Abstract
Sepsis, a lethal inflammatory syndrome, is characterized by organ system dysfunction. In the liver, we have observed decreased expression of genes encoding proteins modulating key processes. These include organic anion and bile acid transport. We hypothesized that the inflammatory mediator IL-6 modulates altered expression of several key hepatic genes in sepsis via induction of the intracellular transcription factor signal transducer and activator of transcription (Stat) 3. Sepsis was induced in IL-6 +/+ and IL-6 -/- mice, and expression of the liver-restricted genes encoding the sodium-taurocholate cotransporter (Ntcp), the multidrug resistant protein (MRP) 2 and the organic anion transporter protein (OATP), was determined. As demonstrated previously, cecal ligation and puncture decreases expression of Ntcp, MRP-2, and OATP in IL-6 +/+ mice. Transcription elongation analysis demonstrated that altered expression resulted from decreased transcription. These changes were not observed in IL-6 -/- animals. Cecal ligation and puncture increased the DNA binding activity of Stat-3 in IL-6 +/+ but not IL-6 -/- mice. Because the promoters of Ntcp, MRP-2, and OATP do not contain Stat-3 binding sites, we postulated that altered Ntcp, MRP-2, and OATP expression resulted from activation of hepatocyte nuclear factor (HNF) 1α, which is IL-6 dependent. Cecal ligation and puncture decreased HNF-1α expression and DNA binding activity in IL-6 +/+ but not IL-6 -/- mice. Recombinant human IL-6 restored the sepsis-induced decrease in Ntcp, MRP-2, OATP, and HNF-1α expression in IL-6 -/- mice. We conclude that sepsis decreases the expression of three key hepatic genes via a transcriptional mechanism that is IL-6, Stat-3, and HNF-1α dependent.
Keywords: Liver, cholestasis, steatosis, fatty liver, transcription factors, gene expression, HNF-1α, sodium taurocholate transporter, multidrug resistant protein 2, organic anion transporter protein
INTRODUCTION
Sepsis and the associated systemic inflammatory response syndrome and multiple organ dysfunction syndrome are the leading causes of death in critically ill patients (1). Because hepatic dysfunction is an important but poorly understood component of these disorders, we have investigated changes in hepatic gene expression, structure, and function after cecal ligation and puncture (CLP), a clinically relevant murine model of sepsis (2-6). Early on, CLP provokes a hyper-inflammatory state. This is characterized by increases in serum levels and in the intrahepatic activity of a number of proinflammatory mediators, especially the cytokines TNF-α, IL-1, and IL-6 (7-9). Because many of these cytokines also affect transcription, CLP is an excellent system to study how the biochemistry of hepatic gene expression is modulated by inflammatory mediators. As a result of early cytokine activity, several cytokine-dependent transcription factors become activated or repressed during sepsis. Among these are the signal transducer and activator of transcription (Stat) 3, nuclear factor (NF)κB, and hepatocyte NF (HNF) 1α (8-10). It is logical to propose that the observed changes in gene expression reflect altered transcription factor activation.
Among the gene products whose expression we found to be altered by CLP are those encoding bile acid and organic anion transporters (11). These include the sodium taurocholate transporter (Ntcp), the multidrug resistant protein (MRP) 2, and the organic anion transporter protein (OATP). The expression of each is reduced shortly after CLP (11). Because sepsis is associated with cholestasis and steatosis (5, 8, 11), we have postulated that the development of these clinically relevant abnormalities might result from altered expression of these genes (11). Studies by others have implicated several cytokines in the development of cholestasis and steatosis and in the expression of Ntcp, MRP-2, and OATP in response to toxic and anatomical insults (11-21). We have shown that CLP increases the activity of the IL-6-dependent transcription factor Stat-3 (8). It is known that the genes encoding Ntcp, MRP-2, and OATP are IL-6/cytokine responsive (12, 13, 17, 20, 22-24). Unfortunately, binding sites for Stat-3 have not been identified in the Ntcp, MRP-2, or OATP promoters. However, these sequences do contain binding sites for HNF-1α, and stimuli that down-regulate bile acid/organic anion transporter expression are associated with decreased HNF-1α DNA binding activity (16). Other studies have indicated that HNF-1α is Stat-3 responsive (23). Therefore, it is reasonable to hypothesize that HNF-1α may play an intermediate role in the regulation of Ntcp, MRP-2, and OATP genes by IL-6 after CLP (16). In this study, we performed CLP on IL-6 +/+ and IL-6 -/- mice to test the hypothesis that sepsis-associated changes in the expression of the bile acid/OATPs Ntcp, MRP-2, and OATP are modulated by IL-6 via HNF-1α.
MATERIALS AND METHODS
Induction of sepsis
All animal protocols were approved by the Penn Institutional Animal Care and Utilization Committee and adhered to National Institutes of Health standards. Study animals were 6- to 8-week-old male wild-type C57BL/6 mice (IL-6 +/+) or mice with a targeted deletion of exon 2 of the IL-6 gene (IL-6 -/-) (Jackson Laboratories, Bar Harbor, Maine). On arrival, experimental animals were housed in a climate-controlled facility and allowed free access to food and water. After acclimation, mice underwent CLP with a 23-gauge needle as previously described (10). Sham-operated animals, subjected to anesthesia, laparotomy, and intestinal manipulation but not CLP, served as controls. Immediately after surgery and every 24 h thereafter, animals were fluid resuscitated with 40 mL/kg saline injected subcutaneously. At the time of CLP, some mice in both the IL-6 +/+ and the IL-6 -/- cohorts had 1 mg/kg recombinant human IL-6 (rhIL-6) (BioSource Int., Camarillo, Calif) dissolved in 0.3 mL of isotonic sodium chloride solution administered into the subcutaneous tissue. In these mice, fluid resuscitation was reduced to assure that they received the same volume of fluid as other animals. At time 0 (unoperated control) and 3, 6, 16, 24, or 48 h after surgery, mice were reanesthetized with 50 mg/kg pentobarbital (i.p.). Blood was obtained for aspartate aminotransferase and alanine aminotransferase determinations, and animals were killed by exsanguination. Liver tissue was harvested for isolation of (1) total RNA for Northern blot hybridization analysis and (2) nuclei for transcription elongation analysis and electrophoretic mobility shift assays.
Northern blot analysis
Total RNA was isolated from liver homogenate using the guanidium acid-phenol method of Chomczynski and Sacchi (25). Northern blotting was performed as previously described using 10 μg of RNA/lane (7). Blots were probed with 32P-labeled cDNAs complementary to the NtcX, the MRP-X, the OATP, β2-macroglobulin (β2M) and HNF-1α. Densitometry was performed, and values for Ntcp, MRP2, and OATP were normalized to those of β2M.
Transcription elongation analysis
Hepatic nuclei were isolated and transcription elongation analysis performed as previously described (7). We used 10 μg of target cDNAs to Ntcp, MRP2, OATP, and β2M. Autoradiography and densitometry were performed, and the signal density for Ntcp, MRP-2, or OATP was normalized to that of β2M.
Probe labeling and purification
Double-stranded oligonucleotides (250 ng) containing consensus HNF-1α or Stat-3 binding sites (Santa Cruz Biotechnology Inc., Santa Cruz, Calif) were incubated in a mixture containing 2 μL of 10× polynucleotide kinase buffer, 9.5 μL of double-deionized water, 2.5 μL (25 μCi) of γ[-32P]adenosine triphosphate, and 1 μL (10 U) of T4 polynucleotide kinase (New England Biolabs, Beverly, Mass) at 37°C for 60 min. The mixture was purified by phenol-chloroform extraction and centrifugation through a G-25 Sephadex column.
Electrophoretic mobility shift assay for HNF-1α
Nuclear protein (7 μg) was incubated on ice in an equal volume of reaction buffer (25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid/KOH, pH 7.9; 50 mM KCl, 0.1 mM EDTA; 1 mM dithiothreitol; and 10% glycerol) and 2 μL of poly (dI-dC) for 1 h. In super shift assays, nuclear protein extracts (7 μg) were incubated on ice with 2 μL of HNF-1α antibody (Santa Cruz Biotechnology) for 1 h. One microliter of oligonucleotide probe (10,000 cpm) was added to each reaction mixture and incubated on ice for 30 min. The samples were loaded on an 8% nondenaturing Tris-boric acid, EDTA polyacrylamide gel for electrophoresis (300 V, 3 h, 4°C). The gels were dried, and autoradiography was performed.
Electrophoretic mobility shift assay for Stat-3
Nuclear protein extracts (5 μg) were incubated at room temperature in an equal volume of reaction buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid/KOH, pH 7.8; 50 mM KCl; 1 mM EDTA; 5 mM MgCl2; 5 mM dithiothreitol; and 10% glycerol) and 1 μL of poly (dI-dC) for 15 min. In the super shift assay, the nuclear protein extract was incubated on ice with 1 μL of antimouse monoclonal phospho-Stat-3 antibody (Santa Cruz Biotechnology) for 1 h. One microliter of oligonucleotide probe (100,000 cpm) was added to each reaction mixture and incubated at room temperature for 15 min. The samples were loaded on a 5% nondenaturing Tris-boric acid, EDTA-polyacrylamide gel for electrophoresis (300 V, 2 h, 4°C). The gels were dried, and autoradiography was performed.
HNF-1α Co-immunoprecipitation
To preclear the samples, nuclear protein extracts (400 μg) were incubated at 4°C with 100 μL (50% slurry in phosphate-buffered saline) of Protein G-conjugated Sepharose (Sigma, St. Louis, Mo) for 30 min. After brief centrifugation, the protein extracts were removed and incubated overnight at 4° C with an antigoat polyclonal HNF-1α antibody (Santa Cruz Biotechnology). Protein G-conjugated Sepharose (100 μL [50% slurry in phosphate-buffered saline]) was added to the samples and incubated at 4°C for 3 h. The protein G Sepharose was briefly centrifuged and washed three times with modified RIPA buffer (50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1% NP-40; 4% Na-deoxycholate; and 1mM EDTA). Seventy-five microliters of 2× sample buffer (125 mM Tris-HCl, pH 6.8; 20% glycerol; 4% sodium dodecyl sulfate, and 0025% Bromo-phenol blue) was added to each sample and incubated at 60°C for 7 min. A negative control was prepared by adding an unrelated antibody in place of the HNF-1α antibody. The samples were loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel for electrophoresis. The gels were then transferred onto polyvinyldifluoride (Biorad, Hercules, Calif) for immunoblotting.
Immunoblotting for HNF-1α and Stat-3
Blots were briefly rinsed in double-deionized water for several minutes after transfer and then incubated at room temperature with blocking solution (5% nonfat dry milk in Tris-buffered saline, pH 7.6, with 001% Tween-20) for 2 h. The blots were incubated at room temperature with either an antimouse monoclonal phospho-Stat-3 antibody (Santa Cruz Biotechnology) or an antigoat polyclonal HNF-1α antibody (Santa Cruz Biotechnology) diluted 1:1,000 with blocking solution for 1.5 h. After a brief wash step, the blots were incubated at room temperature with a species-specific horseradish-peroxidase-conjugated secondary antibody diluted 1:1,000 with blocking solution for 1 h. Resulting protein bands (HNF-1α or Stat-3) were detected by enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, NJ).
Statistics
Differences within and between groups were analyzed using ANOVA with Bonferroni correction.
RESULTS
Steady-state mRNA levels of Ntcp, MRP-2, and OATP after CLP
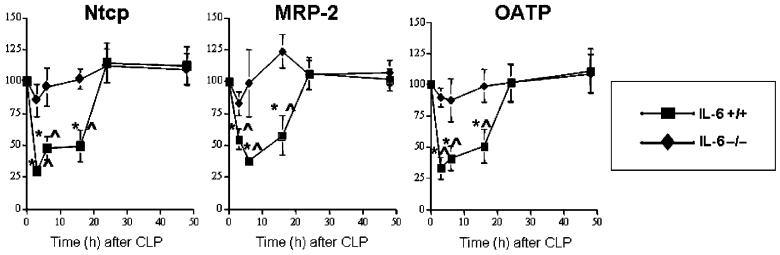
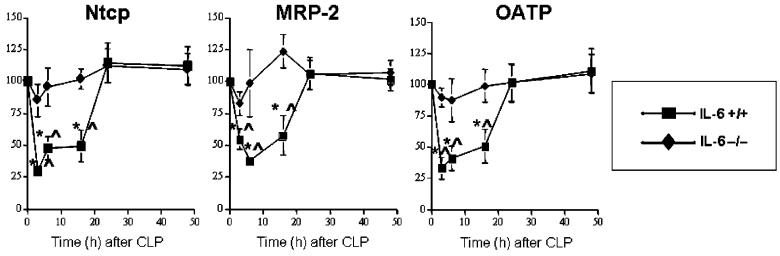
As reported previously, CLP caused an early reduction in transcription of Ntcp and MRP-2 genes in wild-type mice (11). These changes began to resolve by 24 h. Data presented here (Fig. 1) demonstrate that expression of OATP was reduced in a similar manner. Because expression of the genes encoding these proteins is modulated by IL-6 and other cytokines (12, 13, 20, 24, 26-29), we postulated that this change in expression might be altered in IL-6 -/- mice subjected to CLP. Densitometric data derived from Northern blots are depicted in Figure 1. As predicted, CLP decreased steady-state mRNA levels of Ntcp, MRP-2, and OATP in IL-6 +/+ mice. We next evaluated these levels in IL-6 -/- mice after CLP. There was no change in steady-state levels of mRNA encoding Ntcp, MRP-2, or OATP genes after CLP in IL-6 -/- mice. Injection of 1 mg/kg rhIL-6 to the IL-6 -/- mice at the time of CLP resulted in a decrease in steady-state levels of Ntcp, MRP-2, and OATP mRNAs for at least 16 h. These reductions were comparable to those observed after CLP in IL-6 +/+ animals. However, levels in the IL-6 -/- mice returned to baseline at later time points. These data indicate that increased IL-6 activity decreased steady-state levels of the mRNAs encoding Ntcp, MRP-2, and OATP.
Fig. 1. Graphic representation of densitometrically determined steady-state mRNA levels after CLP.
Data as mean ± SD. x axis indicates time after CLP; y axis, normalized density; filled squares, IL-6 +/+ mice; open squares, IL-6 +/+ mice administered rhIL-6; filled diamonds, IL-6 -/- mice; open diamonds, IL-6 -/- mice administered rhIL-6. Values were determined using Northern Blotting (see text). Blots were subjected to autoradiography and densitometry. Data for Ntcp, MRP-2, and OATP normalized to values for the constitutively transcribed β2M at the same time point. Values at time 0 were arbitrarily set at 100. For Ntcp, values for IL-6 +/+ and IL-6 +/+ + rhIL-6 were significantly different than T0 at 3, 6, 16, 24, and 48 h. Values for IL-6 -/- + rhIL-6 were significantly different than values for T0 at 3, 6, 16, and 24 h. Values for IL-6 +/+ and IL-6 +/+ + rhIL-6 were significantly different than values for IL-6 -/- at 3, 6, 16, 24, and 48 h. Values for IL-6 -/- + rhIL-6 were significantly different than values for IL-6 -/- at 3, 6, 16, and 24 h. For MRP-2, values for IL-6 +/+ and IL-6 +/+ + rhIL-6 were significantly different than T0 at 3, 6, 16, 24, and 48 h. Values for IL-6 -/- + rhIL-6 were significantly different than values for T0 at 3, 6, and 16 h. Values for IL-6 +/+ and IL-6 +/+ + rhIL-6 were significantly different than values for IL-6 -/- at 3, 6, 16, 24, and 48 h. Values for IL-6 -/- + rhIL-6 were significantly different than values for IL-6 -/- at 3, 6, and 16 h. For OATP, values for IL-6 +/+ and IL-6 +/+ + rhIL-6 were significantly different than T0 at 3, 6, 16, 24, and 48 h. Values for IL-6 -/- + rhIL-6 were significantly different than values for T0 at 3, 6, and 16 h. Values for IL-6 +/+ and IL-6 +/+ + rhIL-6 were significantly different than values for IL-6 -/- at 3, 6, 16, 24, and 48 h. Values for IL-6 -/- + rhIL-6 were significantly different than values for IL-6 -/- at 3, 6, and 16 h. Differences within and between groups were analyzed using ANOVA with Bonferroni correction.
Transcription of Ntcp, MRP-2 and OATP in IL-6 +/+ and IL-6 -/- mice after CLP
Previous studies have indicated that low steady-state mRNA levels result from decreased transcription (5, 11). However, sepsis-induced changes in mRNA levels also might reflect accelerated degradation. Therefore, we used transcription elongation analysis to examine transcription of the genes encoding Ntcp, MRP-2, and OATP after CLP in IL-6 +/+ and IL-6 -/- mice directly (Fig. 2). Previous studies revealed an initial CLP-induced decrease in Ntcp and MRP-2 transcription rates that resolved within 48 h (11). Use of transcription elongation analysis revealed a decrease in transcription of these two genes and OATP after CLP in IL-6 +/+ mice compared with the T0 controls. No such change was observed in IL-6 -/- mice subjected to CLP (Fig. 2). Ntcp, MRP-2, and OATP transcript levels in IL-6 +/+ mice differed significantly from those in IL-6 -/- mice at 3, 6, and 16 h after CLP. Therefore, the decreased steady-state Ntcp, MRP-2, and OATP mRNA levels can be explained in part by an IL-6-mediated reduction in transcription.
Fig. 2. Graphic representation of densitometrically determined transcription rates after CLP.
Data as mean ± SD. x axis indicates time after CLP; y axis, normalized density; filled squares, IL-6 +/+ mice; filled diamonds, IL-6 -/- mice. Values were determined using blots from transcription elongation analysis (see text). Blots were subjected to autoradiography and densitometry. Data for Ntcp, MRP-2, and OATP were normalized to value for the constitutively transcribed β2M at the same time point. Values at time 0 were arbitrarily set at 100. * indicates significantly different than T0. †Significantly different than IL-6 -/- at same time point. Differences within and between groups were analyzed using ANOVA with Bonferroni correction.
Stat-3 DNA binding activity in IL-6 +/+ and IL-6 -/- mice after CLP
IL-6 activates a signal transduction pathway that culminates in the nuclear translocation and DNA binding of the dimeric transcription factor Stat-3 (30). Previous studies in our laboratory have demonstrated increased Stat-3 DNA binding after CLP in the rat (8). Therefore, we evaluated Stat-3 DNA binding in IL-6 +/+ and IL-6 -/- mice (Fig. 3). Consistent with previous studies, Stat-3 DNA binding peaked at 3 to 6 h after CLP in IL-6 +/+ mice and decreased afterwards, although levels never returned to baseline (Fig. 3A). There was no detectable Stat-3 DNA binding at any time point after either CLP or sham operation (SO) in the IL-6 -/- animals (data not shown). At 16 h after CLP in IL-6 -/- mice given rhIL-6, an increase in Stat-3 DNA binding was noted (Fig. 3B). No increase was detected at later time points or after SO in either IL-6 +/+ or IL-6 -/- mice (data not shown). When coupled with the data presented in Figures 1 and 2, these data suggest that Stat-3 mediates the IL-6-induced inhibition of Ntcp, MRP-2, and OATP expression.
Fig. 3. Representative electrophoretic mobility shift assay for Stat-3 DNA binding in IL-6 +/+ and IL-6 -/- mice.
Animals were subjected to either CLP or SO. Detection with a radiolabeled DNA sequence homologous to a consensus Stat-3 binding site. T0 represents unoperated controls. Subscript indicates the time (T) in hours after CLP or SO. A, Five micrograms of liver nuclear extract from IL-6 +/+ mice was used in this assay. No Stat-3 DNA binding was detected after CLP or SO in IL-6 -/- mice (data not shown). B, Five micrograms of liver nuclear extract from IL-6 +/+ and IL-6 -/- mice was used. Some samples (indicated with “+”) were incubated with a monoclonal antibody to the phosphorylated form of Stat-3 (anti-pStat-3) (Santa Cruz Biotechnology) and, thus, “supershifted,” whereas others (indicated by “-”) were not. IL-6 -/- mice given rhIL-6 at the time of surgery are indicated by “+”.
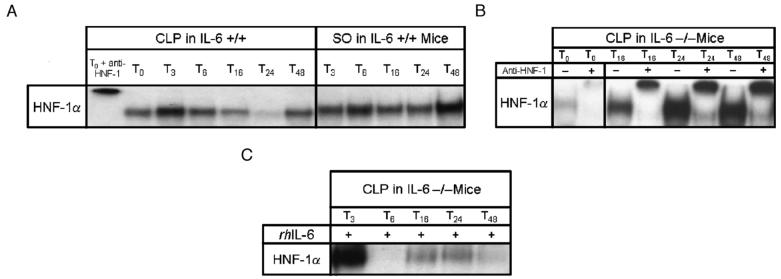
HNF-1α DNA binding activity in IL-6 +/+ and IL-6 -/- mice after CLP
The data presented here suggest that IL-6-induced activation of Stat-3 DNA binding decreases transcription of Ntcp, MRP-2, and OATP. However, this cannot be a direct effect because the promoter regions of Ntcp, MRP-2, and OATP do not contain Stat-3 binding sequences. These promoters do, however, contain HNF-1α binding sites. Therefore, we tested the hypothesis that sepsis altered expression of Ntcp, MRP-2 and OATP by decreasing HNF-1α binding activity. In a previous study, HNF-1α DNA binding was significantly reduced 3, 6, and 16 h after CLP in IL-6 +/+ rats, but levels beyond these time points remained unchanged compared with the T0 controls (10). In the current study, HNF-1α DNA binding was significantly reduced 16, 24, and 48 h after CLP in IL-6 +/+ mice when compared with untreated (T0) and sham-operated mice (Fig. 4A). In contrast, CLP increased HNF-1α DNA binding activity in IL-6 -/- mice relative to T0 controls (Fig. 4B). The time course of reduced HNF-1α DNA binding in IL-6 +/+ animals correlates temporally with the CLP-induced decrease in Ntcp, MRP-2, and OATP transcription as previously reported (10). At 16, 24, and 48 h after CLP in IL-6 -/- mice given rhIL-6, there was a decrease in HNF-1α DNA binding activity compared with T0 controls (Fig. 4C). This pattern is similar to that seen in IL-6 +/+ mice after CLP. These findings were confirmed with immunoprecipitation (Fig. 4C). Data reveal that the amount of HNF-1α immunoprecipitated in IL-6 -/- mice that received rhIL-6 was markedly reduced compared with the T0 control. Our studies indicate that HNF-1α binding is influenced by IL-6 and suggest that this transcription factor is a determinant of Ntcp, MRP-2, and OATP expression.
Fig. 4. Representative electrophoretic mobility shift assay of HNF-1α DNA binding in IL-6 +/+ and IL-6 -/- mice.
Animals were subjected to either CLP or SO. T0 represents unoperated controls. Subscript indicates the time (T) in hours after CLP or SO. A, Seven micrograms of liver nuclear extract from IL-6 +/+ mice was used. The first well was loaded with a T0 sample and “supershifted” via incubation with a polyclonal antibody to HNF-1α (Santa Cruz Biotechnology) as a control. B, Seven micrograms of liver nuclear extract from IL-6 -/- mice was used. Samples indicated with a “+”were incubated with a polyclonal antibody to HNF-1α (Santa Cruz Biotechnology) and, thus, supershifted. Lower bands indicate HNF-1α complex; upper bands, supershifted complex detected after antibody administration. C, Seven micrograms of liver nuclear extract from IL-6 -/- mice was used. “+” indicates that IL-6 -/- mice were injected with rhIL-6 at the time of surgery.
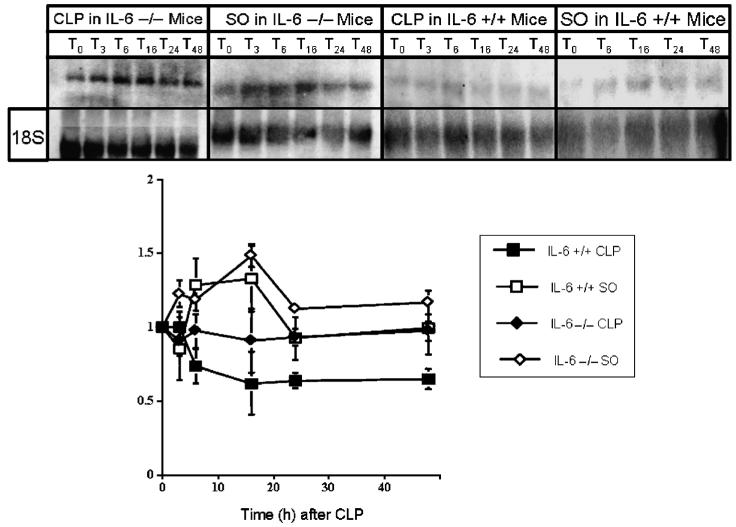
Steady-state HNF-1a mRNA levels in IL-6 +/+ and IL-6 -/- mice after CLP
The absence of a Stat-3 binding site in Ntcp, MRP-2, and OATP promoters makes it unlikely that IL-6/Stat-3 directly affect expression of these genes. However, HNF-1α activity is altered in sepsis, and the promoter of the HNF-1α gene contains a Stat-3 binding site. Therefore, we hypothesized thatIL-6/Stat-3 may exert an effect on Ntcp, MRP-2, and OATP expression by altering expression of HNF-1α. To test this hypothesis, we examined steady-state HNF-1α mRNA levels in IL-6 +/+ and IL-6 -/- mice using Northern blotting. Results are depicted in Figure 5. Hepatocyte nuclear factor 1α mRNA was reduced after CLP in the IL-6 +/+ mice. In contrast, CLP did not alter HNF-1α message levels in IL-6 -/- animals. These data suggest that IL-6/Stat-3 decrease expression of HNF-1α, and this decrease contributed to reduced expression of Ntcp, MRP-2, and OATP.
Fig. 5. Representative autoradiograms of Northern blots to determine steady-state mRNA levels of HNF-1α in IL-6 +/+ and IL-6 -/- mice after CLP and SO (top).
Graphic representation of densitometrically determined steady-state mRNA levels of HNF-1α in IL-6 +/+ and IL-6 -/- mice after CLP and SO (bottom). Data as mean ± SD. x axis indicates time in hours after CLP or SO; y axis, normalized density; solid squares, IL-6 +/+ mice subjected to CLP; solid diamonds, IL-6 -/- mice subjected to CLP; open squares, IL-6 +/+ mice subjected to SO; open diamonds, IL-6 -/- mice subjected to SO. Data for HNF-1α normalized to values for the constitutively expressed 18S ribosomal subunit at same time point. Values at time 0 were arbitrarily set at 100. Values for IL-6 +/+ mice subjected to CLP differed significantly from values for IL-6 -/- subjected to CLP and IL-6 +/+ subjected to SO at 6, 16, 24, and 48 h. Values for IL-6 +/+ subjected to CLP differed significantly from values for IL-6 -/- subjected to SO at 3, 6, 16, 24, and 48 h. Differences within and between groups were analyzed using ANOVA with Bonferroni correction.
Co-immunoprecipitation for HNF-1α and Stat-3 in IL-6 +/+ and IL-6 -/- mice after CLP
The reduction in HNF-1α DNA binding activity may result from decreased gene expression or from a direct interaction with Stat-3 itself. The latter has been reported previously by Leu et al. (23). To test this hypothesis, we used coimmunoprecipitation to determine if there was direct interaction between HNF-1α and Stat-3 in IL-6 +/+ and IL-6 -/- mice. The 16-h time point was chosen because previous studies indicate that Stat-3 is consistently activated at that time point. No HNF-1α/Stat-3 interaction was observed (Fig. 6). These data suggest that the reduction in HNF-1α DNA binding activity results from decreased HNF-1α gene expression and not from a direct HNF-1α/Stat-3 interaction.
Fig. 6. Representative immunoprecipitation/immunoblot of HNF-1α using IL-6 +/+ and IL -/- mice.
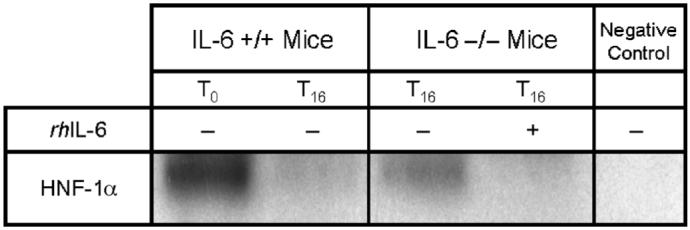
Hepatocyte nuclear factor 1α was immunoprecipitated from 400 μg of liver nuclear extract. Some IL-6 -/- mice received rhIL-6. T0 represents unoperated controls. Subscript indicates the time in hours after CLP. The negative control is a T0 sample that was immunoprecipitated with an antibody not related to HNF-1α.
DISCUSSION
The data presented here suggest a potential mechanism by which IL-6 affects expression of Ntcp, MRP-2, and OATP. Stimulation of the IL-6 signal transduction pathway enhances Stat-3 activity (30). This exerts an inhibitory effect on the expression of the gene encoding the transcription factor HNF-1α. HNF-1α increases transcription of Ntcp, MRP-2, and OATP; therefore, decreased expression of this transcription factor leads to decreased transcription of the bile acid/organic anion genes in question. Our data, whereas not completely conclusive, strongly support this scenario.
Our findings clearly demonstrate that IL-6 influences transcription of the genes encoding Ntcp, MRP-2, and OATP in vivo. Whereas others have reported that IL-6 affects Ntcp, MRP-2, and OATP expression (13, 18, 20-22, 27, 30-32), our data are more complete in that we increased IL-6 activity in a biologically relevant model, demonstrated a change in transcription in that model, and showed that elimination of IL-6, using a targeted knockout of the IL-6 gene in mice, eliminated this change. Use of IL-6 -/- mice assured a complete absence of the cytokine in the tissue in question. This contrasts with strategies where administration of antibodies or naturally occurring antagonists may block cytokine activity incompletely. In addition, administration of rhIL-6 to IL-6 -/- animals restored sepsis-induced decreases in Ntcp, MRP-2, and OATP gene expression. This suggests that IL-6 knockout does not result in a loss of IL-6 receptors.
Sepsis in IL-6 +/+ mice activates the nuclear transcription factor Stat-3 (8). These findings are important because IL-6 exerts its intracellular effects via a cell surface receptor, gp80, which lacks a transmembrane domain. Once ligand is bound to gp80, the combination facilitates dimerization with and activation of the ubiquitous cell surface molecule gp130. Active gp130 interacts with other proteins, undergoes a conformational change, and is able to phosphorylate Stat-3. This allows for Stat-3 dimerization, nuclear transmigration, and modulation of transcription. gp130 can homodimerize in response to other extracellular signaling molecules and also can dimerize with receptors other than gp80 (30). One therefore might anticipate that Stat-3 can be activated even in the absence of IL-6. However, in our studies, no Stat-3 DNA binding activity was detected in septic IL-6 -/- mice. Thus, it seems that activation of Stat-3 in sepsis is, to some degree, dependent on the presence of IL-6 and not other inflammatory mediators. This is confirmed by our demonstration that administration of rhIL-6 partially restored Stat-3 activity in the livers of IL-6 -/- mice. These findings indicate that the IL-6 exerts its effect on Ntcp, MRP-2, and OATP expression via a mechanism that involves Stat-3.
Of additional importance, our data indicate that Stat-3-mediated IL-6 effects reduced Ntcp, MRP-2, and OATP gene expression. The absence of Stat-3 binding domains in the promoters of Ntcp, MRP-2, and OATP indicate that this effect does not occur as a direct result of Stat-3 binding to the Ntcp, MRP-2, and OATP promoters. Two possible indirect effects suggest themselves. The first involves interaction of Stat-3 with a protein that does bind to the Ntcp, MRP-2, and OATP promoters. In this regard, Leu et al. (23) have reported that Stat-3 is capable of forming complexes with a number of transcription factors that may be important in the expression of these genes. Our data do not demonstrate a complex that contains Stat-3 and HNF-1α. Moreover, Leu et al. (23) indicate that the complexes that contain Stat-3 and other transcription factors increase, rather than impair, gene expression. Thus, the relevance of this finding to decreased Ntcp, MRP-2, and OATP transcription remains unclear. Alternatively, expression of each of these gene products is increased by binding of the transcription factor HNF-1α, and the gene encoding HNF-1α contains a Stat-3 binding site. Our data, both in previous studies and as presented here, indicate that sepsis decreased HNF-1α expression and DNA binding activity in IL-6 +/+ mice. This suggests that Stat-3 decreases expression of the HNF-1α gene in IL-6 +/+ mice, limiting increases in protein abundance and DNA binding activity. The correlation of reduced protein levels of HNF-1α and subsequent reduced DNA binding is supported by others (15). In IL-6 -/- animals, HNF-1α expression was unchanged, and DNA binding activity was enhanced slightly. Thus, transcription of the Ntcp, MRP-2 and OATP was not altered. Induction of sepsis in HNF-1α -/- mice would be most useful in validation of our proposed mechanism. However, attempts at generating these animals have been unsuccessful.
It is interesting to note that injection of wild-type mice with rhIL-6 after CLP and sham operated failed to stimulate further Stat-3 DNA binding (data not shown). In the sham-operated animals, this suggests that other cofactors are needed to activate Stat-3 or, as we have suggested previously, that serum levels of IL-6 have little to do with hepatocellular processes (8). A lack of enhanced Stat-3 signaling after rhIL-6 injection in the wild-type mice after CLP suggests either an intracellular signaling defect for Stat-3 inherent to CLP, an inability of the rhIL-6 to reach the liver in adequate concentrations to effect cell signaling, or that serum concentrations of IL-6 do not necessarily influence hepatocellular signaling unless there is a complete deficit of IL-6 as in the knockout animals. Similarly, whereas one might surmise that administration of IL-6 to an otherwise healthy animal should be sufficient to demonstrate the dependence of Ntcp, MRP-2, and OATP expression on this cytokine, this is not the case. In the absence of an insult to stimulate increased hepatic transcellular transport, IL-6 effects are not discernable. This emphasizes the use of the septic model. Furthermore, the responses to IL-6 seen in this study suggest therapeutic possibilities.
In summary, we report that transcription of the genes encoding the key hepatic transport proteins Ntcp, MRP-2, and OATP are repressed by an IL-6-dependent pathway after CLP involving HNF-1α and Stat-3. Although the contribution of these findings to hepatic pathology in sepsis remains unknown, therapeutic possibilities remain.
Acknowledgments
This work was supported in part by NIGMS grant R01 GM059930 (CSD) and T32 GM07612 (CSD, KMA) and the Stavropoulus Sepsis Research Program.
REFERENCES
- 1.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Andrejko KM, Deutschman CS. Altered hepatic gene expression in fecal peritonitis: changes in transcription of gluconeogenic, beta-oxidative, and ureagenic genes. Shock. 1997;7:164–169. [PubMed] [Google Scholar]
- 3.Deutschman CS, Haber BA, Andrejko KM, Cressman DE, Harrison R, Elenko E, Taub R. Increased expression of cytokine-induced neutrophil chemoattractant (CINC) in septic rat liver. Am J Physiol. 1996;271:R593–R600. doi: 10.1152/ajpregu.1996.271.3.R593. [DOI] [PubMed] [Google Scholar]
- 4.Deutschman CS, DeMaio A, Buchman TG, Clemens MG. Sepsis-induced alterations in phosphoenolpyruvate carboxykinase expression: the role of insulin and glucagon. Circ Shock. 1993;40:295–302. [PubMed] [Google Scholar]
- 5.Deutschman CS, Andrejko KM, Haber BA, Elenko E, Harrison R, Taub R. Sepsis-induced depression of rat glucose-6-phosphatase gene expression and activity. Am J Physiol. 1997;273:R1709–R1718. doi: 10.1152/ajpregu.1997.273.5.R1709. [DOI] [PubMed] [Google Scholar]
- 6.Weiss YG, Bouwman AR, Gehan B, Raj N, Deutschman CS. Cecal ligation and double puncture impairs heat shock protein 70 (HSP-70) expression in the lungs of rats. Shock. 2000;13:19–23. doi: 10.1097/00024382-200013010-00004. [DOI] [PubMed] [Google Scholar]
- 7.Andrejko KM, Deutschman CS. Acute-phase gene expression correlates with intrahepatic tumor necrosis factor-alpha abundance but not with plasma tumor necrosis factor concentrations during sepsis/systemic inflammatory response syndrome in the rat. Crit Care Med. 1996;24:1947–1952. doi: 10.1097/00003246-199612000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Andrejko KM, Chen J, Deutschman CS. Intrahepatic STAT3 activation and acute phase gene expression predict outcome after CLP sepsis in the rat. Am J Physiol. 1998;275:G1423–G1429. doi: 10.1152/ajpgi.1998.275.6.G1423. [DOI] [PubMed] [Google Scholar]
- 9.Villa P, Sartor G, Angelini M, Sironi M, Conni M, Gnocchi P, Isetta AM, Grau G, Buurman W, van Tits LJ. Pattern of cytokines and pharmacomodulation in sepsis induced by cecal ligation and puncture compared with that induced by endotoxin. Clin Diagn Lab Immunol. 1995;2:549–553. doi: 10.1128/cdli.2.5.549-553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haaxma CA, Kim PK, Andrejko KM, Raj NR, Deutschman CS. Transcription factors C/EBP-alpha and HNF-1-alpha are associated with decreased expression of liver-specific genes in sepsis. Shock. 2003;19:45–49. doi: 10.1097/00024382-200301000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Kim P, Andrejko KM, Chen J, Raj N, Deutschman CS. Intraabdominal sepsis downregulates transcription of sodium taurocholate cotransporter and multidrug resistance-associated protein in rats. Shock. 2000;14:176–181. doi: 10.1097/00024382-200014020-00017. [DOI] [PubMed] [Google Scholar]
- 12.Gartung C, Ananthanarayanan M, Rahman MA, Schuele S, Nundy S, Soroka CJ, Stolz A, Suchy FJ, Boyer JL. Down-regulation of expression and function of the rat liver Na+/bile acid cotransporter in extrahepatic cholestasis. Gastroenterology. 1996;110:199–209. doi: 10.1053/gast.1996.v110.pm8536857. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann G, Cheung AK, Piquette-Miller M. Inflammatory cytokines, but not bile acids, regulate expression of murine hepatic anion transporters in endotoxemia. J Pharmacol Exp Ther. 2002;303:273–281. doi: 10.1124/jpet.102.039404. [DOI] [PubMed] [Google Scholar]
- 14.Roelofsen H, Van der Veere CN, Ottenhoff R, Schoemaker B, Jansen PL, Oude Elferink RP. Decreased bilirubin transport in the perfused liver of endotoxemic rats. Gastroenterology. 1994;107:1075–1084. doi: 10.1016/0016-5085(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 15.Trauner M, Arrese M, Soroka CJ, Ananthanarayanan M, Koeppel TA, Schlosser SF, Suchy FJ, Keppler D, Boyer JL. The rat canalicular conjugate export pump (Mrp2) is down regulated in intrahepatic and obstructive cholestasis. Gastroenterology. 1997;113:255–264. doi: 10.1016/s0016-5085(97)70103-3. [DOI] [PubMed] [Google Scholar]
- 16.Trauner M, Arrese M, Lee H, Boyer JL, Karpen SJ. Endotoxin downregulates rat hepatic ntcp gene expression via decreased activity of critical transcription factors. J Clin Invest. 1998;101:2092–2100. doi: 10.1172/JCI1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiting JF, Green RM, Rosenbluth AB, Gollan JL. Tumor necrosis factor-alpha decreases hepatocyte bile salt uptake and mediates endotoxin-induced cholestasis. Hepatology. 1995;22:1273–1278. doi: 10.1016/0270-9139(95)90639-8. [DOI] [PubMed] [Google Scholar]
- 18.Li N, Klaussen CD. Lipopolysaccharide-induced down-regulation of organic anion transporting polypeptide 4 (Oatp4; Slc21a10) is independent of tumor necrosis factor-alpha, interleukin-1-beta, interleukin-6, or inducible nitric oxide synthase. Toxicol Sci. 2005;83:197–203. doi: 10.1093/toxsci/kfi003. [DOI] [PubMed] [Google Scholar]
- 19.Bemelmans MH, Gouma DJ, Greve JW, Buurman WA. Cytokines tumor necrosis factor and interleukin-6 in experimental biliary obstruction in mice. Hepatology. 1992;15:1132–1136. doi: 10.1002/hep.1840150626. [DOI] [PubMed] [Google Scholar]
- 20.Moseley RH, Wang W, Takeda H, Lown K, Shick L, Ananthanarayanan M, Suchy FJ. Effect of endotoxin on bile acid transport in rat liver: a potential model for sepsis-associated cholestasis. Am J Physiol. 1996;271:G137–G146. doi: 10.1152/ajpgi.1996.271.1.G137. [DOI] [PubMed] [Google Scholar]
- 21.Green RM, Whiting JF, Rosenbluth AB, Beier D, Gollan JL. Interleukin-6 inhibits hepatocyte taurocholate uptake and sodium-potassium-adenosinetri-phosphatase activity. Am J Physiol. 1994;267:1094–1100. doi: 10.1152/ajpgi.1994.267.6.G1094. [DOI] [PubMed] [Google Scholar]
- 22.Green RM, Beier D, Gollan JL. Regulation of hepatocyte bile salt transporters by endotoxin and inflammatory cytokines in rodents. Gastroenterology. 1996;111:193–198. doi: 10.1053/gast.1996.v111.pm8698199. [DOI] [PubMed] [Google Scholar]
- 23.Leu JL, Crissey MAS, Leu JP, Ciliberto G, Taub R. Interleukin-6-induced STAT3 and AP-1 amplify hepatocyte nuclear factor-1 mediated transactivation of hepatic genes, an adaptive response to liver injury. Mol Cell Biol. 2001;21:414–424. doi: 10.1128/MCB.21.2.414-424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geier A, Dietrich DG, Voight S, Ananthanarayanan M, Lammert F, Schmitz A, Trauner M, Wasmuth HE, Boraschi D, Balasubramaniyan N, et al. Cytokine-dependent regulation of hepatic organic anion transporter gene transactivators in mouse liver. Am J Physiol. 2005;289:G831–G841. doi: 10.1152/ajpgi.00307.2004. [DOI] [PubMed] [Google Scholar]
- 25.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Ann Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 26.Kauffmann HM, Schrenk D. Sequence analysis and functional characterization of the 5′μm-flanking region of the rat multidrug resistance protein 2 (MRP2) gene. Biochem Biophys Res Commun. 1998;245:325–331. doi: 10.1006/bbrc.1998.8340. [DOI] [PubMed] [Google Scholar]
- 27.Dreuw A, Hermanns HM, Heise R, Joussen S, Marquardt Y, Jugert F, Merk HF, Heinrich PC, Baron JM. Interleukin-6 type cytokines upregulate expression of multidrug resistance-associated proteins in NHEK and dermal fibroblasts. J Invest Dermatol. 2005;124:28–37. doi: 10.1111/j.0022-202X.2004.23499.x. [DOI] [PubMed] [Google Scholar]
- 28.Cressman DE, Greenbaum LE, DeAngelis RA, Ciliberto G, Furth EE, Taub R. Liver failure and defective hepatocyte regeneration in Interleukin-6 deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 29.Heinrich PC, Behrman I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin-6 type cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teng S, Piquette-Miller M. The involvement of the pregnane X receptor in hepatic gene regulation during inflammation in mice. J Exp Pharmacol Ther. 2005;312:841–848. doi: 10.1124/jpet.104.076141. [DOI] [PubMed] [Google Scholar]
- 31.Sukhai M, Yong A, Kalitsky J, Piquette-Miller M. Inflammation and interleukin-6 mediate reductions in the hepatic expression and transcription of the mdr1a and mdr1b genes. Mol Cell Biol Res Commun. 2000;4:248–256. doi: 10.1006/mcbr.2001.0288. [DOI] [PubMed] [Google Scholar]
- 32.Siewert E, Dietrich CG, Lammert F, Heinrich PC, Matern S, Gartung C, Geier A. Interleukin-6 regulates hepatic transporters during acute-phase response. Biochem Biophys Res Commun. 2004;322:232–238. doi: 10.1016/j.bbrc.2004.07.102. [DOI] [PubMed] [Google Scholar]