Abstract
The potentially deleterious effects of aberrant mRNA lacking a termination codon (nonstop mRNA) are ameliorated by translation arrest, proteasome-mediated protein destabilization, and rapid mRNA degradation. Because polylysine synthesis via translation of the poly(A) mRNA tail leads to translation arrest and protein degradation by the proteasome, we examined the effects of other amino acid sequences. Insertion of 12 consecutive basic amino acids between GFP and HIS3 reporter genes, but not a stem-loop structure, resulted in degradation of the truncated green fluorescent protein (GFP) products by the proteasome. Translation arrest products derived from GFP-R12-FLAG-HIS3 or GFP-K12-FLAG-HIS3 mRNA were detected in a not4Δ mutant, and MG132 treatment did not affect the levels of the truncated arrest products. Deletion of other components of the Ccr4-Not complex did not increase the levels of the translation arrest products or reporter mRNAs. A L35A substitution in the Not4p RING finger domain, which disrupted its interaction with the Ubc4/Ubc5 E2 enzyme and its activity as an ubiquitin-protein ligase, also abrogated the degradation of arrest products. These results suggest that Not4p, a component of the Ccr4-Not complex, may act as an E3 ubiquitin-protein ligase for translation arrest products. The results let us propose that the interaction between basic amino acid residues and the negatively charged exit tunnel of the ribosome leads to translation arrest followed by Not4p-mediated ubiquitination and protein degradation by the proteasome.
Cells have surveillance systems that recognize and eliminate aberrant mRNAs to prevent the production of potentially harmful protein products. In eukaryotes, an aberrant mRNA lacking a termination codon (nonstop mRNA) may be recognized and eliminated by the quality control system referred to as nonstop decay (1, 2). It has been shown that nonstop mRNA is rapidly degraded by a 3′-to-5′ degradation pathway in yeast (1). We have shown that translation of nonstop mRNA is repressed after the initiation step (3, 4); the level of the product of nonstop mRNA containing a poly(A) tail was reduced by ∼100-fold, and this reduction was due to rapid mRNA degradation, translation repression, and protein destabilization, at least in part, by the proteasome (3, 4). Interestingly, insertion of a poly(A) tract upstream of a termination codon resulted in translation repression and protein destabilization but not rapid mRNA decay (3, 4). Recently, it has been shown that translation of nonstop mRNA also is repressed after initiation in mammalian cells and that the ribosome stalls on poly(A) sequences, a common modification of eukaryotic mRNA that is not normally translated (5). Therefore, translation repression following polylysine synthesis may be conserved among eukaryotes. We propose that ribosomes recognize the translation of poly(A) tails as an aberrant process, resulting in translation arrest and degradation of the protein product by the proteasome.
Translation elongation is an important step of gene regulation; the expression levels of several genes are known to be regulated by translation arrest. In prokaryotes, the regulation of the secM gene, for example, has been extensively analyzed. The interaction between the SecM nascent peptide and the wall of the ribosomal exit tunnel functions as a discriminating gate during translation (6, 7). Several eukaryotic examples of translation arrest also have been described (8, 9). Moreover, the synthesis of polylysine caused by translation of a poly(A) sequence leads to translation arrest (3, 4). Recent measurement of the electrostatic potential of the ribosome exit tunnel using chemical modification methods revealed that the wall of the tunnel is negatively charged in eukaryotes (10, 11). These results suggest that consecutive positively charged amino acid residues in a nascent protein may have a high affinity for the negatively charged ribosomal exit tunnel. A more recent report demonstrated that positively charged residues cause pausing in mammalian ribosomes in vitro and showed how far the positively charged side chains move into the tunnel (12).
The evolutionarily conserved Ccr4-Not complex regulates mRNA biosynthesis at various steps. First, Ccr4p/Caf1p has a deadenylase activity that functions in mRNA deadenylation (13–15). Second, Ccr4p/Caf1p regulates histone H3K4 trimethylation (H3K4me3), which plays a significant role in chromatin organization, gene transcription, and epigenetic regulation via interactions with the proteasome to regulate its recruitment to genes (16). In yeast, the Ccr4-Not core complex is composed of nine components. The Not1–5 proteins are critical for transcriptional repression. It recently was shown that NOT1, NOT2, NOT4, and NOT5 are required for trimethylation of H3K4 (16, 17). Third, the Not4p subunit of the Ccr4-Not complex has an E33 ubiquitin-protein ligase activity for the conserved ribosome-associated heterodimeric EGD complex. This complex consists of the Egd1p and Egd2p subunits in yeast and is named NAC (nascent polypeptide-associated complex) in mammals. Finally, the Not4p displays an Ubc4p-dependent ubiquitination activity in vitro (18, 19), which depends on the RING finger domain of Not4p (18).
In this study, we show that 12 consecutive basic amino acid residues cause translation arrest followed by cotranslational protein degradation by the proteasome. In contrast, a stem-loop structure induces only translation arrest but not degradation of the arrested product. Not4p is required for the degradation of translation arrest products, although Not3p and Not5p of the Ccr4-Not complex are not. In addition, translation arrest products are not degraded by the proteasome in the not4L35A mutant, in which ubiquitination of the EGD complex and the interaction with E2 enzymes are defective. These results strongly suggest that nascent peptide sequences play crucial roles in translation elongation arrest, leading to Not4p-mediated protein degradation by the proteasome. More general roles of endogenous nascent peptides in the regulation of translation and protein degradation also are discussed.
EXPERIMENTAL PROCEDURES
Strains and General Methods—Standard procedures were used to manipulate yeast cells (20). The yeast strains and plasmids used in this study are described in supplemental Table S1. Deletions of NOT3, NOT4, and NOT5 were constructed using a PCR-based method (21). Recombinant DNA procedures were carried out as described previously (22).
Plasmids—Two oligonucleotides (supplemental Table S2) were annealed and inserted into the SpeI site of pSA144 (pGPDp-GFP-FLAG-HIS3-CYC1ter) to create the pGPD-GFP-X-FLAG-HIS3 reporters, which contained sequences encoding various 12-amino acid stretches or a stem-loop structure between GFP and HIS3. A NOT4 XbaI-XhoI fragment was amplified in a PCR using two primers (5′-GGTCTAGACGTATATAATCCAGTCATAATGATG-3′ and 5′-CCCTCGAGGAAAAATATTTAGAGTCGGATTAATTACCGGCGA-3′) and inserted into p415ADH1p to generate p415ADH1p-NOT4. To construct p415ADH1p-NOT4L35A, a mutation was introduced using site-directed mutagenesis. Two oligonucleotides (5′-CTAGAATGGACTACAAGGACGACGATGACAAGGGTCTGGTA-3′ and 5′-CTAGTACCAGACCCTTGTCATCGTCGTCCTTGTAGTCCATT-3′) were annealed and inserted into the XbaI site of p415ADH1p-NOT4 to generate p415ADH1p-FLAG-NOT4.
Determination of the Relative Product Levels Using Western Blotting—Yeast cells were grown on SC-Ura medium, and proteins from cell extracts equivalent to 0.2 A600 were prepared. The products derived from the reporter genes were detected using Western blotting with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare). The primary antibodies used for Western blotting were anti-FLAG antibodies (F1804; Sigma), anti-GFP antibodies (sc-9996; Santa Cruz Biotechnology), and anti-FK2 antibodies (PW8810; BIOMOL). The anti-FK2 antibodies recognize the multiubiquitin chains of polyubiquitinated proteins but not free ubiquitin or the protein moieties (23). The intensities of the bands were quantified with the LAS3000 mini system using Multi-Gauge version 3.0 (Fuji Film, Japan), and the relative levels of the products were determined based on a comparison with a standard curve. When the intensity of the band was outside the range of the standard curve, a series of sample dilutions was prepared. The intensities of the bands for the diluted samples were compared with those from the standard curve, and the relative levels of the products in the samples compared with the control products were determined.
Northern Blotting—Total RNA was separated using 1.2% agarose gel electrophoresis in the presence of formaldehyde and blotted onto Hybond-N+ membrane (Amersham Biosciences). mRNA was visualized using digoxigenin (DIG) reagents and kits for PCR-based nonradioactive nucleic acid labeling detection (Roche Applied Science) according to the manufacturer's instructions. DIG-labeled probes were prepared with the following oligonucleotides: HIS3 (5′-GCTCTAGATGACAGAGCAGAAAGCCCTAG-3′ and 5′-CGGGATCCCATAAGAACACCTTTGGTGGAGG-3′) and GFP (5′-GCTCTAGAGGCCTATGCGGCCGCAGTAAAGGAG-3′ and 5′-CGGGATCCTTTGTATAGTTCATCCATGCC-3′). The band intensities were quantified using the LAS3000 mini system and Multi-Gauge version 3.0 (Fuji Film).
Yeast Extract and Sucrose Gradient Separation—Yeast cells were grown exponentially at 30 °C and harvested by centrifugation. The cells were washed once with lysis buffer, and extracts were prepared as described previously (24). The equivalent of 50 A260 units was then layered onto linear 10–50% sucrose density gradients (10–50% sucrose in 10 mm Tris acetate, pH 7.4, 70 mm ammonium acetate, 4 mm magnesium acetate), prepared in 25 × 89-mm polyallomer tubes (Beckman Coulter) using a gradient master. Crude extracts were layered on top of the sucrose gradients and centrifuged at 27,000 rpm in a P28S rotor (Hitachi Koki) for 3 h at 4 °C. The gradients were then fractionated (TOWA lab, Tsukuba). Polysome profiles were generated with continuous absorbance measurement at 254 nm using a single path UV-1 optical unit (ATTO Biomini UV monitor) connected to a chart recorder (ATTO digital mini-recorder). Equal volume fractions were collected and processed for Western blotting as described above.
RESULTS
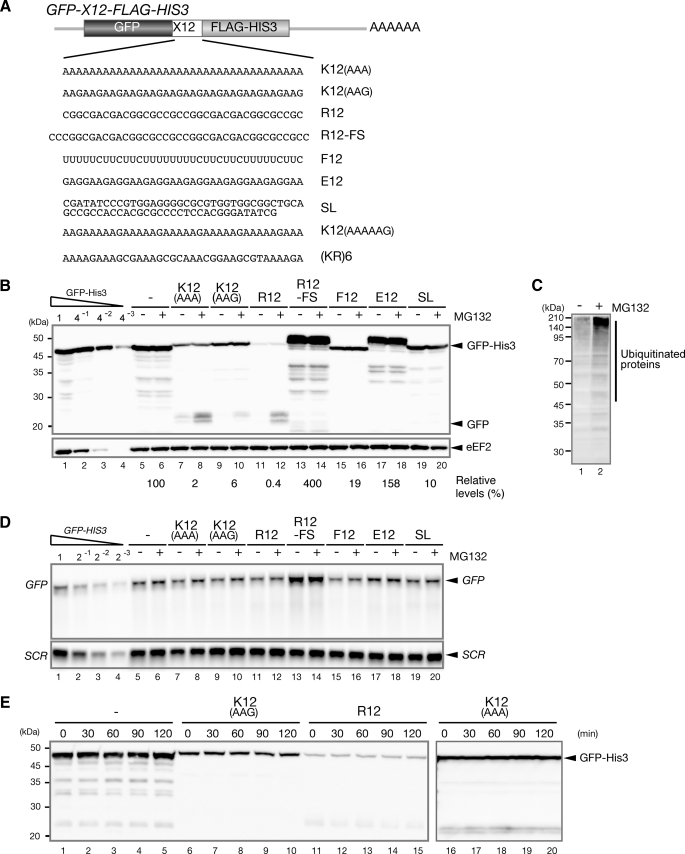
Consecutive Basic Amino Acid Residues Induce Protein Degradation by the Proteasome—It was reported that insertion of a poly(A) tract upstream of a termination codon results in translation repression and protein destabilization but not rapid mRNA decay (3, 4). To determine whether translation is arrested by other amino acid sequences and whether the translation arrest product is degraded by the proteasome, we inserted fragments encoding various amino acid sequences between the GFP and HIS3 genes (Fig. 1A). We hypothesized that translation may be arrested when the ribosome synthesizes polylysine, and the translation arrest product could be detected as a truncated GFP product in the presence of MG132, a proteasome inhibitor. As expected, the level of the full-length product was significantly lower (2–6% of the control product), and in the presence of MG132, a translation arrest product was detected as a truncated GFP product (Fig. 1B, lanes 8 and 10). We also found that insertion of 12 consecutive arginine residues strongly reduced the level of the full-length GFP-R12-FLAG-His3 product (less than 1% of the control sample; Fig. 1B, lanes 11 and 12). The pGPDp-GFP-R12-FS-FLAG-HIS3 construct is identical to the pGPDp-GFP-R12-FLAG-HIS3 construct except for an additional nucleotide just before the sequence encoding polyarginine and two more nucleotides following the sequence. This frameshift mutation completely eliminated translation repression (Fig. 1B, lanes 13 and 14), indicating that the basic amino acid sequence plays a crucial role for translation repression. There was a difference between the translation arrest efficiencies of K12(AAA) and K12(AAG) (Fig. 1B, lanes 7–10). The mRNA levels of GFP-K12(AAA)-FLAG-HIS3 and GFP-K12(AAG)-FLAG-HIS3 were similar, indicating that the nucleotide sequence could play a role in translation repression. One possibility is that Pab1p may bind to poly(A) sequences and inhibit translation elongation. To confirm that MG132 indeed inhibited proteasomes under the test conditions, we performed Western blotting with anti-FK2 antibodies, which recognize monoubiquitin and polyubiquitin but not free ubiquitin. As shown in Fig. 1C, the levels of ubiquitinated proteins were significantly higher in the presence of MG132. In addition, we confirmed that the levels of the truncated products derived from reporters increased in the presence of MG132 in erg6Δ mutant cells, which show an increased uptake of MG132 (supplemental Fig. S1) (25).
FIGURE 1.
Protein degradation by the proteasome in response to translation arrest caused by consecutive basic amino acid residues. A, construction of the pGPDp-GFP-X-FLAG-HIS3 reporter plasmids, which contain various sequences encoding 12 consecutive amino acids inserted between GFP and HIS3. B, consecutive basic amino acid sequences induce translation arrest coupled with protein degradation by the proteasome. Cells harboring the indicated pGPDp-GFP-X-FLAG-HIS3 plasmid were grown on SC-Ura medium, and the samples were analyzed using Western blotting with anti-GFP (top panel) or anti-eEF2 (bottom panel) antibodies. When indicated (+), cell extracts were prepared 2 h after the addition of 0.2 mm MG132. When determining the relative expression levels, we did not include shorter (degraded) products. C, proteasome activity is inhibited in the presence of MG132. W303 cells were grown on SC medium, and cell extracts were prepared 2 h after the addition of 0.2 mm MG132 when indicated (+). The samples were analyzed using Western blotting with anti-FK2 antibodies, which recognize monoubiquitin and polyubiquitin but not free ubiquitin. D, truncated mRNA is not detected in the presence of MG132. W303 cells harboring the indicated pGPDp-GFP-X-FLAG-HIS3 plasmid were grown on SC-Ura medium, and RNA samples were analyzed using Northern blotting with DIG-labeled GFP or SCR probes. E, the stabilities of GFP-K12(AAG)-FLAG-His3, GFP-R12-FLAG-His3, and GFP-K12(AAA)-FLAG-His3 were analyzed using Western blotting. Samples of W303 cells harboring the indicated plasmid were prepared at the indicated time points after the addition of cycloheximide (0.1 mg/ml). The levels of the remaining proteins were determined by Western blotting with anti-GFP antibodies.
We also confirmed that the levels and sizes of the mRNAs derived from GFP-K12(AAA)-FLAG-HIS3, GFP-K12(AAG)-FLAG-HIS3 or GFP-R12-FLAG-HIS3 were not affected by MG132 (Fig. 1D, lanes 7–12), indicating that the truncated proteins were not produced from truncated mRNAs. We found that the levels of the mRNA and protein derived from GFP-R12-FS-FLAG-HIS3 were 2-fold higher than those derived from the control reporter (Fig. 1, B and C, lane 13). MG132, however, did not affect level of mRNA and protein derived from GFP-R12-FS-FLAG-HIS3 (Fig. 1, B and D, lanes 13 and 14). Insertion of polyphenylalanine but not polyglutamic acid significantly reduced the levels of the full-length products, mimicking the effects of polylysine (AAG) (Fig. 1B, lanes 15–18). Truncated product derived from the construct expressing both polyglutamic acid and polyphenylalanine was not detected, however, even in the presence of MG132. These results suggest that sequential basic amino acid residues may induce proteasome-dependent degradation of arrest products.
We also examined whether nascent peptides are degraded by the proteasome when translation is arrested by a stem-loop structure. The level of the intact product expressed from GFP-SL-FLAG-HIS3 mRNA was reduced significantly (10% of the control product) (Fig. 1B, lanes 5 and 19), whereas the mRNA level was not significantly reduced (Fig. 1C, lanes 5 and 19). These results clearly indicate that translation is physically arrested by stable stem-loop structures, as previously described (26). Importantly, MG132 treatment did not affect the expression of the putative small products from translation arrest caused by the stem-loop structure (Fig. 1B, lanes 19 and 20). This strongly suggests that specific nascent peptides, not translation arrest induced by a physical barrier, result in cotranslational protein degradation by the proteasome. To determine whether the truncated products derived from GFP-K12-FLAG-HIS3 or GFP-R12-FLAG-HIS3 may result from protease-mediated cleavage after translation is completed, the proteins remaining after inhibition of translation were examined using Western blotting. The levels of the remaining products were not markedly changed as a function of time after the addition of cycloheximide, indicating that the full-length products were as stable as GFP-His3 (Fig. 1E). Consistently, MG132 treatment did not increase the levels of full-length GFP-K12(AAA)-FLAG-His3, GFP-K12(AAG)-FLAG-His3, or GFP-R12-FLAG-His3 (Fig. 1B, lanes 7–12). These results suggest that the truncated products derived from GFP-FLAG-HIS3, GFP-K12-FLAG-HIS3, and GFP-R12-FLAG-HIS3 were not produced by degradation or the cleavage of full-length products and are consistent with a model in which the truncated product detected in the presence of MG132 is produced by translation arrest. Thus, we propose that specific nascent peptides induce translation arrest, which in turn leads to cotranslational protein degradation by the proteasome in yeast.
Rare Codons Are Not Required for Translation Arrest and Subsequent Protein Degradation—To address the specificity of the amino acid sequences required to induce translation arrest and cotranslational protein degradation, we examined the expression of GFP-X-FLAG-HIS3 reporters containing sequences that encode other 12-amino acid sequences between GFP and HIS3, in the presence or absence of a proteasome inhibitor. We observed significant reductions in the levels of the full-length proteins containing 12 sequential hydrophobic residues, (valine, phenylalanine), although truncated GFP products derived from these reporter genes were not detected, even after MG132 treatment (Fig. 2A, lanes 19, 20, 29, and 30). In contrast, truncated products derived from GFP-W12-FLAG-HIS3 were detected only upon the addition of MG132 (Fig. 2A, lanes 27 and 28), although the observed levels were not as high as those produced by 12 consecutive Lys or Arg residues. Introduction of a frameshift mutation abrogated the arrest product in the presence of MG132 (Fig. 2B, lanes 7 and 8). These results suggest that translation of consecutive 12 basic or Trp residues results in translation arrest, which leads to nascent protein degradation by the proteasome. We also identified truncated products expressed from the GFP-K12(AAAAAG)-FLAG-HIS3 reporter, which contains six repeats of the AAAAAG sequence and encodes polylysine (Fig. 2B, lanes 11 and 12). The levels of truncated products increased in the presence of MG132 and a frameshift mutation again inhibited production of truncated products (Fig. 2B, lanes 13 and 14). These results indicate that polylysine, rather than sequential AAA codons, are required for the translation arrest coupled with protein degradation as previously described (4). We also detected several different truncated GFP products derived from GFP-G12-FLAG-HIS3. The level of full-length GFP-G12-FLAG-His3, however, was similar to that of control GFP-FLAG-His3 product (Fig. 2A, lanes 1 and 15). In addition, the levels of truncated products did not increase in the presence of MG132 (Fig. 2A, lanes 15 and 16). These results strongly suggest that the truncated GFP products are not translation arrest products. We also detected a truncated GFP product derived from GFP-C12-FLAG-HIS3 without MG132 treatment (Fig. 2A, lanes 9 and 10). A truncated His3 product was detected using Western blot analysis with anti-FLAG antibodies, and its level relative to that of the intact GFP-C12-FLAG-His3 product was similar to the results observed for truncated GFP (data not shown). These findings suggest that GFP-C12-FLAG-His3 is cleaved post-translationally independently of the proteasome because MG132 had no effect on the levels of either truncated product (Fig. 2A, lanes 9 and 10).
FIGURE 2.
Consecutive basic amino acid residues rather than rare codons induce translation arrest coupled with protein degradation. A, sequence specificity of the translation arrest-induced protein degradation process mediated by the proteasome. W303 cells harboring the indicated pGPDp-GFP-X-FLAG-HIS3 plasmids were grown on SC-Ura medium, and cell extracts were prepared. When indicated (+), cell extracts were prepared 2 h after the addition of 0.2 mm MG132. The relative levels of the reporter proteins were determined using Western blotting with anti-GFP or anti-eEF2 antibodies. The levels of the products derived from GFP-S12-HIS3, GFP-D12-HIS3, GFP-E12-HIS3, GFP-G12-HIS3, GFP-I12-HIS3, and GFP-P12-HIS3 were higher than that of the product derived from GFP-HIS3. Additionally, the relative levels of those products were determined using a standard curve of samples from cell harboring pGPDp-GFP-S12-HIS3 or pGPDp-GFP-HIS3. B, W303 cells harboring the indicated pGPDp-GFP-X-FLAG-HIS3 plasmid were grown on SC-Ura medium, and the samples were analyzed using Western blotting. When indicated (+), cell extracts were prepared 2 h after the addition of 0.2 mm MG132.
To confirm that 12 consecutive basic amino acid residues induce translation arrest, we constructed a GFP-(KR)6-FLAG-HIS3 reporter that contains a sequence encoding six Lys-Arg repeats between GFP and HIS3. Importantly, the inserted sequence did not contain any rare codons. The truncated product was identified only in the presence of MG132 (Fig. 2B, lanes 9 and 10). These results strongly suggest that consecutive basic amino acids rather than rare codons or pure polylysine or polyarginine sequences are required for translation arrest coupled with protein degradation.
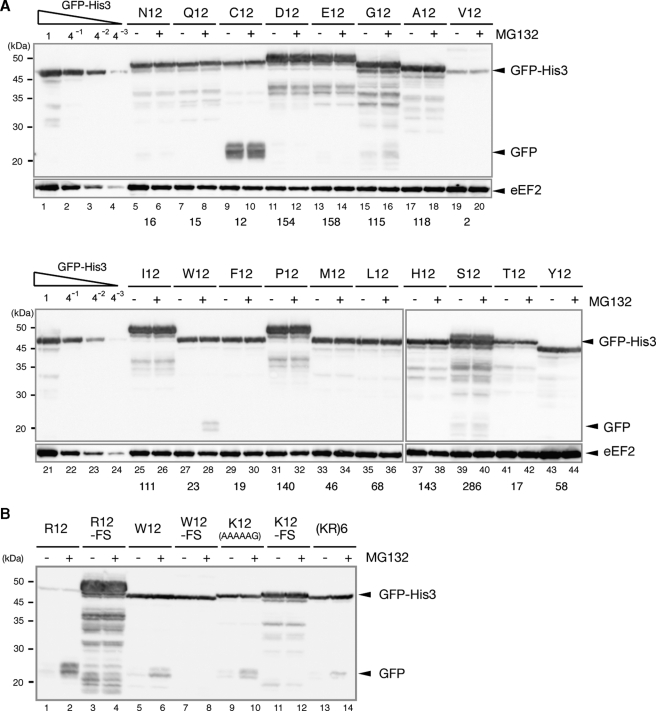
Not4p Is Required for Proteasome-induced Degradation of Translation Arrest Products—Our results indicated that nascent peptide-dependent protein degradation is cotranslational, suggesting that an E3 ubiquitin-protein ligase may be associated with the ribosome. Not4p, a component of the Ccr4-Not complex, has been identified as an E3 ubiquitin-protein ligase for EGD complexes associated with nascent polypeptides (19). Therefore, we examine the expression from GFP-R12-FLAG-HIS3 in a not4Δ mutant. In the not4Δ mutant, the level of the truncated product derived from GFP-R12-FLAG-HIS3 was markedly higher, and, along with the level of GFP-R12-FLAG-HIS3 mRNA, was not affected by the addition of MG132 (Fig. 3, A, lanes 6–9, and B, lanes 5–8). A corresponding truncated mRNA was not detected, even in the presence of MG132 (Fig. 3B, lanes 5–8). We also found that the levels of truncated products derived from GFP-K12(AAA)-FLAG-HIS3 or GFP-K12(AAG)-FLAG-HIS3 also were higher in the not4Δ mutant (Fig. 3C, lanes 2 and 4). These results strongly suggest that Not4p is required for the degradation of translation arrest products by the proteasome. The level of GFP-R12-FLAG-HIS3 mRNA also increased significantly in the not4Δ mutant (Fig. 3B, lanes 5 and 7), although the level of the GFP-R12-FLAG-His3 protein was slightly lower (Fig. 3A, lanes 6 and 8), suggesting that translation arrest of GFP-R12-FLAG-HIS3 mRNA may be enhanced in the not4Δ mutant.
FIGURE 3.
Not4p is required for the degradation of translation arrest products by the proteasome. A, Not4p is required for the proteasome-mediated degradation of the truncated product. W303 or W303not4Δ cells were transformed with the pGPDp-GFP-R12-FLAG-HIS3 plasmid. The cells were grown in SC-Ura, and samples were analyzed using Western blotting with anti-GFP antibodies. When indicated (+), cell extracts were prepared 2 h (W303) or 5 h (W303not4Δ) after the addition of 0.2 mm MG132. B, RNA samples were prepared from cells shown in A and analyzed using Northern blotting with DIG-labeled GFP or SCR probes. C, the level of truncated product derived from GFP-K12-FLAG-HIS3 was higher in the not4Δ mutant. W303 or W303not4Δ cells were transformed with the indicated pGPDp-GFP-X-FLAG-HIS3 plasmid. The cells were grown in SC-Ura, and samples were analyzed using Western blotting. D, W303, W303not4Δ, or W303rpn6–2 (YNK7) cells harboring pGPDp-GFP-R12-FLAG-HIS3 were grown in SC medium at 30 °C. The samples were prepared and analyzed using Western blotting with anti-GFP antibodies (lanes 1–3) or anti-FK2 antibodies that recognize monoubiquitin and polyubiquitin but not free ubiquitin (lanes 4–6). E, the level of the truncated product derived from GFP-R12-FLAG-HIS3 was higher in the not4Δ mutant. W303, W303not3Δ, W303not4Δ, or W303not5Δ cells were transformed with the pGPDp-GFP-R12-FLAG-HIS3 plasmid. The cells were grown in SC-Ura, and the samples were analyzed using Western blotting. F, RNA samples were prepared from the cells shown in D and analyzed using Northern blotting with DIG-labeled GFP or SCR probes. G, the level of truncated product derived from GFP-R12-FLAG-HIS3 was higher in the not4Δ mutant expressing the Not4L35A mutant (mutation in the RING finger domain). W303not4Δ cells containing pGPDp-GFP-R12-FLAG-HIS3 were transformed with pADHp-NOT4 or pADHp-not4L35A. The cells were grown in SC-UraLeu medium, and the samples were analyzed using Western blotting. H, FLAG-Not4p protein was distributed in the polysome fractions. W303 cells were transformed with pADHp-FLAG-NOT4. Cell extracts were prepared, and polysome analysis was performed as described previously (3). Protein samples prepared from each fraction were analyzed using Western blotting with anti-FLAG antibodies. WT, wild type.
Because Not4p is involved in transcriptional regulation via H3K4 trimethylation, the proteasomes in the not4Δ mutant may be generally impaired. To address this possibility, we determined the levels of ubiquitinated proteins using Western blotting with anti-FK2 antibodies. It was shown previously that Rpn6p is a component of the lid of the proteasome and that the activity of the proteasome is impaired in the rpn6–2 temperature-sensitive mutant (27). We found that the levels of ubiquitinated proteins were markedly higher in the rpn6–2 mutant compared with the not4Δ mutant (Fig. 3D, lanes 4–6), indicating that the activity of the proteasome in the rpn6–2 mutant was more severely impaired than in the not4Δ mutant. The increase in the truncated GFP product in the rpn6–2 mutant, however, was smaller than in the not4Δ mutant (Fig. 3D, lanes 1–3). The truncated product in the not3Δ and not5Δ mutants also was not augmented (Fig. 3E, lanes 6 and 8). We obtained similar results for the mRNA levels (Fig. 3F, lanes 6 and 8). Because Not4p and Not5p, rather than Not3p, are required for trimethylation of H3K4 (16, 17), these results suggest that the H3K4me3-mediated defect in transcriptional regulation in the not4Δ mutant may not account for the increased levels of the truncated product. These results are consistent with the idea that Not4p is required for the degradation of truncated products by the proteasome.
The N-terminal domain of Not4p contains a RING finger domain, which mediates its interaction with the E2 ligase Ubc4p/Ubc5p; the L35A mutation disrupted the ubiquitin-protein ligase activity of Not4p (18). The level of the truncated product derived from GFP-R12-HIS3 was markedly higher in the not4Δ mutant harboring the pNOT4-L35A plasmid (L35A) but not in the not4Δ mutant harboring the pNOT4 plasmid (wild type) (Fig. 3G). Interestingly, FLAG-Not4p was distributed mainly in the heavy polysome fractions of sucrose gradients (Fig. 3H), suggesting that Not4p may be associated with ribosome. Thus, Not4p appears to function as an E3 ubiquitin-protein ligase for translation arrest products, which is followed by their proteasome-mediated degradation.
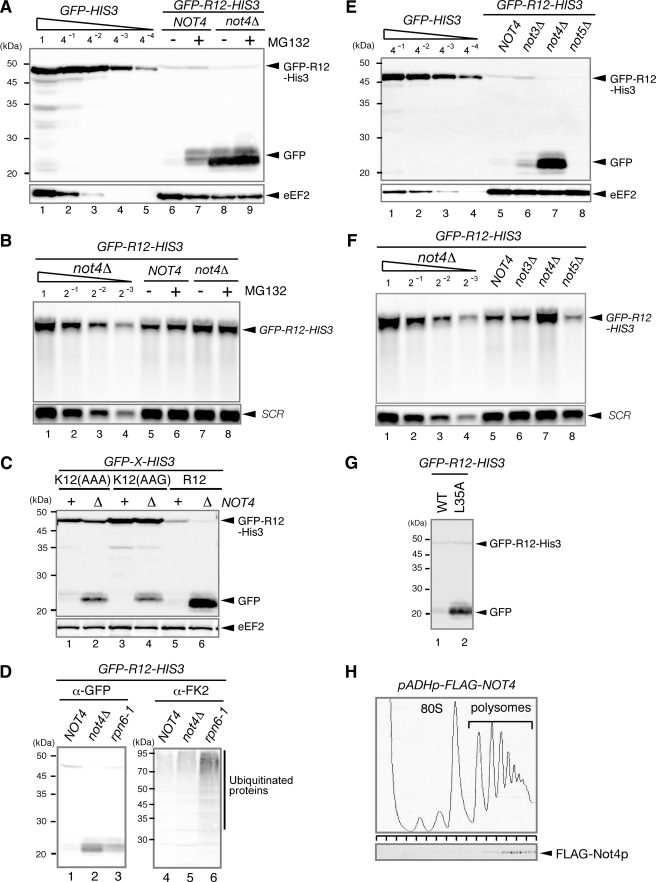
Degradation of Nonstop mRNA Products Is Not4p-independent—We previously demonstrated that translation of a poly(A) tail from nonstop mRNA results in translation arrest and post-translational protein degradation of the nonstop products (3, 4). To investigate the role of Not4p in the degradation of nonstop products, we examined the expression of nonstop products in a not4Δ mutant. The level of the nonstop product derived from GFP-FLAG-HIS3-NS did not increase in the not4Δ mutant (Fig. 4A, lanes 3 and 4), suggesting that Not4p is not involved in the degradation of nonstop products. We also confirmed that the levels of GFP-FLAG-HIS3 and GFP-FLAG-HIS3-NS mRNA were not changed in the not4Δ mutant (Fig. 4B, lanes 5–8), although the level of GFP-FLAG-HIS3-NS mRNA was 4-fold lower than that of GFP-FLAG-HIS3 mRNA as described previously (4). This result is consistent with the role of Not4p as an E3 ubiquitin-protein ligase for translation arrest products but not for aberrant proteins released from the ribosome.
FIGURE 4.
Not4p is not involved in the degradation of nonstop products. W303 or W303not4Δ cells were transformed with the indicated pGPDp-GFP-FLAG-HIS3 (pSA156) or pGPDp-GFP-FLAG-HIS3-NS (pSA157) plasmids (4). A, the cells were grown in SC-Ura, and the samples were analyzed using Western blotting with anti-GFP antibodies. B, the cells were grown in SC-Ura, and the samples were analyzed using Northern blotting with DIG-labeled GFP and SCR probes. WT, wild type.
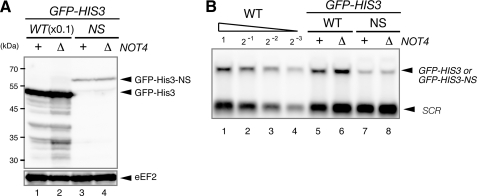
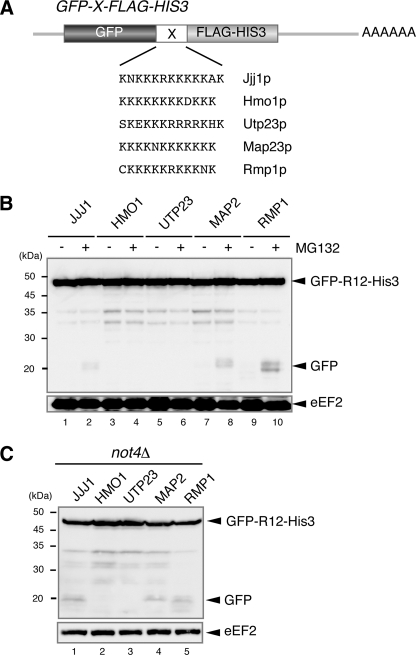
Endogenous mRNA Sequences Induce Translation Arrest Coupled with Protein Degradation—Our results suggest that translation arrest induced by sequences encoding consecutive basic amino acid residues leads to Not4p-mediated protein degradation by the proteasome. To evaluate a possible role for translation arrest in gene regulation, we screened genomic sequences for areas encoding consecutive basic amino acid sequences and identified five genes (Fig. 5A). The five endogenous sequences were inserted between GFP and HIS3 to determine whether they induced translation arrest and associated protein degradation (Fig. 5A). Truncated products derived from the reporter genes were detected in the presence of MG132. The strongest induction of translation arrest and protein degradation was observed with the CKKKKKRKKKNK sequence derived from Rmp1p (Fig. 5B, lanes 9 and 10). Truncated products derived from the reporter genes were also detected in the not4Δ mutant, and the same CKKKKKRKKKNK sequence showed the translation arrest activity (Fig. 5C, lane 5). A possible role for these sequences in gene regulation is currently being investigated.
FIGURE 5.
Endogenous sequences induce translation arrest coupled with protein degradation. A, construction of the pGPDp-GFP-X-FLAG-HIS3 reporter plasmids. These plasmids contain various sequences inserted between GFP and HIS3. The inserted amino acid sequences and original gene names are shown. B, endogenous sequence elements induce translation arrest coupled with protein degradation by the proteasome. W303 cells were transformed with pGPDp-GFP-X-FLAG-HIS3 plasmids that contained sequences from the indicated genes. The cells were grown in SC-Ura, and the samples were analyzed using Western blotting with anti-GFP antibodies. When indicated (+), cell extracts were prepared 2 h after the addition of 0.2 mm MG132. C, translation arrest products in the not4Δ mutant. W303not4Δ cells were transformed with pGPDp-GFP-X-FLAG-HIS3 plasmids that contained sequences from the indicated genes. The cells were grown in SC-Ura, and the samples were analyzed using Western blotting with anti-GFP antibodies.
DISCUSSION
Translation Arrest and Protein Degradation Induced by Consecutive Basic Amino Acids—Gene regulation mediated by the synthesis of specific amino acid sequences and associated translation arrest has been observed in prokaryotes and eukaryotes. Translation arrest of secM mRNA is coupled with protein secretion and plays a crucial role in the autoregulation of SecA protein expression (7, 28). The SecM nascent peptide interacts with the exit tunnel of the ribosome at the discrimination gate, thereby regulating the translation elongation rate of secM mRNA (6, 7). We found that 12 consecutive basic residues caused translation arrest, suggesting that positively charged side chains interact with the ribosome tunnel formed primarily by ribosomal RNA (29). The exit tunnel wall of the mammalian ribosome has a negative electrostatic potential (10) and may interact with peptides that contain positively charged residues (12). The interaction partner for the consecutive positively charged side chains has yet to be determined, although the phosphate rRNA backbone is a good candidate. Recently, it was shown that translation of nonstop mRNA is repressed after initiation and that the ribosome stalls on poly(A) sequences in mammalian cells (5). In addition, translation of poly(A) mRNA was less efficient in an in vitro system using rabbit reticulocyte lysates (5). Therefore, translation repression by consecutive basic amino acids may be conserved among eukaryotes.
Degradation of Translation Arrest Products Depends on Not4p—We found that Not4p is required for the degradation of translation arrest products. Not4p is also involved in transcriptional regulation via trimethylation of H3K4 (16, 17). Therefore, one could argue that Ccr4-Not4 may play a crucial role in the transcription of proteasomal subunits, an activity that may be impaired in the not4Δ mutant, leading to the stabilization of translation arrest products. Two lines of evidence do not support this possibility, however. First, we found that the levels of ubiquitinated proteins were markedly higher in the rpn6–2 mutant compared with the not4Δ mutant, although the increased level of the truncated GFP product was less than that in the not4Δ mutant (Fig. 3D). Second, translation of nonstop mRNA was not enhanced in the not4Δ mutant (Fig. 4A), although the nonstop product was degraded by the proteasome (3, 4). These findings suggest that the reduced degradation of translation arrest products in the not4Δ mutant is not due to a general defect in proteasomal activity.
We found that translation arrest induced by a stable stem-loop does not lead to arrest product degradation by the proteasome (Fig. 1B). This result is consistent with the idea that specific nascent peptide sequences are required for cotranslational degradation of arrest products by the proteasome. What is the mechanism underlying cotranslational degradation of the specific translation arrest products? One possibility is that the stalled ribosome may have a high affinity for Not4p, leading to efficient ubiquitination of the nascent peptides. It has been shown that a SecM nascent peptide modulates the conformation of the ribosome (30), indicating that the interaction between a nascent peptide and the exit tunnel wall can regulate translation by modulating the conformation of the ribosome. We suspect that interactions between certain nascent peptides and the exit tunnel wall lead to changes in the conformation of the ribosome and increase binding between the ribosome and Not4p, thereby regulating protein degradation.
It was shown previously that the interaction between Not4p and Ubc4p, which is dependent on the Not4p RING finger domain, is required for the ubiquitin-protein ligase activity for the EGD complex (18, 19). Ubc4p has recently been shown to interact with the 26 S proteasome in response to hygromycin B-induced translational misreading (18). Results from this study suggest that translation arrest induced by consecutive basic amino acids leads to Not4p-mediated protein degradation by the proteasome. Further, we propose a more general regulatory process in which translation arrest caused by specific nascent peptides leads to Not4p-dependent cotranslational protein degradation by the proteasome. Not4p is associated with ribosomes (Fig. 3H) and may be involved in the ubiquitination of aberrant nascent polypeptides on ribosomes by recruiting the E2 enzymes Ubc4p and Ubc5p as well as the proteasome.
Translation Arrest by Endogenous Basic Amino Acid Sequences—The results of this study led us to hypothesize that nascent peptides may play more general roles in translation elongation, protein stability, and protein expression, probably by modulating the ribosome structure. We found that the CKKKKKRKKKNK sequence from Rmp1p can induce translation arrest (Fig. 5). Rmp1p is a cytoplasmic component of the RNase MRP (31), an endonuclease that contributes to cell cycle-regulated degradation of daughter cell-specific mRNA (32). Because translation arrest leads to endonucleolytic cleavage of mRNA in prokaryotes and eukaryotes (8, 9, 33–36), translation arrest induced by consecutive basic amino acid sequences may lead to an endonucleolytic cleavage of mRNA, and RNase MRP may be responsible for this cleavage activity. It is likely that the cleavage products are degraded from the 3′ end by an exosome or from the 5′ end by Xrn1p and would only be detected in mutant cells. Experiments to investigate this possibility are ongoing.
Supplementary Material
Acknowledgments
We thank Dr. John Hershey for critically reading the manuscript and for valuable comments and suggestions. We thank Dr. Akio Toh-e and Dr. Yasushi Saeki for the rpn6–2 yeast strain. We also thank Dr. Toshiya Endo, Dr. Tohru Yoshihisa, and Dr. Shu-ichi Nishikawa for helpful discussions and materials. We also thank all members of the lab, especially Dr. Sayoko Ito-Harashima for the construction of the mutants and plasmids.
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Naito Foundation (to T. I.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Fig. S1.
Footnotes
The abbreviations used are: E3, ubiquitin-protein isopeptide ligase; E2, ubiquitin carrier protein; GFP, green fluorescent protein; DIG, digoxigenin.
References
- 1.van Hoof, A., Frischmeyer, P. A., Dietz, H. C., and Parker, R. (2002) Science 295 2262–2264 [DOI] [PubMed] [Google Scholar]
- 2.Frischmeyer, P. A., van Hoof, A., O'Donnell, K., Guerrerio, A. L., Parker, R., and Dietz, H. C. (2002) Science 295 2258–2261 [DOI] [PubMed] [Google Scholar]
- 3.Inada, T., and Aiba, H. (2005) EMBO J. 24 1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito-Harashima, S., Kuroha, K., Tatematsu, T., and Inada, T. (2007) Genes Dev. 21 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akimitsu, N., Tanaka, J., and Pelletier, J. (2007) EMBO J. 26 2327–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakatogawa, H., and Ito, K. (2001) Mol. Cell 7 185–192 [DOI] [PubMed] [Google Scholar]
- 7.Nakatogawa, H., and Ito, K. (2002) Cell 108 629–636 [DOI] [PubMed] [Google Scholar]
- 8.Onouchi, H., Lambein, I., Sakurai, R., Suzuki, A., Chiba, Y., and Naito, S. (2004) Biochem. Soc. Trans. 32 597–600 [DOI] [PubMed] [Google Scholar]
- 9.Chiba, Y., Ishikawa, M., Kijima, F., Tyson, R. H., Kim, J., Yamamoto, A., Nambara, E., Leustek, T., Wallsgrove, R. M., and Naito, S. (1999) Science 286 1371–1374 [DOI] [PubMed] [Google Scholar]
- 10.Lu, J., Kobertz, W. R., and Deutsch, C. (2007) J. Mol. Biol. 371 1378–1391 [DOI] [PubMed] [Google Scholar]
- 11.Lu, J., and Deutsch, C. (2005) Nat. Struct. Mol. Biol. 12 1123–1129 [DOI] [PubMed] [Google Scholar]
- 12.Lu, J., and Deutsch, C. (2008) J. Mol. Biol. 384 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tucker, M., Staples, R. R., Valencia-Sanchez, M. A., Muhlrad, D., and Parker, R. (2002) EMBO J. 21 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, J., Rappsilber, J., Chiang, Y. C., Russell, P., Mann, M., and Denis, C. L. (2001) J. Mol. Biol. 314 683–694 [DOI] [PubMed] [Google Scholar]
- 15.Tucker, M., Valencia-Sanchez, M. A., Staples, R. R., Chen, J., Denis, C. L., and Parker, R. (2001) Cell 104 377–386 [DOI] [PubMed] [Google Scholar]
- 16.Laribee, R. N., Shibata, Y., Mersman, D. P., Collins, S. R., Kemmeren, P., Roguev, A., Weissman, J. S., Briggs, S. D., Krogan, N. J., and Strahl, B. D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5836–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulder, K. W., Brenkman, A. B., Inagaki, A., van den Broek, N. J., and Timmers, H. T. (2007) Nucleic Acids Res. 35 2428–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulder, K. W., Inagaki, A., Cameroni, E., Mousson, F., Winkler, G. S., De Virgilio, C., Collart, M. A., and Timmers, H. T. (2007) Genetics 176 181–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panasenko, O., Landrieux, E., Feuermann, M., Finka, A., Paquet, N., and Collart, M. A. (2006) J. Biol. Chem. 281 31389–31398 [DOI] [PubMed] [Google Scholar]
- 20.Amberg, D. C., Burke, D. J., and Strathern, J. N. (2005) Methods in Yeast Genetics, pp. 262–308, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 21.Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998) Yeast 14 953–961 [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., and Russell, D. W. (2006) The Condensed Protocols from Molecular Cloning:A Laboratory Manual, 3rd Ed., pp. 237–356, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 23.Fujimuro, M., Sawada, H., and Yokosawa, H. (1994) FEBS Lett. 349 173–180 [DOI] [PubMed] [Google Scholar]
- 24.Inada, T., Winstall, E., Tarun, S. Z., Jr., Yates, J. R., III, Schieltz, D., and Sachs, A. B. (2002) RNA 8 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, D. H., and Goldberg, A. L. (1996) J. Biol. Chem. 271 27280–27284 [DOI] [PubMed] [Google Scholar]
- 26.Berthelot, K., Muldoon, M., Rajkowitsch, L., Hughes, J., and McCarthy, J. E. (2004) Mol. Microbiol. 51 987–1001 [DOI] [PubMed] [Google Scholar]
- 27.Isono, E., Saito, N., Kamata, N., Saeki, Y., and Toh, E. A. (2005) J. Biol. Chem. 280 6537–6547 [DOI] [PubMed] [Google Scholar]
- 28.Nakatogawa, H., Murakami, A., and Ito, K. (2004) Curr. Opin. Microbiol. 7 145–150 [DOI] [PubMed] [Google Scholar]
- 29.Ban, N., Nissen, P., Hansen, J., Capel, M., Moore, P. B., and Steitz, T. A. (1999) Nature 400 841–847 [DOI] [PubMed] [Google Scholar]
- 30.Mitra, K., Frank, J., and Driessen, A. (2006) Nat. Struct. Mol. Biol. 13 957–964 [DOI] [PubMed] [Google Scholar]
- 31.Salinas, K., Wierzbicki, S., Zhou, L., and Schmitt, M. E. (2005) J. Biol. Chem. 280 11352–11360 [DOI] [PubMed] [Google Scholar]
- 32.Gill, T., Cai, T., Aulds, J., Wierzbicki, S., and Schmitt, M. E. (2004) Mol. Cell. Biol. 24 945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doma, M. K., and Parker, R. (2006) Nature 440 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onouchi, H., Nagami, Y., Haraguchi, Y., Nakamoto, M., Nishimura, Y., Sakurai, R., Nagao, N., Kawasaki, D., Kadokura, Y., and Naito, S. (2005) Genes Dev. 19 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sunohara, T., Jojima, K., Tagami, H., Inada, T., and Aiba, H. (2004) J. Biol. Chem. 279 15368–15375 [DOI] [PubMed] [Google Scholar]
- 36.Sunohara, T., Jojima, K., Yamamoto, Y., Inada, T., and Aiba, H. (2004) RNA 10 378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.