Abstract
IRAK2, a member of the interleukin-1 receptor-associated kinase (IRAK) family, has been implicated in Toll-like receptor (TLR)-mediated signaling. We generated IRAK2-deficient mice to examine its function in detail. These mice are resistant to lipopolysaccharide-induced septic shock, because of impaired TLR4-mediated induction of pro-inflammatory cytokines and chemokines. Although IRAK2 deficiency did not affect TLR4-mediated NFκB activation, a reduction of lipopolysaccharide (LPS)-mediated mRNA stabilization contributed to the reduced cytokine and chemokine production observed in bone marrow-derived macrophages from IRAK2-deficient mice. Furthermore, the ratios of LPS-induced cytokine and chemokine mRNAs in translation-active (polysomal) versus translation-inactive (free ribosomes) pools were reduced in IRAK2-deficient macrophages compared with wild type macrophages. Importantly, LPS-induced phosphorylation of MKK3/6, MNK1, and eIF4E was significantly reduced in IRAK2-deficient macrophages compared with wild type macrophages. Moreover, LPS stimulation induced an interaction of IRAK2 with TRAF6, MKK3/6, and MK2, implicating a critical role for mitogen-activated protein kinase signaling in LPS-induced IRAK2-mediated post-transcriptional control. These results reveal that IRAK2 is required for LPS-mediated post-transcriptional control of cytokine and chemokine expression, which plays an essential role in TLR4-induced septic shock.
Toll-like receptors (TLRs)4 are employed by the innate immune system to recognize conserved molecules associated with invading microorganisms, leading to inflammatory responses and linking to adaptive immunity (1–6). While TLR4 recognizes lipopolysaccharide (LPS) from Gram-negative bacteria, which can cause septic shock (7), TLR2 forms heterodimers with TLR1 and TLR6 to respond to a wide variety of lipid ligands associated with Gram-positive bacteria, mycoplasma, and fungi (8–11). Although TLR5 and TLR11 recognize conserved protein motifs (including those present in bacterial flagellin and a profilin-like molecule of the protozoan parasite Toxoplasma gondii) (12), TLR3, TLR7, TLR8, and TLR9, which are intracellular, detect nucleic acids derived from viruses and bacteria (13, 14). In addition, TLR7 and TLR8 can also recognize synthetic imidazoquinoline-like molecules, which have anti-viral activities (13, 14).
Upon binding their ligands, all of the TLRs except TLR3 recruit the adaptor molecule MyD88 through the Toll/IL-1 receptor domain, mediating MyD88-dependent pathways (15). MyD88 then recruits the serine-threonine kinases IRAK4 and IRAK1. IRAK4 phosphorylates IRAK1, which then mediates the recruitment of TRAF6 to the receptor complex (16). The IRAK1-TRAF6 complex then dissociates from the receptor to interact with and activate TAK1, a member of the MAP kinase kinase kinase (MAPKKK) family (17). The activation of TAK1 eventually leads to the activation of NFκB and JNK (18), which in turn induce transcription of inflammatory cytokine and chemokine genes, such as those encoding TNFα, IL-1β, IL-6, and IL-8. TLR3 and TLR4 also employ a MyD88-independent pathway that uses Toll/IL-1 receptor domain-containing adaptor inducing interferon-β (TRIF) to activate NFκB and IRF3. TLR4-mediated MyD88-independent activities are abolished in mice lacking TRAM, another adaptor in this pathway (19).
While IL-1R and TLRs regulate transcription, they also enable gene expression by stabilizing otherwise unstable mRNAs encoding pro-inflammatory proteins. Many of the mRNAs of cytokines and chemokines have very short half-lives because of AU-rich sequence elements (AREs) located within their 3′-untranslated regions (20). Therefore, the regulation of mRNA stability controls inflammatory gene expression. Degradation of mRNA in the 3′ to 5′ direction, the predominant mechanism in mammalian cells, is carried out through the activation of a large complex of exonucleases termed the exosome. Binding proteins that interact with ARE in mRNAs play essential roles in directing mRNA decay through their interactions with the exosome. The ARE-binding protein, tristetraprolin (TTP), plays a critical role in destabilizing cytokines and chemokines (20). Several signaling pathways modulate the function of these ARE-binding proteins, including the p38 mitogen-activated protein kinase (p38 MAPK) and p38/MAPK-activated protein kinase 2 (MK2), extracellular signal-regulated kinase (ERK), and JNK pathways (20). The MKK3/6-p38-MK2 signaling cascade has been implicated in LPS-mediated mRNA stabilization through the phosphorylation and stabilization of the ARE-binding protein TTP, resulting in a reduction of the affinity of TTP for AREs, relieving ARE-dependent destabilization of TNFα mRNA (21, 22). On the other hand, MAP kinase-interacting serine/threonine kinases (MNKs), substrates of the p38 and ERK pathways, are components of a translational regulatory pathway. Specifically, MNK1 interacts with the translation initiation complex, where it phosphorylates the main CAP-binding protein eIF4E, promoting CAP-dependent translation (23–25).
We recently found that the kinase activity of IRAK4 is essential for IL-1R and TLR-mediated stabilization of cytokine and chemokine mRNAs (26). A reduction of LPS-, R848-, and IL-1-mediated mRNA stability contributed to reduced cytokine and chemokine production in bone marrow (BM)-derived macrophages from IRAK4 kinase-inactive knock-in mice. IRAK2 is another member of the IRAK family. Because the overexpression of IRAK2 activates NFκB, it has been implicated in multiple TLR signaling pathways (27). In a recent study, peritoneal macrophages from IRAK2-deficient mice showed impaired production of cytokines in response to multiple TLR ligands (28). Intriguingly, the IRAK2-deficient macrophages showed normal early but reduced late activation of NFκB in response to the TLR2 ligand Malp-2, suggesting that IRAK2 is critical for the late transcriptional response to the activation of TLR2.
To investigate the role of IRAK2 in TLR-mediated signaling, we also generated IRAK2-deficient mice, which are resistant to lipopolysaccharide-induced septic shock, because of impaired TLR4-mediated induction of proinflammatory cytokines and chemokines. Importantly, a reduction of LPS-mediated mRNA stability and translation initiation contributed to the reduced cytokine and chemokine production is observed in BM-derived macrophages from IRAK2-deficient mice. Together, these results demonstrate a critical role for IRAK2 in regulating post-transcriptional control of LPS-triggered proinflammatory cytokine and chemokine production, an essential step in the development of septic shock.
EXPERIMENTAL PROCEDURES
Reagents—LPS (Escherichia coli 055:B5 (macrophages) and O111:B4 (mice)) was purchased from Sigma-Aldrich. R848 (1-(2-hydroxy-2-methylpropyl)-2-methyl-1H-imidazo[4,5-c]-quinolin-4-amine) was commercially synthesized by GLSynthesis. Antibodies to phospho-MKK3/6, phospho-ERK, phospho-p38, phospho-MNK1, phospho-eIF4E, phospho-IKKα/β, phospho-IκBα, and MK2 were purchased from Cell Signaling. Antibodies to IRAK1, IκBα, and IKKα/β were purchased from Santa Cruz. Antibody to MEKK3 was purchased from BD Biosciences Pharmingen. Antibody to IRAK2 was purchased from Abcam. Antibody to β-actin was purchased from Sigma-Aldrich. Antibody to TTP was a gift from Dr. Perry Blackshear at National Institute of Environmental Health Sciences. cDNA encoding mouse IRAK2 was amplified with primers 5′-CATGGCTTGCTACATCTACC-3′ and 5′-ACGTTTGTCTGTCCAGTTGA-3′ and cloned into TA cloning vector pCR8/GW/TOPO (Invitrogen). A 223-bp fragment, obtained from this construct after cutting with EcoRI and XmaI, was used as a probe for Northern analysis of IRAK2. The oligomer for NF-κB electrophoretic mobility assay was 5′-AGTTGAGGGGACTTTCCCAGGC-3′ from Santa Cruz Biotechnology.
Generation of IRAK2 Knock-out Mice—IRAK2-deficient mice were generated using standard gene targeting technology. A targeting vector was constructed that replaced 5 kb of the Irak2 gene, including 2.7 kb of the 5′ regulatory region, exon 1, and 2.1 kb of intron 1, with a neomycin resistance gene and included the HSV-tk gene in the vector sequence. Successfully targeted ES cells were injected into blastocysts and implanted into pseudopregnant females. Chimeric male offspring were mated to WT C57BL/6 female, and germ line transmission of the mutant Irak2 allele was confirmed by Southern and PCR analyses.
In Vivo Endotoxin Challenges—Adult wild type (129×C57BL6) and knock-out mice (129×C57BL6-Irak 2 KO) were used for all experiments listed in the study, and the studies were approved by the Institutional Animal Care and Use Committee at The University of Texas Southwestern Medical Center. For survival studies adult WT and IRAK2-deficient mice were injected intraperitoneally with LPS (20 mg/kg) and observed for 7 days. Animals in distress or moribund were euthanized and counted as nonsurvivors. For cytokine response, the mice were injected intraperitoneally with saline or LPS (4 mg/kg). At either 1.5 or 3.5 h, the animals were anesthetized, phlebotomized, and euthanized, and the serum was separated and frozen for subsequent cytokine analysis.
Bone Marrow-derived Macrophages—These obtained from bone marrow of tibia and femur by flushing with Dulbecco's minimal essential medium. The bone marrows were cultured in Dulbecco's minimal essential medium with 20% heat-inactivated fetal bovine serum, and 30% L929 cell-conditioned medium for 5 days for differentiation and proliferation of bone marrow-derived macrophages.
Northern Analysis and Quantitative Real Time PCR—Total RNA was isolated by using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Quantitative PCR was performed as described (26). The results were normalized to mouse β-actin. The specific primer sequences used for mouse β-actin, mouse TNF-α, mouse IL-6, mouse KC and mouse glyceraldehyde-3-phosphate dehydrogenase, are as follows: β-actin (133 bp), 5′-GGTCATCACTATTGGCAACG-3′ and 5′-ACGGATGTCAACGTCACACT-3′; TNF-α (103 bp), 5′-CAAAGGGAGAGTGGTCAGGT-3′ and 5′-ATTGCACCTCAGGGAAGAGT-3′; IL-6 (127 bp), 5′-GGACCAAGACCATCCAATTC-3′ and 5′-ACCACAGTGAGGAATGTCCA-3′; KC (125 bp), 5′-TAGGGTGAGGACATGTGTGG-3′ and 5′-AAATGTCCAAGGGAAGCGT-3′; and glyceraldehyde-3-phosphate dehydrogenase (179bp), 5′-GAGCTGAACGGGAAGCTCAC-3′ and 5′-TGTCATACCAGGAAATGAGC-3′.
Electrophoretic Mobility Shift Assay, Co-immunoprecipitation, and Immunoblotting—BM-derived macrophages from IRAK2 wild type and knock-out mice were cultured as indicated above. After stimulation with LPS (1 μg/ml) or R848 (1 μg/ml), the cells were harvested, and an electrophoretic mobility shift assay was performed as described (29). Co-immunoprecipitation and immunoblotting were performed as described (30).
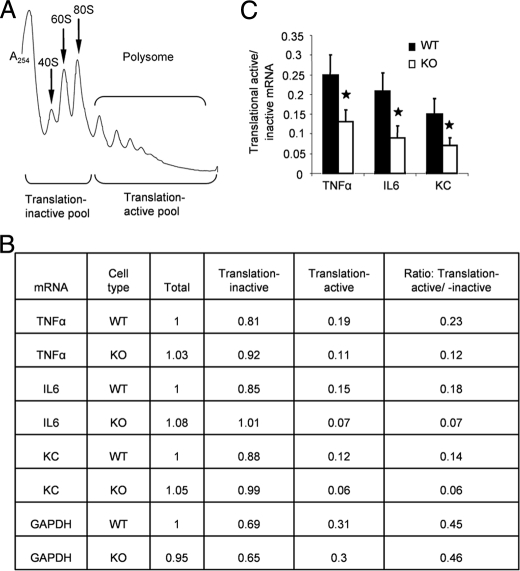
Polysome Fraction Analysis—Cytoplasmic extracts were prepared from BM-derived macrophages stimulated with LPS (1 μg/ml) as described (31). Cytoplasmic exacts were carefully layered over 5–50% linear sucrose gradients in polysome buffer (10 mm HEPES, pH 7.5, 100 mm KCl, 2.5 mm MgCl2, 1 mm dithiothreitol, 50 units of recombinant RNasin (Promega), and 0.1% Igepal-CA630 (Sigma)) and centrifuged at 17,000 rpm in a Beckman SW32.1 Ti rotor for 18 h at 4 °C. Gradients were fractioned by using an ISCO gradient fractionation system equipped with a UA-6 detector. Light RNP fractions, 40, 60, and 80 S and heavy polysome fractions were monitored by the continuous UV absorption profile at A254, and 22 tubes of 750-μl fractions were collected. The fractions collected in the first eight tubes, representing light RNP and free ribosomes, were used to isolate the translation-inactive pool of mRNAs, and the following fractions were used to isolate the translation-active mRNAs. RNAs were isolated from these fractions by extraction with TRIzol.
Statistical Analysis—Survival comparison was analyzed using a Log-Rank test for survivors versus nonsurvivors at day 7. One-way analysis of variance was used to compare serum cytokine and chemokine concentrations from in vivo challenges and macrophages stimulated with TLR ligands. One-way analysis of variance was also used to compare cytokine and chemokine mRNA levels from IRAK2 wild type and deficient macrophages stimulated with TLR ligands. Repeated measures analysis of variance was used to compare LPS-mediated cytokine and chemokine mRNA stabilization in IRAK2 wild type and deficient macrophages.
RESULTS
IRAK2 Is Required for TLR4-induced Cytokine and Chemokine Production—We recently generated IRAK2-deficient mice by deleting the promoter, first exon, including the translation start site, and proximal first intron of the Irak2 gene (Fig. 1). To examine the role of IRAK2 in TLR4-mediated pathways, we compared wild type and IRAK2-deficient mice for their susceptibility to LPS-induced septic shock. Whereas 80% of the wild type mice died within 7 days of LPS challenge, only 10% of the IRAK2-deficient mice died during this period (Fig. 2A). These results demonstrate that IRAK2 is critical for the lethal response to LPS in vivo. Consistent with the lethality curve, IRAK2-deficient mice had lower serum cytokine concentrations compared with wild type mice following LPS challenge (Fig. 2B). Furthermore, cytokine (IL-6 and TNF-α) and chemokine (KC) production were significantly reduced in BM-derived macrophages from IRAK2-deficient mice in response to LPS (TLR4) and R848 (TLR7) stimulation compared with wild type cells (Fig. 3A), indicating that IRAK2 is crucial for TLR-mediated cytokine and chemokine production.
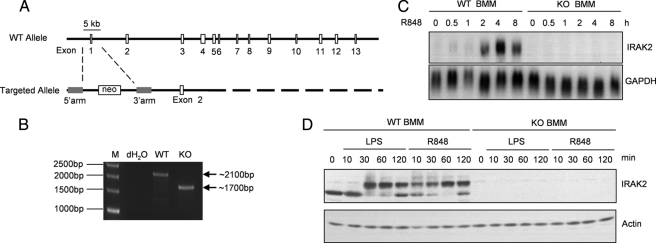
FIGURE 1.
Generation of IRAK2-deficient mice. A, structure of the mouse IRAK2 gene and the targeted IRAK2 gene. B, PCR analysis of IRAK2 genomic DNA in wild type and knock-out mice. KO, knock-out. C, BM-derived macrophages (BMM) from wild type and IRAK2-deficient mice were untreated or treated with R848 (1 μg/ml). Total RNA (10 μg) was analyzed by the Northern method with a probe specific for IRAK2. IRAK2 mRNA was induced by R848 in BM-derived macrophages from wild type but not IRAK2-deficient mice. D, BMM from WT and KO mice were untreated or treated with LPS (1 μg/ml) or R848 (1 μg/ml). Cell lysates were analyzed by Western method with an antibody against IRAK2. Stimulation by either LPS or R848 led to modification of IRAK2 in macrophages from wild type mice. IRAK2 protein was absent in IRAK2-deficient macrophages.
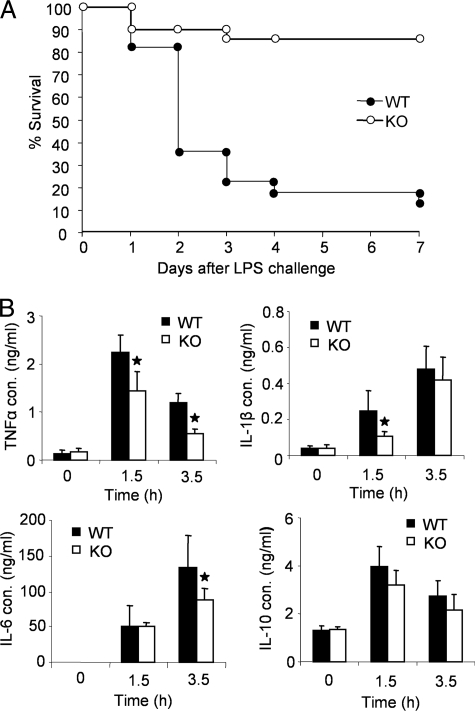
FIGURE 2.
Resistance of IRAK2-deficient mice to LPS-induced septic shock. A, age- and sex-matched wild type (n = 22) and IRAK2-deficient (n = 21) mice were injected intraperitoneally with LPS (20 mg/kg body weight). Survival was monitored for 7 days after LPS challenge. B, concentrations of TNFα, IL1β, IL 6, and IL 10 were measured by enzyme-linked immunosorbent assay in sera from wild type and IRAK2-deficient mice 0, 1.5, and 3.5 h after injection of LPS (4 mg/kg of body weight). The results shown are the means ± S.D. of triplicate determinations. *, p < 0.05.
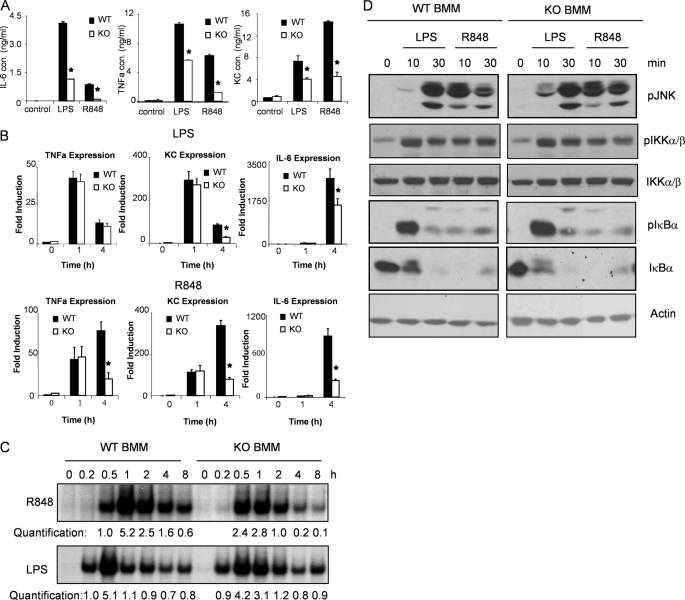
FIGURE 3.
TLR-mediated cytokine and chemokine production and NFκB activation in IRAK2-deficient macrophages. A, WT and IRAK2-deficient (KO) BM-derived macrophages were treated with LPS (1 μg/ml) or R848 (1 μg/ml) for 24 h. IL-6, KC, and TNF-α concentrations in supernatant were measured by enzyme-linked immunosorbent assay. The results shown are the means ± S.D. of triplicate determinations. *, p < 0.05. B, WT or IRAK2-deficient (KO) BM-derived macrophages were treated with LPS (1 μg/ml) and R848 (1 μg/ml) for indicated times, and total RNAs (2 μg) were analyzed by reverse transcription-PCR. *, p < 0.05. C, cell lysates from WT and IRAK2-deficient (KO) BM-derived macrophages, either untreated or treated with LPS (1 μg/ml) or R848 (1 μg/ml) for indicated times analyzed by electrophoresis mobility shift assay with an NFκB-specific probe. The levels of NFκB activation were analyzed by Scion Image 1.62C alias and are presented as fold increases compared with the samples treated for 30 min with R848 or 15 min with LPS. D, WT or IRAK2-deficient (KO) BM-derived macrophages were untreated or treated with LPS (1 μg/ml) or R848 (1 μg/ml). The cell lysates were analyzed by the Western method with antibodies against phospho-JNK, phospho-IKKα/β, total IKKα/β, phospho-IκBα, total IκBα, and actin.
IRAK2 Is Not Required for Either Early or Late TLR4-induced NFκB Activation—One hour after stimulation, IRAK2-deficient BM-derived macrophages showed similar LPS/R848-induced expression of TNFα and KC mRNAs compared with wild type macrophages, indicating that IRAK2 deficiency does not have a significant impact on TLR4/7-induced transcription at early times (Fig. 3B). Consistent with this, LPS/R848 activated the signaling cascade leading to NFκB activation and JNK activation within 30 min after stimulation (Fig. 3, C and D).
Previous studies assigned a critical role to IRAK2 in sustaining TLR2-induced NFκB activation (28), and the loss of this prolonging effect could explain the overall reduction in TLR2-induced cytokine and chemokine production in IRAK2-deficient macrophages. We also observed attenuated Malp2-induced NFκB activation at late times in IRAK2-deficient macrophages (supplemental Fig. S1). Moreover, R848-induced late NFκB activation (mediated by TLR7) was also reduced in IRAK2-deficient macrophages (Fig. 3C). Consistent with this observation, TLR7-dependent induction of TNFα, IL-6, and KC mRNAs was dramatically reduced 4 h after stimulation (Fig. 3B), indicating that IRAK2 was also critical for sustained R848-mediated gene expression.
Surprisingly, IRAK2-deficient and wild type macrophages showed similar LPS-induced NFκB DNA binding activity at both early (within 30 min) and late times (within 1–8 h) of stimulation, indicating that IRAK2 is not essential for sustained NFκB activation in TLR4-dependent signaling (Fig. 3C and supplemental Fig. S1). Whereas the LPS-induced late NFκB activation was normal in IRAK2-deficient macrophages (Fig. 3C), the LPS-mediated expression of IL-6 and KC mRNA was reduced at late times (Fig. 3B). Moreover, although IRAK2-deficient macrophages showed reduced TNFα protein production compared with wild type macrophages after LPS stimulation (Fig. 3A), LPS stimulation induced similar levels of TNFα mRNA both early and late (Fig. 3B). Together, these results indicate that the reduced TLR4-induced cytokine and chemokine production in IRAK2-deficient macrophages, and the resistance of IRAK2-deficient mice to LPS-induced septic shock are probably not simply due to lack of sustained TLR4 signaling (NFκB activation) and gene transcription at late times. Therefore, we propose that IRAK2 has a unique role in mediating LPS-induced cytokine and chemokine production.
IRAK2 Is Required for Stabilization of a Subset of Cytokine and Chemokine mRNAs in Response to LPS—We previously discovered that the kinase activity of IRAK4 is required to stabilize cytokine and chemokine mRNAs in response to TLR4 activation (26), leading us to hypothesize that IRAK2 might be involved in TLR4-induced cytokine and chemokine mRNA stabilization. To test this hypothesis, we investigated cytokine and chemokine mRNA stability in IRAK2-deficient macrophages in response to LPS. IRAK2-deficient and wild type macrophages were first treated with LPS alone for 1.5 h to induce gene expression, followed by treatment with actinomycin D (to block transcription) and LPS (to stabilize mRNAs) for 0.5–4 h. Although KC and IL-6 mRNAs were induced to comparable levels in IRAK2-deficient and wild type macrophages, the decay rate of those mRNAs was accelerated, and the levels of both messages were lower in BM-derived macrophages from IRAK2-deficient mice as compared with wild type cells (Fig. 4). These results demonstrate that, as for IRAK4, IRAK2 is also required for LPS-mediated cytokine and chemokine mRNA stabilization.
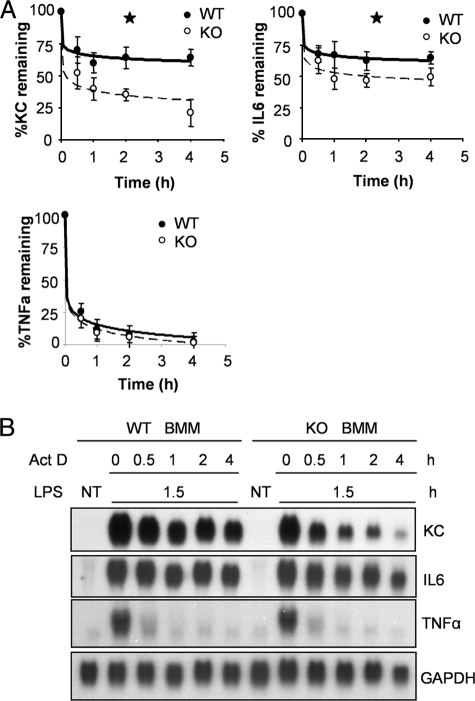
FIGURE 4.
Impaired LPS-mediated mRNA stabilization in IRAK2-deficient macrophages. A, WT and IRAK2-deficient (KO) BM-derived macrophages were pretreated with LPS (1 μg/ml) for 90 min and then treated with actinomycin D (Act D, 5 μg/ml) and LPS (1 μg/ml) for indicated times. KC, IL-6, and TNFα and mRNAs were analyzed by quantitative reverse transcription-PCR, normalized by β-actin, and plotted. The results shown are the mean ± S.D. of three independent experiments. *, p < 0.05. B, WT and IRAK2-deficient (KO) BM-derived macrophages were either untreated (NT) or pretreated with LPS (1 μg/ml) for 90 min and then treated with actinomycin D (5 μg/ml) and LPS (1 μg/ml) for the indicated times. Total RNAs (10 μg) were analyzed by the Northern method.
IRAK2 Plays an Important Role in the Regulation of LPS-mediated Gene Translation—Although the production of TNFα protein was reduced in IRAK2-deficient macrophages in response to LPS (Fig. 3A), the expression of TNFα mRNA was similar to that in wild type macrophages (Fig. 3B). Therefore, we investigated the role of IRAK2 in the regulation of cytokine and chemokine translation. BM-derived macrophages from IRAK2-deficient and wild type mice were treated with LPS for 1.5 h, and a sucrose gradient was used to separate the mRNA translation-inactive pool (free ribosomes) and translation-active pool (polysomal) (Fig. 5A). Compared with wild type macrophages, the IRAK2-deficient macrophages had more cytokine and chemokine mRNAs in the translation-inactive pool than that in the translation-active pool (Fig. 5B). As a result, the ratios of LPS-induced cytokine and chemokine mRNAs in translation-active versus translation-inactive pools were reduced in IRAK2-deficient macrophages compared with wild type macrophages (Fig. 5C). These results suggest that IRAK2 is essential for normal cytokine and chemokine protein production. IRAK2-deficient macrophages showed reduced capacity to stabilize both KC and IL-6 mRNAs but did not modulate the half-life of TNFα. Although prior studies have demonstrated that TNFα mRNA decay is regulated by changes in decay mediated in part through the action of TTP, there is also significant evidence that translation of TNFα mRNA is a more important regulatory control than its decay (21, 32, 33). Even under conditions of increased stability, TNFα mRNA remains highly unstable, consistent with the more significant role of translational control indicated in the present data.
FIGURE 5.
Impaired LPS-mediated protein translation in IRAK2-deficient macrophages. A, translation-active and -inactive mRNAs from LPS-treated BM-derived macrophages were isolated by sucrose gradient fraction. B, BM-derived macrophages from wild type and IRAK2-deficient mice were treated with LPS (1 μg/ml) for 1.5 h. Cytokine, chemokine, and glyceraldehyde-3-phosphate dehydrogenase mRNAs from unfractionated cell lysates, translation-active pools, and translation-inactive pools were analyzed by quantitative reverse transcription-PCR and normalized to β-actin. Similar results were obtained in three independent experiments. One representative is shown. C, the ratios of TNFα, IL-6, and KC mRNA from translation-active and -inactive pools are shown. The results shown are the means ± S.D. of three independent experiments. *, p < 0.05.
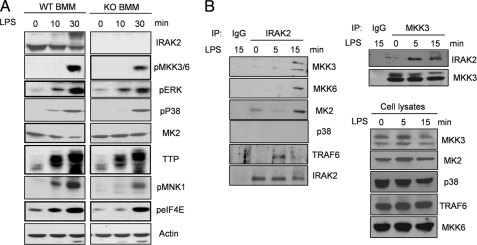
Activation of MAP Kinase Signaling Is Impaired in IRAK2-deficient Macrophages in Response to LPS—To study the role of IRAK2 on TLR4-mediated post-transcriptional control, BM-derived macrophages from wild type and IRAK2-deficient mice were examined for TLR-mediated activation of MAP kinase signaling. Although the phosphorylation of two MAP kinases, p38 and ERK, was slightly reduced in the IRAK2-deficient macrophages in response to LPS (Fig. 6A), the upstream kinase MKK3/6 and downstream effector molecules MNK1 and eIF4E were phosphorylated at lower levels in LPS-treated IRAK2-deficient macrophages (Fig. 6A). Of note, LPS stimulation led to MK2 degradation in wild type macrophages, but not in IRAK2-deficient macrophages, probably reflecting the lack of MK2 activation in the absence of IRAK2 (Fig. 6A). MK2 activation has been shown to play a critical role in LPS-induced TTP modification and stabilization (22). TTP accumulation was reduced in IRAK2-deficient macrophages upon LPS stimulation compared with wild type macrophages, indicating the potential role of IRAK2 in mediating TLR4-induced TTP stabilization (Fig. 6A). Taken together, these results demonstrate the TLR4-mediated activation of MAP kinase signaling is reduced in IRAK2-deficient macrophages, indicating a critical role for MAP kinase signaling in LPS-induced IRAK2-mediated post-transcriptional control (Fig. 7).
FIGURE 6.
Impaired activation of MAP kinase-mediated signaling in IRAK2-deficient macrophages in response to LPS. A, WT or IRAK2-deficient (KO) BM-derived macrophages were untreated or treated with LPS (1 μg/ml). The cell lysates were analyzed by the Western method with antibodies against IRAK2, phospho-MKK3/6, phospho-ERK, phospho-p38, MK2, TTP, phospho-MNK1, phospho-eIF4E, and actin. B, cell lysates from IRAK2 wild type or knock-out BM-derived macrophages, untreated or treated with LPS (1 μg/ml) for the indicated times, were immunoprecipitated (IP) with antibodies against IRAK2 and IgG, followed by Western analysis with antibodies against MKK3, MKK6, p38, MK2, and TRAF6. The cell lysates were also analyzed directly by the Western method with antibodies against MKK3, MKK6, p38, MK2, and TRAF6. The cell lysates from IRAK2 wild type BM-derived macrophages untreated or treated with LPS (1 μg/ml) were immunoprecipitated with antibodies against MKK3 and IgG, followed by Western analysis with antibodies against MKK3 and IRAK2.
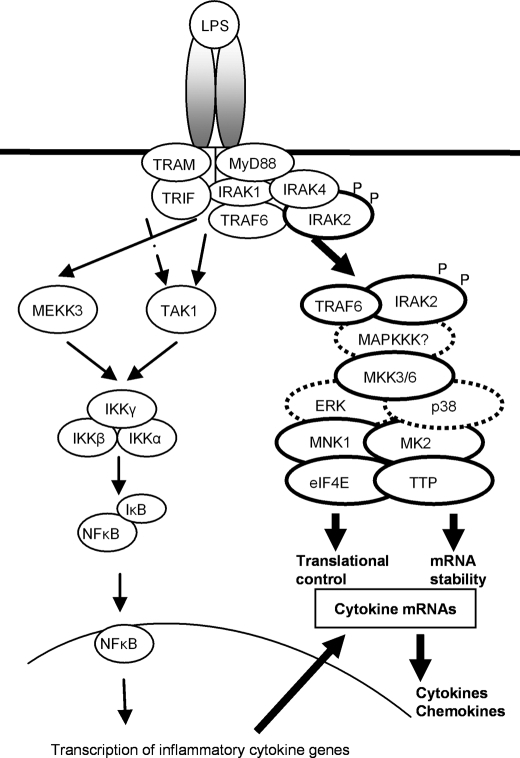
FIGURE 7.
Model of LPS-mediated IRAK2-dependent post-transcriptional control. Upon LPS stimulation in macrophages, IRAK4 is activated, leading to the modification and activation of IRAK1 and IRAK2. While IRAK1 and IRAK2 activate TAK1 and MEKK3 to mediate NFκB activation, the activated IRAK2 recruits kinases (MKK3/6, ERK, p38, MK2, and MNK1) and effector molecules (TRAF6, eIF4E, and TTP) to mediate mRNA stabilization and translation initiation.
To determine whether IRAK2 mediates the activation of MAP kinase signaling through protein-protein interactions, IRAK2 was immunoprecipitated from cell lysates of BM-derived macrophages, with or without LPS treatment, followed by Western analysis with antibodies against TRAF6 and components of MAP kinase pathways (Fig. 6B). TRAF6, MKK3, MKK6, and MK2 formed a complex with IRAK2 in an LPS-dependent manner. Importantly, an LPS-dependent IRAK2-MKK3 interaction was also detected in reciprocal experiments in which cell lysates were immunoprecipitated with anti-MKK3 followed by Western analysis with IRAK2 antibody (Fig. 6B). However, we failed to detect interaction of p38 with IRAK2. Collectively, these results suggest that IRAK2 forms one or more ligand-dependent signaling complexes with the MAP kinases important for mRNA stabilization and translation, thereby exerting its impact on LPS-induced post-transcriptional control (Fig. 7).
DISCUSSION
We report a novel function for IRAK2 in mediating LPS-triggered post-transcriptional control of cytokine and chemokine expression. Although IRAK2 deficiency did not affect the extent of either early or late TLR4-mediated NFκB activation, a reduction in LPS-mediated mRNA stability contributed to the reduced cytokine and chemokine production observed in BM-derived macrophages from IRAK2-deficient mice. Furthermore, the ratios of LPS-induced cytokine and chemokine mRNAs in translation-active versus translation-inactive pools were lower in IRAK2-deficient macrophages compared with wild type macrophages. These data show the requirement of IRAK2 for sustained translation of these mRNAs. Consequently, the IRAK2-deficient mice were resistant to lethal endotoxic shock, because of impaired proinflammatory cytokine and chemokine induction.
A previous study showed that IRAK2 is critical for sustaining NFκB activation at late times during TLR2-mediated signaling (28). In the present study, we observed that IRAK2 was also important for TLR7-induced NFκB activation at late times. Therefore, IRAK2 may play a similar role in TLR2- and TLR7-mediated signaling. However, our data demonstrated that IRAK2 was not required for sustained NFκB activation in TLR4-mediated signaling. Although TLR2 and TLR7 mediate NFκB activation solely through a MyD88-dependent pathway, delayed NFκB activation was observed in MyD88-deficient mice upon TLR4 activation with LPS. The TLR4-induced MyD88-independent pathway uses the adaptor molecule TRIF to recruit RIP1 and TRAF6 to mediate late activation of NFκB in response to LPS. This MyD88-independent pathway may compensate for an IRAK2 requirement in sustained NFκB activation in TLR4-mediated signaling. As a result, IRAK2 plays a more critical role following transcription than during its initiation or continuation in TLR4-dependent signaling. Therefore, the lack of LPS-induced cytokine mRNA stabilization and inefficient translation initiation probably accounts for the resistance of the IRAK2-deficient mice to lethal endotoxic shock.
Because TLR2- and TLR7-mediated late NFκB activation was reduced in IRAK2-deficient macrophages, TLR2/TLR7-induced gene transcription was affected, which significantly contributed to the reduced TLR2/7-mediated cytokine and chemokine production in the IRAK2-deficient macrophages. However, it is important to note that TLR7-induced phosphorylation of MKK3/6, MNK1, and eIF4E was significantly reduced in IRAK2-deficient macrophages compared with wild type macrophages, suggesting the potential role of IRAK2 in mediating the TLR7-induced signaling pathway important for post-transcriptional regulation.5 Future studies are required to uncouple transcription from post-transcriptional regulation by using reporter systems (driven by a constitutive promoter or an unrelated inducible promoter and under the control of 3′-untranslated region of cytokines/chemokines) to measure TLR2/TLR7-mediated post-transcriptional regulation with or without IRAK2.
Because IRAK4 kinase activity is required for IL-1R and TLR-mediated mRNA stability, IRAK4 substrates are likely to be downstream signaling components that mediate mRNA stabilization. We found that although IRAK4 kinase activity is required for LPS-induced modification of both IRAK1 and IRAK2, IRAK1 and IRAK2 do not depend on each other for modification in response to LPS stimulation.6 Although it is possible that IRAK4 phosphorylates IRAK2 directly upon receptor activation, it has recently been shown that IRAK2 itself also has kinase activity (28). Therefore, it is possible that IRAK4 mediates the activation of IRAK2, followed by IRAK2 autophosphorylation. Future studies are required to map the IRAK2 phosphorylation sites, identify the kinases for IRAK2 phosphorylation, and study the functional importance of IRAK2 modification. Although the kinase activity of IRAK2 is critical for TLR2-mediated late NFκB activation, it is important to determine whether IRAK2 kinase activity is necessary for the activation of downstream kinases (such as MAPKKKs and MKK3/6) that are known to modulate post-transcriptional regulation. Furthermore, Keating et al. (27) showed that IRAK2 can promote TRAF6 ubiquitination (27). In support of their study, we detected IRAK2-TRAF6 interaction upon LPS stimulation. Although TRAF6 appears not to be required for IL-1-induced mRNA stabilization in mouse embryonic fibroblasts (34), it is possible that TRAF6 or ubiquitinated TRAF6 plays a role in recruiting MKK3/6 to modulate LPS-induced post-transcriptional control in macrophages.
Co-immunoprecipitation experiments showed that MKK3/6 and MK2 are associated with IRAK2 in response to LPS, suggesting that IRAK2 may mediate cytokine mRNA stabilization through MAP kinase signaling. A MKK3/6-p38-MK2 signaling cascade has been implicated in LPS-mediated mRNA stabilization through phosphorylation and stabilization of the ARE-binding protein TTP (21, 22). However, p38 was not detected in the IRAK2-MKK3/6-MK2 complex. Furthermore, LPS-induced p38 phosphorylation was only slightly reduced, whereas phosphorylation of MKK3/6, MK2 activation and TTP stability were substantially reduced in IRAK2-deficient macrophages compared with wild type macrophages. These results suggest that MAP kinases other than p38 might be also involved in MKK3/6-mediated activation of the MK2 pathway. In support of this idea, LPS induced similar levels of p38 phosphorylation in IRAK4 kinase-inactive knock-in macrophages and wild type macrophages, despite the fact that the kinase activity of IRAK4 is required for LPS-mediated mRNA stabilization (26). On the other hand, LPS-induced phosphorylation of MKK3/6, MK2 activation and TTP stability were significantly reduced in IRAK4 kinase-inactive knock-in macrophages.6
Data from fractionated polysomes showed that the ratios of LPS-induced cytokine and chemokine mRNAs in translation-active versus translation-inactive pools were reduced in IRAK2-deficient macrophages compared with wild type macrophages, indicating an important role for IRAK2 in translational control. Previous studies have shown that MNK1 phosphorylates the main CAP-binding protein eIF4E, promoting CAP-dependent translation initiation (23–25). LPS-induced phosphorylation of MNK1 was indeed reduced in IRAK2-deficient macrophages and IRAK4 kinase-inactive knock-in macrophages. However, although previous studies indicate that MNK1 is a substrate of the p38 and ERK pathways, LPS-induced phosphorylation of these upstream kinases was only modestly reduced in IRAK2-deficient macrophages and not altered in IRAK4 kinase-inactive knock-in macrophages (26), suggesting the possible involvement of additional MAP kinase(s) in the activation of MNK1-mediated translational initiation.
In summary, whereas previous studies have shown redundant functions of IRAK1 and IRAK2 in TLR-induced early NFκB activation, IRAK2 is important for late NFκB activation in response to certain TLR ligands (including TLR2 and TLR7). Importantly, we now identify a novel and important function of IRAK2 in TLR4-mediated mRNA stabilization and translational control in macrophages, resulting in resistance of IRAK2-deficient mice to lethal endotoxic shock. Analysis of IRAK1-deficient mice showed that IRAK1 contributes only modestly to TLR-induced cytokine production in macrophages, and IRAK1 deficiency had marginal impact on LPS-induced septic shock (35). Taken together, these results suggest that IRAK2-mediated post-transcriptional regulation plays a critical role in TLR4-induced cytokine and chemokine production in macrophages (Fig. 7). It is important to note that IRAK1 has been shown to mediate IL-1-induced mRNA stabilization in nonmyeloid cells (34). Interestingly, we found that IL-1-induced mRNA stabilization was greatly reduced in IRAK1-deficient, but only modestly attenuated in IRAK2-deficient mouse embryonic fibroblasts and kidney epithelial cells.5 It is possible that IRAK1 and IRAK2 have cell type-specific roles in TLR/IL-1R-mediated post-transcriptional regulation.
Supplementary Material
Acknowledgments
We thank May Tsen and Junko Kuno for expert technical assistance. We thank Dr. Perry Blackshear for the gift of TTP antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants GM060020 (to X. L.), RO1 AI50019 (to J. T.), P50 GM021681 (to J. T.), and RO1 HL 79164 (to B. M.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: TLR, Toll-like receptor; IL, interleukin; IL-1R, IL-1 receptor; IRAK, IL-1R-associated kinase; LPS, lipopolysaccharide; BM, bone marrow; MAP, mitogen-activated protein kinase; MAPKKK, MAP kinase kinase kinase; TNF, tumor necrosis factor; ARE, AU-rich sequence element; TTP, tristetraprolin; MK2, MAPK-activated protein kinase 2; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; MEKK, MAPK/ERK kinase; WT, wild type; KC, mouse chemokine CXCL1.
J. Thomas and X. Li, unpublished data.
T. W. Kim and X. Li, unpublished data.
References
- 1.Medzhitov, R., Preston-Hurlburt, P., and Janeway, C. A., Jr. (1997) Nature 388 394–397 [DOI] [PubMed] [Google Scholar]
- 2.Rock, F. L., Hardiman, G., Timans, J. C., Kastelein, R. A., and Bazan, J. F. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi, O., Kawai, T., Sanjo, H., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Takeda, K., and Akira, S. (1999) Gene (Amst.) 231 59–65 [DOI] [PubMed] [Google Scholar]
- 4.Chuang, T. H., and Ulevitch, R. J. (2000) Eur. Cytokine Netw. 11 372–378 [PubMed] [Google Scholar]
- 5.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K., and Akira, S. (2000) Nature 408 740–745 [DOI] [PubMed] [Google Scholar]
- 6.Zhang, D., Zhang, G., Hayden, M. S., Greenblatt, M. B., Bussey, C., Flavell, R. A., and Ghosh, S. (2004) Science 303 1522–1526 [DOI] [PubMed] [Google Scholar]
- 7.Poltorak, A., He, X., Smirnova, I., Liu, M. Y., Huffel, C. V., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., Freudenberg, M., Ricciardi-Castagnoli, P., Layton, B., and Beutler, B. (1998) Science 282 2085–2088 [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi, O., Hoshino, K., Kawai, T., Sanjo, H., Takada, H., Ogawa, T., Takeda, K., and Akira, S. (1999) Immunity 11 443–451 [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi, O., Kaufmann, A., Grote, K., Kawai, T., Hoshino, K., Morr, M., Muhlradt, P. F., and Akira, S. (2000) J. Immunol. 164 554–557 [DOI] [PubMed] [Google Scholar]
- 10.Underhill, D. M., Ozinsky, A., Hajjar, A. M., Stevens, A., Wilson, C. B., Bassetti, M., and Aderem, A. (1999) Nature 401 811–815 [DOI] [PubMed] [Google Scholar]
- 11.Underhill, D. M., Ozinsky, A., Smith, K. D., and Aderem, A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 14459–14463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., Eng, J. K., Akira, S., Underhill, D. M., and Aderem, A. (2001) Nature 410 1099–1103 [DOI] [PubMed] [Google Scholar]
- 13.Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S., and Reis e Sousa, C. (2004) Science 303 1529–1531 [DOI] [PubMed] [Google Scholar]
- 14.Heil, F., Hemmi, H., Hochrein, H., Ampenberger, F., Kirschning, C., Akira, S., Lipford, G., Wagner, H., and Bauer, S. (2004) Science 303 1526–1529 [DOI] [PubMed] [Google Scholar]
- 15.Akira, S., Takeda, K., and Kaisho, T. (2001) Nat. Immunol. 2 675–680 [DOI] [PubMed] [Google Scholar]
- 16.Cao, Z., Xiong, J., Takeuchi, M., Kurama, T., and Goeddel, D. V. (1996) Nature 383 443–446 [DOI] [PubMed] [Google Scholar]
- 17.Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C., and Chen, Z. J. (2000) Cell 103 351–361 [DOI] [PubMed] [Google Scholar]
- 18.Ninomiya-Tsuji, J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z., and Matsumoto, K. (1999) Nature 398 252–256 [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto, M., Sato, S., Hemmi, H., Uematsu, S., Hoshino, K., Kaisho, T., Takeuchi, O., Takeda, K., and Akira, S. (2003) Nat. Immunol. 4 1144–1150 [DOI] [PubMed] [Google Scholar]
- 20.Anderson, P. (2008) Nat. Immunol. 9 353–359 [DOI] [PubMed] [Google Scholar]
- 21.Hitti, E., Iakovleva, T., Brook, M., Deppenmeier, S., Gruber, A. D., Radzioch, D., Clark, A. R., Blackshear, P. J., Kotlyarov, A., and Gaestel, M. (2006) Mol. Cell Biol. 26 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronkina, N., Kotlyarov, A., ttrich-Breiholz, O., Kracht, M., Hitti, E., Milarski, K., Askew, R., Marusic, S., Lin, L. L., Gaestel, M., and Telliez, J. B. (2007) Mol. Cell Biol. 27 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukunaga, R., and Hunter, T. (1997) EMBO J. 16 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda, T., Watanabe-Fukunaga, R., Fukuyama, H., Nagata, S., and Fukunaga, R. (2004) Mol. Cell Biol. 24 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowlett, R. M., Chrestensen, C. A., Nyce, M., Harp, M. G., Pelo, J. W., Cominelli, F., Ernst, P. B., Pizarro, T. T., Sturgill, T. W., and Worthington, M. T. (2008) Am. J. Physiol. 294 G452–G459 [DOI] [PubMed] [Google Scholar]
- 26.Kim, T. W., Staschke, K., Bulek, K., Yao, J., Peters, K., Oh, K. H., Vandenburg, Y., Xiao, H., Qian, W., Hamilton, T., Min, B., Sen, G., Gilmour, R., and Li, X. (2007) J. Exp. Med. 204 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keating, S. E., Maloney, G. M., Moran, E. M., and Bowie, A. G. (2007) J. Biol. Chem. 282 33435–33443 [DOI] [PubMed] [Google Scholar]
- 28.Kawagoe, T., Sato, S., Matsushita, K., Kato, H., Matsui, K., Kumagai, Y., Saitoh, T., Kawai, T., Takeuchi, O., and Akira, S. (2008) Nat. Immunol. 9 684–691 [DOI] [PubMed] [Google Scholar]
- 29.Qin, J., Jiang, Z., Qian, Y., Casanova, J. L., and Li, X. (2004) J. Biol. Chem. 279 26748–26753 [DOI] [PubMed] [Google Scholar]
- 30.Qin, J., Yao, J., Cui, G., Xiao, H., Kim, T. W., Fraczek, J., Wightman, P., Sato, S., Akira, S., Puel, A., Casanova, J., Su, B., and Li, X. (2006) J. Biol. Chem. 281 21013–21021 [DOI] [PubMed] [Google Scholar]
- 31.Chaudhuri, S., Vyas, K., Kapasi, P., Komar, A. A., Dinman, J. D., Barik, S., and Mazumder, B. (2007) RNA 13 2224–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson, K., and Sundler, R. (2006) Cytokine 33 52–57 [DOI] [PubMed] [Google Scholar]
- 33.Buxade, M., Parra, J. L., Rousseau, S., Shpiro, N., Marquez, R., Morrice, N., Bain, J., Espel, E., and Proud, C. G. (2005) Immunity 23 177–189 [DOI] [PubMed] [Google Scholar]
- 34.Hartupee, J., Li, X., Hamilton, T. (2008) J. Biol. Chem. 283 15689–15693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swantek, J. L., Tsen, M. F., Cobb, M. H., Thomas, J. A. (2000) J. Immunol. 164 4301–4306 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.