Abstract
Chromosome segregation at mitosis depends critically on the accurate assembly of kinetochores and their stable attachment to microtubules. Analysis of Saccharomyces cerevisiae kinetochores has shown that they are complex structures containing ≥50 protein components. Many of these yeast proteins have orthologs in animal cells, suggesting that key aspects of kinetochore structure have been conserved through evolution, despite the remarkable differences between the 125-base pair centromeres of budding yeast and the Mb centromeres of animal cells. We describe here an analysis of S. cerevisiae Ndc10p, one of the four protein components of the CBF3 complex. CBF3 binds to the CDEIII element of centromeric DNA and initiates kinetochore assembly. Whereas CDEIII binding by Ndc10p requires the other components of CBF3, Ndc10p can bind on its own to CDEII, a region of centromeric DNA with no known binding partners. Ndc10p-CDEII binding involves a dispersed set of sequence-selective and -nonselective contacts over ∼80 base pairs of DNA, suggesting formation of a multimeric structure. CDEII-like sites, active in Ndc10p binding, are also present along chromosome arms. We propose that a polymeric Ndc10p complex formed on CDEII and CDEIII DNA is the foundation for recruiting microtubule attachment proteins to kinetochores. A similar type of polymeric structure on chromosome arms may mediate other chromosome–spindle interactions.
INTRODUCTION
Accurate chromosome segregation depends on the attachment of microtubules to kinetochores, protein–DNA complexes assembled on centromeric DNA. During mitosis, each sister chromatid must assemble one and only one kinetochore. Chromatids lacking kinetochores cannot be pulled into daughter cells at anaphase, whereas those with two or more kinetochores are in danger of being pulled in two directions at once and becoming torn. Correct chromosome segregation therefore requires proper regulation of kinetochore assembly.
Saccharomyces cerevisiae is an attractive organism in which to study kinetochore assembly because it contains particularly short centromeres. A 125-base pair CEN sequence is necessary and sufficient to mediate accurate chromosome segregation during mitosis and meiosis (Clarke and Carbon, 1980; Cottarel et al., 1989). In contrast, Schizosaccharomyces pombe centromeres are 40–100 kb in length, and human centromeres span megabases (Bloom, 1993). Despite the dramatic difference in the complexity of centromeric DNA among different eukaryotes, there is good evidence that many kinetochore proteins have been conserved through evolution (Dobie et al., 1999). It is therefore reasonable to expect that lessons learned in yeast will be of general importance for the study of chromosome segregation in other organisms.
The 16 centromeres in S. cerevisiae contain three conserved DNA elements: CDEI, CDEII, and CDEIII (reviewed in Hegemann and Fleig, 1993). CDEI is not essential for centromere function, but both CDEII and CDEIII are. CDEI is the binding site for the Cbf1 protein (Cai and Davis, 1989; Jiang and Philippsen, 1989; Mellor et al., 1990) and CDEIII is the binding site for the four-protein CBF3 complex (Ng and Carbon, 1987; Lechner and Carbon, 1991). Both CDEI and CDEIII contain highly conserved bases in which point mutations impair protein binding and centromere function (Niedenthal et al., 1991; McGrew et al., 1986; Ng and Carbon, 1987; Hegemann et al., 1988). CDEII has no known binding partners but it has a conserved length (78–84 bases) and high A-T composition (>90%; Clarke and Carbon, 1980).
CBF3 is the most extensively studied kinetochore complex in budding yeast (Lechner and Carbon, 1991; Goh and Kilmartin, 1993; Sorger et al., 1995; Strunnikov et al., 1995; Connelly and Hieter, 1996; Espelin et al., 1997) and seems to be required for the initiation of kinetochore assembly (Kaplan et al., 1997; Russell et al., 1999). S. cerevisiae kinetochores are characterized by a hierarchy of protein–DNA and protein–protein contacts, and all known kinetochore proteins require CBF3 activity for in vivo association with centromeric DNA. CBF3 contains four protein subunits: Ndc10p, Cep3p, Ctf13p, and Skp1p, all of which are necessary for DNA binding and for cell viability. Three CBF3 proteins, Ndc10p, Cep3p, and Ctf13p are in direct contact with DNA, as judged by DNA cross-linking in vitro (Espelin et al., 1997). The fourth, Skp1p, mediates the phosphorylation-dependent activation of Ctf13p (Kaplan et al., 1997).
Although CDEII is essential (Panzeri et al., 1985; Gaudet and Fitzgerald-Hayes, 1987), it has no previously identified protein ligands. The experiments described here show that Ndc10p is a sequence-specific CDEII DNA binding protein. Although CDEIII binding by Ndc10p requires the three other members of the CBF3 protein complex, CDEII binding does not. Linker-scanning mutagenesis of CDEII reveals that sequences across a roughly 80-base pair region are involved in protein–DNA recognition with a particularly important site near the center. Mutations in this sequence significantly decrease the fidelity of chromosome segregation. We therefore propose that Ndc10p has two functions at kinetochores, one as an essential component of CBF3 and a second as a CDEII-binding factor.
MATERIALS AND METHODS
Protein Expression and Purification
Ndc10p, Ctf13p/Skp1p, and Cep3p-containing extracts were prepared from nuclear lysates of baculovirus-infected Hi5 insect cells (Kaplan et al., 1997). These extracts were analyzed in bandshift assays with radiolabeled probes, as described previously (Sorger et al., 1995). To purify Ndc10p, Hi5 cell lysates were prepared 36 h postinfection, centrifuged to remove particulate material, and applied to a cation exchange POROS HQ column (Applied Biosystems, Foster City, CA) in 150 mM KCl, 10 mM HEPES pH 8.0, 20 mM β-glycerophosphate, 20 mM NaF, 10% glycerol, 0.1% β-mercaptoethanol and eluted with 600 mM KCl in the same buffer. Fractions containing Ndc10p were collected and the KCl concentration adjusted to 150 mM. The pooled fractions were then applied to an anion-exchange POROS HS column (Applied Biosystems) and eluted with 600 mM KCl. Fractions were again pooled on the basis of Ndc10p amount as judged by SDS-PAGE. Anti-myc antibody (9E10) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids and Probes
Bandshift probes were generated as described previously (Espelin et al., 1997) from subcloned fragments in plasmids or directly from genomic DNA. CEN3 mutants were generated from pRN505 as follows: the CDEIM probe includes a CDEI mutation from CACATG to CATTATG and CDEIIIM probes delete the central CCG of CDEIII (Sorger et al., 1995). Probes for the experiments in Figure 2 were generated using PCR “mega-priming” with the final products subcloned into vectors and sequenced. A fragment of the tetracycline resistance gene from pBR322 was used as a source of “random” DNA in substitution and linker scanning mutations. Chromatin immunoprecipitation (ChIP) was performed as described previously (Megee et al., 1999; Tanaka et al., 1999).
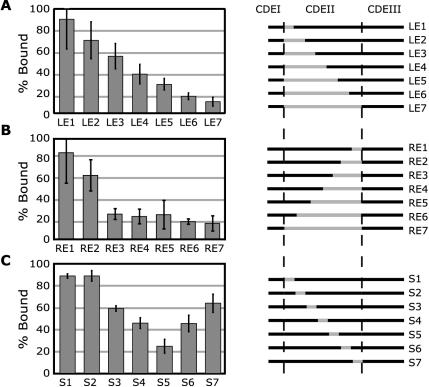
Figure 2.
Mapping the Ndc10p binding site in CDEII by substitution and linker scanning mutagenesis. The amount of binding of each 184-base pair probe to 2 pmol purified Ndc10p is shown as a percentage of binding to a wild-type CEN3 control. In the diagrams to the right of each bar graph, wild-type CEN3 sequences are denoted in black and fragments of pBR322 or linker DNA with gray. (A) Substitutions from the left (CDEI; Left Extension) side of CEN3 in 12-base pair steps. (B) Substitutions from the right (CDEIII; Right Extension) side of CEN3 in 12-base pair steps. (C) Linker-scanning mutations in successive 12-base pair steps across CDEII (Scanning).
Strains and Chromosome Loss Assay
Yeast strains were derived from W303 (MATa, ade2-1, trp1-1, can1-100, leu2-3112, his3-11,15, ura3, GAL, psi+). The strain harboring Ndc10p-myc6 was produced by transformation of W303 by NDC10-myc6::TRP1 (gift of S. Piatti, Universita degli Studi di Milano-Bicocca, Italy). The Mcd1p-myc18 strain was a gift of the Nasmyth laboratory (K6565: W303 MATa, ade2-1, can1-100, leu2-3112, GAL, psi+, MCD1-myc18::TRP1). Strains for chromosome loss assays were generated as described previously (Clarke et al., 1983) by transforming W303 haploid cells with constructs containing variant CEN sequence and a linked URA3 marker and subsequent mating with cells of the opposite mating type. Correct integrations were confirmed by PCR genotyping. The chromosome loss assay was performed as follows: cells were grown in selective media (SD-URA) and then plated immediately (t = 0) or grown under nonselective conditions for 7 or 13 generations in YPD before plating on SD-URA and YPD. Results shown are the average of three separate cultures and 200–400 colonies analyzed for each strain. Generations were monitored by OD600 and cell number. Cells were counted by hemacytometer for plating. The fraction of Ura+ cells was determined from the ratio of colonies obtained by plating on SD-URA and YPD. (Analysis was also performed using 5-fluoro-orotic acid SD-complete plates with similar results. PCR analysis of several randomly chosen colonies from 5-fluoro-orotic acid plates confirmed loss of the chromosome carrying the CDEII mutation; our unpublished data).
Pattern Identification Program
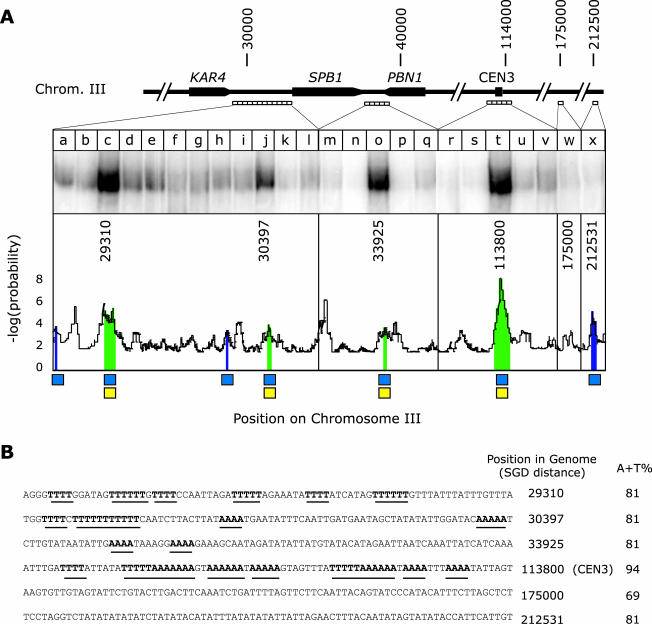
To analyze CDEII sequences, a program was written that applies a sliding, nonoverlapping window of 80 nucleotides (similar in length to CDEII) to the sequence of Chromosome III; this analysis uncovered 71 sites of high A-T composition (>80%). Four of these sites were selected at random for bandshift analysis (Figure 3, lanes c, j, o, and x) along with a randomly selected chromosomal site as a control (Figure 3, lane w). Bandshift probes were 200 bases in length and covered the 80-base pair CDEII-matched property window along with 60 flanking bases on both sides. The probability of a particular A + T composition is derived from the distribution observed in the yeast genome. Regions of DNA containing stretches of poly-A or poly-T were compared with each other by using simulation: for a particular content of adenine, the sequence was randomized and number of stretches counted; from this distribution probabilities were determined. We compute that CDEII-like sites occur on average every ∼5 kb.
Figure 3.
Binding of recombinant Ndc10p to sequences on Chromosome III arms and in CEN3. (A) DNA probes in 200-base pair increments were generated across the indicated regions in the KAR4-PBN1 interval and analyzed for binding to purified recombinant Ndc10p. Distances are indicated in nucleotides from the left telomere of Chromosome III. The graph shows the product of the log probability of the A-T composition in successive 80-base pair intervals and the log probability of the number of stretches containing four or more adenines or thymines in a row. To find potential Ndc10p binding sites, sequences were sought in which the A-T composition exceeded 80% (blue boxes) and the number of poly-A/poly-T stretches was significantly above average for the genome (yellow boxes). Ndc10p binding was predicted to occur on sequences in which both of these thresholds had been exceeded (green regions). (B) Sequences of six 80-base pair windows in Chromosome III analyzed for Ndc10p binding in vitro. Position refers to the number of the first nucleotide in the window relative to the left telomere of Chromosome III (Cherry et al., 1997) and “A + T” % to the percentage of adenine plus thymine. Stretches of A and T are emphasized in bold and underlined. Sequence 113800 is CDEII of CEN3; 175000 (lane w) represents a randomly chosen genomic location; 212531 is a site of high A-T content but lacking poly-A/poly-T stretches.
RESULTS
Ndc10p Binds to CDEII
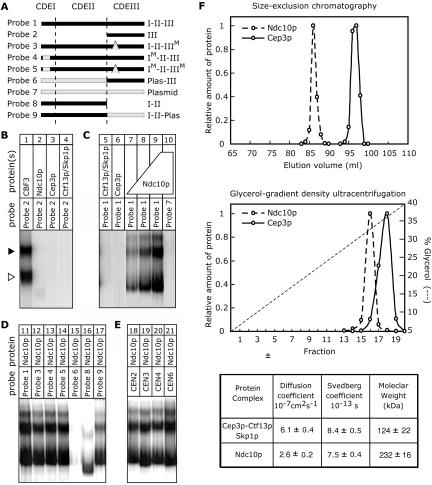
To identify proteins that associate specifically with CDEII DNA, we performed in vitro binding experiments using wild-type and variant CEN3 probes that cover different regions of the S. cerevisiae centromere (Figure 1A). Both whole-cell lysates from S. cerevisiae and recombinant kinetochore proteins expressed in insect cells were used as sources of potential centromere-binding proteins. Consistent with our previous findings, when insect cell extracts containing recombinant Ndc10p, Cep3p or Ctf13p-Skp1p were mixed and incubated with an 88-base pair CDEIII probe (probe 2), we observed a rapidly migrating CBF3 species and a second more slowly migrating species (Figure 1B, lane 1; Espelin et al., 1997). Both of these complexes contain all four CBF3 subunits but the fast-migrating species has a single Ndc10p dimer, whereas the slow-migrating species has two dimers. As expected, we saw no complexes when extracts containing only one or two CBF3 proteins were present in the binding reaction (Figure 1B, lanes 2–4; Kaplan et al., 1997). A strikingly different result was obtained with a 184-base pair probe spanning all of CEN3 (probe 1). When Ndc10p alone was mixed with probe 1, several distinct DNA–protein complexes formed (Figure 1C, lanes 7–9) but none was detected with Ctf13p-Skp1p alone or Cep3 alone (Figure 1C, lanes 5 and 6). Moreover, complexes did not form when Ndc10p was incubated with a randomly selected fragment of pUC19 (Figure 1C, lane 10), suggesting that the Ndc10p–probe 1 complex is sequence selective. To measure this sequence selectivity, mixed sequence salmon sperm DNA was titrated into Ndc10p-probe 1 binding reactions. Binding of Ndc10p to CEN3 DNA (50 fM) was 50% inhibited at a salmon sperm DNA concentration of 20 mg/ml (our unpublished data). Thus, mixed sequence DNA can compete with probe DNA for Ndc10p binding only at very high concentrations. The microscopic sequence selectivity for Ndc10p binding to CEN3, relative to mixed sequence DNA, is ∼2 × 105. This is 10-fold lower than that of CBF3 for CDEIII (Espelin et al., 1997) but typical for many sequence-specific DNA binding proteins. From these data, we conclude that Ndc10p binds in a sequence-selective manner to intact CEN3 DNA in the absence of other proteins but does not bind to DNA containing only CDEIII, the binding site for CBF3.
Figure 1.
Binding of CBF3 and Ndc10p to CDEII and CDEIII centromeric DNA. Nuclear extracts from insect cells expressing recombinant proteins were incubated with radiolabeled DNA fragments and complexes resolved on nondenaturing Bandshift gels (see MATERIALS AND METHODS). Free probe has run off the bottom of the gel. (A) Diagram of CEN3-derived probes used in this study and their names, as used in the text. Dashed lines denote approximate boundaries between CDEI, CDEII, and CDEIII. “M” indicates a point or linker-scanning mutation as described in MATERIALS AND METHODS. Probe 2 is 88 base pairs in length, probe 8 is 96 base pairs, and all others are 184 base pairs. (B) Binding of recombinant CBF3 proteins to CDEIII DNA (probe 2). “CBF3” (lane 1) denotes a binding reaction containing a mixture of three nuclear extracts from Hi5 cells expressing Ndc10p, Cep3p, or Ctf13p/Skp1p. Binding reactions in lanes 2–4 contain only one extract with the proteins indicated. The open arrowhead marks the position of the CBF3-CDEIII core complex and the solid arrowhead the extended CBF3 complex (Espelin et al., 1997). (C) Binding of recombinant CBF3 proteins to intact centromeric DNA (probe 1) and a plasmid control (probe 7). Ndc10p was purified from insect cell lysates by ion exchange chromatography and added at 0.4 pmol (lanes 7), 1 pmol (lane 8), or 2 pmol (lane 9–10). Binding of 2 pmol of purified Ndc10p to the indicated probes (D) and to 184-base pair fragments of CEN2, CEN3, CEN4, and CEN6 (E) that span CDEI-II-III (probe 1). (F) Analysis of whole-cell yeast extracts was performed using a Sephacryl S-500 HR column (Amersham Biosciences, Piscataway, NJ) and glycerol velocity gradients as described previously (Russell et al., 1999) followed by immunoblotting with anti-Cep3p antibodies and anti-myc antibodies (for Ndc10p-myc6). The table shows estimated hydrodynamic properties (see Russell et al., 1999 for details).
Point mutations in CDEI or CDEIII disrupt association with Cbf1p and CBF3, respectively (McGrew et al., 1986; Ng and Carbon, 1987; Baker and Masison, 1990; Cai and Davis, 1990; Neidenthal et al., 1991). Incorporation of these mutations into CEN3 either singly (probes 3 and 4) or in combination (probe 5) had no detectable effect on Ndc10p binding (Figure 1D, lanes 12–14). However, when an 85-base pair fragment of plasmid DNA was inserted in place of 85-base pairs of CDEII, Ndc10p binding was abolished (Figure 1D, lane 15). This result suggests that Ndc10p binds to sequences in CDEII in a sequence-specific manner. To investigate whether CDEII is sufficient for Ndc10p binding, we used a 127-base pair sequence comprising only wild-type CDEI + II of CEN3 (probe 8). Ndc10p bound to this DNA fragment, but more weakly than full-length CEN3 (probe 1). Suspecting that diminished Ndc10p binding might reflect the shorter length of probe 8 relative to intact CEN, we generated a 184-base pair probe (probe 9) in which 88 base pairs of CDEIII were substituted with an equal length fragment of plasmid DNA while maintaining wild-type sequence for CDEI and CDEII. Ndc10p bound tightly to probe 9 (complex formation was ∼75% as efficient as for probe 1), leading us to conclude that DNA flanking CDEII is involved in nonsequence-specific Ndc10p binding. CDEII binding by Ndc10p is not a phenomenon limited to CEN3 because a similar binding pattern was obtained with probes (probe 1) derived from four different centromeres (Figure 1E). Thus, binding to CDEII by Ndc10p seems to be a conserved feature of yeast kinetochores.
These findings with recombinant proteins indicate that Ndc10p is capable of existing independently of the other CBF3 components and that this Ndc10p is active in CDEII binding. To demonstrate that this situation exists within a cell, whole-cell protein extracts were prepared from wild-type yeast and subjected to hydrodynamic and bandshift analysis. Sizing columns and glycerol velocity gradients demonstrate that Ndc10p exists as a distinct species in yeast extract (Figure 1F). When incubated with CDEII-containing DNA (Probe 9), these extracts gave rise to set of complexes whose electrophoretic mobilities were indistinguishable from those of purified recombinant Ndc10p (our unpublished data). Combined with our analysis of recombinant proteins also showing that Ndc10p can exist both as a homodimer on its own and as part of CBF3 (Russell et al., 1999; our unpublished observations), we conclude that unbound Ndc10p exists as a dimer and Cep3p is part of a 185-kDa complex containing two Cep3p, one Ctf13p and one Skp1p subunits. From these observations we conclude that Ndc10p dimers represent the primary CDEII-binding activity that can be detected in yeast extracts by using bandshift gels.
Mapping Ndc10p Binding Sites in CDEII
To delineate the Ndc10p binding site in CDEII, we constructed a series of CEN3 variants by substituting progressively larger regions of CDEII with plasmid DNA while maintaining the wild-type sequence of CDEI and CDEIII and the spacing between them (Figure 2). When a substitution series from the left (starting at CDEI; LE1–7) was examined, Ndc10p-DNA binding decreased progressively (Figure 2A). When substitutions from the right (starting at CDEIII; RE1–7) were examined, the most dramatic effects occurred with mutations covering the first 36 base pairs (Figure 2B; compare RE2 and RE3). From experiments with a set of linker-scanning mutations (S1–7), the CDEII region ∼36 base pairs to the left of CDEIII seems to be the most critical for Ndc10p binding (Figure 2C, S5), but no single 12-base pair mutation abolished binding completely. We conclude from these data that Ndc10p-CDEII binding interactions are dispersed across the length of CDEII with particularly important contacts near the middle. We propose that Ndc10p makes extended DNA contacts and that multiple Ndc10p dimers participate in CDEII binding. The latter suggestion is consistent with the detection bandshift of more than one distinct Ndc10p–DNA complex in our experiments (Figure 1C). We have previously come to similar conclusions about the capacity of Ndc10p to form higher order complexes on CDEIII, in association with CBF3 (Espelin et al., 1997).
Properties of Ndc10p Binding Sites
Does CDEII contain a series of Ndc10p sites in tandem? The region of CDEII most critical for Ndc10p association (bases 49–60 of CDEII; Figure 2C probe S5) does not represent a motif found elsewhere in CDEII. It therefore seems likely that Ndc10p binds to a family of related but nonidentical AT-rich sequences. To capture this sequence selectivity computationally, we used a supervised learning algorithm to derive a pattern that extracts the characteristic features of CDEII in all 16 S. cerevisiae centromeres. The yeast genome has an overall A + T composition of 61% but the distribution of A-T bases is nonrandom. When an 80-base pair window (the approximate length of CDEII) is used to scan the genome, more regions with very high or very low A + T content are found than expected by chance (∼100 times more frequent). Moreover, for any particular A-T composition, stretches of poly-adenine or poly-thymine are also more common than expected. The 16 CDEII sequences in S. cerevisiae are characterized both by high A-T content (averaging 92%), by the presence of 4–6 poly-A and poly-T stretches, and by a length of 78–84 base pairs.
Are there sequences at locations in the genome other than centromeres that match the CDEII pattern? When we scanned Chromosome III for matches to the CDEII pattern, 71 sites were found with a mean spacing of 4.5 kb (a similar density was found elsewhere in the genome; see MATERIALS AND METHODS). A scan for the CDEIII pattern yielded only the single centromere present on each chromosome. Within the selected region of the Chromosome III arm shown in Figure 3, four matches to the CDEII pattern are found (including CEN3; Figure 3, A and B). These sites are characterized both by high A-T content (marked with blue squares) and poly-A/poly-T stretches (yellow squares). A highly AT-rich sequence is also present but it lacks poly-A and poly-T stretches (Figure 3; fragment SGD 212531). To determine whether Ndc10p can bind to these noncentromeric sequences, we generated 25 successive 200-base pair probes spanning the identified sequences and flanking DNA and analyzed Ndc10p binding to the probes on nondenaturing gels. Ndc10p associated efficiently with the three CDEII-like arm sequences (Figure 3, lanes c, j, and o) and to CEN3-CDEII (Figure 3, lane t), but not to intervening DNA, a random genomic sequence (Figure 3 lane w) or to the control AT-rich site (Figure 3, lane x). In addition, the DNA–protein complexes formed on arm and CDEII sequences had similar electrophoretic mobilities and the two classes of sequence (arms and CEN) cross-competed with one another (our unpublished data). Thus, the CDEII pattern generated by computer learning captures the DNA features responsible for Ndc10p binding, even though the pattern does not correspond to a simple consensus. In summary, our results support the conclusion that Ndc10p binds in the absence of other CBF3 proteins, specifically to a subset of AT-rich, poly-A–, and poly-T–containing sequences similar to CDEII. The CDEII pattern is present considerably more often in the genome (∼1000 times more frequent) than would be predicted by chance and Ndc10p binding sites may therefore be present along chromosome arms away from centromeres.
Increased Chromosome Loss Associated with Mutations in Ndc10p Binding Sites
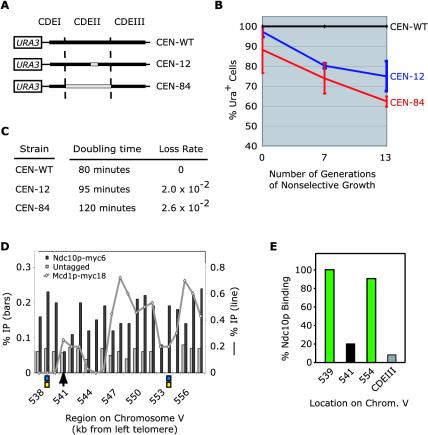
To begin to establish a biological function for the binding of Ndc10p to CDEII, we asked whether mutations that affect Ndc10p-CDEII binding in vitro also affect centromere function in vivo. A subset of CEN3 constructs containing wild-type or mutant sequences in combination with the URA3 gene were recombined into Chromosome III in diploid cells, replacing the natural centromere on one chromosome with a modified sequence while maintaining the wild-type centromeric sequence on the other chromosome (Figure 4A). We then determined the percentage of URA+ cells by plating on SD-URA and YPD after 0, 7, and 13 generations of nonselective growth in YPD (Figure 4B) and determined the loss rate of the mutated chromosomes (Figure 4C). The loss rate of wild-type centromeres was essentially zero after 13 generations of nonselective growth (Figure 4, B and C). Alteration of bases “49–60” in the middle of CDEII, the mutation with the greatest reduction in Ndc10p binding in vitro, drastically decreased the mitotic stability of Chromosome III, resulting in a loss rate of 2 × 10-2 per generation (Figure 4, B and C). When CDEII was replaced in its entirety with plasmid (Tetr) DNA, mitotic chromosome segregation fidelity was again severely compromised, with an estimated loss rate of 2.6 × 10-2 per generation observed (Figure 4, B and C). Centromere function in Chromosome III could be substantially rescued, although not up to wild-type levels, by inserting a DNA sequence from an Ndc10p binding site from the Chromosome III arm, whereas an AT-rich sequence that lacks Ndc10p binding activity in vitro rescued CEN function much less efficiently (our unpublished data). However, these data must be interpreted with some caution. The presence of chromosomes with CDEII mutations causes a substantial increase in cell-cycle doubling time (Figure 4C), apparently as a consequence of a mitotic delay that is Mad2 dependent. Moreover, starting cultures seem to be quite aneuploid, probably as a consequence of chromosome nondisjunction. Both of these problems are much more severe in our hands with CDEII mutations than with CDEIII mutations, and CDEII mutant chromosomes clearly warrant further investigation by live cell analysis (He et al., 2001). Nevertheless, our data do show that CDEII sequences that bind Ndc10p in vitro are important for centromere function in vivo and that Ndc10p binding sites from chromosome arms can function at the centromere. We have therefore established a correlation between Ndc10p activity in vitro and CDEII function in vivo (see DISCUSSION).
Figure 4.
In vivo analysis of Ndc10p–DNA interactions. (A) Schematic of centromeric sequences carrying CDEII mutations and URA3 used to evaluate chromosome loss. Wild-type regions are shown in black and mutant regions in gray. (B) Chromosome loss in cells carrying mutant centromeres on a URA3-marked chromosome after 0, 7, and 13 generations of nonselective growth in YPD. % Ura+ cells represents the ratio of colonies on SD-URA and YPD plates for each time point. Values represent the average of three individual cultures of each strain along with the SE of the mean. (C) Doubling time under nonselective conditions (YPD); chromosome loss rates determined from B. (D) Binding of Ndc10p and the cohesin subunit Mcd1p to a region of Chromosome V previously subjected to detailed analysis (Tanaka et al., 1999) by ChIP in 200-base pair DNA fragments spaced every 1 kb. A wild-type strain lacking a tagged protein (Untagged) serves as negative control for the level of background cross-linking. Percentage of cross-linking (relative to total input DNA) of Ndc10p-myc6 and the wild-type (Untagged) to Chromosome V are graphed against the left axis (vertical bars); percentage of cross-linking of Mcd1p-myc18 is graphed against the right axis (line) (separate axes were used because absolute ChIP values differed). The sites indicated by the boxes and arrow correspond to sites used in the bandshift assay in E. (E) In vitro binding of Ndc10p to Chromosome V sites with high and low levels of ChIP cross-linking. CDEIII serves as a negative control.
Ndc10p Association with Chromosome Arms
Does Ndc10p bind to chromosome arms in vivo as suggested by our in vitro data? To investigate this question, we performed ChIP analysis on a region of Chromosome V that had previously been examined in detail for the binding of cohesins (Tanaka et al., 1999). The cohesin proteins Mcd1/Scc1p, Scc3p, Smc1p, and Smc3p (Strunnikov et al., 1993; Guacci et al., 1997; Michaelis et al., 1997; Skibbens et al., 1999; Toth et al., 1999) link sister chromatids together during metaphase and associate preferentially with AT-rich DNA (Blat and Kleckner, 1999; Megee et al., 1999; Tanaka et al., 1999). It therefore seemed possible that the cohesins and Ndc10p might bind to the same arm sequences. When the Chromosome V region was scanned for potential Ndc10p binding sites, several strong matches were identified. To examine the region for cohesin and Ndc10p binding in vivo by ChIP, yeast cells expressing either Mcd1p-myc18 or Ndc10p-myc6 in place of the wild-type proteins were cross-linked with formaldehyde, extracts prepared, DNA sheared to an average length of 500 base pairs, and the myc-tagged proteins isolated by immunoprecipitation. Extract from cells lacking a myc-tagged protein (untagged) served as a negative control. Cross-links were hydrolyzed and DNA sequences associated with Ndc10p-myc6 and Mcd1p-myc18 were detected by PCR (method of Megee et al., 1999; Chromosome V oligos used by Tanaka et al., 1999). We observed that Ndc10p-myc6 cross-linked strongly to CEN5 DNA (our unpublished data), as expected, and to different extents to DNA along the Chromosome V arms (Figure 4D). The peaks of cross-linking were significantly above background levels observed with an untagged strain and corresponded to sequences that were positive for Ndc10p binding in vitro (Figure 4E). The Ndc10p cross-linking peaks did not correspond (or alternate with) peaks of Mcd1p binding, however, arguing against a connection between Ndc10p and cohesin (Figure 4D). Furthermore, binding of Ndc10p to arm sites was at best three- to fourfold above the untagged control and 5- to 10-fold lower than the level of Ndc10p cross-linking to centromeres. We have repeated this experiment with many variations using synchronized and asynchronous cultures and in each case obtained similar results: Ndc10p-myc6 cross-links more strongly to some arm sites than others, cross-linking is well above background levels in untagged strains, but the absolute level of cross-linking is low. One explanation for the low but reproducible signal along arms is that the epitope tag used for Ndc10 immunoprecipitation is less accessible on arms than at centromeres. A second, more likely explanation, is low fractional occupancy of Ndc10p on arm sites. Consistent with this idea, the on-off rate of Ndc10p at CDEII in vitro is high (seconds per minute as opposed to hours for CBF3 on CDEIII; our unpublished data). In summary, ChIP data are consistent with noncentromeric binding by Ndc10p but short of conclusive.
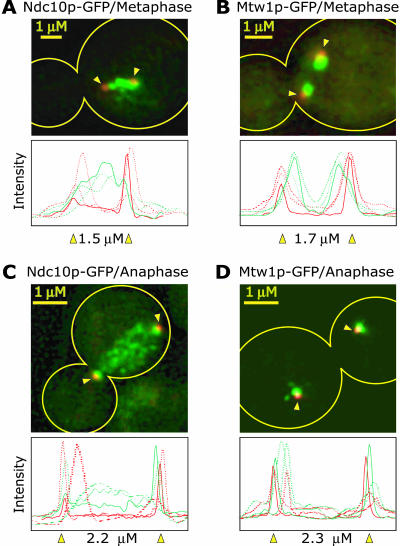
We therefore sought, using high-resolution imaging, to obtain independent confirmation that Ndc10p is present at cellular locations other than centromeres. Asynchronous cultures of cells carrying Ndc10-GFP were compared with cells in which known kinetochore proteins were similarly tagged. To determine the position of the spindle as well as verify cell cycle status, the spindle poles were marked with Spc42-CFP and three-dimensional images were obtained from live and fixed cells by using deconvolution microscopy. In other work from this laboratory, high-resolution imaging has shown that kinetochore proteins localize during metaphase to two lobes on either side of the spindle mid-zone (Goshima and Yanagida, 2000; He et al., 2001). These lobes correspond to the average positions of congressed sister kinetochores, and thus to the budding yeast metaphase plate. The localization of one of these kinetochore-specific proteins, Mtw1p-GFP, is shown in Figure 5, B and D (Goshima and Yanagida, 2000; He et al., 2001). The apparent overlap of Mtw1p-GFP lobes and spindle poles is an artifact of projecting a three-dimensional distribution onto two dimensions (He et al., 2000). Some kinetochore proteins have a more complex distribution than Mtw1p, being localized not only to lobes of congressed kinetochores but also to nuclear microtubules, spindle pole bodies, and cytoplasmic structures (Bik1p, Stu2p, and Ipl1p; He et al., 2001; Tanaka et al., 2002).
Figure 5.
Localization of Ndc10-GFP and Mtw1p-GFP in metaphase and anaphase cells. Ndc10p and Mtw1p, a known kinetochore protein (Goshima and Yanagida, 2000), were tagged with GFP (green) and the spindle pole component Spc42p was tagged with CFP (red; indicated by yellow arrowheads). Maximum intensity projections of three-dimensional image stacks containing 10 to 20 0.2-μm sections are shown representing typical images. The outline of the cell is indicated in yellow. All cells were exposed similarly and images have been adjusted to give the most accurate comparison. The graphs show the GFP and CFP fluorescence (raw pixel intensities) integrated along the spindle axis for three different representative cells with the solid line and indicated spindle length representing data from the image shown.
Among the localization patterns, we have observed for ∼25 kinetochore proteins examined thus far, Ndc10p is unique. Ndc10p is found both in a region of the nucleus consistent with congressed kinetochores and is also present along much of the chromatin (Figure 5A). In early anaphase spindles, Ndc10p-GFP fluorescence is visible in a broad distribution similar to that of DAPI-stained DNA (Figure 5C; colocalization of Ndc10p and DAPI has not been possible because of the much greater brightness of the DAPI-stained DNA, but the pattern of Ndc10p localization is very similar). Somewhat later in anaphase, Ndc10p is also visible along the interpole microtubules, a pattern that has been noticed by others (our unpublished observations; Zeng et al., 1999). In contrast, in control cells carrying only tagged spindle poles and imaged under identical conditions, neither chromatin nor microtubule fluorescence are visible, confirming that they are specific to Ndc10p-GFP (our unpublished observations). These data argue strongly that Ndc10p is present in nuclei not only at centromeres but also at other structures that probably correspond to chromosome arms.
DISCUSSION
Homologies between kinetochore proteins in higher and lower eukaryotes argue for substantial evolutionary conservation in kinetochore architecture. Centromeric DNA is strikingly different among different species, however, consisting of as few as 125-base pairs of specific DNA in budding yeast and as many as several megabases in humans. One of the central mysteries in mitosis is the molecular basis of this radical divergence in centromere organization within the context of substantial conservation in kinetochore composition. One way to clarify this mystery is to uncover the molecular principles of yeast centromere organization and to use these principles to study complex animal cell centromeres. The least well understood of the three sequence elements in S. cerevisiae centromeres is CDEII. Cbf1p has been shown to bind CDEI (Jiang and Philippsen, 1989; Cai and Davis, 1990; Mellor et al., 1990) and CBF3 to bind CDEIII (Lechner and Carbon, 1991; Espelin et al., 1997), but no CDEII-interacting proteins have been identified thus far. A key finding from a large number of studies is that the association of all known kinetochore proteins with centromeric DNA depends on functional Ndc10p (Ortiz et al., 1999; He et al., 2001; Janke et al., 2001). In this work, we propose that the critical role for Ndc10p is a consequence of its sequence-specific binding to both CDEII and CDEIII.
DNA Binding Activities of Ndc10p
Previous analysis of Ndc10p focused on its binding to CDEIII DNA as part of a CBF3 core complex containing a homodimer of Ndc10p, a homodimer of Cep3p, and a heterodimer of Skp1p and Ctf13p. When assembled into CBF3, Ndc10p, Ctf13p, and Cep3p are in direct contact with CDEIII DNA, as judged by DNA–protein cross-linking, but none of these proteins can bind CDEIII on its own (Figure 6; Espelin et al., 1997). We now show that Ndc10p interacts with CDEII in the absence of other CBF3 proteins. This binding includes an extended series of sequence-specific contacts more than ∼80 base pairs of CDEII as well as sequence-independent contacts with flanking DNA. In addition, three or more CDEII–Ndc10p complexes differing in electrophoretic mobility form in vitro (depending on Ndc10p concentration), probably representing different numbers of Ndc10p dimers on each DNA molecule. It is further notable that the in vivo loss rate of chromosomes carrying a specific 12 base pair mutation in CDEII, which markedly reduces in vitro binding of Ndc10p, is similar to the loss rate observed by replacing all of CDEII with plasmid DNA. This suggests that we have disrupted a key function of CDEII, which we believe is the ability to bind Ndc10p. Together, these data suggest that Ndc10p forms a multimeric structure and makes dispersed contacts throughout the length of CDEII. A similar type of binding by Ndc10p also occurs in CDEIII. Although there is only one Ndc10p dimer in the CBF3 core (Russell et al., 1999), a second dimer associates with the core and flanking DNA to generate an extended CBF3 complex (Espelin et al., 1997).
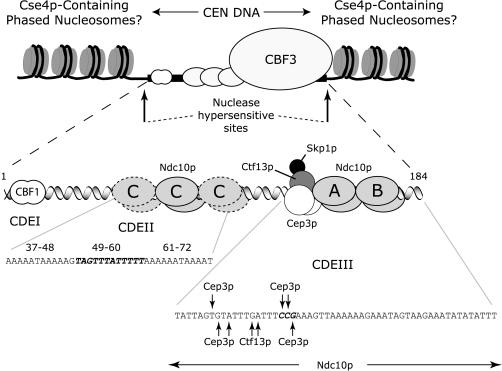
Figure 6.
Speculative model for the interaction of Ndc10p, CBF3, and nucleosomes with yeast centromeres. Three different modes of Ndc10p–DNA interaction are shown. The extended CBF3 complex on CDEIII involves sequence-specific binding by one Ndc10p dimer (marked A) and sequence-independent binding by a second dimer (marked B; Espelin et al., 1997; Russell et al., 1999). We propose that multiple Ndc10p dimers bind sequence-selectively to CDEII (marked C), with particularly important binding by one dimer (C, solid outline) to positions 49–60 of CDEII (Figure 2). The sites of cross-linking by CBF3 subunits to bases in CDEIII are indicated by arrows (Espelin et al., 1997). This CBF3–Ndc10p–CEN DNA complex is proposed to be flanked by nuclease hypersensitive sites and embedded in a region of phased nucleosomes that contain the specialized H3 protein Cse4p (Bloom and Carbon, 1982; Bloom et al., 1983; Funk et al., 1989). No attempt has been made to speculate on the overall folding of centromeric chromatin, but it seems likely from studies in other organisms that it adopts a special structure.
Our current model is that Ndc10p has multiple functions in kinetochore assembly involving three modes of DNA binding (Figure 6): sequence-selective binding to CDEII in the absence of other proteins (this work), sequence-selective binding to CDEIII in the context of a CBF3 core complex (Espelin et al., 1997), and sequence-independent but CBF3-dependent binding to centromere-distal sequences in the context of the extended CBF3 complex (Espelin et al., 1997). We have generated a large series of Ndc10p truncation and point mutations and tested them for binding to both CDEII and CDEIII but have found no separation-of-function mutations that retain one type of binding but eliminate another (our unpublished data). We therefore believe that all three binding modes involve a single DNA binding domain, rather than multiple distinct domains. There are many precedents for the binding of a protein to DNA in more than one context. Transcription factors such as activator protein-1 can associate with a variety of other DNA-binding proteins on complex regulatory sites (Shaulian and Karin, 2002). The yeast transcription factors, Ste12p and Pho4p, can bind DNA as homomultimers and bind as cooperative complexes with other proteins (Baur et al., 1997; Magbanua et al., 1997). Consistent with binding in several modes, Ndc10p exist as a free dimer in whole-cell yeast extracts.
Potential CDEII Binding Proteins
Our data argue that Ndc10p binds to both CDEII and CDEIII DNA in vivo, but proteins other than Ndc10p have previously been proposed to be CDEII interactors. These proteins include ubiquitous AT-binding proteins and the kinetochore-specific proteins Mif2p, the yeast homolog of mammalian CENP-C (Meluh and Koshland, 1995; Meluh and Koshland, 1997), and Cse4p, the yeast homolog of the variant mammalian histone H3 CENP-A (Pluta et al., 1995; Stoler et al., 1995; Smith et al., 1996; Meluh et al., 1998). Although it is possible, and even likely, that more than one kinetochore protein has CDEII-binding activity, it is informative to compare the candidates. A variety of ubiquitous proteins bind to A-T tracts, including DATIN (Winter and Varshavsky, 1989), CMBF (Horn et al., 1999), and HMG-I(Y) (Reeves and Nissen, 1993), but none have been shown to have a specific role in kinetochore function. DATIN specifically has been shown to be incapable of CDEII binding in vitro (Winter and Varshavsky, 1989). The discussion below therefore focuses on Ndc10p, Mif2p, and Cse4p. In considering the evidence discussed below it is important to note that none of these three proteins has been shown to be capable of binding to CDEII in vivo in the absence of a functional CDEIII sequence. It therefore seems that the binding of all kinetochore proteins (including the proposed CDEII-bound Ndc10p polymer) requires that CBF3 be associated with CDEIII. Based on our current understanding of transcription, it seems likely that one important function of CDEIII is to disperse nucleosomes that might otherwise make centromeric DNA inaccessible to other binding factors.
We have obtained strong biochemical evidence that Ndc10p binds in a sequence-specific manner to CDEII in vitro but cannot yet determine the precise function of this binding in vivo. By comparison, the function of the CDEIII–CBF3 interaction has been firmly established by multiple in vitro and in vivo experiments. CDEIII is a relatively short and well-conserved sequence in which single point mutations abolish activity. When a range of point mutations can be made in a sequence and activities in vitro and in vivo correlated, a strong argument for the biological function of a particular DNA–protein interaction can be made. In the case of CDEII, however, the distributed nature of a site in which no single linker-scanning block mutation eliminates binding, makes it hard to draw as tight a link between biochemical and cellular activities. However, we have uncovered some of the key features of Ndc10p–CDEII association. CDEII mutations that impair Ndc10p binding in vitro impair centromere function in vivo. An algorithm that extracts the sequence features of CDEII identifies noncentromeric DNA that is active in Ndc10p binding in vitro, and at least partially active in substituting for CDEII at centromeres in vivo. Additionally, we have attempted to use ChIP to map Ndc10p binding to CDEII and CDEIII in vivo and to evaluate the effects of CDEII mutations on Ndc10p-CEN association. However, the difficulty in resolving immediately adjacent sequences by ChIP has made it impossible to distinguish between binding to CDEII and to CDEIII. Nevertheless, we conclude that a biological role for Ndc10p in CDEII binding is likely.
The argument that Mif2p binds to CDEII is largely based on its homology to the mammalian centromere-binding protein CENP-C (Meluh and Koshland, 1995) and the presence of an “AT-hook” motif proposed to assist in binding DNA (Brown et al., 1993). However, the region responsible for DNA binding in CENP-C (Politi et al., 2002) is not conserved in Mif2p, and we have found that Mif2p lacking the AT-hook seems to be fully functional in vivo (our unpublished data). We have also examined recombinant Mif2p in vitro, but have not detected binding to CEN DNA, either in the presence or absence of CBF3. Furthermore, Mif2p has been shown to be incapable of binding CDEI+II DNA in vivo as determined by ChIP (Meluh and Koshland, 1997). Thus, although it is clear that Mif2p is bound to centromeric DNA (Meluh and Koshland, 1997), the evidence is not strong that this involves a specific interaction with CDEII DNA as opposed to association with other kinetochore components.
Cse4p, a specialized histone H3, has also been proposed to be a CDEII-binding protein. This proposal is based on interpretation of ChIP results (Meluh et al., 1998) and the existence of genetic synergy between cis-acting mutations in CDEII and trans-acting mutations in CSE4 (Smith et al., 1996; Keith and Fitzgerald-Hayes, 2000), but no direct interaction has been demonstrated. Given the small size of the yeast centromere and the proximity of CDEII and CDEIII relative to the resolution of ChIP (∼200–500 base pairs), it is difficult, if not impossible to determine that Cse4p is binding CDEII using this method. Enhanced cross-linking of Cse4p to the center of CEN DNA (Meluh et al., 1998) would be likely if Cse4p-containing nucleosomes flank the centromere on both sides. Furthermore, Ortiz et al. (1999) have shown that Cse4p can be cross-linked by ChIP to a CEN3 fragment that includes CDEIII without CDEII, showing that CDEII is not necessary for Cse4p recruitment to centromeres. Classic experiments describing the nature of nucleosomes (Kornberg and Klug, 1981) dictate that changes in the number of nucleotide bases alter the phasing of DNA. Were core centromeric DNA sequences wrapped around a nucleosome, changes in the length of CDEII would have a dramatic effect on the ability of kinetochore proteins to make specific DNA contacts and would thus alter their orientation and interaction with other kinetochore proteins. Previous data demonstrate binding of the kinetochore proteins Cep3p, Ctf13p, Ndc10p, and Cbf1p (Espelin et al., 1997; Cai and Davis, 1990) to both sides of the DNA, making it a steric improbability that kinetochore proteins could bind on the face of the nucleosome and not be affected by the length of CDEII (Bloom et al., 1989; Meluh et al., 1998; Cheeseman et al., 2002). Nucleosomes containing Cse4p should also be similar in their binding specificities to conventional nucleosomes because the Cse4p-specific sequences, particularly the extended N terminus, project away from the DNA (Luger et al., 1997). Conventional nucleosomes are known to bind poorly to sequences containing stretches of Adenine followed by stretches of Thymine (Kunkel and Martinson 1981; Prunell 1982). Although not excluded from nucleosomes, AT-rich DNA tends to be found at the ends of DNA wrapped around histones (Satchwell et al., 1986), and CDEII would therefore seem to be a poor nucleosome binding sequence.
These considerations raise questions about the widely held assumption that Cse4p binds specifically to CDEII (Keith and Fitzgerald-Hayes, 2000; Cheeseman et al., 2002). Instead, we put forward the hypothesis that CDEII is bound by a polymeric Ndc10p complex; that phased nucleosomes reside on either side of the central CEN DNA-kinetochore structure, as originally suggested by Bloom and Carbon (1982); and that it is these nucleosomes that contain Cse4p. The evidence in favor of this model is by no means definitive, but we believe it should be seriously considered as an alternative to the “nucleosome”-centric view.
Noncentromeric Roles for Ndc10p
Using the sixteen CDEII sequences in S. cerevisiae as a guide, we have identified sequences approximately every 5 kb along chromosome arms that match the CDEII pattern and are high-affinity Ndc10p binding sites in vitro. The AT-rich, poly-A/poly-T pattern characteristic of these sites occurs 1000-fold more frequently in the yeast genome than would be expected by chance, implying a possible biological function. The Ndc10p–DNA complexes that form on arm sites are similar in electrophoretic mobility to complexes that form on CDEII, and arm sites cross-complete with CDEII for Ndc10p binding. Because the arm sites are not near CDEIII sequences, it is possible to probe the extent of Ndc10p binding in vivo by using ChIP. Ndc10p is observed to cross-link to arm sites at levels well above background but below the levels observed at centromeres. The relatively low efficiency of cross-linking makes the result less than totally convincing but may reflect low occupancy of the sites in vivo. However, the localization of Ndc10p to structures other than kinetochores has been confirmed by three-dimensional deconvolution microscopy (this study) and by other methods (Goh and Kilmartin, 1993; Zeng et al., 1999). The function of extracentromeric Ndc10p is not yet known, but may reflect the assembly of structures that transiently associate with microtubules, perhaps to generate a yeast analog of polar ejection forces (Rieder et al., 1986).
Summary
In summary, the data in this article support the conclusion that, in addition to binding to CDEIII as an essential component of CBF3, Ndc10p also binds to CDEII and to sites along chromosome arms. We have not yet distinguished the functions of Ndc10p at CDEII and CDEIII, but it may not be meaningful to separate the kinetochore structure formed on CDEII from the assembly on CDEIII because proteins bound to each region of DNA likely interact. In fact, inversion of CDEIII, relative to CDEII, results in complete loss of centromere function (Murphy et al., 1991). ChIP experiments demonstrate that kinetochore proteins do not associate in vivo with CDEI or CDEII in the absence of a proximal CDEIII sequence, not even Cbf1p, which recognizes a discrete site at CDEI in vitro (Meluh and Koshland, 1995). A more detailed knowledge of Ndc10p is required to resolve these issues, and we have therefore embarked on a high-resolution crystallographic analysis of Ndc10p complexes as well as electron microscopy of CEN DNA with the CBF3 protein complex, Ndc10p- and Cse4p-containing nucleosomes. We nevertheless propose that kinetochores contain a polymeric Ndc10p assembly covering CDEII and CDEIII that forms a platform onto which the microtubule-binding components of kinetochores are recruited. The presence of an Ndc10p polymer may help to solve a compliance problem encountered in firmly anchoring 13 microtubule protofilaments to a single DNA sequence.
Acknowledgments
We thank Peter DeWulf for the data in Figure 1F, Ken Kaplan for help with early stages of this work, Simonetta Piatti for the Ndc10p-myc6 plasmid, Paul Megee and Doug Koshland for help with ChIP, Iain Russell for reagents, and Dan Rines for assistance with microscopy. All strains and plasmids are available from the authors upon request. This work was supported by National Institutes of Health grant GM-51464 to P.K.S. K.T.S. was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship. S.C.H. is an investigator in the Howard Hughes Medical Institute.
References
- Baker, R.E., and Masison, D.C. (1990). Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell. Biol. 10, 2458-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur, M., Esch, R.K., and Errede, B. (1997). Cooperative binding interactions required for function of the Ty1 sterile responsive element. Mol. Cell. Biol. 17, 4330-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat, Y., and Kleckner, N. (1999). Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98, 249-259. [DOI] [PubMed] [Google Scholar]
- Bloom, K. (1993). The centromere frontier: kinetochore components, microtubule-based motility, and the CEN-value paradox. Cell 73, 621-624. [DOI] [PubMed] [Google Scholar]
- Bloom, K.S., and Carbon, J. (1982). Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29, 305-317. [DOI] [PubMed] [Google Scholar]
- Bloom, K.S., Fitzgerald-Hayes, M., and Carbon, J. (1983). Structural analysis and sequence organization of yeast centromeres. Cold Spring Harb. Symp. Quant. Biol. 47, 1175-1185. [DOI] [PubMed] [Google Scholar]
- Bloom, K.S., Hill, A., Kenna, M., and Saunders, M. (1989). The structure of a primitive kinetochore. Trends Biochem. Sci. 14, 223-227. [DOI] [PubMed] [Google Scholar]
- Brown, M.T., Goetsch, L., and Hartwell, L.H. (1993). MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J. Cell Biol. 123, 387-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, M., and Davis, R.W. (1990). Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell 61, 437-446. [DOI] [PubMed] [Google Scholar]
- Cai, M.J., and Davis, R.W. (1989). Purification of a yeast centromere-binding protein that is able to distinguish single base-pair mutations in its recognition site. Mol. Cell. Biol. 9, 2544-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Drubin, D.G., and Barnes, G. (2002). Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157, 199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.M., et al. (1997). Genetic and physical maps of Saccharomyces cerevisiae. Nature 387, 67-73. [PMC free article] [PubMed] [Google Scholar]
- Clarke, L., and Carbon, J. (1980). Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287, 504-509. [DOI] [PubMed] [Google Scholar]
- Clarke, L., Hsiao, C.L., and Carbon, J. (1983). Selection procedure for isolation of centromere DNAs from Saccharomyces cerevisiae. Methods Enzymol. 101, 300-307. [DOI] [PubMed] [Google Scholar]
- Connelly, C., and Hieter, P. (1996). Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell, 86, 275-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottarel, G., Shero, J.H., Hieter, P., and Hegemann, J.H. (1989). A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol. Cell. Biol. 9, 3342-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie, K.W., Hari, K.L., Maggert, K.A., and Karpen, G.H. (1999). Centromere proteins and chromosome inheritance: a complex affair. Curr. Opin. Genet. Dev. 9, 206-217. [DOI] [PubMed] [Google Scholar]
- Espelin, C.W., Kaplan, K.B., and Sorger, P.K. (1997). Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 139, 1383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet, A., and Fitzgerald-Hayes, M. (1987). Alterations in the adenineplus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol. Cell. Biol., 7, 68-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, P.Y., and Kilmartin, J.V. (1993). NDC 10, a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121, 503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Yanagida, M. (2000). Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100, 619-633. [DOI] [PubMed] [Google Scholar]
- Guacci, V., Hogan, E., and Koshland, D. (1997). Centromere position in budding yeast: evidence for anaphase A. Mol. Biol. Cell 8, 957-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Asthana, S., and Sorger, P.K. (2000). Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101, 763-775. [DOI] [PubMed] [Google Scholar]
- He, X., Rines, D.R., Espelin, C.W., and Sorger, P.K. (2001). Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106, 195-206. [DOI] [PubMed] [Google Scholar]
- Hegemann, J.H., and Fleig, U.N. (1993). The centromere of budding yeast. Bioessays 15, 451-460. [DOI] [PubMed] [Google Scholar]
- Hegemann, J.H., Shero, J.H., Cottarel, G., Philippsen, P., Hieter, P. (1988). Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 2523-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, J., Dietz-Schmidt, A., Zundorf, I., Garin, J., Dingermann, T., and Winckler, T. (1999). A Dictyostelium protein binds to distinct oligo(dA) x oligo(dT) DNA sequences in the C-module of the retrotransposable element DRE. Eur. J. Biochem. 265, 441-448. [DOI] [PubMed] [Google Scholar]
- Janke, C., Ortiz, J., Lechner, J., Shevchenko, A., Magiera, M.M., Schramm, C., and Schiebel, E. (2001). The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20, 777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W.D., and Philippsen, P. (1989). Purification of a protein binding to the CDEI subregion of Saccharomyces cerevisiae centromere DNA. Mol. Cell. Biol. 9, 5585-5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, K.B., Hyman, A.A., and Sorger, P.K. (1997). Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91, 491-500. [DOI] [PubMed] [Google Scholar]
- Keith, K.C., and Fitzgerald-Hayes, M. (2000). CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a cse4p variant nucleosome. Genetics 156, 973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg, R.D., and Klug, A. (1981). The nucleosome. Sci. Am. 244, 52-64. [DOI] [PubMed] [Google Scholar]
- Kunkel, G., and Martinson, H. (1981). Nucleosomes will not form on double-stranded RNA or over poly(dA):poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 9, 6869-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, J., and Carbon, J. (1991). A 240 kD multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64, 717-725. [DOI] [PubMed] [Google Scholar]
- Luger, K., Rechsteiner, T.J., Flaus, A.J., Waye, M.M., and Richmond, T.J. (1997). Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272, 301-311. [DOI] [PubMed] [Google Scholar]
- Magbanua, J.P., Fujisawa, K., Ogawa, N., and Oshima, Y. (1997). The homeodomain protein Pho2p binds at an A/T-rich segment flanking the binding site of the basic-helix-loop-helix protein Pho4p in the yeast PHO promoters. Yeast 13, 1299-1308. [DOI] [PubMed] [Google Scholar]
- McGrew, J., Diehl, B., and Fitzgerald-Hayes, M. (1986). Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 6, 530-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee, P.C., Mistrot, C., Guacci, V., and Koshland, D. (1999). The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 4, 445-450. [DOI] [PubMed] [Google Scholar]
- Mellor, J., Jiang, W., Funk, M., Rathjen, J., Barnes, C.A., Hinz, T., Hegemann, J.H., and Philippsen, P. (1990). CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9, 4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and Koshland, D. (1995). Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6, 793-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and Koshland, D. (1997). Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes. Dev. 11, 3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., Yang, P., Glowczewski, L., Koshland, D., and Smith, M.M. (1998). Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94, 607-613. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., Ciosk, R., and Nasmyth, K. (1997). Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91, 35-45. [DOI] [PubMed] [Google Scholar]
- Murphy, M.R., Fowlkes, D.M., Fitzgerald-Hayes, M. (1991). Analysis of centromere function in Saccharomyces cerevisiae using synthetic centromere mutants. Chromosoma 101, 189-197. [DOI] [PubMed] [Google Scholar]
- Ng, R., and Carbon, J. (1987). Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 4522-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal, R., Stoll, R., Hegemann, J.H. (1991). In vivo characterization of the Saccharomyces cerevisiae centromere DNA element I, a binding site for the helix-loop-helix protein CPF1. Mol. Cell. Biol. 11, 3545-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal, R., Stoll, R., and Hegemann, J.H. (1991). In vivo characterization of the Saccharomyces cerevisiae centromere DNA element I, a binding site for the helix-loop-helix protein CPF1. Mol. Cell. Biol. 11, 3545-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, J., Stemmann, O., Rank, S., and Lechner, J. (1999). A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13, 1140-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeri, L., Landonio, L., Stotz, A., Philippsen, P. (1985). Role of conserved sequence elements in yeast centromere DNA. EMBO J. 4, 1867-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta, A.F., Mackay, A.M., Ainsztein, A.M., Goldberg, I.G., and Earnshaw, W.C. (1995). The centromere: hub of chromosomal activities. Science 270, 1591-1594. [DOI] [PubMed] [Google Scholar]
- Politi, V., Perini, G., Trazzi, S., Pliss, A., Raska, I., Earnshaw, W.C., and Valle, G.D. (2002). CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 115, 2317-2327. [DOI] [PubMed] [Google Scholar]
- Prunell, A. (1982). Nucleosome reconstitution on plasmid-inserted poly(dA)*poly(dT) EMBO J. 1, 173-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, R., and Nissen, M.S. (1993). Interaction of high mobility group-I (Y) nonhistone proteins with nucleosome core particles. J. Biol. Chem. 268, 21137-21146. [PubMed] [Google Scholar]
- Rieder, C.L., Davison, E.A., Jensen, L.C., Cassimeris, L., and Salmon, E.D. (1986). Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103, 581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, I.D., Grancell, A.S., and Sorger, P.K. (1999). The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145, 933-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell, S.C., Drew, H.R., and Travers, A.A. (1986). Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 191, 659-675. [DOI] [PubMed] [Google Scholar]
- Shaulian, E., and Karin, M. (2002). AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4, E131-E136. [DOI] [PubMed] [Google Scholar]
- Skibbens, R.V., Corson, L.B., Koshland, D., and Hieter, P. (1999). Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13, 307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.M., Yang, P., Santisteban, M.S., Boone, P.W., Goldstein, A.T., and Megee, P.C. (1996). A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16, 1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger, P.K., Doheny, K.F., Hieter, P., Kopski, K.M., Huffaker, T.C., and Hyman, A.A. (1995). Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc. Natl. Acad. Sci. USA 92, 12026-12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler, S., Keith, K.C., Curnick, K.E., and Fitzgerald-Hayes, M. (1995). A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9, 573-586. [DOI] [PubMed] [Google Scholar]
- Strunnikov, A.V., Kingsbury, J., and Koshland, D. (1995). CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 128, 749-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov, A.V., Larionov, V.L., and Koshland, D. (1993). SMC 1, an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 123, 1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., Cosma, M.P., Wirth, K., and Nasmyth, K. (1999). Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98, 847-858. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M.J., and Nasmyth, K. (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317-329. [DOI] [PubMed] [Google Scholar]
- Toth, A., Ciosk, R., Uhlmann, F., Galova, M., Schleiffer, A., and Nasmyth, K. (1999). Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13, 320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, E., and Varshavsky, A. (1989). A DNA binding protein that recognizes oligo(dA). oligo(dT) tracts. EMBO J. 8, 1867-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., Kahana, J.A., Silver, P.A., Morphew, M.K., McIntosh, J.R., Fitch, I.T., Carbon, J., and Saunders, W.S. (1999). Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146, 415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]