Abstract
Aberrant coregulator expression that occurs during prostate cancer (PCa) progression correlates with poor prognosis and aggressive disease. This has been attributed to the ability to regulate androgen receptor-mediated transcription. We have shown previously that the androgenic milieu regulates the expression of the coactivators p300 and FHL2, with severe consequences for PCa cell proliferation and androgen receptor transcriptional activity. To determine the extent of androgen dependency of coregulator genes, we designed a cDNA-mediated annealing, selection, extension, and ligation RNA profiling array that probes the expression of 186 coregulators. Using this assay, we demonstrated androgen control over approximately 30% of coregulator genes in PCa cells. For a subset of 15 functionally diverse coregulators, androgen regulation was confirmed using real-time RT-PCR and immunoblotting. The extent, dose dependency, and kinetics by which androgens affect coregulator expression differed widely, indicating diverse molecular mechanisms underlying these effects. Moreover, differences in coregulator expression were observed between isogenic androgen-dependent and castration-recurrent PCa cells. Small interfering RNA-mediated changes in coregulator expression had profound effects on cell proliferation, which were most pronounced in castration-recurrent cells. Taken together, our integrated approach combining expression profiling, characterization of androgen-dependent coregulator expression, and validation of the importance of altered coregulator expression for cell proliferation identified several potential novel therapeutic targets for PCa treatment.
Androgen control over coregulator expression in prostate cancer cells was demonstrated using a custom assay and revealed consequences for cell proliferation and androgen receptor activity.
Androgen receptor (AR) signaling is critical for prostate cancer (PCa) cell proliferation (1,2,3,4). Patients with locally advanced and metastatic PCa that do not benefit from surgery or radiation therapy are normally treated with androgen ablation therapies. These therapies inhibit the production of androgens, the natural ligands for the AR, and/or prevent binding of androgens to AR by administration of an excess of AR antagonists (5). Despite castrate levels of circulating androgens in these patients, the AR is increasingly considered as an attractive therapeutic target in castration-recurrent (CR) PCas that emerge when androgen ablation therapies fail (3,6,7).
To assemble a productive transcriptional complex, the AR relies on functional interactions with multiple coregulatory proteins (8,9). Coregulators recruited by the AR either enhance (i.e. coactivators) or reduce (i.e. corepressors) AR transactivation without significantly altering its basal transcription rate and without binding directly to DNA. Instead, coregulators act at AR target gene promoter and/or enhancer regions to facilitate DNA occupancy, induce chromatin remodeling and/or recruit general transcription factors associated with RNA polymerase II. Alternatively, they assure that the AR is able to affect target gene expression by modulating folding of the AR, ensuring AR stability or directing the subcellular localization of the AR. To date, approximately 170 proteins have been identified as AR coregulators (9). The remarkable functional diversity displayed by these proteins and the number of cellular pathways with which they are involved offer a glimpse of the extraordinary level of complexity of protein-protein interactions involved in generating an AR-mediated transcriptional response. AR transcriptional activity is further regulated by several specific transcription factors, such as Foxa1, Oct1, GATA binding protein 2 (GATA2), ETS-1 [v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)], and activator protein 1 (AP1), which bind to cognate DNA sequences that are interspersed between, or in close proximity to, AR binding sites (10,11,12).
Recently, AR-associated coregulators have been suggested as potential targets for therapeutic intervention in PCa disease (13,14,15). Several coregulatory proteins are aberrantly expressed during PCa progression, where their expression often correlates with a poor prognosis. Moreover, changes in coregulator expression have been demonstrated to substantially contribute to (CR) AR activity. To better understand the mechanism of AR action in PCa cells and to gain insights that are urgently needed to generate novel therapeutic approaches, our laboratory has been investigating the cues and signal transduction pathways that drive altered expression of these critical cofactors in PCa.
Using p300 and FHL2, two AR-associated coactivators with distinct functions (histone acetylation and signal integration, respectively) as model genes, we demonstrated that the androgenic milieu of PCa cells can have a profound impact on the expression level of coregulators (16,17). We showed that androgen-induced changes in these coactivators have severe repercussions on PCa cell proliferation and provided evidence for the existence of feedback loops that establish androgenic control over AR transcriptional activity. In addition, our work exploring the molecular mechanism that underlies androgen regulation of these coactivators suggested that the activity of the transcription factors, serum response factor and nuclear factor-κB, constitute a novel, indirect means of targeting AR signaling in PCa cells (16,17).
In view of the large number of proteins that have been identified as AR coregulators, which interact to form the AR transcriptional complex (8,9,14), we hypothesized that androgen control over coregulator expression is not limited to p300 and FHL2. To explore the extent to which other cofactors may be affected by androgens, we designed a cDNA-mediated annealing, selection, extension, and ligation (DASL) array that assesses expression of the AR, 186 coregulator proteins, and 42 genes under androgen control. Using this array, we showed that more than 30% of coregulators under investigation (64/186) are subject to androgen regulation. The extent, dose dependency, and kinetics by which androgens affect coregulator expression indicate that diverse molecular mechanisms underlie these events. Moreover, changes in coregulator expression occurred during progression to the CR state, which profoundly affect PCa cell viability. Taken together, our integrated approach confirms the overall validity of coregulators as therapeutic targets and identifies several cofactors that may hold promise for such therapeutic intervention.
Results
Design of a custom DASL array
To coordinately interrogate the expression of multiple coregulators, we designed a DASL gene expression profiling array (Illumina) (18,19). The strength of the DASL assay lies within its ability to concurrently query the expression of up to 1536 sequence targets (typically 512 genes, targeted in three different regions) in a highly sensitive manner. Probes are designed to target 50-bp exonic gene regions that display specificity within the transcriptome and the genome. DASL assays have been extensively characterized and typically show very low false-positive rates (<0.3%), high reproducibility (routinely R2 of 0.99 between replicates (Refs. 18 and 19 and data not shown) and a sensitivity that allows correct classification of samples with transcripts differing by less than 1.10-fold (18,19). These features reflect that DASL arrays were designed to generate profiling data from partially degraded RNA derived from formalin-fixed, paraffin-embedded (FFPE) tissues. These arrays are also well suited to assess expression levels of transcripts that are present in a cell in limited amounts, such as coregulators, which are often difficult to profile using conventional oligonucleotide arrays. Moreover, the scope of the assay, which is limited compared with traditional microarray approaches (∼500 genes compared with tens of thousands) allows one to design an assay to explore and answer a specific question. Thus, we designed a custom DASL array to query AR function in PCa cells. To this end, we included probe groups targeting the expression of the AR, 42 androgen-regulated genes, and 186 coregulators, 100 of which have been shown to structurally or functionally interact with the AR (Tables 1–3). The remaining genes targeted by our array (list available upon request) have been suggested to play important roles in PCa cell biology in gene profiling studies performed in our institution or available in the public domain (20,21,22,23,24,25,26) or represent loading control genes the expression of which is not expected to be subject to androgen regulation.
Table 1.
Effect of androgen treatment on DASL expression profile: androgen target genes
| Gene symbol | Fold induction
|
|
|---|---|---|
| 0.1 nm | 1.0 nm | |
| KLK2 | 6.552 | 9.633 |
| MSMB | 4.231 | 3.538 |
| ACPP | 0.649 | 1.100 |
| TMPRSS2 | 1.924 | 3.408 |
| ODC1 | 1.051 | 2.181 |
| AMD1 | 1.100 | 1.179 |
| SMS | 1.166 | 1.198 |
| SAT | 1.056 | 1.251 |
| ACACA | 1.400 | 1.452 |
| ACACB | 2.336 | 0.813 |
| ME1 | 1.317 | 1.699 |
| FASN | 0.880 | 3.797 |
| HMGCS1 | 1.120 | 1.308 |
| HMGCS2 | 1.516 | 1.862 |
| HMGCR | 1.097 | 2.052 |
| DHCR24 | 1.783 | 4.296 |
| FDPS | 1.221 | 1.209 |
| SREBF1 | 0.684 | 1.202 |
| SREBF2 | 1.072 | 1.553 |
| SCAP | 1.094 | 1.294 |
| INSIG1 | 1.310 | 2.351 |
| INSIG2 | 0.721 | 0.604 |
| ACTA2 | 1.327 | 4.317 |
| ACTG2 | 2.180 | 5.963 |
| MYL9 | 0.518 | 3.074 |
| MYLK | 0.275 | 0.294 |
| TPM1 | 0.775 | 1.328 |
| FN1 | 0.753 | 0.221 |
| NKX3-1 | 2.133 | 2.749 |
| CDK8 | 0.612 | 0.414 |
| BCL2 | 1.207 | 0.769 |
| CASP2 | 1.060 | 0.837 |
| PIK3R3 | 0.917 | 0.467 |
| RAB4A | 1.072 | 1.333 |
| LIFR | 1.156 | 3.006 |
| HERC3 | 1.413 | 3.013 |
| VEGFA | 1.025 | 3.006 |
| VEGFC | 2.357 | 0.904 |
| NDRG1 | 1.358 | 5.378 |
| NFKBIA | 0.869 | 1.167 |
| BCHE | 0.763 | 0.306 |
| AR | 0.526 | 0.407 |
The order of the genes listed corresponds to the order in which the genes are listed and grouped in the heatmap shown in supplemental Fig. 1. LNCaP cells were seeded in medium supplemented with charcoal-stripped serum. Medium was changed 3 d later, and cells were treated with 0, 0.1 or 1 nm of R1881. Cells were harvested 48 h later, and RNA was isolated and subjected to DASL analysis. Values represent the average fold change in gene expression after treatment with 0.1 or 1 nm of R1881 from three biological replicates each.
Table 2.
Effect of androgen treatment on DASL expression profile: control genes
| Gene symbol | Fold induction
|
|
|---|---|---|
| 0.1 nm | 1.0 nm | |
| GAPDH | 1.064 | 0.955 |
| PGK1 | 1.080 | 1.120 |
| YWHAZ | 1.021 | 1.081 |
LNCaP cells were seeded in medium supplemented with charcoal-stripped serum. Medium was changed 3 d later, and cells were treated with 0, 0.1 or 1 nm of R1881. Cells were harvested 48 h later, and RNA was isolated and subjected to DASL analysis. Values represent the average fold change in gene expression after treatment with 0.1 or 1 nm of R1881 from three biological replicates each.
Table 3.
Effect of androgen treatment on DASL expression profile: coregulator genes
| Gene symbol | Fold induction
|
Gene symbol | Fold induction
|
Gene symbol | Fold induction
|
|||
|---|---|---|---|---|---|---|---|---|
| 0.1 nm | 1.0 nm | 0.1 nm | 1.0 nm | 0.1 nm | 1.0 nm | |||
| BAZ1B | 0.974 | 0.843 | SFPQa | 0.812 | 0.904 | DDCa | 1.565 | 0.675 |
| PB1 | 0.886 | 0.833 | SNW1 | 0.937 | 0.918 | DJBPa | 0.963 | 0.647 |
| SMARCA2a | 0.765 | 0.740 | RNPC2 | 1.005 | 0.934 | EDF1 | 0.938 | 0.964 |
| SMARCA4a | 0.795 | 0.748 | WDR77a | 0.970 | 1.081 | GMEB1 | 1.281 | 0.932 |
| SMARCB1 | 0.800 | 0.890 | BRCA1a | 1.576 | 0.904 | GMEB2 | 2.184 | 0.709 |
| SMARCC1a | 1.074 | 1.109 | BRCA2a | 1.603 | 1.132 | HIPK3 | 0.879 | 0.815 |
| SMARCC2 iso1 | 0.797 | 0.726 | MGMT | 0.539 | 0.715 | HR | 0.522 | 1.432 |
| SMARCC2 iso2 | 0.716 | 0.576 | MMS19L | 0.754 | 1.026 | HTATIP2 | 1.042 | 1.154 |
| SMARCD1 | 0.916 | 0.804 | PRKDCa | 1.188 | 0.816 | IQWD1a | 1.017 | 1.269 |
| SMARCD2 | 0.880 | 1.061 | RAD9Aa | 0.938 | 0.808 | JDP2 | 0.946 | 0.498 |
| SMARCD3 iso1 | 0.388 | 0.302 | XRCC5a | 1.077 | 1.168 | MAGEA11a | 1.344 | 1.552 |
| SMARCD3 iso2 | 1.265 | 0.877 | XRCC6a | 0.899 | 1.073 | MLL2 | 0.829 | 0.846 |
| SMARCE1a | 0.947 | 0.868 | BAG1a | 0.891 | 0.935 | MLL3 | 0.586 | 0.437 |
| SRCAPa | 1.165 | 0.644 | CDC37a | 0.990 | 1.202 | MLR2 | 0.889 | 0.961 |
| CREBBPa | 0.978 | 0.663 | DNAJA1a | 1.022 | 0.968 | MTA1 | 0.754 | 0.684 |
| EP300a | 0.788 | 0.889 | FKBP5a | 2.924 | 6.008 | NCOA4a | 1.039 | 1.032 |
| GCN5L2 | 1.044 | 0.915 | ACTL6A | 1.201 | 1.006 | NCOA5 | 0.538 | 0.721 |
| HDAC1a | 0.748 | 0.759 | APPBP2a | 1.078 | 1.896 | NCOA6a | 0.835 | 0.888 |
| HDAC3 | 0.998 | 1.071 | FLNAa | 0.575 | 1.110 | NCOA7 | 0.616 | 0.397 |
| HTATIPa | 0.749 | 0.828 | GSNa | 1.392 | 0.977 | NCOR1a | 0.949 | 0.986 |
| MYST2a | 1.078 | 0.772 | SVILaiso1 | 0.781 | 0.600 | NCOR2a | 0.496 | 0.846 |
| NCOA1a | 0.796 | 0.760 | SVILaiso2 | 1.297 | 0.525 | NRIP1a | 0.826 | 0.660 |
| NCOA2a | 0.816 | 0.657 | APPLa | 0.806 | 0.816 | NRIP2 | 2.362 | 0.857 |
| NCOA3a | 0.789 | 0.505 | CAV1a | 1.461 | 0.909 | PARK7a | 1.114 | 1.032 |
| PCAFa | 1.115 | 0.901 | GAKa | 0.532 | 0.945 | PHB2 | 0.985 | 0.928 |
| SIN3B | 0.862 | 1.006 | COPS5 | 0.887 | 0.905 | PLAGL1a | 2.043 | 0.970 |
| SIRT1a | 0.945 | 0.860 | CTNNB1a | 0.796 | 0.851 | PNRC1a | 0.802 | 0.888 |
| AOF2a | 0.958 | 0.851 | FHL2a | 2.278 | 3.119 | PNRC2 | 0.748 | 0.760 |
| CARM1a | 1.238 | 0.997 | GNB2L1a | 1.014 | 0.947 | PPARBPa | 0.889 | 0.943 |
| HRMT1L1 | 0.893 | 1.054 | HEY1a | 1.330 | 0.864 | PPARGC1A | 1.748 | 0.776 |
| HRMT1L2a | 1.065 | 0.996 | ITGB3BP | 1.209 | 1.015 | PPARGC1B | 2.020 | 0.981 |
| NSD1a | 0.824 | 0.694 | PA2G4a | 1.224 | 1.048 | PRIC285a | 0.614 | 0.887 |
| EDD1 | 0.904 | 0.683 | PAK6a | 0.695 | 1.042 | RAD54L2 | 1.929 | 0.863 |
| NEDD4 | 1.287 | 0.870 | PELP1a | 1.565 | 0.943 | RAP80 | 1.288 | 1.094 |
| PSMC3 | 1.221 | 1.006 | PXNa | 0.813 | 1.094 | RBM14 | 1.059 | 0.908 |
| PSMC4 | 0.877 | 1.009 | RANa | 1.162 | 0.945 | RING1 | 0.680 | 0.646 |
| PSMC5 | 0.967 | 0.961 | RANBP9a | 0.790 | 0.929 | SAFB | 1.008 | 0.792 |
| RCHY1a | 0.891 | 0.929 | SMAD3a | 0.765 | 0.461 | SAFB2 | 1.150 | 0.945 |
| RNF4a | 1.091 | 1.203 | SQSTM1 | 0.586 | 1.005 | SCAND1 | 0.543 | 0.916 |
| RNF14a | 1.032 | 1.303 | TGFB1l1 | 1.301 | 1.117 | SIX3 | 0.343 | 0.234 |
| STUB1a | 0.577 | 1.049 | TRIP6 | 0.640 | 1.202 | SPEN | 1.032 | 0.849 |
| TSG101a | 0.824 | 0.916 | ANP32Aa | 0.996 | 0.987 | SRA1a | 1.826 | 0.705 |
| UBE3Aa | 0.967 | 0.883 | CDK7 | 0.964 | 1.023 | TADA2L | 1.536 | 0.863 |
| PIAS1a | 0.872 | 1.218 | CCND1a | 0.877 | 0.930 | TADA3L | 0.908 | 1.059 |
| PIAS2aiso1 | 0.756 | 0.685 | CCNE1a | 1.421 | 0.782 | TBL1X | 0.965 | 1.095 |
| PIAS2aiso2 | 0.950 | 0.888 | RB1a | 0.715 | 0.893 | TBL1XR1 | 0.933 | 0.974 |
| PIAS3a | 0.829 | 0.992 | RBAKa | 0.801 | 0.770 | TBPIPa | 2.511 | 0.979 |
| PIAS4a | 0.622 | 0.927 | AESa | 0.955 | 0.965 | TGIFa | 1.040 | 1.061 |
| UBE1Ca | 0.900 | 0.809 | ARID1Aa | 0.708 | 0.875 | THRAP4a | 1.052 | 0.903 |
| UBE2Ia | 0.943 | 0.979 | BCL3 | 0.539 | 1.148 | TLE1 | 1.032 | 0.956 |
| ZIMP7a | 0.446 | 1.039 | BCL11A | 1.106 | 0.930 | TMF1a | 0.953 | 1.064 |
| ANT-1a | 0.726 | 0.943 | BCL11B | 0.598 | 1.091 | TRA16 | 1.427 | 1.227 |
| DDX5 | 0.844 | 0.918 | BLOC1S1 | 0.859 | 0.988 | TRIM24a | 0.997 | 1.087 |
| DDX17 | 0.703 | 0.858 | BRD8 | 1.166 | 0.888 | TRIP4 | 1.410 | 0.889 |
| DDX20 | 1.395 | 0.908 | C1D | 1.073 | 1.053 | TRIP11 | 1.114 | 0.775 |
| DDX54 | 1.042 | 1.234 | COCOAa | 0.799 | 0.569 | TRRAP | 0.930 | 0.880 |
| DHX9 | 1.081 | 0.828 | CITED1 | 1.153 | 1.515 | TSC2 | 0.665 | 0.981 |
| FUS | 0.955 | 0.551 | CMTM2a | 2.020 | 0.937 | UXTa | 0.982 | 1.011 |
| HNRPU | 1.026 | 0.966 | COPS2a | 1.060 | 0.875 | ZNF278a | 1.195 | 0.665 |
| (Continued) | ||||||||
Table 3A.
Continued
| Gene symbol | Fold induction
|
Gene symbol | Fold induction
|
Gene symbol | Fold induction
|
|||
|---|---|---|---|---|---|---|---|---|
| 0.1 nm | 1.0 nm | 0.1 nm | 1.0 nm | 0.1 nm | 1.0 nm | |||
| NCOA6IP | 1.075 | 1.005 | CRABP2 | 0.660 | 0.391 | ZNF335 | 1.022 | 1.020 |
| NONOa | 0.923 | 0.872 | CRSP2a | 0.783 | 0.887 | ZNF653 | 0.705 | 1.025 |
| RBM9 | 0.791 | 0.661 | CTBP1 | 0.900 | 0.523 | ZNHIT3 | 0.866 | 0.955 |
The order of the genes listed corresponds to the order in which the genes are listed and grouped in the heatmap shown in supplemental Fig. 2. LNCaP cells were seeded in medium supplemented with charcoal-stripped serum. Medium was changed 3 d later, and cells were treated with 0, 0.1 or 1 nm of R1881. Cells were harvested 48 h later, and RNA was isolated and subjected to DASL analysis. Values represent the average fold change in gene expression after treatment with 0.1 or 1 nm of R1881 from three biological replicates each.
Coregulator that has been shown to interact with the AR. Iso, Coregulator isoform.
DASL analysis reveals widespread androgen regulation of coregulator expression
To explore the effect of androgen stimulation on coregulator expression, androgen-responsive LNCaP PCa cells were cultured for 48 h in the presence of 0, 0.1, or 1.0 nm of the synthetic androgen R1881. RNA prepared from biological triplicates was analyzed using our custom DASL array. Tables 1–3 show the average fold change in gene expression after androgen treatment compared with the vehicle control. Absolute expression values, standard deviations, and statistical significance are listed in supplemental Table 1 published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org. Supplemental Figs. 1 and 2 represent expression data for all samples as heatmaps and include functional classification of coregulators and positive control genes. In supplemental Table 2, the data, which are classified according to function in Tables 1–3, are listed alphabetically. As shown in Table 1, the expression of the 42 previously described androgen-responsive genes, which were included as positive controls, was regulated in a manner that is in line with published reports. Consistent with data obtained from conventional oligoarray experiments, the extent of the androgen-induced effect observed in our array was less pronounced as compared with that observed by other quantification methods. This may also reflect the particular nature of the DASL assay, in which the intensity of a given transcript may be influenced by a change in expression of another transcript. Nonetheless, with the exception of acid phosphatase, prostate (ACPP) and vascular endothelial growth factor C (VEGFC), the expression of all positive control genes showed an androgen-dependent fold change of at least 1.15-fold, which is within the sensitivity range of the assay. Importantly, none of the loading control genes displayed changes in expression exceeding this level of regulation. These observations suggest that a 15% difference in DASL expression levels is indicative of actual changes in expression. Using this value as a guideline, the expression of approximately one third (64/186) of coregulators was subject to androgen regulation.
Validation of DASL results
To confirm our hypothesis and to validate the array expression data, we studied the expression of a subset of 15 coregulators in more detail. For this, we selected coregulators that display different androgen dependency on the DASL array and that function in distinct cellular processes and pathways (see supplemental Fig. S2). Androgen regulation of coregulator expression suggests the existence of feed-forward and feedback mechanisms in which androgens, through modulation of regulators of their cognate receptor, ensure strong activation of androgen-responsive genes. Therefore, we first examined the impact of changes in coregulator expression on the full androgen induction of a set of endogenously expressed genes containing well-characterized androgen response elements (AREs) (10,11,27,28,29,30,31). To this end, LNCaP cells were transfected with siRNAs targeting coregulator expression or control siRNAs. Forty-two hours after transfection, when coregulator levels had declined substantially in cells exposed to specific siRNA (data not shown), cells were treated with R1881 or vehicle. Forty-eight hours later, marked changes in the androgen-responsiveness of the genes encoding prostate-specific antigen (PSA), kallikrein-related peptidase 2 (KLK2), transmembrane protease serine 2 (TMPRSS2), sterol regulatory element binding transcription factor cleavage activating protein (SCAP), maspin and fibronectin-1 (FN1) were noted in cells where coregulator expression was targeted (Fig. 1). It is significant that loss of a coregulator could differentially impact the androgen responsiveness of AR target genes, thus suggesting specificity in the target genes controlled by coregulators. Transcriptionally regulatory effects were observed also for cofactors not previously known to affect AR-mediated transcription such as PB1 and NCoA7. These data confirm the relevance of the coregulators that were chosen for follow-up for AR target gene expression.
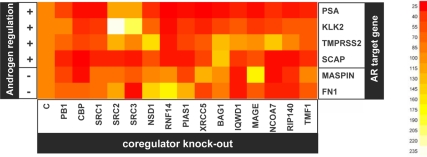
Figure 1.
Importance of coregulator expression for androgen responsiveness of AR target genes. LNCaP cells were transfected with siRNAs targeting coregulators or nonspecific control siRNAs (c). Forty-two hours after transfection, cells were treated with 5 nm R1881 or ethanol vehicle. Forty-eight hours later, androgen responsiveness of expression of ARE-driven genes that are up-regulated [(+), PSA, KLK2, TMPRSS2 and SCAP] or down-regulated [(−), maspin and fibronectin-1 (FN1)] after androgen exposure was evaluated by real-time RT-PCR. AR target gene mRNA levels were normalized with the values obtained from glyceraldehyde-3-phosphate dehydrogenase expression and are expressed as relative expression values, taking the value obtained from one of the vehicle-treated control transfected conditions as 1. Values represent relative fold induction of mRNA expression after R1881 treatment and correspond to the means of values obtained from biological triplicate samples. The heatmap representing the results was generated using R. Full (100%) androgen induction of AR target gene expression obtained in cells transfected with nontargeting control (c) siRNA is represented by orange. Yellow and white indicate an increase (>100%) in androgen responsiveness; red represents decreases (<100%) in androgen-responsiveness of AR target genes upon loss of a specific coregulator. Right, Scale numerically matching color shades with extent of the androgen regulation of AR target gene expression. MAGE, MAGEA11. SRC1, SRC2, and SRC3 correspond to DASL nomenclature NCOA1, NCOA2 and NCOA3 in Tables 1–3.
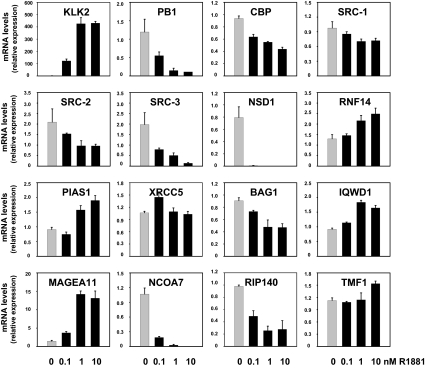
To validate the results from the DASL array, real-time RT-PCR was performed on cDNA derived from the same RNA samples that were used for DASL analysis. The ability of the primers used to detect specific coregulator mRNA expression was verified by experiments using specific siRNAs (supplemental Fig. S3). Androgen responsiveness of the cells was confirmed by measuring the androgen induction of KLK2 expression (Fig. 2). Figure 2 shows that, overall, the androgen-dependent changes in coregulator expression observed by DASL analysis are confirmed by real-time RT-PCR. With the exception of XRCC5, which displays a modest increase in expression at 1 nm on DASL array, androgen modulation of the expression of 14 of 15 coregulators chosen for follow-up is seen also in the RT-PCR data. For BCL2-associated athanogene (BAG1), a trend toward down-regulation observed on the array became more pronounced by real-time RT-PCR analysis. Except for TATA element modulatory factor 1 (TMF1), exposure to higher doses of R1881 did not or only slightly increased the magnitude of androgen regulation of coregulator genes. In addition, data presented in Fig. 2 confirm that coregulator expression can be either up-regulated or down-regulated after stimulation with R1881. Obvious patterns of coregulator regulation or classification could not be discerned. Moreover, the extent of the androgen effect and the dose dependency of the effect varied widely between different coregulators, suggesting multiple molecular mechanisms underlying these events. To evaluate the involvement of the AR in the effects of androgens on coregulator expression, LNCaP cells were treated with R1881 (1 nm) in the absence or presence of an excess of the antiandrogen Casodex (bicalutamide, 10 μm). As shown in supplemental Fig. 4, the addition of Casodex counteracted, either partially or completely, the effects of androgen treatment.
Figure 2.
Real-time RT-PCR validation of DASL results. LNCaP cells were seeded in medium supplemented with charcoal-stripped serum. Medium was changed 2 d later, and cells were treated with 0.1, 1, or 10 nm of the synthetic androgen R1881 (black bars) or ethanol vehicle (gray bars) for 48 h. RNA was isolated and converted into cDNA. Real-time RT-PCR was performed with the primer pairs listed in supplemental Table 3. Coregulator mRNA levels were normalized with the values obtained from glyceraldehyde-3-phosphate dehydrogenase . Values are expressed as relative expression levels, taking the value obtained from one of the untreated samples as 1. Columns indicate means of values obtained from three independent biological replicates; bars, sem values.
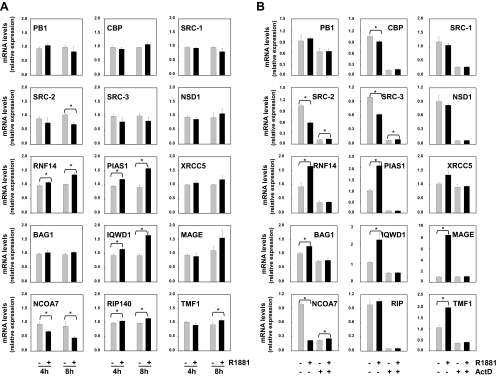
To distinguish between direct, ARE-driven, and indirect mechanisms of androgen action, we performed a time course in which LNCaP cells were treated for 4 and 8 h in the presence or the absence of 1 nm R1881. At these time points, mRNA expression of PSA, a direct target gene for the AR, was markedly induced (data not shown). Expression of steroid receptor coactivator (SRC)-2 and IQ motif and WD repeats 1 (IQWD1), which have recently proposed to be regulated by AREs (32,33), were also affected. Data shown in Fig. 3a suggest that a similar direct mechanism of AR action could underlie the androgen regulation of SRC-3, RNF14, protein inhibitor of activated STAT 1 (PIAS1), and NCoA7 as well. Melanoma antigen family A, 11 (MAGEA11), on the other hand, seems to be under control of an indirect mechanism. Androgen modulation of other coregulator genes could not be observed at these time points and is therefore likely to be mediated by indirect mechanisms of androgen action.
Figure 3.
Characterization of mechanism(s) underlying androgen regulation of coregulator expression. A, Short-term time course. LNCaP cells were treated with 1 nm R1881 (black bars) or ethanol (gray bars) for 4 or 8 h. B, Coregulator expression was analyzed by real-time RT-PCR as described. LNCaP cells were treated for 30 min with actinomycin D (ActD, 5 μg/ml) or vehicle. R1881 (5 nm) (black bars) or ethanol vehicle (gray bars) was added. After 16 h, coregulator expression was assessed by real-time RT-PCR as described. Columns indicate mean values from samples in triplicate; bars, sem values; *, statistically significant changes (P <0,05) as determined by t test.
To investigate whether androgen regulation of coregulator gene mRNA involves active transcription, LNCaP cells were pretreated for 30 min with the transcriptional inhibitor actinomycin D before androgen treatment. Sixteen hours later, coregulator expression was evaluated by real-time RT-PCR. As shown in Fig. 3B, actinomycin D completely prevented androgen modulation of coregulator expression, suggesting involvement of active transcription. Actinomycin D also affected basal levels of most coregulators. Remarkably, at this time point, expression of XRCC5 and BAG1 appeared to be slightly and transiently induced by androgen treatment.
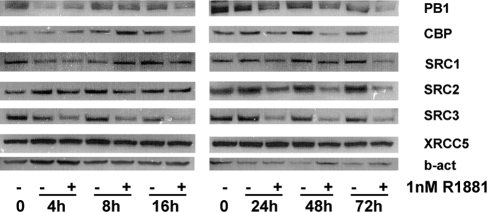
To further characterize and validate the data derived from the DASL array, a more extensive time course was performed in which LNCaP cells were treated for up to 72 h with 1 nm R1881 or vehicle. For those coregulators for which reliable antibodies could be obtained [i.e. PB1, cAMP response element-binding protein (CREB)-binding protein (CBP) (gene symbol CREBBP), SRC-1 (gene symbol NCOA1), SRC-2 (gene symbol NCOA2), SRC-3 (gene symbol NCOA3) and XRCC5], cell lysates were analyzed by Western blot. The specificity of the antibodies was verified using siRNAs targeting coregulator expression (supplemental Fig. S5). As shown in Fig. 4, and rogen regulation of coregulator expression was confirmed at the protein level. The kinetics by which R1881 exposure affects coregulator protein levels were markedly different, which underscores the complexity underlying coregulator gene regulation. Except for SRC-2 and PB-1, androgen-induced changes in coregulator expression at the protein level were preceded by alterations at the mRNA level. Moreover, for some coregulators (e.g. CBP), the magnitude of the androgenic control was more pronounced at the protein level. These observations indicate that additional androgenic regulation of coregulators occurs at the level of protein translation, stabilization, and/or turnover.
Figure 4.
Time course of the effects of androgens on coregulator protein expression. LNCaP cells were seeded in charcoal-stripped serum medium. Medium was changed 2 d later, and cells were treated with 1 nm of R1881 (+) or vehicle (−) for the indicated periods of time. Total protein extracts were prepared, and equal amounts of protein were analyzed by Western blotting using antibodies directed against PB1, CBP, SRC1, SRC2, SRC3, or XRCC5. To evaluate potential loading differences, blots were stripped and reprobed with an antibody against β-actin (b-act).
Differential coregulator expression in isogenic PCa cells
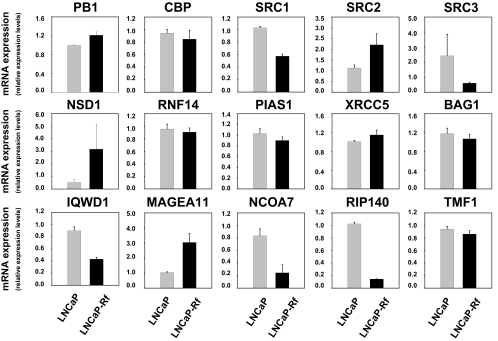
Having established that androgenic stimulation can modulate the expression of multiple coregulators, we explored whether androgen deprivation, which is therapeutically relevant, impacts coregulator expression. To this end, we evaluated the expression levels of the subset of 15 coregulators chosen for validation in LNCaP cells and its isogenic cell line LNCaP-Rf. The LNCaP-Rf cell line has been established in our laboratory by long-term androgen ablation of LNCaP cells and is considered to be a valuable model for the study of CR PCa (34). Real-time RT-PCR analysis showed elevated mRNA expression levels of PB1, SRC-2, nuclear receptor binding SET domain protein 1 (NSD1), and MAGEA11 in LNCaP-Rf cells, whereas expression of SRC-1, SRC-3, IQWD1, NCoA7, and RIP140 (gene symbol NRIP1) was decreased compared with its parental line. Expression of other cofactors (CBP, RNF14, PIAS1, XRCC5, BAG1, TMF1) was similar between LNCaP and LNCaP-Rf cells (Fig. 5). These observations were confirmed at the protein level for PB1, SRC-1, SRC-2, SRC-3, and XRCC5 (Fig. 6). CBP protein levels, on the other hand, were higher in LNCaP-Rf than in LNCaP cells, whereas CBP mRNA expression levels in LNCaP and LNCaP-Rf cells did not differ. Short-term androgen deprivation had similar effects, and readministration of androgen to LNCaP-Rf cells restored androgen responsiveness to coregulator expression, albeit with slower kinetics (data not shown).
Figure 5.
Real-time RT-PCR evaluation of differential expression of coregulators in isogenic LNCaP cells. LNCaP (gray bars) and LNCaP-Rf cells (black bars) were seeded in their regular medium. Cells were harvested 3 d later, and total RNA was isolated. Real-time RT-PCR was performed as described. Values are expressed as relative expression levels, taking the value obtained from one of the LNCaP-derived samples as 1. Columns represent means of values obtained from three independent biological replicates; bars, sem values.
Figure 6.
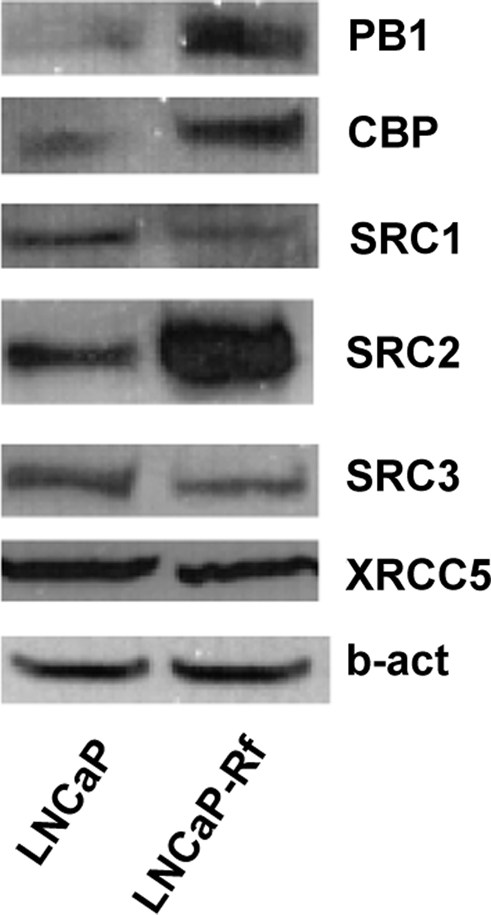
Western blot analysis of differential expression of coregulators in isogenic LNCaP cells. LNCaP and LNCaP-Rf cells were seeded in their regular medium. Total protein extracts were prepared 3 d later, and equal amounts of protein were analyzed by Western blotting using antibodies directed against PB1, CBP, SRC1, SRC2, SRC3, or XRCC5. To evaluate potential loading differences, blots were stripped and reprobed with an antibody against β-actin (b-act).
Changes in coregulator expression affect PCa cell proliferation
To assess the importance of changes in coregulator expression for PCa cell survival during disease progression, we compared the dependency of the isogenic cell lines LNCaP and LNCaP-Rf on the expression of each of the 15 coregulators for cell viability. Cells were transfected with siRNAs targeting each coregulator or nontargeting control siRNAs. One to six days after transfection, cell viability was evaluated by an MTS [3-(4,5- dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H tetrazolium)] assay. In LNCaP cells, down-regulation of PB1, CBP, NSD1, RNF14, BAG1, MAGEA11, and NCOA7 expression led to relatively minor decreases in cell viability that were obvious at different times after transfection (Fig. 7). Loss of PIAS1 had a more profound negative effect on cell viability, whereas the knockdown of XRCC5, IQWD1, and TMF1 led to increases in the number of viable cells, again with different kinetics. Changes in the expression of SRC-1, SRC-2, SRC-3, or RIP140 did not notably affect LNCaP cell viability (Fig. 7). CR LNCaP-Rf cells tended to rely more on normal coregulator expression levels to maintain cell viability. This was particularly true for coregulators such as PB1, CBP, SRC-2, NSD1, and MAGEA11, which exhibited increased expression in LNCaP-Rf cells compared with LNCaP cells. The loss of RNF14, BAG1, and TMF1, as well as NCoA7, led to decreases in the number of viable LNCaP-Rf cells that were greater than those observed for LNCaP. In line with observations in LNCaP cells, depletion of PIAS1 markedly inhibited LNCaP-Rf cell viability. Remarkably, targeting the expression of the core coregulators SRC-1 and SRC-3 did not affect the number of viable LNCaP-Rf cells. Loss of XRCC5 or IQWD1, which led to increased cell viability of the parental line LNCaP, had the opposite effect on LNCaP-Rf cells or did not affect them at all.
Figure 7.
Dependency of isogenic LNCaP cells lines on coregulator expression for cell proliferation. LNCaP and LNCaP-Rf cells were transfected with siRNAs targeting the expression of a particular coregulator (black bars) or control siRNAs (gray bars). Medium was changed 12–16 h after transfection. Cells were harvested 1, 2, 3, 4, 5, and 6 d later, and cell proliferation was assessed by means of an MTS assay reading absorbance at 490 nm. Columns represent mean values from five individual measurements, bars, sem values.
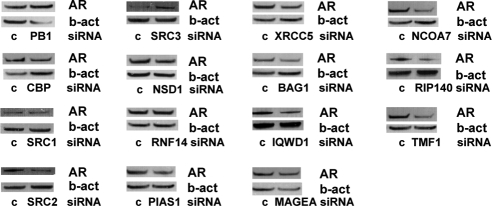
The results presented in Fig. 7 could not be attributed to marked differences in the efficiency of coregulator knockdown between the LNCaP and LNCaP-Rf cell lines (Refs. 16 and 17 and data not shown). Consistent with previous studies in our laboratory in which the expression of other cofactors were modulated (16,17), changes in cell viability upon loss of coregulator expression appeared to be due to decreased cell proliferation rather than increased apoptosis. Cells did not show morphological characteristics consistent with apoptosis after transfection with siRNAs targeting coregulator expression (data not shown), nor did cleavage of poly(ADP-ribose)polymerase, a widely recognized marker for cells undergoing apoptosis, occur (supplemental Fig. S6). Because posttranslational modification of the AR by several of the coregulators studied here can affect AR activity and/or stability (8,9,33), we also investigated the impact of the loss of each coregulator on AR expression levels. As shown in Fig. 8, loss of coregulator expression did not result in major changes in AR levels. Taken together, our data indicate that the changes in number of viable cells after transfection with siRNAs targeting several coregulators are due to changes in cell proliferation, rather than apoptosis or AR depletion.
Figure 8.
Loss of coregulator expression does not markedly affect AR protein expression. LNCaP cells were transfected with specific siRNAs targeting coregulator expression or control (c) siRNAs. Medium was changed 12–16 h after transfection. Proteins were isolated 96 h later, and Western blotting was performed using an antibody directed against the AR. To assess potential intersample loading differences, blots were stripped and reprobed with antibodies recognizing β-actin (b-act).
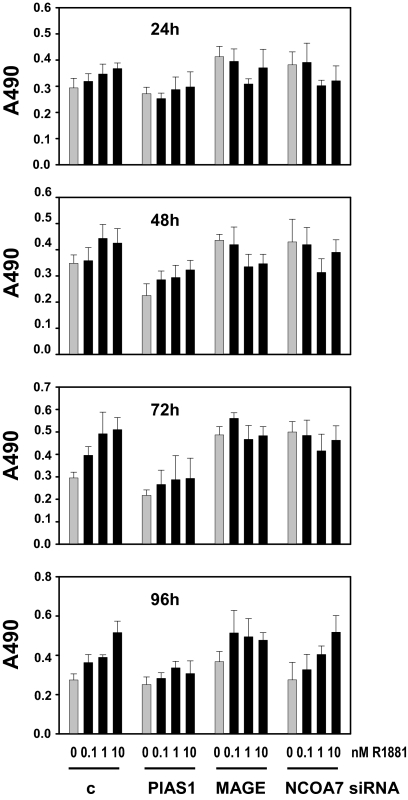
To assess the relevance of regulation of coregulator expression for androgen-dependent PCa cell viability, we performed MTS studies combining androgen treatment with siRNA-mediated silencing of several coregulators that showed promise as therapeutic targets in the experiments shown in Fig. 7 (PIAS1, NCOA7, and MAGEA11). To this end, LNCaP cells were transfected with siRNAs targeting PIAS1, MAGEA11, and NCOA7 or nontargeting control siRNAs. Cells were treated, 16 h after transfection, with 0, 0.1, 1, or 10 nm R1881 and MTS assays were done 1–4 d later. As shown in Fig. 9, silencing of PIAS1 blunted androgen-dependent increases in the number of viable LNCaP cells at all time points studied. Similar observations were done for MAGEA11 and NCOA7, although cell viability appeared either partially or almost completely restored, respectively, after 96 h of androgen stimulation. These data indicate that androgen regulation of coregulator expression is important for PCa proliferation and may point to differences in the timing when specific coregulators are most important for cell viability. In line with data shown in Fig. 8, loss of PIAS1, NCOA7, or MAGEA11 did not affect androgen regulation of AR levels (data not shown).
Figure 9.
Role of coregulator expression in androgen-dependent cell proliferation. LNCaP cells were transfected with siRNAs targeting the expression of PIAS1, MAGEA11 (MAGE) or NCOA7, or control siRNAs (c). Cells were treated 16 h after transfection with 0.1, 1 or 10 nm R1881 (black bars) or vehicle bars (gray bars). Cell proliferation was assessed 24, 48, 72, and 96 h later by means of an MTS assay reading absorbance at 490 nm. Columns represent mean values from five individual measurements; bars, sem values.
Discussion
Deregulated coregulator expression during PCa progression has been associated with a poor prognosis and aggressive disease and has been suggested as an attractive target for therapeutic intervention (13,14,15). Whereas some of the effects of coregulators on PCa cells may be independent of the AR, most are likely due to their ability to modulate AR transcriptional output. As such, coregulator expression and/or action could represent an indirect means of targeting the activity of the AR, which is emerging as a critical determinant for PCa cell survival (1,2,3,4). Thus, a better understanding of the signals and signaling mechanisms that govern coregulator expression could lead to novel therapeutic approaches, which are urgently needed. Here, we designed a gene expression-profiling assay to evaluate AR signaling and, more specifically, to comprehensively and simultaneously evaluate the expression patterns of a large set of coregulators. After validation and calibration of our DASL assay, we showed that 30% of coregulators included in our array display some level of androgen regulation. Overall, general trends and patterns of androgen regulation of coregulator expression were hard to discern. For example, the expression of coregulators that fulfil roles in chromatin remodeling and histone (de)acetylation or (de)methylation all trended toward down-regulation. Conversely, cofactors belonging to other functional groups (such as those with roles in sumoylation, RNA metabolism, and chaperone action) could either be repressed or induced by androgens. Noteworthy, apart from LNCaP cells [as evidenced by our data, SRC-2 (32), CBP (35)], androgen-dependent coregulator expression was also observed in AR-positive VCaP and LAPC4 PCa cells but not in other malignant or normal androgen-responsive models systems (Boorjian S. A., H. V. Heemers, I. Frank, S. Farmer, L. J. Schmidt, T. J. Sebo, and D. J. Tindall; and Heemers, H. V., E. Kidd, K. A. Raclaw, L. J. Schmidt, K. M. Regan, T. J. Sebo, and D. J. Tindall, manuscripts in preparation), suggesting PCa cell specificity.
The kinetics by which androgens affect coregulator expression at the mRNA and protein level, in combination with studies using the transcriptional inhibitor actinomycin D, point toward diverse molecular machineries driving these events. Our results are in agreement with recent reports of ARE-driven expression of SRC-2 and IQWD1 (32,33) and suggest that a similar direct mechanism of androgen action regulates expression of the genes encoding SRC-3, RNF14, PIAS1, and NCoA7. Conversely, characterization of the androgen regulation of the gene encoding MAGEA11 indicates the involvement of transcriptional events that are indirectly regulated by the AR, which is reminiscent of the manner by which androgens modulate FHL2 expression (17). Indirect mechanisms are likely to play a role also in the androgen modulation of other coregulator genes (e.g. NSD1). Moreover, at least for some coregulators, an additional level of androgen regulation occurs at the level of protein translation, turnover, or stability. A thorough examination of these mechanisms has been hampered by the lack of reliable antibodies. Other means by which androgens could affect coregulator expression that were not addressed by our studies include effects on coregulator mRNA and/or protein half-lives or alterations in the posttranslational modification status of cofactor proteins. The importance of the latter mechanism has been demonstrated for SRC-3. Androgen stimulation induces several site-specific phosphorylations, which are critical for SRC-3 to coactivate the AR and determine the proteins with which it interacts (36).
Regardless of the mechanism(s) responsible for androgen control over coregulator expression, our data indicate that such regulation results in feedback and feed-forward loops that impact the composition of the AR transcriptional complex and, consequently, the transcriptional output by the AR. We have reported previously a similar feed-forward mechanism for FHL2, the expression of which is induced by androgens (17). In line with previous observations for BRM and SRC-2 (37,32), our data provide compelling evidence for specificity in the AR target genes controlled by coregulators. Additional mechanistic studies will reveal whether the kinetics by which androgens affect individual coregulator expression are either: 1) reflected in the temporal composition of the AR transcriptional complex at regulatory regions of target genes, and/or 2) impose a preference or selectivity toward transcription of a particular set of target genes at a given time after androgen stimulation. Ongoing efforts to map genome-wide recruitment of the AR, in combination with gene expression profiling of PCa specimens, will help focus these studies to those AR target genes and coregulators that are clinically relevant.
Because transcription factors compete for limited amounts of coregulators within the cell, androgen modulation of coregulators may also affect the transcriptional program of transcription factors other than the AR and induce positive or negative cross talk between the AR and other transacting factors. Androgenic modulation of several members of the PIAS family of coregulator proteins could, for instance, impart steroid regulation on Stat-mediated signaling. Such transcriptional interference may be particularly relevant for those cofactors that have not (yet) been shown to directly associate with the AR.
In line with observations in patient samples (13,14), we observed alterations in coregulator expression in an isogenic cell model that mimics the transition toward CR PCa. These changes appear to have important consequences for PCa proliferation because CR LNCaP-Rf cells tend to rely more on those coregulators that are overexpressed to maintain cell viability when compared with the parental LNCaP cells. On the other hand, even though NCoA7 mRNA expression levels are lower in LNCaP-Rf, maintenance of normal expression levels of NCoA7 were more essential for proliferation of these cells than for LNCaP cells, suggesting that NCoA7 is a potential target for the treatment of CR PCa disease. Similarly, our work indicates that PIAS1 may hold promise as a target for therapeutic intervention because PIAS1 was essential for cell proliferation in both the LNCaP and LNCaP-Rf cells despite a modest level of androgen regulation and the lack of notable differences in PIAS mRNA expression in LNCaP and LNCaP-Rf cells. As for NCoA7, additional regulation of PIAS1 stability, turnover, or posttranslational status could not be assessed because of the lack of an antibody. Our observations of deregulated coregulator expression in the isogenic LNCaP model system are reminiscent of those reported for the CWR22 xenograft model and those observed for the LUCAP 35/35V and LUCAP23.1/23.1 AI models in our laboratory (Ref. 38 and data not shown).
Taken together, our work illustrates the importance of combining and integrating different experimental approaches to identify a coregulator as a valid therapeutic target. The data generated by our current study will provide the foundation for follow-up studies aimed at unraveling the signaling events that govern the expression of clinically relevant coregulators. Eventually, these approaches may lead to the development of alternative therapeutic options to target AR action in PCa.
Materials and Methods
Cell culture
LNCaP and LNCaP-Rf cells were maintained as described elsewhere (17).
Reagents
Methyltrienolone (R1881) was purchased from DuPont (Boston, MA). Staurosporine and actinomycin D were obtained from Sigma (St. Louis, MO). Casodex was kindly provided by Zeneca Pharmaceuticals (Wilmington, DE). Antibodies (AR, CBP) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); β-actin and poly(ADP-ribose)polymerase were obtained from Cell Signaling Technology (Beverly, MA); SRC-1, SRC-2, and SRC-3 were supplied by BD Transduction Laboratories (San Jose, CA); and XRCC5 was obtained from Novus Biologicals (Littleton, CO). The antibody recognizing PB-1 was a kind gift from Dr. Wang (39).
Generation of DASL data
A custom DASL array was designed according to the manufacturer’s instructions (Illumina, San Diego, CA). Our custom DASL gene list consists of genes encoding the AR, androgen-regulated control genes, coregulator genes, loading control genes, and genes relevant to PCa biology (see Tables 1–3; complete gene list available upon request). Total RNA isolated from biological triplicates (200 ng) was used for DASL analysis. RNA samples were subjected to quality control and processed for DASL analysis by the microarray core facility at Mayo Clinic’s Advanced Genomics Technology Center. Illumina BeadStudio version 3 was used to transform the scanned arrays into summarized nonnormalized average signal expression values for 1536 probes representing 532 genes. These average signal expression values were imported to the software R. The nine experimental samples were normalized together using the R-package fastlo (40). The probe-level expression values for each gene were summarized using the average of expression values for all probes associated with each gene. Differential expression between control and androgen-treated samples was assessed using the R-package called limma (41). The function lmFit (42) was used to fit a full-rank model, and contrasts fit was used to obtain coefficients and ses for the two-group contrasts of interest. The function eBayes was used to rank genes in order of evidence for differential expression using the log odds. This function uses an empirical Bayes method to shrink the probe-wise sample variances toward a common value and to augment the degrees of freedom for the individual variances. This empirical Bayes method calculates a moderated t-statistic test, which tests each individual contrast equal to zero. The function top table was used to summarize results for each contrast and compute an adjusted P value based on the less conservative method of Benjamini and Hochberg (43) that controls the false discovery rate (FDR). This FDR is the expected proportion of false discoveries among the rejected hypotheses. Summary statistics were calculated for each gene within the control and each experimental group. The average and sd were calculated for each group. For each of the two comparisons the fold change, P value, rank order of the P value, and the FDR were tabulated for each gene. The maximum of the two P value ranks for each gene was determined as the maximum overall rank. Specific sets of genes were analyzed using heatmap. The heatmap were generated using R. Color intensity shown in each heatmap corresponds to the relative level of expression for a gene across all nine samples. The lighter the color the higher the relative expression is for that sample within that gene.
Western blotting
Whole-cell lysates were prepared and analyzed as described previously (17).
Real-time RT-PCR
RNA was isolated and real-time RT-PCR was performed as described elsewhere (17). The primers used are listed in supplemental Table 3.
Cell proliferation assay
LNCaP and LNCaP-Rf cells were seeded in 96-well tissue culture plates at a density of 6 × 103 cells per well in their regular medium without added antibiotics. siRNA transfections and cell proliferation assays were carried out as described (17).
Supplementary Material
Acknowledgments
We thank Drs. Scott Dehm and Lucas Nacusi for helpful discussions.
Footnotes
This work was supported by National Institutes of Health Grants CA121277, CA91956, CA15083, CA125747, and DK65236, the T.J. Martell Foundation, the Charlotte Geyer Foundation, and the Belgian-American Educational Foundation (BAEF) (to H.V.H.).
Disclosure Summary: H.V.H., K.M.R., L.J.S., S.K.A., K.V.B., and D.J.T. have nothing to declare.
First Published Online January 22, 2009
Abbreviations: AR, Androgen receptor; ARE, androgen response element; BAG1, BCL2-associated athanogene; CBP, cAMP response element-binding protein (CREB)-binding protein; CR, castration-recurrent; DASL, cDNA-mediated annealing, selection, extension, and ligation; FDR, false discovery rate; IQWD1, IQ motif and WD repeats 1; KLK2, kallikrein-related peptidase 2; MAGEA11, melanoma antigen family A, 11; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H tetrazolium; NSD1, nuclear receptor binding SET domain protein 1; PCa, prostate cancer; PIAS, protein inhibitor of activated STAT; PSA, prostate-specific antigen; SRC, steroid receptor coactivator; TMF1, TATA element modulatory factor 1.
References
- Grossmann ME, Huang H, Tindall DJ 2001 Androgen receptor signaling in androgen-refractory prostate cancer. J Natl Cancer Inst 93:1687–1697 [DOI] [PubMed] [Google Scholar]
- Litvinov IV, De Marzo AM, Isaacs JT 2003 Is the Achilles’ heel for prostate cancer therapy a gain of function in androgen receptor signaling? J Clin Endocrinol Metab 88:2972–2982 [DOI] [PubMed] [Google Scholar]
- Debes JD, Tindall DJ 2004 Mechanisms of androgen-refractory prostate cancer. N Engl J Med 351:1488–1490 [DOI] [PubMed] [Google Scholar]
- Balk SP, Knudsen KE 2008 AR, the cell cycle, and prostate cancer. Nucl Recept Signal 6:e001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto H, Messing EM, Chang C 2004 Androgen deprivation therapy for prostate cancer: current status and future prospects. Prostate 61:332–353 [DOI] [PubMed] [Google Scholar]
- Roy-Burman P, Tindall DJ, Robins DM, Greenberg NM, Hendrix MJ, Mohla S, Getzenberg RH, Isaacs JT, Pienta KJ 2005 Androgens and prostate cancer: are the descriptors valid? Cancer Biol Ther 4:4–5 [DOI] [PubMed] [Google Scholar]
- Mohler JL 2008 Castration-recurrent prostate cancer is not androgen-independent. Adv Exp Med Biol 617:223–234 [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C 2002 Androgen receptor (AR) coregulators: an overview. Endocr Rev 23:175–200 [DOI] [PubMed] [Google Scholar]
- Heemers HV, Tindall DJ 2007 Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev 28:778–808 [DOI] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Jänne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M 2007 A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton EC, So AY, Chaivorapol C, Ha CM, Li H, Yamamoto KR 2007 Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev 21:2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, Mills IG 2007 New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep 8:871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemers HV, Tindall DJ 2005 Androgen receptor coregulatory proteins as potential therapeutic targets in the treatment of prostate cancer. Curr Cancer Ther Rev 1:175–186 [Google Scholar]
- Chmelar R, Buchanan G, Need EF, Tilley W, Greenberg NM 2007 Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int J Cancer 120:719–733 [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Weigel NL 2008 Androgen receptor coactivators and prostate cancer. Adv Exp Med Biol 617:245–255 [DOI] [PubMed] [Google Scholar]
- Heemers HV, Sebo TJ, Debes JD, Regan KM, Raclaw KA, Murphy LM, Hobisch A, Culig Z, Tindall DJ 2007 Androgen deprivation increases p300 expression in prostate cancer cells. Cancer Res 67:3422–3430 [DOI] [PubMed] [Google Scholar]
- Heemers HV, Regan KM, Dehm SM, Tindall DJ 2007 Androgen induction of the androgen receptor coactivator four and a half LIM domain protein-2: evidence for a role for serum response factor in prostate cancer. Cancer Res 67:10592–10599 [DOI] [PubMed] [Google Scholar]
- Fan JB, Yeakley JM, Bibikova M, Chudin E, Wickham E, Chen J, Doucet D, Rigault P, Zhang B, Shen R, McBride C, Li HR, Fu XD, Oliphant A, Barker DL, Chee MS 2004 A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res 14:878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Talantov D, Chudin E, Yeakley JM, Chen J, Doucet D, Wickham E, Atkins D, Barker D, Chee M, Wang Y, Fan JB 2004 Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol 165:1799–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghray A, Feroze F, Schober MS, Yao F, Wood C, Puravs E, Krause M, Hanash S, Chen YQ 2001 Identification of androgen-regulated genes in the prostate cancer cell line LNCaP by serial analysis of gene expression and proteomic analysis. Proteomics 1:1327–1338 [DOI] [PubMed] [Google Scholar]
- DePrimo SE, Diehn M, Nelson JB, Reiter RE, Matese J, Fero M, Tibshirani R, Brown PO, Brooks JD 2002 Transcriptional programs activated by exposure of human prostate cancer cells to androgen. Genome Biol 3:RESEARCH0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B 2002 The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci USA 99:11890–11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg N, Eroglu B, Ferguson C, Arnold H, Moorman A, Nelson PS 2002 Digital expression profiles of the prostate androgen-response program. J Steroid Biochem Mol Biol 80:13–23 [DOI] [PubMed] [Google Scholar]
- Segawa T, Nau ME, Xu LL, Chilukuri RN, Makarem M, Zhang W, Petrovics G, Sesterhenn IA, McLeod DG, Moul JW, Vahey M, Srivastava S 2002 Androgen-induced expression of endoplasmic reticulum (ER) stress response genes in prostate cancer cells. Oncogene 21:8749–8758 [DOI] [PubMed] [Google Scholar]
- Dehm SM, Tindall DJ 2006 Molecular regulation of androgen action in prostate cancer. J Cell Biochem 99:333–344 [DOI] [PubMed] [Google Scholar]
- Velasco AM, Gillis KA, Li Y, Brown EL, Sadler TM, Achilleos M, Greenberger LM, Frost P, Bai W, Zhang Y 2004 Identification and validation of novel androgen-regulated genes in prostate cancer. Endocrinology 145:3913–3924 [DOI] [PubMed] [Google Scholar]
- Cleutjens KB, van Eekelen CC, van der Korput HA, Brinkmann AO, Trapman J 1996 Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J Biol Chem 271:6379–6388 [DOI] [PubMed] [Google Scholar]
- Cleutjens KB, van der Korput HA, van Eekelen CC, van Rooij HC, Faber PW, Trapman J 1997 An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol Endocrinol 11:148–161 [DOI] [PubMed] [Google Scholar]
- Mitchell SH, Murtha PE, Zhang S, Zhu W, Young CY 2000 An androgen response element mediates LNCaP cell dependent androgen induction of the hK2 gene. Mol Cell Endocrinol 168:89–99 [DOI] [PubMed] [Google Scholar]
- Heemers H, Verrijdt G, Organe S, Claessens F, Heyns W, Verhoeven G, Swinnen JV 2004 Identification of an androgen response element in intron 8 of the sterol regulatory element-binding protein cleavage-activating protein gene allowing direct regulation by the androgen receptor. J Biol Chem 279:30880–30887 [DOI] [PubMed] [Google Scholar]
- He ML, Jiang AL, Zhang PJ, Hu XY, Liu ZF, Yuan HQ, Zhang JY 2005 Identification of androgen-responsive element ARE and Sp1 element in the maspin promoter. Chin J Physiol 48:160–166 [PubMed] [Google Scholar]
- Agoulnik IU, Vaid A, Nakka M, Alvarado M, Bingman III WE, Erdem H, Frolov A, Smith CL, Ayala GE, Ittmann MM, Weigel NL 2006 Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res 66:10594–10602 [DOI] [PubMed] [Google Scholar]
- Chen PH, Tsao YP, Wang CC, Chen SL 2008 Nuclear receptor interaction protein, a coactivator of androgen receptors (AR), is regulated by AR and Sp1 to feed forward and activate its own gene expression through AR protein stability. Nucleic Acids Res 36:51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo H, Huang H, Schmidt LJ, Smith DI, Tindall DJ 2001 Role of PI3K signaling in survival and progression of LNCaP prostate cancer cells to the androgen refractory state. Endocrinology 142:4795–4805 [DOI] [PubMed] [Google Scholar]
- Comuzzi B, Nemes C, Schmidt S, Jasarevic Z, Lodde M, Pycha A, Bartsch G, Offner F, Culig Z, Hobisch A 2004 The androgen receptor co-activator CBP is up-regulated following androgen withdrawal and is highly expressed in advanced prostate cancer. J Pathol 204:159–166 [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW 2004 Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell 15:937–949 [DOI] [PubMed] [Google Scholar]
- Marshall TW, Link KA, Petre-Draviam CE, Knudsen KE 2003 Differential requirement of SWI/SNF for androgen receptor activity. J Biol Chem 278:30605–30613 [DOI] [PubMed] [Google Scholar]
- Gregory CW, He B, Johnson RT, Ford OH, Mohler JL, French FS, Wilson EM 2001 A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res 61:4315- 4319 [PubMed] [Google Scholar]
- Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, Young MK, Salmon ED, Wang W 2000 The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci USA 97:13015–13020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballman KV, Grill DE, Oberg AL, Therneau TM 2004 Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics 20:2778–2786 [DOI] [PubMed] [Google Scholar]
- Smyth GK 2004 Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article 3 [DOI] [PubMed] [Google Scholar]
- Smyth GK 2005 Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer; 397–420 [Google Scholar]
- Benjamini Y, Hochberg Y 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.