Abstract
SUV39H1 is a histone H3K9-specific methyltransferase important for heterochromatin formation, regulation of gene expression, and induction of senescence in premalignant cells. SUV39H1 forms a complex with SirT1, and its activity is stimulated by SirT1 binding. Here we present evidence that the product of the DBC1 (deleted in breast cancer 1) gene disrupts the SUV39H1-SirT1 complex. Furthermore, DBC1 binds to the SUV39H1 catalytic domain and inhibits its ability to methylate histone H3 in vitro and in vivo. Knockdown of endogenous DBC1 increased the level of cellular H3K9 methylation. As expected, DBC1 also binds to SirT1 and inhibits the deacetylase activity of SirT1. These results identify DBC1 as a novel cellular inhibitor of SUV39H1 activity. DBC1 may be an important regulator of heterochromatin formation and genomic stability by disrupting the SUV39H1-SirT1 complex and inactivating both enzymes.
SUV39H1 is the human homolog of the Drosophila Su(var)3-9 histone methyltransferase that specifically mediates trimethylation of histone H3 lysine 9 (1). A mouse knock-out experiment showed that SUV39H1 and its testis-specific homolog, SUV39H2, are required for most of the H3K9 methylation in heterochromatin regions (2). SUV39H1 has been shown to form a complex with pRb and plays a role in the inhibition of E2F1 by methylation of E2F1 target promoters (3). SUV39H1/H2-double-null mice have reduced viability during embryonic development and reduced growth as adult animals and are infertile due to abnormal chromosome segregation during spermatogenesis (2). SUV39H1-null mice are viable but predisposed to spontaneous B cell lymphomas (2). Furthermore, SUV39H1 deficiency blocks ras-induced premature senescence and promotes the development of T cell lymphoma when crossed into the Eμ-N-ras transgenic mouse (4). These observations suggest that SUV39H1 is involved in tumor suppressor pathways.
Sir2 (silent information regulator 2) functions as an NAD-dependent deacetylase and regulates chromatin silencing in Saccharomyces cerevisiae (5). Increased SIR2 gene dosage results in the extension of life span in yeast (6). The mammalian homolog of Sir2 (SirT1) regulates glucose homeostasis in mice by deacetylating and activating the transcription factor PGCα (7). Transgenic mice overexpressing SirT1 in pancreatic beta cells showed improved glucose tolerance and increased insulin secretion in response to glucose (8, 9). Furthermore, pharmacological activators of SirT1 such as resveratrol can mimic the anti-aging effects of calorie restriction in lower organisms, reduce insulin resistance in mice fed a high fat diet, and prolong survival (10–12). A more potent activator of SirT1 has shown therapeutic potential in the treatment of type 2 diabetes in animal models by improving insulin sensitivity and lowering plasma glucose levels (13).
SirT1 functions through regulating histone acetylation and heterochromatin formation (5). When targeted to promoters by other proteins, SirT1 deacetylates histones H4K16 and H3K9 and recruits H1 to promote the establishment of repressed chromatin (14). SirT1 also deacetylates many nonhistone proteins such as p53, Foxo, and Ku70 to regulate sensitivity to apoptosis (15–19). Recent studies suggest that SirT1 interacts with other chromatin-modifying enzymes to form multimeric complexes in which different enzymes act in a sequential or coordinated fashion. An interesting example is the interaction between SirT1 and SUV39H1 (20, 21). SirT1 interacts with the N-terminal chromodomain of SUV39H1 and deacetylates SUV39H1 on Lys266 to stimulate its methyltransferase activity. Furthermore, SirT1 binding alone also activates SUV39H1 through additional mechanisms independent of its deacetylase activity. Importantly, SirT1-null mouse embryo fibroblasts showed a significant loss of H3K9me3 in heterochromatin regions (21). Therefore, the SUV39H1-SirT1 complex may coordinately promote H3K9 methylation using the deacetylase activity of SirT1 and the enhanced methylase activity of SUV39H1.
Recent studies also revealed that SirT1 interacts with DBC1 (deleted in breast cancer 1) (22, 23). DBC1 was initially identified by its localization to a region of chromosome 8p21 that was homozygously deleted in human breast cancer (24). However, DBC1 was not considered to be the primary target of the deletion, and its role in cancer development remains to be determined. DBC1 is a large nuclear protein of 923 residues with no clear functional domains. During tumor necrosis factor α-induced apoptosis, DBC1 undergoes protease cleavage and cytoplasmic translocation, which may play a role in the cell death process (25). DBC1 uses an N-terminal putative leucine zipper to bind to the SirT1 catalytic domain and to inhibit its deacetylase activity (22). DBC1 knockdown increases SirT1 activity and protects cells from DNA damage-induced apoptosis. In this study, we present evidence that in addition to binding and inhibiting SirT1, DBC1 also binds to the catalytic domain of SUV39H1 and causes its complete inactivation. Furthermore, DBC1 expression disrupts the interaction between SUV39H1 and SirT1. Our results suggest that DBC1 may regulate heterochromatin formation by targeting both subunits of the SUV39H1-SirT1 complex.
MATERIALS AND METHODS
Cell Lines, Plasmids, and Reagents—H1299 and 293T cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. Plasmids expressing SirT1 were provided by Dr. Wei Gu (15). FLAG-tagged murine DBC1 was obtained by PCR amplification of cDNA. FLAG-tagged DBC1 deletion mutants were generated using the murine cDNA construct. Hemagglutinin (HA)2-tagged human DBC1 was a kind gift from Dr. Junjie Chen. Anti-SirT1 monoclonal antibody 10E4 was generated by immunization using SirT1-(470–747). To inhibit DBC1 expression by RNA interference, an ON-TARGETplus siRNA pool for DBC1 was purchased from Dharmacon. H1299 cells were transfected with 100 nm siRNA and Lipofectamine 2000 reagent (Invitrogen) for 48 h.
Western Blotting—Cells were lysed in lysis buffer (50 mm Tris-HCl, pH 8.0, 5 mm EDTA, 150 mm NaCl, 0.5% Nonidet P-40, and 1 mm phenylmethylsulfonyl fluoride) and centrifuged for 15 min at 14,000 × g, and the supernatant was used for immunoprecipitation and Western blotting. Insoluble nuclear pellet was washed once with lysis buffer and extracted with 0.4 m HCl for histones. Histones were then neutralized with NaOH and Tris-HCl buffer. Cell lysate (10–50 μg of protein) and histones (∼1 μg of protein) were fractionated by SDS-PAGE and transferred to Immobilon-P filters (Millipore). The filter was blocked for 1 h with phosphate-buffered saline containing 5% nonfat dry milk and 0.1% Tween 20. Endogenous human SirT1 was detected by Western blotting of whole cell extract using monoclonal antibody 10E4. DBC1 was detected using an antibody from Bethyl Laboratories. The filter was developed using SuperSignal (Pierce) or ECL Plus reagent (Amersham Biosciences).
Luciferase Reporter Assay—Cells (50,000/well) were cultured in 24-well plates and transfected with a mixture containing 10 ng of Gal4-TK-luciferase reporter, 10 ng of cytomegalovirus lacZ, 0.8 ng of Gal4-E2F1-C, 5–20 ng of SUV39H1, 5–10 ng of Rb, and 30 ng of DBC1 plasmids. Transfection was achieved using Lipofectamine 2000 reagent, and cells were analyzed for luciferase and β-galactosidase expression after 24–48 h. The luciferase/β-galactosidase activity ratio was used as an indicator of transcriptional activity.
In Vitro Methylation Assay—H1299 cells were transfected with plasmids encoding Myc-SUV39H1, FLAG-DBC1, and FLAG-DBC1 deletion mutants. SUV39H1 was immunoprecipitated with anti-Myc antibody and protein G beads, whereas DBC1 was immunoprecipitated with M2 beads (Sigma). The beads (∼20-μl bed volume) were mixed with 15 μl of methylation buffer (50 mm Tris-HCl, pH 8.5, 5 mm MgCl2, and 4 mm dithiothreitol), 10 μg of core histones (Sigma), and 2 μCi of S-[3H]adenosylmethionine (15 Ci/mmol; Amersham Biosciences) and incubated for 1 h at 30 °C with mixing. The sample was boiled in Laemmli sample buffer, fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, sprayed with EN3HANCE™, and exposed to film for 24–48 h at –80 °C. After obtaining a 3H-labeled histone autoradiograph, the filter was washed in methanol and used for Western blotting to detect protein expression levels.
Deacetylase Assay—Plasmids encoding SirT1, SirT1-363A, and FLAG-DBC1 were transfected into H1299 cells. Cells were lysed in 50 mm Tris-HCl, pH 8.0, 5 mm EDTA, 150 mm NaCl, and 1 mm phenylmethylsulfonyl fluoride by sonication and centrifuged for 15 min at 14,000 × g, and the insoluble debris was discarded. Cell lysate containing 10 μg of protein was used for deacetylation assay. A 3H labeling histone deacetylase kit (Upstate) was used for testing SirT1 activity. Trichostatin A (200 ng/ml) was added to the reaction to suppress NAD+-independent histone deacetylases in the cell lysate. Histone deacetylase activities (3H counts) were normalized by relative SirT1 concentrations in the sample, which were determined by SirT1 enzyme-linked immunosorbent assay. In the SirT1 enzyme-linked immunosorbent assay, antibody 10E4 was immobilized on the enzyme-linked immunosorbent assay plate, followed by incubation with cell lysate. The bound SirT1 was detected with a rabbit anti-SirT1 antibody.
In Vitro Binding Assay—GST-DBC1 truncation mutants were expressed in Escherichia coli and bound to glutathione beads. SUV39H1 was translated with a TnT translation kit in the presence of [35S]methionine (Promega). GST-DBC1 beads were incubated with 35S-labeled SUV39H1 at 4 °C for 1 h. The beads were washed with 5% sucrose, 50 mm Tris-HCl, pH 7.4, 5 mm EDTA, 0.1% Nonidet P-40, and 500 mm NaCl. Bound proteins were eluted with 15 mm reduced glutathione and analyzed by SDS-PAGE and autoradiography.
RESULTS
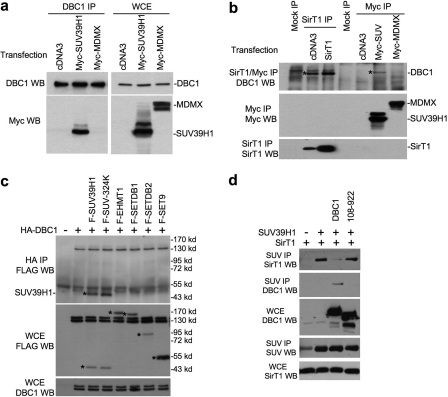
DBC1 Interacts Specifically with SUV39H1—Recent studies revealed that DBC1 binds to SirT1 and inhibits its deacetylase activity (22, 23). Furthermore, SirT1 interacts with SUV39H1 and stimulates its methyltransferase activity (21). In testing the effects of DBC1 on the SUV39H1-SirT1 complex, we found that DBC1 also interacts strongly with SUV39H1. In a cotransfection assay, immunoprecipitation (IP) of FLAG-DBC1 specifically coprecipitated Myc-SUV39H1 (Fig. 1a), and no binding was detected between DBC1 and the negative control Myc-MDMX. Because of the very low levels of endogenous SUV39H1, the interaction between endogenous SUV39H1 and DBC1 was not detectable. However, when cells were transfected with Myc-SUV39H1, IP of Myc-SUV39H1 (but not Myc-MDMX) coprecipitated endogenous DBC1 (Fig. 1b). As expected, IP of endogenous or transfected SirT1 also coprecipitated endogenous DBC1 (Fig. 1b). SUV39H1-DBC1 coprecipitation was not enhanced by expression of SirT1 (data not shown), suggesting that the interaction was not due to the bridging effect of SirT1 (see below). In an in vitro binding assay, GST-DBC1 efficiently pulled down in vitro-translated SUV39H1 (Fig. 2b), suggesting that the binding is a direct interaction. These results demonstrate that DBC1 is a novel and specific binding partner of SUV39H1.
FIGURE 1.
Interaction between DBC1 and SUV39H1. a, H1299 cells were transiently transfected with the indicated plasmids for 48 h. Endogenous DBC1 was immunoprecipitated with anti-DBC1 antibody. Coprecipitated SUV39H1 was detected by anti-Myc Western blotting (WB). DBC1, Myc-MDMX, and Myc-SUV39H1 expression levels were confirmed in WCE by anti-DBC1 and anti-Myc antibodies. b, SirT1 was immunoprecipitated with antibody 10E4, whereas SUV and MDMX were immunoprecipitated with anti-Myc antibody. Coprecipitated DBC1 was detected by anti-DBC1 Western blotting. SUV39H1 and MDMX were detected by anti-Myc Western blotting. c, HA-DBC1 was cotransfected into H1299 cells with FLAG-tagged histone methyltransferases and immunoprecipitated with anti-HA antibody. Coprecipitated enzymes were detected by anti-FLAG Western blotting. Expression of HA-DBC1 in the WCE was detected with anti-DBC1 antibody. d, H1299 cells were transiently transfected with the indicated plasmids. SUV39H1 was immunoprecipitated with mouse anti-SUV39H1 antibody. Coprecipitated SirT1 and DBC1 were detected with antibody 10E4 and anti-DBC1 antibody, respectively. SUV39H1 was detected by reprobing with rabbit anti-SUV antibody.
FIGURE 2.
Mapping of DBC1 domain involved in SUV39H1-DBC1 and SirT1-DBC1 interactions. H1299 cells were transiently transfected with the indicated plasmids for 48 h. a, DBC1 was immunoprecipitated with anti-FLAG antibody. SUV39H1 coprecipitation was detected by anti-SUV39H1 Western blotting (WB). WT, wild-type. b, in vitro-translated SUV39H1 was incubated with GST-DBC1 truncation mutants bound to glutathione beads. The captured SUV39H1 was detected by SDS-PAGE and autoradiography. c, DBC1 and truncation mutants were immunoprecipitated with M2 beads. SirT1 coprecipitation was detected by rabbit anti-SirT1 antibody, whereas DBC1 was detected by rabbit anti-FLAG antibody. d, the diagram summarize results in a–c. NLS, nuclear localization signal; LZ, leucine zipper.
To further test the specificity of the SUV39H1-DBC1 interaction, DBC1 was cotransfected with several FLAG-tagged histone methyltransferases (EHMT1, Setdb1, Setdb2, and Set9). The common FLAG epitope allowed us to compare the binding efficiency with FLAG-SUV39H1. The result indicated that only SUV39H1 showed detectable binding to DBC1 (Fig. 1c). Furthermore, the enzymatically inactive SUV39H1-324K mutant showed identical binding to DBC1 compared with wild-type SUV39H1 (Fig. 1c). Therefore, SUV39H1-DBC1 binding is highly specific among the histone H3K9 methyltransferases.
SUV39H1 forms a complex with SirT1 and is activated by SirT1 (21). To test the effect of DBC1 on the SUV39H1-SirT1 interaction, cells transfected with combinations of SUV39H1, SirT1, and DBC1 were analyzed by IP/Western blotting. The results showed efficient SUV39H1-SirT1 binding as reported previously (21). Interestingly, DBC1 expression strongly inhibited SUV39H1-SirT1 complex formation, accompanied by the formation of the SUV39H1-DBC1 complex (Fig. 1d). A DBC1 mutant defective for SirT1 and SUV39H1 binding (residues 108–922) did not affect the SUV39H1-SirT1 complex. Therefore, DBC1 binding to SirT1 or SUV39H1 disrupts the SUV39H1-SirT1 complex. This result also ruled out SirT1 as a mediator of the SUV39H1-DBC1 interaction.
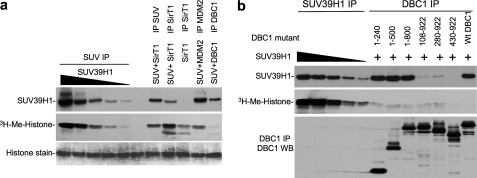
Mapping of Binding Sites on SUV39H1 and DBC1—To identify the domains on DBC1 and SUV39H1 that mediate the interaction, deletion mutants were generated for SUV39H1 and DBC1. Co-IP of transfected FLAG-DBC1 mutants with Myc-SUV39H1 also showed that the N-terminal residues 1–240 of DBC1 were necessary and sufficient for binding SUV39H1 (Fig. 2, a and d). In an in vitro binding assay, GST-DBC1-(100–240) was sufficient for binding in vitro-translated SUV39H1 (Fig. 2b). Therefore, it appears that DBC1 uses a sizable domain (residues 100–240) rather than a small peptide region to interact with SUV39H1. The first eight amino acid residues (positions 100–107) of this domain seem to be important for binding because mutant 108–922 was unable to bind SUV39H1 (Fig. 2a). Deletion mapping using SUV39H1 mutants revealed that a region between positions 220 and 300 was sufficient for binding to DBC1 (Fig. 3, a and b). Interestingly, this region is part of the SET domain that encodes methyltransferase activity. This interaction may be responsible for the inhibition of SUV39H1 activity by DBC1 (see below).
FIGURE 3.
Mapping of SUV39H1 domain involved in SUV39H1-DBC1 interaction. a, DBC1 was immunoprecipitated with anti-HA antibody. SUV39H1 coprecipitation was detected by rabbit anti-FLAG antibody. WT, wild-type; WB, Western blot. b, diagrams summarizing the results are shown. Chromo, chromodomain.
DBC1 binds to both SUV39H1 and SirT1 but disrupts the SUV39H1-SirT1 complex rather than bridging their interaction (Fig. 1d). This suggests that the SUV39H1- and SirT1-binding sites on DBC1 may overlap, precluding the formation of a trimeric complex. However, the leucine zipper region (positions 243–264) of DBC1 has been reported as the binding site for SirT1 (22), which does not overlap with the SUV39H1-binding site we identified (positions 1–240). To address this question, we tested the DBC1 deletion mutants for SirT1 binding by cotransfection and IP/Western blotting. The result showed that deleting the N-terminal residues 1–108 of DBC1 abrogated much of the SirT1 binding, whereas the DBC1-(1–240) fragment retained full activity for SirT1 binding (Fig. 2c). DBC1-(1–240) was able to abrogate SirT1 suppression of p53 tran-scriptional activity in a reporter gene assay (data not shown). Examination of published results also revealed that the DBC1-(Δ1–230) mutant lost most of its SirT1 binding (22). Therefore, we conclude that DBC1 uses the same N-terminal domain (amino acids 1–240) to bind SUV39H1 and SirT1. This is consistent with its ability to disrupt the SUV39H1-SirT1 complex rather than forming a trimeric complex.
DBC1 Inhibits Methyltransferase Activity of SUV39H1—To test the functional effect of DBC1 on SUV39H1, purified core histones were used as substrate to measure the methyltransferase activity of SUV39H1. Myc-SUV39H1 was transfected into cells and immunopurified by anti-Myc IP. This enzyme preparation showed strong activity in methylating histone H3 in the presence of S-[3H]adenosylmethionine. However, when Myc-SUV39H1 was purified by coprecipitation with FLAG-DBC1, the complex showed almost no methyltransferase activity (Fig. 4a). In contrast, SUV39H1 copurified as a complex with SirT1 showed stronger activity than SUV39H1 alone as reported previously (21). In a second control, SUV39H1 copurified by binding to MDM23 also remained active. Therefore, DBC1 binding to SUV39H1 inactivates its histone methylase function, possibly by blocking the catalytic SET domain.
FIGURE 4.
DBC1 inhibits SUV39H1 methyltransferase activity with its N-terminal domain. H1299 cells were transiently transfected with the indicated plasmids for 48 h. a, different amounts of SUV39H1 (relative levels 1×, 2×, 4×, 8×, and 16×) were immunoprecipitated with anti-Myc antibody. SirT1 was immunoprecipitated with antibody 10E4; MDM2 and SUV39H1 were immunoprecipitated with anti-FLAG antibody. The beads were used for in vitro methylation of histone H3 and detected by 3H autoradiography. SUV39H1 was detected by Western blotting. Histone H3 on the membrane was revealed by Coomassie staining. b, SUV39H1 was immunoprecipitated in the same manner as indicated in a. DBC1 and deletion mutants were immunoprecipitated with anti-FLAG antibody. The beads were used for in vitro methylation of histone H3. Expression levels of DBC1 mutants and coprecipitation of SUV39H1 were confirmed by Western blotting (WB). Wt, wild-type.
Because DBC1 is a large protein but uses only a small N-terminal region to bind SUV39H1, we tested whether the N-terminal domain alone is sufficient to inactivate SUV39H1. When DBC1 deletion mutants were used to copurify SUV39H1 after cotransfection, SUV39H1 captured by the smallest DBC1 fragment (residues 1–240), was inactive (Fig. 4b). Therefore, the N-terminal domain of DBC1 is necessary and sufficient to bind and inhibit SUV39H1.
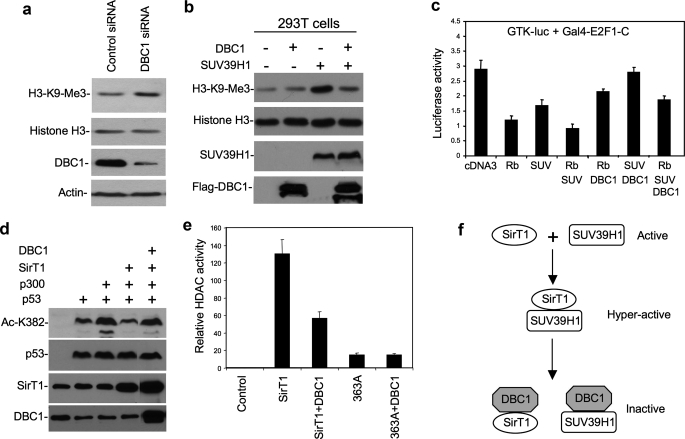
DBC1 Inhibition of SUV39H1 in Vivo—To test whether DBC1 has a role in regulating SUV39H1 in vivo, H1299 cells were treated with DBC1 siRNA to inhibit its expression. The level of global H3K9 trimethylation was analyzed by Western blotting of acid-extracted histones. The results showed that partial knockdown of DBC1 resulted in a moderate but reproducible increase in total K9me3 levels (Fig. 5a). Transient transfection of H1299 cells with SUV39H1 induced a significant increase in total K9me3 levels. Coexpression of DBC1 abrogated the induction of K9me3 by SUV39H1 (Fig. 5b). SUV39H1 is recruited by Rb to suppress E2F1 activity (3, 26). In a reporter gene assay, SUV39H1 cooperated with Rb to inhibit the ability of Gal4-E2F1-C (the E2F1 activation domain) to activate the Gal4-TK-luciferase reporter. Expression of DBC1 partially neutralized the repression by Rb and SUV39H1 (Fig. 5c). These results showed that the activity of SUV39H1 in vivo is regulated by DBC1.
FIGURE 5.
DBC1 inhibits SUV39H1 in vivo. a, H1299 cells were transfected with DBC1 siRNA or control siRNA. Histones were extracted, and the H3K9me3 level was detected with anti-H3K9me3 antibody and reprobing with anti-H3 antibody. DBC1 in the WCE was detected with anti-DBC1 antibody. b, 293T cells were transiently transfected with the indicated plasmids for 48 h. Histone was extracted, and H3K9me3 was detected with anti-H3K9me3 antibody. DBC1 in the WCE was detected with anti-DBC1 antibody. SUV39H1 in the WCE was detected with anti-Myc antibody. c, H1299 cells were transfected with the indicated plasmids for 48 h. Relative luciferase activity was determined and normalized by cotransfected lacZ control. d, H1299 cells were transiently transfected with the indicated plasmids for 48 h. Acetylation of p53 Lys382 was detected with anti-Ac-K382 antibody and reprobed with p53 antibody DO1. e, H1299 was transfected with the indicated plasmids for 48 h. Cell lysate was analyzed for histone deacetylase (HDAC) activity using a 3H-labeled acetylated H3 peptide as substrate. f, a model of DBC1 regulation of the SUV39H1-SirT1 complex is shown. Error bars represent S.D. from three experiments.
Recent studies suggested that DBC1 inhibits SirT1 deacetylase activity. Our results confirmed this observation. Cotransfection of DBC1 blocked the ability of SirT1 to deacetylate p53 Lys382 in vivo (Fig. 5d). Furthermore, SirT1 coexpressed with DBC1 showed reduced deacetylase activity in vitro when assayed using acetylated histone H3 peptide as substrate (Fig. 5e). Therefore, DBC1 is an effective regulator of both SirT1 and SUV39H1 by disrupting complex formation and also inactivating the enzymatic functions of both proteins (Fig. 5f).
DISCUSSION
The results described above identified the first cellular inhibitor for SUV39H1. Formation of the SUV39H1-SirT1 complex not only results in deacetylation and functional activation of SUV39H1 but may also allow the two enzymes to act cooperatively and sequentially to promote histone methylation and heterochromatin formation (21). By disrupting SUV39H1-SirT1 binding and inactivating both SUV39H1 and SirT1, DBC1 has the potential to act as an efficient regulator of chromatin modifications.
SUV39H1 is responsible for most of the H3K9 trimethylation in heterochromatin regions (2). SUV39H1 deficiency causes predisposition to spontaneous B cell lymphomas and failure to induce premature senescence in response to the activated ras oncogene (4). Growth regulation by the retinoblastoma protein Rb also involves recruitment of SUV39H1. Therefore, SUV39H1 has an overall biological function as a tumor suppressor.
Despite its ability to inhibit apoptosis during acute DNA damage, SirT1 also has features of a growth suppressor. SirT1-deficient mouse embryo fibroblasts show increased proliferation and spontaneous immortalization (27). SirT1 knockdown increases the proliferation of human fibroblasts (28). Furthermore, transgenic overexpression of SirT1 inhibits the development of intestinal neoplasia in the Adenomatosis polyposis coli mutant mouse (29). SirT1 also inhibits E2F1 and causes cell cycle arrest after overexpression (30). SirT1 deficiency abrogates the formation of heterochromatin regions in mouse embryo fibroblasts (21). SUV39H1 and SirT1 interaction is also important for repression of rDNA transcription during glucose starvation (20). These observations suggest that SirT1 and SUV39H1 function in a common pathway. Formation of the SUV39H1-SirT1 complex and activation of SUV39H1 in such a complex provide a molecular basis for functional cooperation and synergism. The ability of DBC1 to disrupt and inactivate both members of the SUV39H1-SirT1 complex suggests that DBC1 may be an important regulator of heterochromatin formation, gene expression, genomic stability, and cell proliferation.
Acknowledgments
We thank the Moffitt Molecular Biology Core for DNA sequence analysis. We also thank Drs. Junjie Chen and Wei Gu for constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant CA121291 (to J. C.).
Footnotes
The abbreviations used are: HA, hemagglutinin; siRNA, small interfering RNA; GST, glutathione S-transferase; IP, immunoprecipitation; WCE, whole cell extract; MDMX, murine double-minute gene X.
L. Chen, Z. Li, and J. Chen, unpublished data.
References
- 1.Aagaard, L., Laible, G., Selenko, P., Schmid, M., Dorn, R., Schotta, G., Kuhfittig, S., Wolf, A., Lebersorger, A., Singh, P. B., Reuter, G., and Jenuwein, T. (1999) EMBO J. 18 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., Opravil, S., Doyle, M., Sibilia, M., and Jenuwein, T. (2001) Cell 107 323–337 [DOI] [PubMed] [Google Scholar]
- 3.Nielsen, S. J., Schneider, R., Bauer, U. M., Bannister, A. J., Morrison, A., O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R. E., and Kouzarides, T. (2001) Nature 412 561–565 [DOI] [PubMed] [Google Scholar]
- 4.Braig, M., Lee, S., Loddenkemper, C., Rudolph, C., Peters, A. H., Schlegelberger, B., Stein, H., Dorken, B., Jenuwein, T., and Schmitt, C. A. (2005) Nature 436 660–665 [DOI] [PubMed] [Google Scholar]
- 5.Imai, S., Armstrong, C. M., Kaeberlein, M., and Guarente, L. (2000) Nature 403 795–800 [DOI] [PubMed] [Google Scholar]
- 6.Haigis, M. C., and Guarente, L. P. (2006) Genes Dev. 20 2913–2921 [DOI] [PubMed] [Google Scholar]
- 7.Rodgers, J. T., Lerin, C., Haas, W., Gygi, S. P., Spiegelman, B. M., and Puigserver, P. (2005) Nature 434 113–118 [DOI] [PubMed] [Google Scholar]
- 8.Moynihan, K. A., Grimm, A. A., Plueger, M. M., Bernal-Mizrachi, E., Ford, E., Cras-Meneur, C., Permutt, M. A., and Imai, S. (2005) Cell Metab. 2 105–117 [DOI] [PubMed] [Google Scholar]
- 9.Bordone, L., Motta, M. C., Picard, F., Robinson, A., Jhala, U. S., Apfeld, J., McDonagh, T., Lemieux, M., McBurney, M., Szilvasi, A., Easlon, E. J., Lin, S. J., and Guarente, L. (2006) PLoS Biol. 4 e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howitz, K. T., Bitterman, K. J., Cohen, H. Y., Lamming, D. W., Lavu, S., Wood, J. G., Zipkin, R. E., Chung, P., Kisielewski, A., Zhang, L. L., Scherer, B., and Sinclair, D. A. (2003) Nature 425 191–196 [DOI] [PubMed] [Google Scholar]
- 11.Lagouge, M., Argmann, C., Gerhart-Hines, Z., Meziane, H., Lerin, C., Daussin, F., Messadeq, N., Milne, J., Lambert, P., Elliott, P., Geny, B., Laakso, M., Puigserver, P., and Auwerx, J. (2006) Cell 127 1109–1122 [DOI] [PubMed] [Google Scholar]
- 12.Baur, J. A., Pearson, K. J., Price, N. L., Jamieson, H. A., Lerin, C., Kalra, A., Prabhu, V. V., Allard, J. S., Lopez-Lluch, G., Lewis, K., Pistell, P. J., Poosala, S., Becker, K. G., Boss, O., Gwinn, D., Wang, M., Ramaswamy, S., Fishbein, K. W., Spencer, R. G., Lakatta, E. G., Le Couteur, D., Shaw, R. J., Navas, P., Puigserver, P., Ingram, D. K., de Cabo, R., and Sinclair, D. A. (2006) Nature 444 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milne, J. C., Lambert, P. D., Schenk, S., Carney, D. P., Smith, J. J., Gagne, D. J., Jin, L., Boss, O., Perni, R. B., Vu, C. B., Bemis, J. E., Xie, R., Disch, J. S., Ng, P. Y., Nunes, J. J., Lynch, A. V., Yang, H., Galonek, H., Israelian, K., Choy, W., Iffland, A., Lavu, S., Medvedik, O., Sinclair, D. A., Olefsky, J. M., Jirousek, M. R., Elliott, P. J., and Westphal, C. H. (2007) Nature 450 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaquero, A., Scher, M., Lee, D., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. (2004) Mol. Cell 16 93–105 [DOI] [PubMed] [Google Scholar]
- 15.Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., and Gu, W. (2001) Cell 107 137–148 [DOI] [PubMed] [Google Scholar]
- 16.Vaziri, H., Dessain, S. K., Ng Eaton, E., Imai, S. I., Frye, R. A., Pandita, T. K., Guarente, L., and Weinberg, R. A. (2001) Cell 107 149–159 [DOI] [PubMed] [Google Scholar]
- 17.Cohen, H. Y., Miller, C., Bitterman, K. J., Wall, N. R., Hekking, B., Kessler, B., Howitz, K. T., Gorospe, M., de Cabo, R., and Sinclair, D. A. (2004) Science 305 390–392 [DOI] [PubMed] [Google Scholar]
- 18.Motta, M. C., Divecha, N., Lemieux, M., Kamel, C., Chen, D., Gu, W., Bultsma, Y., McBurney, M., and Guarente, L. (2004) Cell 116 551–563 [DOI] [PubMed] [Google Scholar]
- 19.Yuan, Z., Zhang, X., Sengupta, N., Lane, W. S., and Seto, E. (2007) Mol. Cell 27 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murayama, A., Ohmori, K., Fujimura, A., Minami, H., Yasuzawa-Tanaka, K., Kuroda, T., Oie, S., Daitoku, H., Okuwaki, M., Nagata, K., Fukamizu, A., Kimura, K., Shimizu, T., and Yanagisawa, J. (2008) Cell 133 627–639 [DOI] [PubMed] [Google Scholar]
- 21.Vaquero, A., Scher, M., Erdjument-Bromage, H., Tempst, P., Serrano, L., and Reinberg, D. (2007) Nature 450 440–444 [DOI] [PubMed] [Google Scholar]
- 22.Zhao, W., Kruse, J. P., Tang, Y., Jung, S. Y., Qin, J., and Gu, W. (2008) Nature 451 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J. E., Chen, J., and Lou, Z. (2008) Nature 451 583–586 [DOI] [PubMed] [Google Scholar]
- 24.Hamaguchi, M., Meth, J. L., von Klitzing, C., Wei, W., Esposito, D., Rodgers, L., Walsh, T., Welcsh, P., King, M. C., and Wigler, M. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13647–13652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundararajan, R., Chen, G., Mukherjee, C., and White, E. (2005) Oncogene 24 4908–4920 [DOI] [PubMed] [Google Scholar]
- 26.Vandel, L., Nicolas, E., Vaute, O., Ferreira, R., Ait-Si-Ali, S., and Trouche, D. (2001) Mol. Cell. Biol. 21 6484–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chua, K. F., Mostoslavsky, R., Lombard, D. B., Pang, W. W., Saito, S., Franco, S., Kaushal, D., Cheng, H. L., Fischer, M. R., Stokes, N., Murphy, M. M., Appella, E., and Alt, F. W. (2005) Cell Metab. 2 67–76 [DOI] [PubMed] [Google Scholar]
- 28.Abdelmohsen, K., Pullmann, R., Jr., Lal, A., Kim, H. H., Galban, S., Yang, X., Blethrow, J. D., Walker, M., Shubert, J., Gillespie, D. A., Furneaux, H., and Gorospe, M. (2007) Mol. Cell 25 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Firestein, R., Blander, G., Michan, S., Oberdoerffer, P., Ogino, S., Campbell, J., Bhimavarapu, A., Luikenhuis, S., de Cabo, R., Fuchs, C., Hahn, W. C., Guarente, L. P., and Sinclair, D. A. (2008) PLoS ONE 3 e2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, C., Chen, L., Hou, X., Li, Z., Kabra, N., Ma, Y., Nemoto, S., Finkel, T., Gu, W., Cress, W. D., and Chen, J. (2006) Nat. Cell Biol. 8 1025–1031 [DOI] [PubMed] [Google Scholar]