However, remarkably, Esp1's protease activity seems not to be required for its mitotic exit function (Sullivan and Uhl-mann, 2003). The FEAR network component Slk19 localizes to kinetochores and the spindle mid-zone during anaphase and is a substrate of Esp1 (Zeng et al., 1999; Sullivan et al., 2001). Spo12 encodes a protein of unknown function.
CDC5, ESP1, SLK19, and SPO12 have been grouped together into a network based on the observation that they are required for Cdc14 release from the nucleolus during early anaphase. Whether these genes function in a linear pathway or in parallel to bring about Cdc14 release from the nucleolus during early anaphase has not been determined. Furthermore, how the FEAR network and the MEN promote the dissociation of Cdc14 from Cfi1/Net1 is not known. Here, we investigate the relationship among FEAR network components and propose an order in which the four identified network components function. ESP1 functions upstream of SLK19 and SPO12 acts in parallel to ESP1, SLK19, and CDC5. We further address how Cdc5 promotes Cdc14 release from the nucleolus. We show that high levels of Cdc5 cause Cdc14 release from the nucleolus in S phase-arrested cells. Furthermore, we find that Cdc5 induces Cdc14 and Cfi1/Net1 phosphorylation. Phosphorylation of Cfi1/Net1 induced by Cdc5 depends on an active mitotic exit network whereas phosphorylation of Cdc14 does not. Finally we present evidence to suggest that Cdc14 release from the nucleolus only occurs when both Cdc14 and Cfi1/Net1 are phosphorylated.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
All strains were derivatives of strain W303 (K699). The GAL-CDC5-3MYC, CDC14-HA, CFI1-MYC, GAL-CDC15[1-974] and GAL-CDC15[1-750] fusions were described in Taylor et al. (1997), Charles et al. (1998), Visintin et al. (1999), and Bardin et al. (2003) respectively. The CFI1-GFP and DBF2-3HA fusion were constructed by using the polymerase chain reaction-based method described in Longtine et al. (1998). Growth conditions for individual experiments are described in the figure legends.
Immunoblot Analysis and Dbf2 Kinase Assay
To prepare protein extracts for Western blot analysis cells were incubated for 10 min in 10% trichloroacetic acid at 4°C, pelleted, and then washed with acetone. Cells were broken in 50 mM Tris pH 7.5, 1 mM EDTA, 1 mM p-nitrophenyl phosphate, 50 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 2 μg/ml pepstatin with glass beads for 40 min and boiled in 1× sample buffer. Immunoblot analysis of the total amount of Clb2, Cdc14-HA, Cfi1-MYC, Cdc5-MYC, Dbf2-HA, and Kar2 was performed as described in Cohen-Fix et al. (1996). Dbf2 kinase activity was assayed as described in Visintin and Amon (2001).
Fluorescence Microscopy
Indirect in situ immunofluorescence methods and antibody concentrations were as described in Visintin et al. (1999). Cells were analyzed on an Axioplan 2 microscope (Carl Zeiss, Thornwood, NY), and images were captured with a camera controller (Hamamatsu, Bridgewater, NJ). Openlab 3.0.2.software was used to process immunofluorescence images.
RESULTS
Overexpression of SPO12 or CDC5 Abolishes the Need for SLK19 and ESP1 in Releasing Cdc14 from the Nucleolus
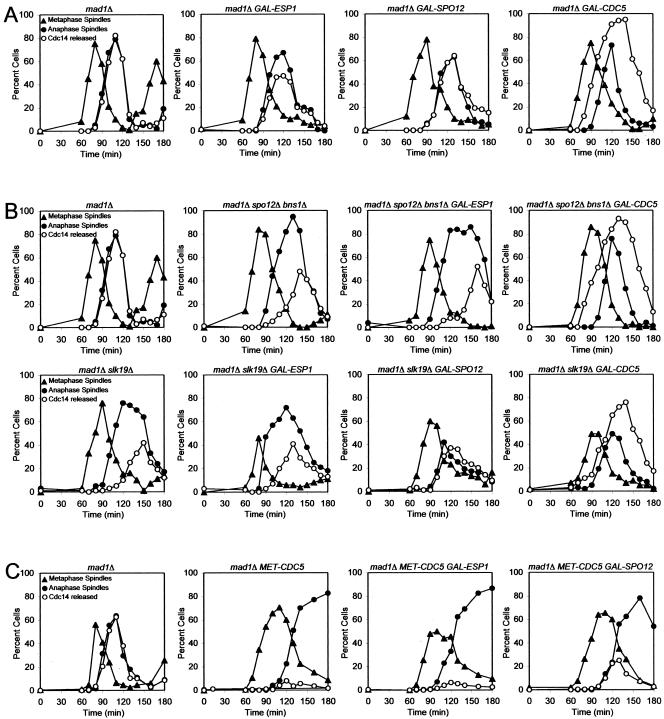
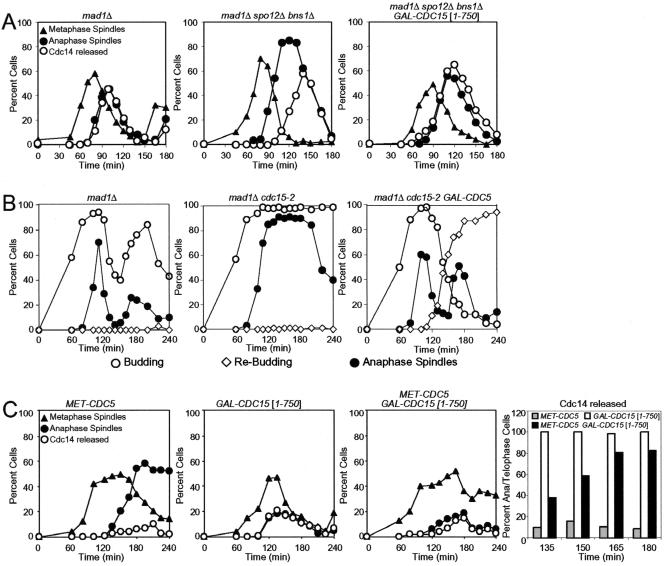
Several components of the FEAR network have been identified but the order in which they function to promote Cdc14 release from the nucleolus is not known. To investigate the epistatic relationship among FEAR network components, we determined whether overexpression of one FEAR network component from the galactose-inducible GAL1-10 promoter could restore Cdc14 release from the nucleolus in cells lacking other FEAR network components. To this end, we first characterized the effects of overexpressing various FEAR network components on Cdc14 localization in otherwise wild-type cells. We conducted this analysis in cells lacking MAD1, a component of the spindle checkpoint, to avoid indirect effects on FEAR network activity due to activation of the mitotic spindle checkpoint (Stegmeier et al., 2002; Yoshida et al., 2002). Overexpression of SPO12 did not significantly affect the kinetics of Cdc14 release from the nucleolus (Figure 1A). Overexpression of CDC5 caused premature release of Cdc14 from the nucleolus during metaphase, whereas, surprisingly, overexpression of ESP1 slightly inhibited Cdc14 release from the nucleolus (Figure 1A). We were not able to examine the consequences of overexpressing SLK19 because high levels of Slk19 caused spindle abnormalities precluding the reliable identification of cells with anaphase spindles (our unpublished data).
Figure 1.
Epistatic relationships among FEAR network components. (A) mad1Δ (A2853), mad1Δ GAL-ESP1 (A7619), mad1Δ GAL-SPO12 (A6490), and mad1Δ GAL-CDC5 (A7297) cells carrying a CDC14-3HA fusion were arrested in G1 in YEP medium containing 2% raffinose (YEPR) with α-factor (5 μg/ml) for 3 h. Cells were then released into YEPR medium containing 2% galactose (YEPRG) lacking pheromone at 25°C. The percentage of cells with metaphase spindles (close triangles), anaphase/telophase spindles (close circles) as well as the percentage of cells with Cdc14-HA released from the nucleolus (open circles) was determined at the indicated times. (B) mad1Δ (A2853) mad1Δ spo12Δ bns1Δ (A5408), mad1Δ spo12Δ bns1Δ GAL-ESP1 (A7619) mad1Δ spo12Δ bns1Δ GAL-CDC5 (A7578), mad1Δ slk19Δ (A4302), mad1Δ slk19Δ GAL-ESP1 (A7302), mad1Δ slk19Δ GAL-SPO12 (A7581), mad1Δ slk19Δ GAL-CDC5 (A8032) cells carrying a CDC14-3HA fusion were grown and analyzed as described in (A). (C) mad1Δ (A2853), mad1Δ MET-CDC5 (A6560), mad1Δ MET-CDC5 GAL-ESP1 (A6562), and mad1Δ MET-CDC5 GAL-SPO12 (A6630) cells carrying a CDC14-3HA fusion were grown in medium lacking methionine and arrested in G1 with α-factor (5 μg/ml). Galactose (2%) was added 1 h before release, and cells were released into YEPRG medium containing 8 mM methionine at 25°C. The percentage of cells with metaphase spindles (closed triangles), anaphase/telophase spindles (closed circles) as well as the percentage of cells with Cdc14-HA released from the nucleolus (open circles) was determined at the indicated times.
Having established the consequences of overexpressing various FEAR network components on Cdc14 localization in otherwise wild-type cells, we determined whether overexpression of CDC5, ESP1, or SPO12 could restore Cdc14 release from the nucleolus to slk19Δ mutants during early anaphase. In slk19Δ cells, Cdc14 release from the nucleolus, in contrast to wild-type cells, occurs only during late anaphase (Figure 1B; Stegmeier et al., 2002). Overexpression of ESP1 did not restore release of Cdc14 from the nucleolus during early anaphase but overexpression of CDC5 and SPO12 did (Figure 1B). These findings raised the possibility that CDC5 and SPO12 functioned downstream of or in parallel to SLK19 and that ESP1 functioned upstream of SLK19. We could indeed show that ESP1 functioned upstream of SLK19 when we examined the effects of high levels of Esp1 in nocodazole arrested cells. In the nocodazole arrest, high levels of Esp1 led to premature release of Cdc14 from the nucleolus, which depended on SLK19 (Sullivan and Uhl-mann, 2003; Supplemental Figure 1).
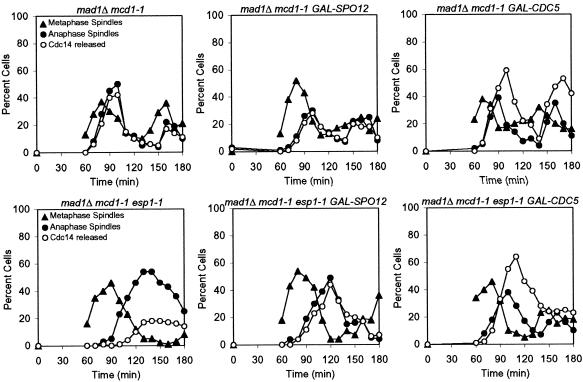
Next, we determined whether overexpression of FEAR network components rendered ESP1 dispensable for Cdc14 release from the nucleolus during early anaphase. Because ESP1 is also required for sister-chromatid separation (Ciosk et al., 1998), we examined the kinetics of Cdc14 release from the nucleolus in esp1-1 mutants carrying a temperature-sensitive mcd1-1 mutation, which makes ESP1 dispensable for chromosome segregation and spindle elongation (Guacci et al., 1997). Overexpression of CDC5 or SPO12 restored release of Cdc14 from the nucleolus during early anaphase in esp1-1 mcd1-1 mutants (Figure 2). These results indicate that Cdc5 and Spo12, when overproduced, do not require SLK19 or ESP1 function to promote Cdc14 release from the nucleolus during early anaphase.
Figure 2.
High levels of Cdc5 and Spo12 allow esp1-1 to release Cdc14 from the nucleolus during early anaphase. mad1Δ mcd1-1 (A2784), mad1Δ mcd1-1 GAL-SPO12 (A6528), mad1Δ mcd1-1 GAL-CDC5 (A7300), mad1Δ mcd1-1 esp1-1 (A3023), mad1Δ mcd1-1 esp1-1 GAL-SPO12 (A7711), and mad1Δ mcd1-1 esp1-1 GAL-CDC5 (A7580) all carrying the CDC14-3HA fusion were grown and analyzed as described in Figure 1A, except cells were released from the G1 block at 37°C.
High Levels of Cdc5 Suppress the Requirement for SPO12 in Promoting Release of Cdc14 during Early Anaphase
To further investigate the relationship between FEAR network components, we determined whether overexpression of CDC5 or ESP1 could restore Cdc14 release from the nucleolus to spo12Δ bns1Δ double mutants during early anaphase. BNS1 shares 55% similarity with SPO12 (Grether and Herskowitz, 1999) and when inactivated further impairs the release of Cdc14 from the nucleolus during early anaphase in spo12Δ cells (Supplemental Figure 2). Thus, BNS1 and SPO12 perform at least partially overlapping functions in promoting Cdc14 release from the nucleolus. Overexpression of ESP1 did not promote release of Cdc14 from the nucleolus during early anaphase in spo12Δ bns1Δ double mutants but overexpression of CDC5 restored Cdc14 release to the extent observed in wild-type cells overexpressing CDC5 (Figure 1B). This finding indicates that high levels of Cdc5 can promote Cdc14 release from the nucleolus independently of SPO12 and BNS1 but that Esp1 cannot.
Overexpression of SPO12 Partially Restores Cdc14 Release from the Nucleolus in CDC5-depleted Cells
To determine whether overexpression of FEAR network components can bypass the requirement for CDC5 in promoting Cdc14 release from the nucleolus during early anaphase, we overexpressed various FEAR network components in cells depleted for Cdc5. We chose to inactivate Cdc5 by depletion rather than by using a temperature sensitive cdc5 allele to ensure complete inactivation of CDC5 function. CDC5 was cloned under the control of the methionine-repressible MET3 promoter (Mountain and Korch, 1991). Because Cdc5 is unstable during G1 (Charles et al., 1998; Shirayama et al., 1998), the protein is depleted from cells during a pheromone-induced G1 arrest. MET-CDC5 strains were arrested in G1 by using α-factor pheromone and transcription of CDC5 was repressed by methionine addition 1 h before release of cells from the G1 block. Under these conditions, cells arrested in telophase with Cdc14 sequestered in the nucleolus (Figure 1C). Furthermore, the telophase arrest of Cdc5-depeleted cells was bypassed by the TAB6-1 mutation, a dominant allele of CDC14 (Supplemental Figure 3; Shou et al., 1999), indicating that promoting Cdc14 release from the nucleolus is CDC5's essential function during exit from mitosis.
Overexpression of ESP1 did not restore Cdc14 release from the nucleolus during early anaphase, but overexpression of SPO12 did to some extent (Figure 1C). We also observed that 30% of Cdc5-depleted cells overexpressing SPO12 disassembled their mitotic spindles and entered a new cell cycle, as judged by their ability to form a new bud (Supplemental Figure 4). However these cells failed to form colonies, perhaps due to the severe cytokinesis defects observed in these cells (our unpublished data). Our findings suggest that SPO12 can to some extent promote Cdc14 release from the nucleolus in CDC5-depleted cells. This suppression could be due to overproduced Spo12 being able to promote Cdc14 release from the nucleolus in the absence of CDC5 function. However, we cannot exclude the possibility that Cdc5-depleted cells contain residual amounts of Cdc5, which though unable to promote Cdc14 release from the nucleolus and exit from mitosis together with high levels of Spo12 promote Cdc14 release from the nucleolus and exit from mitosis.
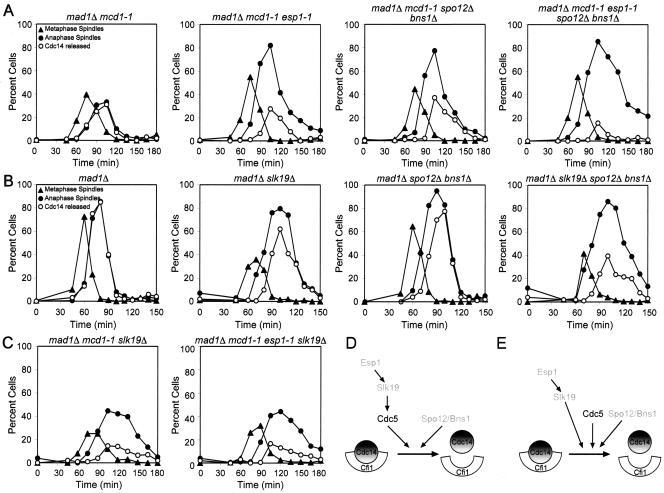
Inactivation of SPO12 and BNS1 Enhances the Mitotic Exit Defect of esp1-1 and slk19Δ Mutants
The observation that high levels of SPO12 at least partially suppress the defect in Cdc14 release from the nucleolus caused by inactivation of ESP1, SLK19, or CDC5 indicates that SPO12 either functions downstream of or in parallel to these genes. To distinguish between these possibilities, we compared the kinetics of Cdc14 release from the nucleolus and mitotic exit of esp1-1 or slk19Δ mutants with that of esp1-1 or slk19Δ mutants lacking SPO12 and BNS1. We could not test whether deletion of SPO12 and BNS1 enhanced the phenotype of cells lacking CDC5 as Cdc14 release from the nucleolus is completely abolished in such cells (Pereira et al., 2002; Stegmeier et al., 2002; Yoshida et al., 2002). Inactivation of SPO12 and BNS1 enhanced the defect in Cdc14 release from the nucleolus and mitotic spindle disassembly of, both, esp1-1 and slk19Δ mutants (Figure 3, A and B). In contrast, the mitotic exit defect of esp1-1 slk19Δ double mutants was similar to that of the slk19Δ and esp1-1 single mutants (Figure 3C).
Figure 3.
SPO12 and BNS1 function in parallel to ESP1 and SLK19. (A) mad1Δ mcd1-1 (A2784), mad1Δ mcd1-1 esp1-1 (A3550), mad1Δ mcd1-1 spo12Δ bns1Δ (A8129), and mad1Δ mcd1-1 esp1-1 spo12Δ bns1Δ (A8130) cells carrying a CDC14-3HA fusion were arrested in G1 in YEPD medium with α-factor (5 μg/ml) and subsequently released into YEPD lacking pheromone at 37°C. α-Factor was readded 90 min after release to prevent entry into a subsequent cell cycle. The percentage of cells with metaphase spindles (closed triangles), anaphase/telophase spindles (closed circles) as well as the percentage of cells with Cdc14-HA released from the nucleolus (open circles) was determined at the indicated times. (B) mad1Δ (A2853), mad1Δ slk19Δ (A4302), mad1Δ spo12Δ bns1Δ (A5408), and mad1Δ spo12Δ bns1Δ slk19Δ (A5515) cells all carrying a CDC14-3HA fusion were arrested in G1 in YEPD medium with α-factor (5 μg/ml) and subsequently released in a synchronous manner into YEPD lacking pheromone at 25°C. α-Factor pheromone was readded 80 min after release to prevent entry into a subsequent cell-cycle. Cells were analyzed as described in (A). (C) mad1Δ mcd1-1 slk19Δ (A8132) and mad1Δ mcd1-1 esp1-1 slk19Δ (A8132) cells carrying a CDC14-3HA grown and analyzed as described in A. (D and E) Possible order of function of FEAR network components. SLK19 functions downstream of ESP1 (D, E). SPO12 and BNS1 function in parallel to ESP1, SLK19, and CDC5 (D and E). CDC5 could function in either of two ways in the FEAR network. CDC5 could act downstream of ESP1 and SLK19 (D) or in parallel to the two genes (E).
In summary our epistasis analyses indicate that ESP1 and SLK19 function in the same pathway and that SLK19 functions downstream of ESP1 (Figure 3, D and E; Sullivan and Uhlmann, 2003). CDC5 either acts downstream of and/or in parallel to ESP1 and SLK19 and in parallel to SPO12/BNS1 (Figure 3, D and E) because overexpression of CDC5 suppresses the defect in Cdc14 release from the nucleolus of esp1-1 or slk19Δ or spo12Δ bns1Δ mutants as well as of spo12Δ bns1Δ slk19Δ triple mutants (Figures 1 and 2; Supplemental Figure 5). The findings that 1) inactivation of SPO12 and BNS1 enhances the mitotic exit defect of slk19Δ and esp1-1 mutants; 2) overexpression of SPO12 at least partially bypasses the requirement of CDC5, ESP1, and SLK19 in Cdc14 release from the nucleolus; and 3) overexpression of CDC5 can promote Cdc14 release from the nucleolus during early anaphase in spo12Δ bns1Δ mutants further suggests that SPO12 and presumably BNS1 function in a branch of the FEAR network that is parallel to that of CDC5, ESP1, and SLK19 (Figure 3, D and E).
Hyperactivation of the MEN Suppresses the Defect of FEAR Network Mutants
Our epistasis analysis thus far has not been able to distinguish between the possibilities that CDC5 functions down-stream of or in parallel to Esp1 and Slk19. Cdc5, besides being a FEAR network component, is also a member of the Mitotic Exit Network (Shou et al., 1999; Visintin et al., 1999; Hu et al., 2001; Lee et al., 2001) that functions in parallel to the FEAR network to promote Cdc14 release from the nucleolus (Pereira et al., 2002; Stegmeier et al., 2002; Yoshida et al., 2002). The finding that overexpression of CDC5 led to ectopic activation of Dbf2 kinase (Figure 7A) raised the possibility that the suppression of FEAR network mutants by high levels of Cdc5 was due to Cdc5's parallel activating function in the MEN. We, therefore, asked whether overactivation of the MEN could suppress the defect of FEAR network mutants in releasing Cdc14 from the nucleolus during early anaphase. Overexpression of a truncated version of CDC15 from the GAL1-10 promoter (GAL-CDC15 [1-750]) leads to pronounced hyperactivation of the MEN (Bardin et al., 2003) and suppressed the defect in Cdc14 release from the nucleolus in spo12Δ bns1Δ mutants (Figure 4A). This finding suggests that hyperactivation of the MEN can suppress the need for the FEAR network in Cdc14 release during early anaphase.
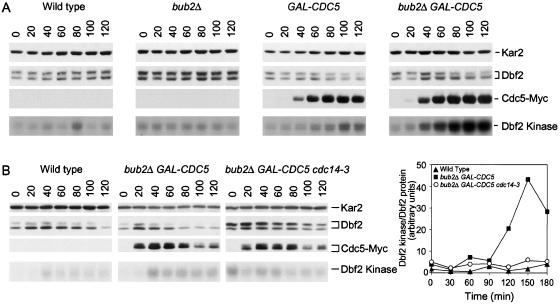
Figure 7.
Overexpression of CDC5 stimulates MEN activity. (A) Wild-type (A3912), bub2Δ (A6243), GAL-CDC5 (A6207), and GAL-CDC5 bub2Δ (A6210) cells all carrying a DBF2-3HA fusion were grown as described in Figure 5. Samples were taken at the indicated times to analyze Dbf2 and Cdc5 protein levels and their mobility in SDS PAGE as well as Dbf2 associated Kinase activity. (B) Wild-type (A3912), GAL-CDC5 bub2Δ (A6210) and GAL-CDC5 bub2Δ cdc14-3 (A7462) cells all carrying a DBF2-3HA fusion were arrested with 10 mg/ml HU at 25°C. Galactose was then added and cells were shifted to 37°C. The specific activity of Dbf2 (Dbf2 kinase/Dbf2 protein) was analyzed at the indicated times.
Figure 4.
Hyperactivation of the MEN can suppress the mitotic exit defect of FEAR network mutants. (A) mad1Δ (A2853), mad1Δ spo12Δ bns1Δ (A5408), and mad1Δ spo12Δ bns1Δ GAL-CDC15[1-750] (A8708) cells carrying a CDC14-3HA fusion were arrested in G1 in YEPR with α-factor (5 μg/ml) followed by released in YEPRG lacking pheromone. The percentage of cells with metaphase (closed triangles), anaphase/telophase spindles (closed circles) as well as the percentage of cells with Cdc14-HA released from the nucleolus (open circles) was determined at the indicated times. (B) mad1Δ (A2853), mad1Δ cdc15-2 (A4300), and mad1Δ cdc15-2 GAL-CDC5 (A5516) cells carrying a CDC14-3HA fusion were arrested in G1 in YEPR with α-factor (5 μg/ml) followed by released in YEPRG lacking pheromone at the restrictive temperature 37°C. The percentage of budded cells (open circles), rebudded cells (open diamonds), which indicates that cells have entered the next cell cycle as well as the percentage of anaphase/telophase spindles (closed circles) was determined at the indicated times. (C) MET-CDC5 (A9006), GAL-CDC15[1-750] (A8459), and MET-CDC5 GAL-CDC15[1-750] (A9007) cells carrying a CDC14-3HA fusion were grown in medium lacking methionine and arrested in G1 with α-factor (5 μg/ml). Methionine (8 mM) was added 1 h before release, and cells were released into YEPRG medium containing 8 mM methionine at 25°C. The percentage of cells with metaphase (closed triangles), anaphase/telophase spindles (closed circles) as well as the percentage of cells with Cdc14-HA released from the nucleolus (open circles) was determined at the indicated times. The histogram on the right shows the percentage of Cdc14 release in anaphase/telophase cells. One hundred cells were scored for each time point.
Does overexpression of CDC5 only activate the MEN or does it also activate the FEAR network? If high levels of Cdc5 were solely activating the MEN, inactivation of this pathway should preclude Cdc14 release from the nucleolus in GAL-CDC5 cells. However, GAL-CDC5 cdc15-2 mutants exited mitosis with almost the same kinetics as wild-type cells (Figure 4B). Suppression of the mitotic exit defect of the cdc15-2 mutant by overexpressed CDC5 was not due to CDC5 functioning downstream of CDC15 in the MEN. Hyperactivation of the MEN caused by overexpression of a truncated version of CDC15 from the GAL1-10 promoter (GAL-CDC15 [1-750]; Bardin et al., 2003) promoted Cdc14 release from the nucleolus during late stages of anaphase in the absence of CDC5 function (Figure 4C; note that Cdc14 release from the nucleolus did not occur in early anaphase, consistent with Cdc5's role in the FEAR network). We conclude that overexpression of CDC5 can promote Cdc14 release from the nucleolus and exit from mitosis in the absence of the MEN, suggesting that high levels of Cdc5 also cause hyperactivation of the FEAR network. However, owing to the dual role of Cdc5 in the FEAR network and the MEN, it was not possible to place Cdc5 within the FEAR network. Thus, CDC5 either functions downstream of and/or in parallel to ESP1 and SLK19 and in parallel to SPO12/BNS1. This parallel function of CDC5 could be in the MEN (Figure 3, D and E).
Overexpression of CDC5 Causes Premature Release of Cdc14 from the Nucleolus
A key question concerning the regulation of Cdc14 activity is how Cdc14 release from its inhibitor is regulated at the molecular level. Cdc5 seemed to play a central role in promoting the release of Cdc14 release from the nucleolus because CDC5 was the only gene, which when overexpressed, caused premature release of Cdc14 from the nucleolus during metaphase (Shou et al., 2002; Figure 1). We, therefore, investigated Cdc5's role in promoting Cdc14 release from the nucleolus in more detail and asked whether overexpression of CDC5 could induce Cdc14 release from the nucleolus in cell cycle stages other than anaphase. Overexpression of CDC5 caused release of Cdc14 from the nucleolus in S phase- and metaphase-arrested cells (Figures 5A and 6C), suggesting that CDC5 when present at high levels is sufficient to induce release of Cdc14 from the nucleolus. However, the kinetics of this release was sluggish, suggesting that other factors were necessary to promote efficient release of Cdc14 from the nucleolus. To identify factors that aid Cdc5 in promoting Cdc14 release from the nucleolus, we first determined whether Spo12, which functions in parallel to Cdc5 in the FEAR network could enhance Cdc5's Cdc14 release-promoting activity. Overexpression of SPO12 did not enhance the ability of CDC5 to promote release of Cdc14 from the nucleolus nor did it cause release of Cdc14 from the nucleolus in S phase- or metaphase-arrested cells on its own (Supplemental Figure 6; our unpublished data).
Figure 5.
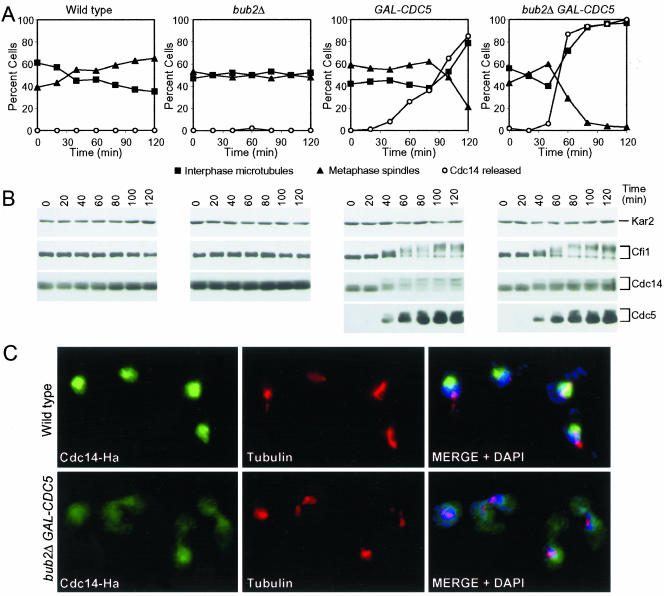
Overexpression of CDC5 causes release of Cdc14 from the nucleolus in hydroxyurea-arrested cells, which is enhanced by the deletion of BUB2. Wild-type (A1681), bub2Δ (A4451), GAL-CDC5 (A4450), and GAL-CDC5 bub2Δ (A4453) cells carrying a CDC14-3HA and a CFI1-3MYC fusion were arrested in early S phase with HU (10 mg/ml) in YEPR medium at 25°C. After 2.5 h 5 mg/ml HU and galactose (2%) were added. Samples were taken at the indicated times to analyze the percentage of cells with metaphase spindles (closed triangles; A) interphase microtubules (closed squares; A), with Cdc14 released from the nucleolus (open circles; A) and to analyze Cfi1/Net1, Cdc14, and Cdc5 protein levels and their mobility (B). Kar2 was used as an internal loading control in Western blots. The pictures in (C) show Cdc14 localization in wild-type and GAL-CDC5 bub2Δ cells 80 min after galactose addition. Cdc14 is shown in green, microtubules in red and nucleus in blue.
Figure 6.
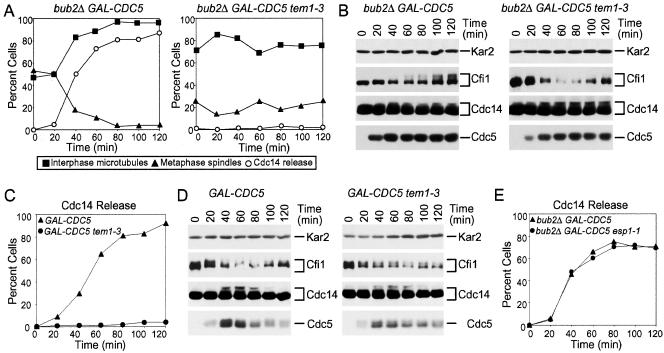
The ectopic release of Cdc14 from the nucleolus brought about by GAL-CDC5 and deletion of BUB2 depends on TEM1. (A, B) GAL-CDC5 bub2Δ (A4453) and GAL-CDC5 bub2Δ tem1-3 (A4549) cells carrying a CDC14-3HA and a CFI1-3MYC fusion were grown and analyzed as described in Figure 5 except, cells were shifted to 37°C at the time of galactose addition. (C, D) GAL-CDC5 (A4450) and GAL-CDC5 tem1-3 (A4547) cells all carrying a CDC14-3HA and a CFI1-3MYC fusion were arrested in early mitosis with the microtubule depolymerazing drug nocodazole (15 μg/ml) in YEPR at 25°C. After 2.5 h, cells were shifted to 37°C in the presence of nocodazole. Samples were taken at the indicated times to analyze the percentage of cells with Cdc14 released from the nucleolus (C) and Cfi1/Net1, Cdc14 and Cdc5 protein levels and mobility in SDS-PAGE (D). (E) GAL-CDC5 bub2Δ (A4453) and GAL-CDC5 bub2Δ esp1-1 (A4282) cells carrying a CDC14-3HA and a CFI1-3MYC fusion were arrested with 10 mg/ml HU at 25°C. Galactose was then added and cells were shifted to 37°C and Cdc14 localization was analyzed at the indicated times.
Next, we determined whether activation of the MEN enhanced Cdc5's ability to promote Cdc14 release from the nucleolus in S phase-arrested cells. Deletion of BUB2 has been shown to cause a slight hyperactivation of the MEN kinase Dbf2 (Fesquet et al., 1999; Lee et al., 2001; Visintin and Amon, 2001). Cells deleted for BUB2 did not release Cdc14 from the nucleolus in a hydroxyurea (HU)-arrest, but inactivation of BUB2 enhanced the ability of CDC5 to promote Cdc14 release from the nucleolus (Figure 5A). In GAL-CDC5 bub2Δ cells, Cdc14 release from the nucleolus occurred very efficiently and as soon as Cdc5 protein accumulated, 40 min after galactose addition (Figure 5, A and C). The release of Cdc14 from the nucleolus was not due to disruption of this organelle, or the release of the Cdc14-Cfi1/Net1 complex from the nucleolus, as the localization of both the nucleolar protein Nop1 and Cfi1/Net1 was not affected by high levels of Cdc5 and inactivation of BUB2 (Supplemental Figure 7). Overexpression of other MEN or FEAR network components did not cause Cdc14 release from the nucleolus in S phase-arrested cells (our unpublished data). Our results show that overexpression of CDC5 and the simultaneous overactivation of the MEN is sufficient to cause efficient Cdc14 release from the nucleolus and that Cdc5 is unique in its ability to promote Cdc14 release from the nucleolus in all cell cycle stages analyzed.
The MEN Is Required for CDC5 to Promote Ectopic Release of Cdc14 from the Nucleolus
High levels of Cdc5 can only bring about efficient ectopic release of Cdc14 from the nucleolus when BUB2 is deleted indicating that CDC5 requires MEN function to effectively release Cdc14 from the nucleolus. Consistent with this idea we found that inactivation of TEM1 prevented the ectopic release of Cdc14 from the nucleolus in S phase-arrested GAL-CDC5 bub2Δ cells or nocodazole-arrested GAL-CDC5 cells (Figure 6, A and C). In contrast, inactivation of the FEAR network component ESP1 did not hamper Cdc5's ability to promote Cdc14 release from the nucleolus (Figure 6E). These data indicate that the activity of other FEAR network components is not necessary for overexpressed CDC5 to bring about Cdc14 release from the nucleolus but MEN activity is. This need for the MEN was, however, not universal. During anaphase, high levels of Cdc5 could promote exit from mitosis in the absence of an active MEN. Overexpression of CDC5 restored Cdc14 release from the nucleolus during anaphase and exit from mitosis in cdc15-2 cells (Figure 4A). Perhaps FEAR network components such as SPO12 aid CDC5 during anaphase in promoting Cdc14 release from the nucleolus but cannot do so in other stages of the cell cycle. Consistent with this idea we found that deletion of SPO12 abolished the ability of overexpressed CDC5 to promote exit from mitosis in cdc15-2 mutants (our unpublished data).
Dbf2 Kinase Is Activated in GAL-CDC5 bub2Δ Cells in a CDC14-dependent Manner
Previous studies suggested that Cdc5 activated the MEN kinase Dbf2 in a BUB2-independent manner (Fesquet et al., 1999, Lee et al., 2001). We, therefore, examined the effects of overexpressing CDC5 and deleting BUB2 on Dbf2 kinase activity. Overexpression of CDC5 slightly stimulated Dbf2 kinase activity but deletion of BUB2 together with the overexpression of CDC5 led to high levels of Dbf2 kinase activity (Figure 7A). To determine the mechanism whereby Dbf2 was activated in GAL-CDC5 bub2Δ cells, we determined whether CDC14 was required for this activation to occur. We examined this possibility because one function of Cdc14 released by the FEAR network is to stimulate MEN activity (Jaspersen and Morgan, 2000; Stegmeier et al., 2002). Inactivation of CDC14 prevented Dbf2 kinase activation in GAL-CDC5 bub2Δ cells as judged by the low specific activity of Dbf2 kinase in GAL-CDC5 bub2Δ cells carrying a cdc14-3 mutation (Figure 7B). Our results show that BUB2-independent activation of the MEN by high levels of Cdc5 is mostly if not solely due to Cdc5's function in the FEAR network.
Overexpression of CDC5 Induces Phosphorylation of Cdc14 and Cfi1/Net1
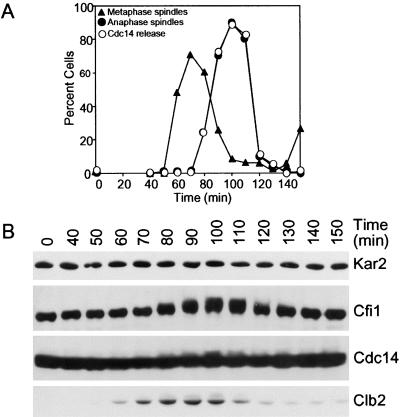
Release of Cdc14 from the nucleolus brought about by high levels of Cdc5 was accompanied by the appearance of slower migrating forms of Cdc14 and Cfi1/Net1 on Western blots (Figure 5B). These slower migrating forms of Cdc14 and Cfi1/Net1 represent phosphorylated forms of the proteins as treatment of both proteins with alkaline phosphatase led to their disappearance (Shou et al., 1999; Shou et al., 2002; our unpublished data). Thus, overexpression of CDC5 either directly or indirectly, induces phosphorylation of Cdc14 and Cfi1/Net1 and this phosphorylation occurs concomitantly with the release of Cdc14 from the nucleolus. A correlation between phosphorylation of Cdc14 and Cfi1/Net1 and Cdc14 release from the nucleolus also exists in wild-type cells progressing through the cell cycle in a synchronous manner (Figure 8). At the time of Cdc14 release from the nucleolus, Cfi1/Net1 and Cdc14 became phosphorylated as judged by the appearance of slower migrating forms of the proteins, although phosphorylation of Cdc14 was less pronounced than that seen in HU-arrested cells overexpressing CDC5 (Figure 8B), perhaps reflecting the transient nature of this phosphorylation. We conclude that Cfi1/Net1 and Cdc14 are phosphorylated at the time when Cdc14 is released from the nucleolus and that these modifications can be brought about by the overexpression of CDC5.
Figure 8.
Cdc14 and Cfi1/Net1 are phosphorylated during mitosis. Cells carrying a CDC14-3HA and a CFI1-3MYC fusion (A1681) were arrested in G1 in YEPD medium with α-factor (5 μg/ml) followed by release into YEPD medium lacking pheromone at 25°C. The percentage of cells with metaphase (closed triangles, A) and anaphase/telophase spindles (closed circles; A) as well as the percentage of cells with Cdc14-HA released from the nucleolus (open circles; A) was determined at the indicated times. Cdc14 and Cfi1/Net1 protein levels as well as their migration in SDS-PAGE were analyzed by Western Blot analysis (B). Clb2 protein levels were examined to determine when entry and exit from mitosis occur.
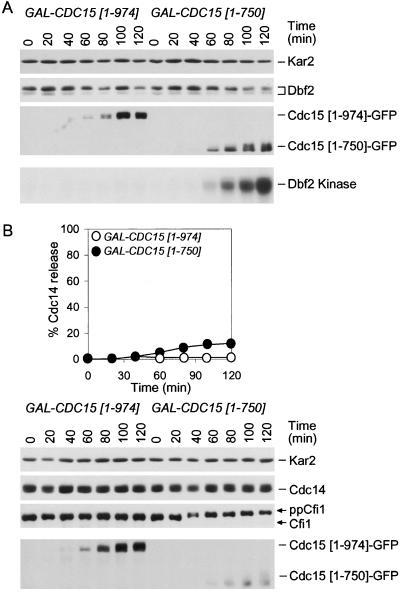
Cdc5 Induces Cfi1/Net1 Phosphorylation by Activating the MEN
Inactivation of TEM1 prevented the release of Cdc14 from the nucleolus in S phase-arrested GAL-CDC5 bub2Δ cells. To determine whether MEN activity also influenced Cdc14 and Cfi1/Net1 phosphorylation, we analyzed the mobility of the two proteins in GAL-CDC5 bub2Δ cells carrying a tem1-3 mutation. Inactivation of TEM1 had little effect on Cdc14 phosphorylation induced by high levels of Cdc5 (Figure 6, B and D), phosphorylation of Cfi1/Net1 was greatly reduced in GAL-CDC5 bub2Δ tem1-3 cells compared with GAL-CDC5 bub2Δ cells (Figure 6, B and D). Similar results were obtained when the MEN was inactivated using a cdc15-2 mutation (our unpublished data), suggesting that the bulk of Cfi1/Net1 phosphorylation induced by high levels of Cdc5 was largely due to MEN activation. To further test this possibility, we asked whether activation of the MEN would induce Cfi1/Net1 phosphorylation. Indeed, overexpression of CDC15[1-750] caused hyperphosphorylation of Cfi1/Net1 in S phase-arrested cells but it did not induce Cdc14 phosphorylation (Figure 9). Phosphorylation of Cfi1/Net1 was independent of CDC5 (our unpublished data), indicating that Cdc5 was not responsible for the phosphorylation of Cfi1/Net1 under these conditions. Remarkably, overexpression of CDC15[1-750] did not promote Cdc14 release from the nucleolus in S phase-arrested cells (Figure 9B). Our data indicate that phosphorylation of Cfi1/Net1 brought about by high levels of Cdc15[1-750] is not sufficient to bring about Cdc14 release from the nucleolus. Conversely, Cdc14 phosphorylation seemed not to be sufficient to cause Cdc14 release from the nucleolus in S phase-arrested cells, as Cdc14 is hyperphosphorylated in GAL-CDC5 bub2Δ tem1-3 mutants, yet Cdc14 release from the nucleolus does not occur (Figure 6). Thus, it seems that only when both, Cdc14 and Cfi1/Net1 are phosphorylated can the phosphatase be released from its inhibitor.
Figure 9.
Hyperactivation of MEN is not sufficient to promote Cdc14 release in hydroxyurea arrested cells. (A) GAL-CDC15[1-940] (A5568) and GAL-CDC15[1-750] (A6130) cells carrying a DBF2-3MYC fusion were arrested in early S phase with hydroxyurea (HU; 10 mg/ml) in YEPR medium at 25°C. After 2.5 h 5 mg/ml HU and galactose (2%) were added. Dbf2 and Cdc15 protein levels and their mobility as well as Dbf2 kinase activity was analyzed at the indicated times. Kar2 was used as an internal loading control in Western blots. (B) GAL-CDC15[1-940] (A6460) and GAL-CDC15[1-750] (A6459) cells carrying a CDC14-3HA fusion were arrested as described in Figure 9A. Samples were taken at the indicated times to analyze the percentage of cells with Cdc14 released from the nucleolus (open circles, GAL-CDC15[1-940]; closed circles, GAL-CDC15[1-750]) and to analyze Cfi1/Net1, Cdc14, and Cdc15 protein levels and their mobility. Kar2 was used as an internal loading control in Western blots.
DISCUSSION
Relationship among FEAR Network Components
Determining whether overexpression of a particular FEAR network component alleviates the need for another component to promote Cdc14 release from the nucleolus during early anaphase allowed us to characterize the epistatic relationship among FEAR network components. We propose that ESP1 and SLK19 function in the same branch of the FEAR network as esp1-1 slk19Δ double mutants exhibit the same defect in Cdc14 release from the nucleolus as slk19Δ single mutants. We further suggest that SLK19 functions downstream of ESP1 as SLK19 is required for ectopic release of Cdc14 from the nucleolus in nocodazole-arrested cells brought about by high levels of Esp1 (Supplemental Figure 1; Sullivan and Uhlmann, 2003).
All our data indicate that SPO12 and BNS1 constitute a separate branch of the FEAR network (Figure 3, D and E). Overexpression of SPO12 restored Cdc14 release from the nucleolus to slk19Δ and esp1-1 mutants and inactivation of SPO12 and BNS1 enhanced the mitotic exit defect of esp1-1 and slk19Δ mutants. The placing of SPO12 and BNS1 in parallel to CDC5 is based on the observations that overexpression of SPO12 partially suppressed the defect in Cdc14 release from the nucleolus in Cdc5-depleted cells and that high levels of CDC5 alleviated the requirement for SPO12 in promoting Cdc14 release from the nucleolus during early anaphase. We were not able to conclusively place CDC5 within the FEAR network. This was due to the fact that high levels of Cdc5 activated the FEAR network and to some extent the MEN. This leads us to propose that CDC5 either functions downstream of and/or in parallel to ESP1 and SLK19 and in parallel to SPO12/BNS1 and that this parallel function of CDC5 could be in the MEN.
Esp1's and Slk19's Role in the FEAR Network
Esp1 is best known for its function in promoting sister-chromatid separation through cleavage of Scc1/Mcd1 (reviewed in Nasmyth, 2001) but the protease is also required for the timely exit from mitosis (Cohen-Fix and Koshland, 1999; Tinker-Kulberg and Morgan, 1999; Stegmeier et al., 2002). The Esp1 substrate Slk19 is also necessary for exit from mitosis but cleavage of Slk19 by Esp1, does not seem to be necessary for Cdc14 release from the nucleolus (Stegmeier et al., 2002). Furthermore, a recent report by Sullivan and Uhlmann (2003) indicates that Esp1's protease is not required for its mitotic exit promoting activity.
We observed in many independent experiments that overexpression of ESP1 in cells progressing through the cell cycle in a synchronous manner did not promote Cdc14 release from the nucleolus but in fact slightly inhibited it. In nocodazole-arrested cells, however, overexpression of ESP1 causes ectopic release of Cdc14 from the nucleolus and exit from mitosis, although the kinetics with which this release occurred is slower than that seen in cells overexpressing CDC5 (Supplemental Figure 1; Sullivan and Uhlmann, 2003). This observation, however, allowed us to order SLK19 with respect to ESP1 function. Our results and that of Sullivan and Uhlmann (2003) suggest that Slk19 functions down-stream of Esp1 to promote Cdc14 release from the nucleolus. Why high levels of Esp1 promote Cdc14 release from the nucleolus in one stage of the cell cycle (metaphase) but slightly inhibit release in others (anaphase) is unknown. We speculate that ESP1 is not only required to promote Cdc14 release from its inhibitor but is also necessary for Cdc14 resequestration after exit from mitosis has been completed.
Spo12's Role in the FEAR Network
Inactivation of the SPO12 homolog BNS1, although causing no obvious defects in Cdc14 release from the nucleolus and exit from mitosis on its own, enhances the defects associated with a deletion of SPO12 thus adding this gene to the list of FEAR network components. However, even inactivation of both SPO12 and BNS1 only caused a mild defect in Cdc14 release from the nucleolus and exit from mitosis. It, thus, comes as a surprise that Spo12 is a potent promoter of Cdc14 release from the nucleolus and exit from mitosis when overexpressed during anaphase. High levels of Spo12 allow temperature-sensitive tem1-3, cdc15-2, cdc5-1, and dbf2-2 mutants to grow with almost wild-type kinetics at the restrictive temperature (Parkes and Johnston, 1992; Jaspersen et al., 1998; Shah et al., 2001). To further complicate the Spo12 puzzle, despite overproduced Spo12 effectively promoting Cdc14 release during anaphase even in the absence of a functional MEN, Spo12 fails to induce Cdc14 release from the nucleolus in stages of the cell cycle when Cdc14 is normally sequestered. The mechanisms that cause Spo12 to effectively promote Cdc14 release during anaphase but not other cell cycle stages remain obscure.
Multiple Roles of Cdc5 in Mitotic Exit
Cdc5 is the only member of the FEAR network and the MEN that, when overexpressed, promoted Cdc14 release from the nucleolus in S phase-arrested cells. In contrast, several MEN and FEAR network components can promote Cdc14 release from the nucleolus in metaphase-arrested cells (Shou et al., 2002; Yoshida et al., 2002; Bardin et al., 2003; Sullivan and Uhlmann, 2003; Visintin, unpublished observations). Our data show that Cdc5 has the unique ability to activate both the FEAR network and the MEN and this is the likely reason why Cdc5 can promote the dissociation of Cdc14 from its inhibitor in S phase-arrested cells. In metaphase-arrested cells and during anaphase, Cdc5 is active (Charles et al., 1998; Shirayama et al., 1998), which may be the reason why other FEAR network and MEN components can promote Cdc14 release from the nucleolus in this cell cycle stage.
Previous data and our results implicate Cdc5 in multiple aspects of mitotic exit. Cdc5 as a component of the FEAR network promotes Cdc14 release from the nucleolus (Pereira et al., 2002; Stegmeier et al., 2002). Cdc5 has also been shown to regulate the MEN. Cdc5 phosphorylation inhibits Bub2-Bfa1 (Hu et al., 2001; Hu and Elledge, 2002, Geymonat et al., 2003) and activates Dbf2 activity independently of Bub2-Bfa1 (Lee et al., 2001). How Cdc5 controlled the activity of Dbf2 was not known. We show that the ability of overexpressed CDC5 to activate Dbf2 in the absence of BUB2 depends on CDC14. Our data further indicate that Cdc5 does not function downstream of CDC15. Thus, our findings together with that of others suggest that Cdc5 functions in two ways to promote the activation of the MEN. Cdc5 directly activates the MEN by inhibiting Bub2-Bfa1 (Hu et al., 2001; Hu and Elledge, 2002; Geymonat et al., 2003). Cdc5's BUB2-independent role in promoting MEN activation is through releasing Cdc14 from the nucleolus as a component of the FEAR network (Pereira et al., 2002; Stegmeier et al., 2002; Yoshida et al., 2002; this study).
A Model for How Cdc14 Is Released from Its Inhibitor Cfi1/Net1 during Anaphase
In vitro studies suggest that phosphorylation is responsible for the dissociation of Cdc14 from its inhibitor Cfi1/Net1 (Shou et al., 1999; Shou et al., 2002). Furthermore, in vivo phosphorylation of Cfi1/Net1 and Cdc14 correlates with the release of Cdc14 from the nucleolus. Two observations suggest that Cdc5 directly phosphorylates Cdc14. Overexpression of CDC5 induced Cdc14 phosphorylation. Furthermore, Cdc5 can phosphorylate Cdc14 in vitro (Visintin, unpublished observations). Previous studies suggested that Cfi1/Net1 is a direct target of Cdc5. Cfi1/Net1 is a substrate of the protein kinase in vitro (Shou et al., 2002), Cfi1/Net1 phosphorylation depends on CDC5 activity in vivo (Shou et al., 2002; Yoshida and Toh-e, 2002) and high levels of Cdc5 induce Cfi1/Net1 phosphorylation (Figure 5). However, at least in S phase-arrested cells, the bulk of Cfi1/Net1 phosphorylation induced by CDC5 depended on an active MEN, indicating that Cdc5 induced Cfi1/Net1 phosphorylation indirectly, by activating the MEN. Consistent with this idea is the finding that Cfi1/Net1 phosphorylation depends on the MEN kinase Dbf2 (Visintin, unpublished observations). Thus, although it is possible that Cdc5 directly phosphorylates Cfi1/Net1, the bulk phosphorylation of the protein that occurs in vivo seems to be mediated by the MEN.
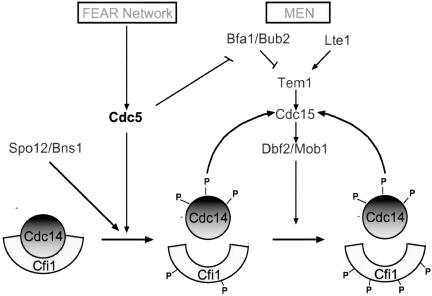
Our studies on Cdc14 and Cfi1/Net1 phosphorylation also revealed that the dissociation of Cdc14 from its inhibitor correlated only with the phosphorylation of both proteins. In GAL-CDC5 tem1-3 mutants, Cdc14 was hyperphosphorylated but Cfi1/Net1 was not and Cdc14 was not released from the nucleolus. In cells overexpressing CDC15[1-750] Cfi1/Net1 was hyperphosphorylated but Cdc14 was not and no Cdc14 release from the nucleolus was observed. Based on this correlation and results described above, we propose a two-step model for how Cdc14 release from the nucleolus is controlled. During early anaphase Cdc5 phosphorylates Cdc14 and perhaps to some extent Cfi1/Net1. We speculate that Spo12/Bns1 then aids in the dissociation of phosphorylated Cdc14 from Cfi1/Net1 (Figure 10). Activation of the MEN, in part through Cdc5 and Cdc14 then leads to phosphorylation of Cfi1/Net1. These phosphorylation events are necessary for maintaining Cdc14 in the released state during late stages of anaphase and telophase. Thus, Cdc5 initiates a positive feedback loop that brings about the rapid activation of Cdc14 at the end of mitosis.
Figure 10.
A model for how Cdc5 controls the release of Cdc14 from the nucleolus. At the onset of anaphase, activation of the FEAR network causes, by an unknown mechanism, Cdc5 to phosphorylate Cdc14 and Cfi1/Net1. Release of Cdc14 then promotes MEN activity by dephosphorylating Cdc15 and perhaps other proteins. Activation of the MEN then leads to further phosphorylation of Cfi1/Net1. Cdc5 also phosphorylates Bfa1 and Bub2, thereby contributing to full activation of the MEN.
Supplementary Material
Acknowledgments
We are grateful to Frank Uhlmann for strains overexpressing ESP1 from the GAL1-10 promoter and for communicating results before publication and to Ray Deshaies for providing the TAB6-1 allele. We thank Frank Solomon and members of the Amon laboratory for critical reading of the manuscript. R.V. is a postdoctoral fellow of the Merck foundation and F.S. is a predoctoral fellow of the National Science Foundation. This research was supported by a National Institute of Health grant GM-56800 to A.A. A.A is an investigator of the Howard Hughes Medical Institute.
Online version of this article contains supplemental material for some figures. Online version is available at www.molbiolcell.org.
References
- Bardin, A.J., and Amon, A. (2001). Men and sin: what's the difference? Nat. Rev. Mol. Cell. Biol. 2, 815-826. [DOI] [PubMed] [Google Scholar]
- Bardin, A.J., Boselli, M.G., and Amon, A. (2003). Mitotic exit regulation through distinct domains within the protein kinase Cdc15. Mol. Biol. Cell 23, 5018-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles, J.F., Jaspersen, S.L., Tinker-Kulberg, R.L., Hwang, L., Szidon, A., and Morgan, D.O. (1998). The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 8, 497-507. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., Zachariae, W., Michaelis, C., Shevchenko, A., Mann, M., Nasmyth, K. (1998). An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93, 1067-1076. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O., and Koshland, D. (1999). Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 13, 1950-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix, O., Peters, J.M., Kirschner, M.W., Koshland, D. (1996). Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10, 3081-3093. [DOI] [PubMed] [Google Scholar]
- Fesquet, D., Fitzpatrick, P.J., Johnson, A.L., Kramer, K.M., Toyn, J.H., and Johnston, L.H. (1999). A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 18, 2424-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat, M., Spanos, A., Walker, P.A., Johnston, L.H., and Sedgwick, S.G. (2003). In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 278, 14591-14594. [DOI] [PubMed] [Google Scholar]
- Grether, M.E., and Herskowitz, I. (1999). Genetic and biochemical characterization of the yeast spo12 protein. Mol. Biol. Cell 10, 3689-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg, U., Campbell, K., Simpson, C., Grindlay, J., and Schiebel, E. (2000). Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 19, 6475-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci, V., Koshland, D., and Strunnikov, A. (1997). A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91, 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, F., and Elledge, S.J. (2002). Bub2 is a cell cycle regulated phospho-protein controlled by multiple checkpoints. Cell Cycle 1, 351-355. [PubMed] [Google Scholar]
- Hu, F., Wang, Y., Liu, D., Li, Y., Qin, J., and Elledge, S.J. (2001). Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107, 655-665. [DOI] [PubMed] [Google Scholar]
- Jaspersen, S.L., Charles, J.F., Tinker-Kulberg, R.L., and Morgan, D.O. (1998). A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 9, 2803-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S.L., and Morgan, D.O. (2000). Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr. Biol. 10, 615-618. [DOI] [PubMed] [Google Scholar]
- Lee, S.E., Frenz, L.M., Wells, N.J., Johnson, A.L., and Johnston, L.H. (2001). Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 11, 784-788. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Mah, A.S., Jang, J., and Deshaies, R.J. (2001). Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98, 7325-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum, D., and Gould, K.L. (2001). Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 11, 89-95. [DOI] [PubMed] [Google Scholar]
- Morgan, D.O. (1999). Regulation of the APC and the exit from mitosis. Nat. Cell Biol. 1, E47-E53. [DOI] [PubMed] [Google Scholar]
- Mountain, H.A., and Korch, C. (1991). TDH2 is linked to MET3 on chromosome X of Saccharomyces cerevisiae. Yeast 7, 873-880. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. (2001). Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35, 673-745. [DOI] [PubMed] [Google Scholar]
- Parkes, V., and Johnston, L.H. (1992). SPO12 and SIT4 suppress mutations in DBF2, which encodes a cell cycle protein kinase that is periodically expressed. Nucleic Acids Res. 20, 5617-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., Manson, C., Grindlay, J., and Schiebel, E. (2002). Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 157, 367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, R., Jensen, S., Frenz, L.M., Johnson, A.L., and Johnston, L.H. (2001). The Spo12 protein of Saccharomyces cerevisiae: a regulator of mitotic exit whose cell cycle-dependent degradation is mediated by the anaphase-promoting complex. Genetics 159, 965-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama, M., Zachariae, W., Ciosk, R., and Nasmyth, K. (1998). The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17, 1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., Azzam, R., Chen, S.L., Huddleston, M.J., Baskerville, C., Charbonneau, H., Annan, R.S., Carr, S.A., and Deshaies, R.J. (2002). Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex. BMC Mol. Biol. 3, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou, W., Seol, J.H., Shevchenko, A., Baskerville, C., Moazed, D., Chen, Z.W., Jang, J., Charbonneau, H., and Deshaies, R.J. (1999). Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97, 233-244. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., Visintin, R., and Amon, A. (2002). Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108, 207-220. [DOI] [PubMed] [Google Scholar]
- Sullivan, M., Lehane, C., and Uhlmann, F. (2001). Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat. Cell Biol 3, 771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M., and Uhlmann, F. (2003). A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol 5, 249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, G.S., Liu, Y., Baskerville, C., and Charbonneau, H. (1997). The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J. Biol. Chem. 272, 24054-24063. [DOI] [PubMed] [Google Scholar]
- Tinker-Kulberg, R.L., and Morgan, D.O. (1999). Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 13, 1936-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso, E.E., Baskerville, C., Liu, Y., Shou, W., James, P., Deshaies, R.J., and Charbonneau, H. (2001). Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J. Biol. Chem. 276, 21924-21931. [DOI] [PubMed] [Google Scholar]
- Uhlmann, F. (2001). Secured cutting: controlling separase at the metaphase to anaphase transition. EMBO Rep. 2, 487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., and Amon, A. (2001). Regulation of the mitotic exit pathway protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12, 2961-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., Hwang, E.S., and Amon, A. (1999). Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398, 818-823. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., Asakawa, K., and Toh-e, A. (2002). Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr. Biol. 12, 944-950. [DOI] [PubMed] [Google Scholar]
- Yoshida, S., and Toh-e, A. (2002). Budding yeast Cdc5 phosphorylates Net1 and assists Cdc14 release from the nucleolus. Biochem. Biophys. Res. Commun. 294, 687-691. [DOI] [PubMed] [Google Scholar]
- Zeng, X., Kahana, J.A., Silver, P.A., Morphew, M.K., McIntosh, J.R., Fitch, I.T., Carbon, J., and Saunders, W.S. (1999). Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146, 415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.