Abstract
Pdi1p (protein-disulfide isomerase) is a folding assistant of the endoplasmic reticulum (ER) that catalyzes disulfide formation and the isomerization of incorrect disulfides. Its disulfide forming activity is its essential function in Saccharomyces cerevisiae. A truncation mutant (Pdi1a′) that is competent in disulfide formation but deficient in catalyzing isomerization has only a small effect on growth, although the maturation of isomerase-requiring substrates (carboxypeptidase Y) is impaired (Xiao, R., Wilkinson, B., Solovyov, A., Winther, J. R., Holmgren, A., Lundstrom-Ljung, J., and Gilbert, H. F. (2004) J. Biol. Chem. 279, 49780–49786). We show here that there are multiple ways to compensate for defects in disulfide formation and isomerization in the ER. Genes of the unfolded protein response are induced, and deletions of the nonessential IRE1 or HAC1 genes are synthetically lethal. Diploid synthetic lethality analysis by microarray (dSLAM) using PDIa′ and a temperature-sensitive mutant of PDIa′ as query mutations reveals a group of 130 synthetically lethal genes. Only 10 of these correspond to genes clearly associated with the unfolded protein response. More than half are involved in vesicle traffic, not only out of and into the ER but anterograde and retrograde traffic from most cellular compartments. This suggests that defects in protein maturation in one intracellular compartment may be compensated for by adjusting vesicular traffic patterns throughout the cell.
Eukaryotic proteins destined for extracellular locations are processed in the endoplasmic reticulum (ER),2 where they fold and acquire the proper three-dimensional structure before exiting the ER to be delivered to their correct location (1). For those proteins that require disulfide formation, the oxidizing environment of the ER and the presence of chaperones and folding catalysts, such as protein-disulfide isomerase (PDI), ensure that the proper cysteines are connected with disulfide bonds. PDI is an essential ER folding assistant present in all eukaryotes. PDI has two catalytic activities, oxidation of protein sulfhydryls into disulfides (oxidase activity) and the rearrangement of incorrectly formed disulfides (isomerase activity) (2). PDI is composed of four structural domains, all with thioredoxin folds (3, 4). The active sites (CGHC) are in the N- and C-terminal thioredoxin domains (a and a′), separated by two structural/binding domains (b and b′). On an active site basis, the isolated a and a′ domains are capable of catalyzing disulfide formation in substrates at the same rate as the full-length protein, but a multidomain structure is required for catalyzing disulfide isomerization (5, 6).
Because PDI1 is an essential gene in Saccharomyces cerevisiae and is the only protein in yeast with significant disulfide isomerase activity, we were surprised to find that the a′ domain by itself expressed from the PDI1 promoter was sufficient to rescue the lethal pdi1Δ mutation (7). In addition, the amount of PDIa′ required to support nearly wild-type growth rates was within 2-fold of that of full-length PDI. The maturation of CPY, a yeast vacuolar protein that requires disulfide isomerization to rapidly gain its function, was significantly compromised (∼2-fold) despite the nearly normal growth rate. Although these observations confirm that the disulfide-forming activity of PDI (oxidase activity) is its essential function, they suggest that either essential yeast proteins do not require disulfide isomerization to fold correctly (CPY is a nonessential gene) or that there are unknown compensation mechanism at work. An obvious compensation mechanism would involve the up-regulation of the other ER homologues of PDI in yeast, Mpd1p, Mpd2p, Eug1p, or Eps1p. However, PDIa′ could still rescue the lethal PDI1 deletion with nearly normal growth even in a strain that had genomic deletions of all of the Pdi1p homologues (8).
Disulfide isomerization is part of an elaborate ER quality control system that ensures that proteins that cannot reach their native structure are retained in the ER until folding and post-translational modifications are completed or the misfolded protein is degraded (9). When proper folding is compromised, part of the cell's response is to induce the unfolded protein response, a signaling pathway from the ER to the nucleus that is responsible for modifying expression to increase the supply of ER chaperones and folding catalysts, including PDI, to help mediate the consequences of accumulating unfolded proteins in the ER (10).
To explore mechanisms that might be required to compensate for defects in the isomerase activity of Pdi1p, we examined the induction of the PDI1 promoter, the mRNA levels of the PDI1 homologues, and the state of induction of the unfolded protein response. The normally nonessential unfolded protein response (UPR) becomes essential when the isomerase activity of Pdi1p is deficient. To detect other synthetically lethal genes, we applied the diploid synthetic lethality array method (dSLAM), where the competitive growth of a mixture of ∼6000 bar-coded individual deletion strains is examined by microarray analysis before and after replacing full-length PDI with the single catalytic domain, PDIa′, or a temperature-sensitive mutant of the PDIa′ domain (tsPDIa′). In addition to a number of genes involved in stress responses, including the UPR, the synthetically lethal genes we identified are involved in almost all aspects vesicular trafficking, including exocytic and endocytic pathways, and genes involved in regulating cell growth. Although limiting cell growth would diminish the requirement for the ER providing new membrane proteins, the other group of genes may be involved in regulating vesicle trafficking, possibly in an effort to balance the appearance and disappearance of external membrane proteins due to a defect in the ER maturation. Experiments with the vital dye FM4-64 showed that compromising the maturation of ER proteins resulted in a decrease in the rate of endocytosis from the plasma membrane, consistent with the proposal of mechanisms to coordinate vesicle traffic pathways within the cell.
EXPERIMENTAL PROCEDURES
Media, Strains, and Plasmids—Antibiotic-resistant strains were maintained in YPD with either 200 μg/ml G418 (Invitrogen) or 50 μg/ml nourseothricin (ClonNAT) (Werner Bioagents). Sporulation medium contained 1% (w/v) potassium acetate, 0.05% (w/v) zinc acetate, and 0.3 mm l-histidine. For transformation with plasmids (URA3, cen) containing wtPDI, PDIa′, or tsPDIa′, transformants were selected on plates of synthetic complete medium without uracil (SC–Ura). Haploid selection magic medium (MM) plates contained 20 g/liter dextrose; 1.7 g/liter Difco yeast nitrogen base without amino acids and ammonium sulfate; 2 g/liter synthetic complete medium without leucine, histidine, uracil, and arginine (SC–Leu–His–Ura–Arg drop out); 1 g/liter sodium glutamate; 0.2 g/liter G418; 60 mg/liter l-canavanine with/without 50 μg/ml nourseothricin (ClonNAT). Plasmids used include YCplac33 (URA3, cen) (11) and Y33-14 (PDI promoter and terminator in YCplac33). Plasmids expressing PDI or PDIa′ (residues 375–485) were derived from YCplac33 by inserting the PDI coding sequence behind 543 bp of the PDI1 promoter in the HindIII/BamHI site of YCplac33.
β-Galactosidase Reporter Expression—The 879-bp fragment of the PDI promoter from YPP414-YPS (8) was digested by KpnI and EcoRI and inserted into the vector pYC-Z110 (12) digested by the same restriction enzymes. The resulting plasmid was transformed into pdi1Δ yeast strains M4174 (pdi1Δ) and M4477 (pdi1Δ, mpd1Δ, mpd2Δ, eps1Δ, eug1Δ) (13). In both strains, the deletion of PDI1 was complemented with a plasmid expressing Pdi1p or PDIa′. The β-galactosidase activity of yeast culture was assayed following the method of Amberg (14). Western blotting was performed as described previously (8), using purified Pdi1p or PDIa′ as standards against customer-made yeast PDIa′ antiserum (Pacific Immunology Corp.) and anti-PGK (Invitrogen) as a control. The protein concentration was analyzed using the program Scion (available on the World Wide Web).
Quantiative PCR—RNA isolation and cDNA synthesis were performed using the RNeasy minikit (Qiagen) and TaqMan RT reagents (Applied Biosystems) (15). The cDNA was synthesized from 2 μg of total RNA in a 50-μl volume. The reaction was incubated at 25 °C for 10 min and at 37 °C for 2 h and then inactivated at 95 °C for 5 min. Real time PCR analysis was performed on an MJ real time PCR using the following primers: PDI1 and PDIa′, aaatc gtcaa cgacc caaag (sense) and caatt acgac gcctc tgaca (antisense); MPD1, tttcc gtggt gttgt tttcc (sense) and ctgtc gctgt tcagg aatga (antisense); MPD2, tggca aagaa tcaag acgtg (sense) and tgcaa ttcct agaat catcg aa (antisense); KAR2, tgaca accaa ccaac cgtta (sense) and tctgt ggcag acacc ttcag (antisense); EUG1, accgt gaagg tactg ccaag (sense) and ttcgt aaata ggcgc aaacc (antisense); EPS1, tcgca agttg ggagt atgaa (sense) and gggaa tctct tcacg cattt (antisense); ERO1, tccca gctgg acaca aaaat (sense) and gcagt tgcgt aaccg gtagt (antisense); actin, gttac gtcgc cttgg acttc (sense) and ccaaa cccaa aacag aagga (antisense). To avoid amplification of genomic DNA, primers for each gene were chosen from different exons. A 2-μl aliquot from the cDNA synthesis from 40 ng of RNA/μl of reaction was used in 20 μl of PCR buffer containing Brilliant SYBR Green (Stratagene) with a 0.1 nm concentration of each primer. Amplification was performed in triplicate using actin, as a control. Denaturation at 95 °C for 7 min was followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 59 °C for 40 s, and extension at 72 °C for 50 s. The change in fluorescence SYBR Green in every cycle was monitored by the MJ system software.
Temperature-sensitive Mutations—Random mutagenesis of the PDI1a′ fragment was performed using error-prone PCR with biased nucleotide composition, high Mg2+, and the addition of Mn2+ (16). Reaction solutions contained 0.23 mm dATP, 0.20 mm dCTP, 0.57 mm dGTP, 4.0 mm dTTP, 0.5 mm MnCl2, 5.2 mm MgCl2, 10 mm Tris (pH 8.3), 50 mm KCl, 5 ng of bovine serum albumin/50 μl, 5 units/50 μl of Mutazyme II and Taq polymerase mix (Stratagene), and a 1 μm concentration of each primer (GATTC CTCTG TCTTC CAATTG and GACGT CGAAG TGACC GTTTT C for PDI1a′). The plasmid YPP414YPS-Ap containing PDI1 a′ with the PDI promoter and terminator was used as a template. PCR conditions were 94 °C for 2 min, 30 cycles at 94 °C for 1 min, 59 °C for 1 min, 72 °C for 80 s, and, finally, 72 °C for 10 min. The error-prone PCR product and vector, Y33-14 (URA3, cen), were digested with NotI and were co-transformed into the PDI1 haploid mutant strain 219A5 (MATa, PDI/pdiΔ::kanMX, CAN1/can1Δ::[LEU2-MFA1pr::HIS3]). To select temperature-sensitive alleles of PDIa′, the 219A5 (PDI/pdi1Δ::kanMX) strain containing the error-prone PDIa′ library was sporulated for 5 days at 25 °C and then plated on “magic medium” plates (MM; SC–Ura–Leu–His–Arg + Canavanine + G418) to select haploid pdi1Δ mutants with a viable PDIa′ plasmid (17). Replica plates were made, and after 2 days of incubation at 25 °C, they were incubated at either 25 or 37 °C. Temperature-sensitive alleles of PDIa′ were selected and sequenced to identify the mutation site, and the isolated plasmids were transformed back into the PDI1 haploid mutant strain 219A5 (PDI/pdi1Δ::kanMX) to confirm the temperature-sensitive phenotype. Individual tsPDIa′ plasmids were screened for growth at intermediate temperatures, and the allele selected for the synthetic lethality screen (D436N) was chosen based on growth characteristics at intermediate temperatures.
Synthetic Lethality Screens—To construct the pool of genome-wide yeast knockouts with a deleted PDI1 query gene and carrying a plasmid source of PDI or PDIa′, the genomic PDI1 gene and 1500 base pairs of 5′- and 3′-flanking sequence were replaced with a PCR-amplified pdiΔ::natMX cassette conferring ClonNAT (nourseothricin) resistance. The pdiΔ::natMX cassette was constructed using NotI-digested pAG25 (18), the natMX coding region, and the translational elongation factor gene promoter and terminator by transformation into the PDI1 diploid mutant 219A5 (PDI/pdi1Δ::kanMX) strain. The pAG25 transformant containing pdiΔ::natMX was sporulated for 5 days, and the haploid, 219A5nat, containing the pdiΔ::natMX cassette, was selected on MM plates containing 50 μg/ml ClonNAT.
The linear pdiΔ::natMX cassette was amplified from selected haploid transformant, 219A5nat, using PCR with primers (556f, TCAAC TTGTC ACGCA ACTCC; 5142r, CGCAC GGTAC AACTA AGCAA), which are complement at almost 1.5 kb up- and downstream of PDI1. The linear PCR-amplified pdiΔ::natMX cassette (10 μg) was used to transform the collection of heterozygote diploid strains according to the procedure as described (17) to produce a collection of heterozygous strains with one of the two PDI1 genes deleted. Briefly, the haploid-convertible heterozygous diploid yeast knockout pool in 50 ml of YPD liquid (starting at 0.13 A600 nm/ml) was shaken at 30°C for 5 h to an absorbance of 0.5 A600 nm/ml and transformed with 10 μg of the pdiΔ::natMX cassette. After transformation, the suspended cells were transferred to 50 ml of YPD and shaken at 30 °C for 2–3 h to allow expression of the ClonNAT resistance gene and then plated on 150 × 25-mm plates containing YPD plus 50 μg/ml ClonNAT. Dilutions were plated in parallel to calculate transformant yield as described (16). A good transformation will give rise to ∼106 transformants. After 2 days on ClonNAT plates, the pool of diploid strains was transferred to 50 ml of YPD medium and transformed with 5 μg of a Y33-14 (URA3, cen) plasmid containing wtPDI1, wtPDIa′, or tsPDIa′.
The transformed diploid strains were selected on SC–Ura media and subsequently sporulated. The haploid pools were then plated on MM/–Ura with 0.2 g/liter G418 to select for strains that contain a disruption in a single gene (xxxΔ::kanMX, gPDI1, and xxxΔ::kanMX, pdiΔ::natMX). After selection on G418, one-half of the strains in the haploid pool contained a genomic copy of PDI1, a PDI-expressing plasmid, and one other bar-coded gene knock-out (xxxΔ::kanMX, gPDI1, plasmid-PDI). The other half of the pool contained the pdiΔ mutation, a PDI-expressing plasmid, and one other bar-coded gene knockout (xxxΔ::kanMX, pdiΔ::natMX, plasmid-PDI). Control genomic DNA was isolated from this pool of mixed haploid strains to represent the genomic DNA of cells surviving with a genomic copy of PDI1. To produce the experimental genomic DNA, the mixed diploid pool was then plated on MM/–Ura + ClonNAT (50 μg/ml) to select for cells containing plasmid PDI and a genomic knock-out PDI1 (xxxΔ::kanMX, pdiΔ::natMX). Genomic DNA was isolated from at least 106 cells, as described (17). To confirm the genotype of selected cells, PCR was performed with primers for PDI genomic DNA and the kanMX marker that replaced PDI by recombination in the PDI1 deletion strains. The expression of PDIa′ in haploid tsPDIa′ and wtPDIa′ strains carrying the tsPDIA′ gene or wtPDIA′ gene was confirmed with Western blots.
TAG Array PCR—For TAG array hybridization, TAGs from the genomic DNA of control or experimental transformants were amplified using the universal UP-TAG or DOWN-TAG primers U1 or D1 and U2 or D2, which were labeled with fluorescent cyanine dyes (Cy5 and Cy3), as described (17). Reaction mixtures (20 μl/tube) for UP-TAG or DOWN-TAG PCR contained 10 μl of 2× ExTaq Premix (Takara), 0.5 μm primer 1 (U1 or D1), 5.0 μm primer 2 (U2 or D2, 5′-labeled with either Cy3 or Cy5 dye), and 200 ng of genomic DNA as template. UP-TAGs and DOWN-TAGs were amplified separately. PCR cycling parameters were as follows: 94 °C for 3 min; 95 °C for 10 s, 50 °C for 20 s, 72 °C for 20 s for 50 cycles; 4 °C. For control samples, the U2 and D2 primers were labeled with Cy5, whereas the experimental samples were amplified using primers labeled with Cy3. The TAG array hybridization is mediated by specific 20-bp bar code TAGs that lie in middle of the 56-bp TAG PCR products. To prevent reannealing of the amplified DNA during TAG array hybridization, an asymmetric PCR method was used where the Cy5- and Cy3-labeled primers were used. The primer sequences were as follows: U1, 5′-GATGT CCACG AGGTC TCT-3′; U2, 5′-(Cy5 or Cy3)-GTCGA CCTGC AGCGT ACG-3′; D1, 5′-CGGTG TCGGT CTCGT AG-3′; D2, 5′-(Cy5 or Cy3)-CGAGC TCGAA TTCAT CGAT-3′. To mask the common priming sites on TAG PCR products, blocking oligonucleotides (complementary to U1 and U2) were added at 10 μm and heat-denatured at 100 °C for 1 min and kept on ice.
TAG Array Hybridization and Scanning—Hybridization was performed in 7.5 ml of SSTE (0.5% Triton X-100, 1 m NaCl, and 100 mm Tris-HCl, pH 7.5) with freshly added dithiothreitol (DTT) (1 mm) and preblocked TAG PCR products. TAG array hybridization was overnight at 40 °C with gentle rocking. The hybridized TAG array was removed and dipped 3–5 times into 50 ml of wash buffer, 6× SSPE (0.9 m NaCl, 60 mm NaH2PO4, 6 mm EDTA), containing 0.05% Triton X-100 and 1 mm DTT and then washed in 50 ml of 0.06× SSPE containing 1 mm DDT and 50 ml of 0.06× SSPE.
Microarray Data Analysis—Microarrays were scanned using a GenePix 4000B (Axon Instruments), and image files were analyzed using Genepix 4.0 to collect the median raw signal intensity for each hybridization feature. For each feature, local background was subtracted, and the signal intensity was normalized according to an arbitrary total signal intensity of 108 for each fluorescence signal (UP and DOWN signals were normalized separately) with normalization factors varying between 1.0 and 3.6. A control/experiment (Cy5/Cy3) ratio was obtained for each feature. Only features with a normalized background-subtracted signal intensity median of ≥1000 (∼3-fold over background and 2.5 S.D. values above the average intensity for all signals with an intensity below 1000) were analyzed further. Because the efficiency of PCR may vary for each bar code, control/experiment ratios from the UP and DOWN primers may be different. Therefore, candidate positives were considered when either the UP or DOWN control/experiment ratio was >2.0, indicating more than a 2-fold reduction in growth rate when PDI1 is replaced by PDIa′. TAGs giving rise to systematic false positive results across multiple experiments were also discarded from further analysis (17).
Confirmation of Candidate Positive Genes—For confirmation of the individual synthetically lethal genes identified from the dSLAM screen, the corresponding individual heterozygote diploid double mutant strain (xxxΔ::kanMX, PDI1Δ::natMX) was constructed by a sequential transformation with the pdiΔ::natMX cassette, followed by transformation with a plasmid (URA3, cen) expressing tsPDIa′ as described above. The strain was subsequently sporulated by transferring to solid sporulation medium at room temperature (22–25 °C) for 5 days. After sporulation, transformants were plated in serial 10-fold dilutions on plates containing MM/–Ura/kan–Ura medium to select for haploid cells containing the individual gene knock-out, and an identical aliquot was placed on MM/–Ura/kan/natMX–Ura + ClonNAT plates to select haploids with the PDI1 gene replaced by the plasmid version of tsPDIa′ deleted. Plates were grown for 2 days at 30 °C before photographing.
Staining with FM4-64—FM4-64 (Molecular Probes, Inc., Carlsbad, CA) was dissolved in DMSO at 1 mg/ml and diluted 1:50 with YPD medium. Strains growing in logarithmic phase were resuspended in YPD medium with 0.2 mg/ml FM4-64 for 30 min on ice. To remove excess FM4-64, the cells were washed three times using ice-cold YPD. At time 0, cells were diluted into YPD and incubated with shaking at 30 °C for various times. A DeltaVision deconvolution fluorescence microscope (Applied Precision Inc., Seattle, WA) with an environment chamber maintained at 30 °C was used for cell imaging. FM4-64 fluorescence was excited at 622 nm (rhodamine), and emission was observed at 572 nm (TRITC). Images were processed using SoftWoRx or ImageJ software. Individual images displayed in the figures are representative single central slices of an optical sectioning image stack. Images were cropped to remove blank space, and the overall brightness and contrast were adjusted uniformly using ImageJ (19).
RESULTS AND DISCUSSION
The Unfolded Protein Response—Despite the fact that PDI1 is an essential yeast gene, only its ability to catalyze disulfide formation (oxidase activity) is required. A single catalytic domain (PDIa′, amino acid residues 375–485) with less than 5% of the isomerase activity of Pdi1p will support a near wild-type growth rate (6) despite a 2-fold decrease in the rate at which a protein that requires disulfide isomerization, CPY (carboxypeptidase Y), exits the ER. Its slower processing by a PDIa′ strain shows that disulfide isomerization is impaired and ∼2-fold slower than in a wild-type strain (8). All known disulfide isomerases rely on thioredoxin domains with a CXXC motif required in the catalytic site. There are five such proteins in the yeast ER (MPD1, MPD2, EUG1, and EPS1) (13) that might compensate for the missing isomerase activity. However, in a strain with all of the PDI homologues deleted, PDIa′ will still support nearly wild-type growth, although the rate of CPY exit from the ER is now ∼6-fold slower than wild-type (8).
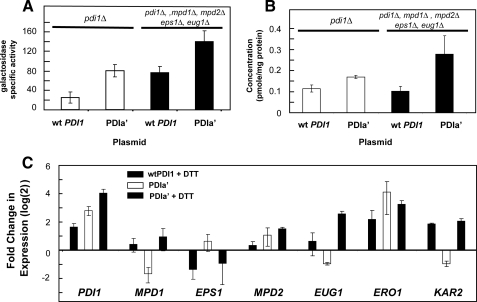
When unfolded proteins accumulate in the ER, the expression of the PDI1 gene and other chaperones and folding catalysts is induced through the UPR (20). The increased expression of PDI1 and other UPR-regulated genes may help compensate for misfolding due to low isomerase activity. Part of the compensation mechanism may involve increased expression of Pdi1p or PDIa′ from the PDI1 promoter. To determine the extent of the activity of the PDI1 promoter in the presence of an isomerase deficiency, promoter activity was examined in yeast strains supported by plasmids expressing PDI1 or PDIa′ using a β-galactosidase reporter driven by the PDI1 promoter. As shown in Fig. 1A, the PDI1 promoter is clearly activated (∼4-fold) when Pdi1p is replaced by PDIa′. The increased expression from the PDI1 promoter also results in a small but significant increase in the level of PDIa′ protein (1.5-fold; p < 0.05, compared with wild-type Pdi1p) as shown by quantitative Western blotting (Fig. 1B). Removing the ER homologs of PDI1 leads to a further increase in the amount of PDIa′ protein expressed (2.4-fold; p < 0.05, compared with wild-type PDI1) (Fig. 1B). Despite the increased expression of PDIa′ compared with full-length Pdi1p, the level of isomerase activity in vivo is still insufficient to produce wild-type rates of CPY maturation (8). In a strain with deletions of all of the ER PDI homologues (13), the PDI1 promoter was also up-regulated even when wild-type PDI1 was provided and even further up-regulated when the PDI1 on the plasmid was replaced by PDIa′. Because the PDIa′ protein exhibits <5% of the isomerase activity of Pdi1p, the additional isomerization capacity provided by this up-regulation would still be insufficient to maintain nearly normal growth rates, since significant growth reduction is observed when the wild-type PDI1 levels are reduced by 50% (7).
FIGURE 1.
Activity of the PDI1 promoter in strains supported with the isomerase-deficient PDIa′. A, β-galactosidase reporter expression. A reporter construct expressing β-galactosidase driven by the PDI1 promoter was introduced into yeast strains M4174 (pdi1Δ) and M4477 (pdi1Δ, mpd1Δ, mpd2Δ, eps1Δ, eug1Δ), where the essential PDI1 function is supplied by a plasmid expressing either wild-type PDI1 or PDIa′ expressed from the PDI1 promoter (8) B, Western blotting to quantitate the expression of PDI1 and PDIa′. Western blots calibrated by standards of purified yeast Pdi1 and PDIa′ were used to quantitate the amount of Pdi1p and PDIa′ protein expressed under the same conditions as in A. The data shown are the averages of eight replicates, and the error bars represent the S.D. values. C, expression (mRNA) of PDI1 homologues when PDI1 is replaced by PDIa′. Quantitative reverse transcription-PCR was used to measure the effect of isomerase deficiency on the mRNA levels of PDI homologues (MPD1, MPD2, EPS1, and EUG1), the ER oxidase responsible for oxidizing Pdi1p (ERO1), and the ER hsp70 chaperone, KAR2. DTT treatment (2.5 mm, 4 h) was used to induce the unfolded protein response in strains supported by Pdi1p or PDIa′. Levels are normalized to an actin loading control and plotted (log2) relative to the expression of the gene level observed in the wild-type strain.
Quantitative real time PCR was also used to determine the effect of decreasing isomerase activity on the expression of the ER homologues of PDI (MPD1, MPD2, EPS1, and EUG1), along with ERO1, the ER oxidase that converts Pdi1p from its reduced to oxidized form, and KAR2, an essential ER chaperone that is responsive to induction of the UPR (20). Consistent with the β-galactosidase reporter and Western blot results shown in Fig. 1, A and B, replacing wild-type Pdi1p with PDIa′ resulted in a significant increase in expression from the PDI1 promoter, as shown by quantitative PCR (Fig. 1C). Treating the strain expressing PDIa′ with DTT, which is known to induce the unfolded protein response (20), resulted in a further increase in expression. By comparison, the mRNA expression levels of the PDI1 homologues (MPD1, MPD2, EPS1, and EUG1) were only modestly affected by replacing Pdi1p with PDIa′. However, stressing the PDIa′-expressing strains with DTT did stimulate transcription further, indicating that the unfolded protein response was not fully induced in the isomerase-deficient strains. The expression of the ER oxidase, ERO1, was significantly induced when PDIa′ was present, as was the expression of the ER hsp70 homologue, KAR2. Of the genes we examined that were identified as part of the canonical unfolded protein response (PDI1, MPD1, MPD2, EUG1, and ERO1) (20), replacing wtPDI1 with PDIa′ induced significant expression of only PDI1, ERO1, and KAR2. For these genes, DTT induced expression comparable with that observed previously (20). However, the induction of the PDI1 homologues, MPD1, MPD2, EPS1, and EUG1, was not as significant. Thus, perturbations in disulfide isomerization in the presence of PDIa′ may not result in a classical unfolded protein response, although clearly some UPR genes are induced.
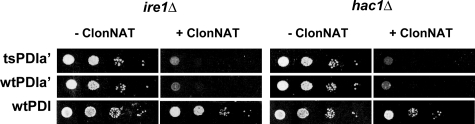
To directly determine if the unfolded protein response is required for growth when PDI1 is replaced with PDIa′, yeast strains were constructed (see “Experimental Procedures”) in which the nonessential genes IRE1 or HAC1 were deleted. IRE1 and HAC1 provide the signal transduction mechanism for the unfolded protein response (20). The deletion constructs were generated from individual diploid yeast strains using the strategy of the dSLAM method (16). The method begins with individual heterozygous diploid strains in which either the IRE1 gene or the HAC1 gene is replaced with a kanamycin resistance marker (kan). One of the two PDI1 genes in the diploid is then replaced with a nourseothricin (ClonNAT) resistance marker (natMX), and the resulting heterozygous diploid strains are transformed with a plasmid supplying either PDI1 or PDIa′ (URA). After sporulation and selection for kanamycin resistance and the presence of the PDI plasmid (URA), the two resulting haploid strains will have genotypes (pdi1Δ::natMX, ire1Δ(or hac1Δ)::kan, and PDI1, ire1Δ(or hac1Δ)::kan) containing the plasmid expressing Pdi1p or PDIa′. After selection for nourseothricin resistance, only strains with a PDI1 deletion will survive but only if the other deleted genes (IRE1 or HAC1) are not required. The results (Fig. 2) clearly show that deletion of either IRE1 or HAC1 is lethal when the only PDI species present is PDIa′, but neither gene is essential when wild-type PDI1 is present. Thus, both HAC1 and IRE1 (and the unfolded protein response) are synthetically lethal, with the loss of the PDI isomerase activity indicating that one or more UPR-responsive genes are required to compensate for defects in disulfide isomerization.
FIGURE 2.
Synthetic lethality of HAC1 or IRE1 deletions with the loss of PDI1 isomerase activity in the PDIa′ mutant. Individual strains were constructed (see “Experimental Procedures”) containing an IRE1 or HAC1 deletion (kan) and a disruption of genomic PDI1 (ClonNAT) in combination with a plasmid (URA) bearing a PDI1 or PDIa′ gene driven by the PDI1 promoter. Strains were spotted in 10-fold serial dilutions onto selection medium (kan, –uracil) to select for IRE1 or HAC1 deletion and the presence of the PDI-expressing plasmid (left panel of each pair). The same number of cells was spotted onto the same selection medium with the addition of ClonNAT to select for the loss of the genomic PDI1 gene (right panel of each pair). The PDI1 gene was first deleted with a natMX marker (pdi1Δ::natMX) in haploid-convertible HAC1/hac1Δ::kanMX and IRE1/ire1Δ::kanMX heterozygous diploid mutants. The resultant double mutants were subsequently transformed with a plasmid that expresses either full-length Pdi1p or PDIa′ and sporulated. The spores were spotted onto MM/–Ura at a 10-fold serial dilution both with and without ClonNAT. Cells were grown at 30 °C and photographed. The entire experimental procedure is shown in supplemental Fig. 1.
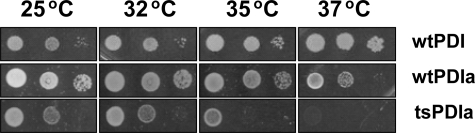
Other Genes Synthetically Lethal with the Loss of PDI—To determine if there are other genes, including those of the UPR response, that become essential when PDI is isomerase-deficient, the dSLAM method (16) was applied to detect synthetically lethal mutants on a genome-wide scale. To help generate a wider range of defects in PDI1p activity, a temperature-sensitive mutation was generated in PDIa′ (tsPDIa′) error-prone PCR. This mutant displayed a growth rate comparable with that from wtPDIa′ at the permissive temperature (25 °C) but failed to grow at the restrictive temperature (37 °C) (Fig. 3). At 30 °C, its growth rate was approximately half that supported by wild type Pdi1p or PDIa′ (Table 1). DNA sequencing of the entire open reading frame revealed a single mutation (D436N) at a site that is located near the surface of the a′ domain in the β-sheet region of the thioredoxin fold and ∼12 Å from the active site cysteines.
FIGURE 3.
Temperature-sensitive PDIa′ strain. Error-prone PCR was used to generate a temperature-sensitive mutation in PDIa′ (D436N). From left to right, the wild-type and temperature-sensitive strains are plated in 10-fold serial dilutions onto MM plates, as described under “Experimental Procedures.”
TABLE 1.
Growth rates of pdi1Δ supported by plasmids expressing wild-type PDI1, PDIa′, and tsPDIa′
Growth rates were measured at 30 °C in exponentially growing cultures in either YPD or minimal medium.
| PDI species | Doubling time, YPDa | Doubling time, minimal mediuma |
|---|---|---|
| h | h | |
| Wild-type Pdi1p | 2.4 (0.1) | 2.8 (0.14) |
| Wild-type Pdia′ | 2.9 (0.3) | 4.2 (0.18)b |
| tsPdia′ | 4.0 (0.2)b | 5.7 (0.20)b |
Averages of at least three measurements.
Significantly different from wild type by Student's t test with p < 0.05.
To construct a pool of the ∼6000 individual deletions used in the dSLAM method, the PDI1 gene was replaced in the pool of strains with natMX (PDI1::natMX), and the desired PDI species was introduced via a URA3 plasmid containing one of the three PDI constructs (wtPDI, PDIa′, or tsPDIa′) under the control of the endogenous PDI1 promoter. After sporulation, four pools of haploid strains were selected either in the presence and absence of ClonNAT (Table 2). The designation wtPDI represents a pool of double deletion mutants (xxxΔ::kanMX pdi1Δ) that contains the full-length wild-type PDI1 on the plasmid. Designations wtPDIa′ and tsPDIa′ represent similar double mutant pools that contain a plasmid expressing wild-type PDIa′ or tsPDIa′, respectively. The designation gPDI represents a pool of strains that contains the wild-type full length PDI1 at its genomic locus. Genomic DNA samples were prepared from the resultant haploid pools and analyzed by microarray to identify gene deletions that were underrepresented in the absence of the chromosomal PDI1 gene. A schematic illustration of the dSLAM experimental protocol is shown in supplemental Fig. 1.
TABLE 2.
Comparison of synthetic lethality candidates in microarray experiments
The total number of significant depletions of genes in the experimental cDNA pool (PDIa′ or tsPDIa′) compared with the control (wild-type or genomic PDI) are shown along the diagonal. Off-diagonal entries indicate the number of common hits between two experiments. The final row provides information about positives that were found in experiments with tsPDIa′ and all three experiments.
|
Microarray experiment B
|
Microarray experiment A
|
||||
|---|---|---|---|---|---|
| wtPDI-wtPDIa′ | wtPDI-tsPDIa′ | gPDI-tsPDIa′ | |||
| wtPDI-wtPDIa′ | Total hits: 186 | Common hits: 134 | Common hits: 38 | ||
| wtPDI-tsPDIa′ | Total hits: 274 | Common hits:131 | |||
| gPDI-tsPDIa′ | Total hits: 622 | ||||
| Common hits in all experiments: 34; GOAT cluster: 19 | Common to tsPDIa′ not in wtPDIa′: 96; GOAT cluster: 70 | ||||
The four pools were compared in three independent experiments using microarray analysis to determine the relative representation of each barcode in different samples. Each experiment is identified using a two-part description. The left part of the description represents control (C) genomic DNA that was isolated from a population of strains that has a wild-type PDI1 gene, either in the genome (gPDI1) or provided by a plasmid (wtPDI1). The right part represents the experimental genomic DNA isolated from the pool of strains remaining after removal of the wtPDI gene (experiment, E). The three experiments were wtPDI1/wtPDa′, wtPDI1/tsPDa′, and gPDI1/tsPDIa′. After hybridization to the TAG microarray and subtraction of background fluorescence at each wavelength (635 and 532 nm), the total fluorescence intensity in each channel was normalized. Because the efficiency of PCR varies for each bar code (for each disrupted gene), ratios from the UP-TAG and DOWN-TAG of each strain may be significantly different. Candidate synthetic lethality genes were selected for subsequent analysis only if the normalized fluorescence intensity was greater than 3-fold above the background signal and the UP or DOWN ratio was >2.0, indicating more than a 2-fold reduction in growth rate when PDI1 was replaced by PDIa′ or tsPDIa′. For each of the experiments, the number of candidates selected using these criteria are shown in Table 2 along with the number of common hits among multiple experiments.
The number of synthetically lethal dSLAM positives when compared with wild-type full-length PDI1 is greater for the tsPDIa′ construct than PDIa′ (Table 2), suggesting that many more genes become synthetically lethal when the tsPDIa′ gene is present at the semipermissive temperature (30 °C). To focus attention on those synthetically lethal genes that may have common biological roles, the positive hits from each experiment were compared with the 34 identified genes that were synthetically lethal in all three experiments. There were 96 additional genes that were synthetically lethal in both of the tsPDIa′ experiments. All 130 genes identified as potentially synthetic lethal are shown in supplemental Table 1.
Synthetically Lethal Gene Ontology Clusters—To help identify those genes products with related function, the list of 130 synthetically lethal genes and the list of 34 genes observed in all three experiments were analyzed independently using the Gene Ontology Analysis Tool (21). This analysis clusters genes in the hit lists using the S. cerevisiae gene ontology (22, 23) and calculates the probability of randomly selecting multiple genes from each ontology cluster based on the total number of genes that have been assigned that clustered GO term and the number of candidate gene selections. A total of 89 GOAT clustered genes were found of the 130 candidate genes initially identified by dSLAM. Of the 34 genes that were observed in all three experiments, all except for DAL81 still clustered significantly (p < 0.05) even with the reduced size of the hit list. A complete list of all 130 synthetically lethal genes along with notations as to their occurrence in other high throughput experiments and a more complete description of known function can be found in supplemental Table 1.
A large proportion (55%) of the clustered genes participate in some aspect of vesicle trafficking, not only into and out of the ER but involving essentially every cellular compartment where ER-synthesized proteins are directed (Table 3). The other genes identified are involved with growth or growth regulation, metabolism, stress responses, and protein degradation (supplemental Table 1). Of the 50 genes involved in some aspect of vesicle trafficking, a large proportion (∼50%) serve in some regulatory capacity. In addition, the genes involved in vesicle trafficking represented virtually all trafficking pathways, including anterograde and retrograde transport involving the ER, Golgi, plasma membrane, and vacuole (Table 3).
TABLE 3.
Genes involved in vesicle trafficking pathways that become essential when disulfide formation/isomerization is compromised by expression of PDIa′ or tsPDIa′ in pdi1Δ strains
Genes (open reading frames) are grouped by the portion of the vesicular trafficking process in which they are involved. The process and function/complex notations were summarized from the description in the SGD (Saccharomyces Genome Database) project (available on the World Wide Web). A listing of all synthetically lethal deletions along with the full description of known function can be found in the supplemental material. All genes in this table showed statistically significant (p < 0.05) clustering by gene ontology component, function, or biological process.
| Open reading frame | Process | Function/Complexa | Verifiedb | In three experimentsc |
|---|---|---|---|---|
| CCZ1 (YBR131W) | Vacuole sorting | Autophagy | ++ | Y |
| APS3 (YJL024C) | Vacuole sorting | Clathrin-associated adaptor | ++ | Y |
| APM3 (YBR288C) | Vacuole sorting | Clathrin-associated, AP3 | ++ | |
| RAV2 (YDR202C) | Vacuole sorting | V-ATPase assembly, RAVE | ++ | |
| VTC1 (YER072W) | Vacuole sorting | V-ATPase assembly, VTC complex | ++ | |
| ARL1 (YBR164C) | Golgi-PM | ARF complex | ++ | Y |
| AVL9P (YLR114C) | Golgi-PM | Unknown | ++ | |
| COG8 (YML071C) | Golgi fusion | COG tethering complex | + | |
| GYP1 (YOR070C) | Golgi fusion | RAB GAP | ++ | |
| SEC28 (YIL076W) | Golgi-ER | COPI vesicles, coatomer | ++ | Y |
| ARR4/GET3 (YDL100C) | Golgi-ER | GET complex | ++ | |
| INP53 (YOR109W) | Golgi-endosome | PIP phosphatase | ++ | |
| KHA1 (YJL094C) | Golgi component | Cation homeostasis | ++ | |
| RGP1 (YDR137W) | Golgi component | GEF, Ypt6p | ++ | |
| ALG8 (YOR067C) | Golgi component | Glucosyltransferase | ++ | |
| GEF1 (YJR040W) | Golgi component | Ion transport | ++ | Y |
| PSD2 (YGR170W) | Golgi component | Lipid metabolism | ++ | Y |
| YUR1 (YJL139C) | Golgi component | N-Linked glycosylation | ++ | |
| SYS1 (YJL004C) | Golgi component | Targeting of ARF complex | ++ | |
| SHR3 (YDL212W) | ER-PM | COPII | ++ | Y |
| GCS1 (YDL226C) | ER-Golgi | ARF GAP | + | Y |
| ERV46 (YAL042W) | ER-Golgi | COPII vesicle fusion | ++ | |
| ERV41 (YML067C) | ER-Golgi | COPII vesicles | -- | Y |
| SPF1 (YEL031W) | ER component | ATPase involved in Ca2+ homeostasis | -- | |
| SCJ1 (YMR214W) | ER component | DnaJ-like chaperone | ++ | |
| HRD1 (YOL013C) | ER component | ER degradation, ERAD | ++ | |
| YER087C-A | ER component | ER translocation, Sec61 complex | ++ | |
| SEC66 (YBR171W) | ER component | ER translocation, Sec63 complex | ++ | |
| YLR404W | ER component | Lipid droplet morphology | ++ | Y |
| PKR1 (YMR123W) | ER component | V-ATPase assembly | ++ | |
| SNF8 (YPL002C) | Endosome-vacuole | ESCRT-II-multivesicular body pathway | ++ | |
| VPS36 (YLR417W) | Endosome-vacuole | ESCRT-II-multivesicular body pathway | ++ | |
| VPS25 (YJR102C) | Endosome-vacuole | ESCRT-II-multivesicular body pathway | ++ | |
| VPS20 (YMR077C) | Endosome-vacuole | ESCRT-III-multivesicular body pathway | ++ | |
| VPS21 (YOR089C) | Endosome-vacuole | GTPase, CORVET complex | ++ | |
| VPS8 (YAL002W) | Endosome-vacuole | GTPase, CORVET complex | ++ | |
| VPS29 (YHR012W) | Endosome-Golgi | Recycling of endocytosed proteins to Golgi-retromer complex | ++ | |
| VPS38 (YLR360W) | Endosome-Golgi | Recycling of endocytosed proteins to Golgi-retromer complex | ++ | |
| RCY1 (YJL204C) | Endosome-Golgi | Recycling of endocytosed proteins to PM | ++ | Y |
| VPS35 (YJL154C) | Endosome-Golgi | Recycling of endocytosed proteins to PM-retromer complex | ++ | |
| VPS17 (YOR132W) | Endosome-Golgi | Recycling of endocytosed proteins to PM-retromer complex | ++ | Y |
| VPS1 (YKR001C) | Endosome sorting | Dynamin-like GTPase | ++ | Y |
| VPS13 (YLL040C) | Endosome sorting | Unknown | ++ | |
| RAV1 (YJR033C) | Endosome sorting | VATPase assembly, RAVE | ++ | |
| CDC50 (YCR094W) | Endosome component | Cell polarity, polarized growth | ++ | |
| NHX1 (YDR456W) | Endosome component | Ion transport, osmotolerance | ++ | |
| RVS167 (YDR388W) | Endocytosis | Actin-associated protein | ++ | |
| YAP1801 (YHR161C) | Endocytosis | Clathrin assembly | None | |
| SWA2 (YDR320C) | Endocytosis | Clathrin uncoating | ++ | |
| SUR7 (YML052W) | Endocytosis | Eisosome | ++ |
Summarized from the description in the SGD (Saccharomyces Genome Database) project.
Independent verification of individual deletion strains, as shown in Fig 4. ++, synthetic lethality phenotype verified by growth before selection for ClonNAT resistance and the absence of growth after selection for ClonNAT resistance, which selects for strains with a deletion in the genomic PDI1 gene; --, the target gene did not display a synthetically lethal phenotype; None, no colonies even in the colonies plated in the absence of ClonNAT selection; ND, not determined.
Y indicates that the indicated gene deletion was scored as a synthetically lethal hit in all three microarray experiments.
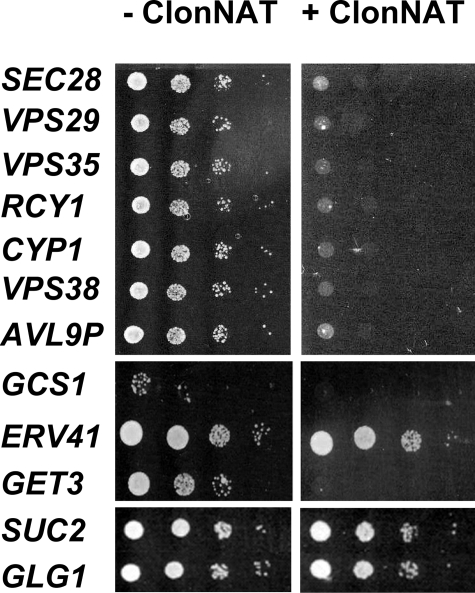
The synthetic lethality of all of the genes involved in vesicle traffic (see below) was also verified directly in individual strains (Table 3 and Fig. 4). In addition, other selected genes, including positive controls, were also tested directly for synthetic lethality (supplemental Table 1). A total of 69 different genes were examined individually. Of those, 61 (88%) showed the expected synthetically lethal phenotype. The false positive rate was 6% overall, and only one gene knock-out repeatedly failed to grow in the absence of ClonNAT selection.
FIGURE 4.
Verification of phenotype for synthetically lethal genes initially identified by dSLAM and GOAT analysis. The PDI1 gene was first deleted (pdi1Δ::natMX) in individual haploid-convertible heterozygous diploid mutants of interest. The resultant heterozygous diploid double mutants were subsequently transformed with a plasmid harboring tsPDIa′ and sporulated. After sporulation and transformation to delete the PDI1 gene and insert the tsPDIa′ plasmid, the spores were spotted at a 10-fold serial dilution between spots onto MM/–Ura/kan with and without ClonNAT to compare the growth phenotype before and after removing genomic PDI1. Photographs were taken after 2 days growth at 30 °C). Photographs have been uniformly adjusted for brightness and contrast. The entire set of verification experiments are shown in the supplemental material.
Notably absent from the dSLAM analysis is the signal transduction pathway of the unfolded protein response, IRE1 and HAC1 (10). Each of these genes had a sufficiently intense signal in the control experiment with wtPDI1 from either the UP-TAG or DOWN-TAG to be analyzed; however, the change in signal upon replacement of the wtPDI1 gene was less than 2-fold in all three experiments, suggesting that they are not required when the isomerase activity of Pdi1p is impaired. These two genes were examined directly for a synthetically lethal phenotype (Fig. 1), suggesting that these two genes are false negatives under the conditions of our dSLAM experiments. Weissman and co-workers (24) have performed a high-throughput experiment for both positive and negative epistasis for 556 specific genes involved in various aspects of ER quality control when the expression of the PDI1 gene is lowered by destabilizing the PDI1 mRNA by a 3′-UTR disruption. For the 130 genes that we identified by dSLAM (before GOAT clustering), the more focused E-MAP experiment considered 26 of the same genes. Of those, nine showed negative epistasis in both the dSLAM and E-MAP experiments. The reasons for the lack of higher complementarity in these two experimental approaches are not known; however, the rather large drop in PDI1 expression in the E-MAP experiment could possibly have queried genes that affected growth under even more stressful conditions than those caused by the PDI species introduced here.
Relation of the dSLAM Synthetically Lethal Genes to the Unfolded Protein Response—The induction of the unfolded protein response is required to compensate for defects in disulfide isomerization. However, there is only a small amount of overlap between the 130 genes identified by dSLAM in these experiments and the 381 genes that display expression patters that have been used to define the unfolded protein response based on their HAC1- and IRE1-dependent induction when protein folding in the ER is disturbed by reduction (DTT) or inhibition of glycosylation (tunicamycin) (20). Only 10 of the 130 genes we have identified here are considered part of the canonical UPR. In addition, 64 of the dSLAM genes are not responsive to DTT or tunicamycin (supplemental Table 1), suggesting that although the unfolded protein response is necessary to compensate for defects in isomerase-dependent folding, the genes of the UPR are not sufficient to rescue growth when the Pdi1p isomerase activity is deficient.
Vesicle Trafficking Pathways—The observation of a cluster of synthetically lethal genes involved in such a large variety of vesicle trafficking processes, and particularly the relatively large fraction that are involved in regulation, suggests the possibility that one mechanism to help compensate for a slower exit of PDI substrates, such as CPY (8), from the ER would be to decrease the rate of retrograde vesicle traffic. For example, decreased endocytosis would decrease the demand for PDI substrates that function in the plasma membrane and compensate for a lower rate of their exit from the ER. Notably, there were dSLAM hits in multiple components of both the retromer complex and the ESCRT complex. These complexes comprise opposing activities in the recycling of plasma membrane proteins from the endosomal system to the plasma membrane (retromer) and the ubiquitin-dependent degradation of plasma membrane proteins from endosomal vesicles (ESCRT) (25).
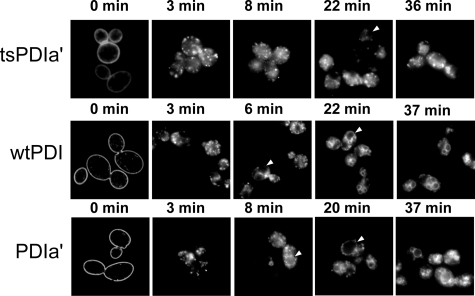
The internalization of the vital dye FM4-64 is energy-dependent and dependent on endocytosis. Incubating yeast cells with FM4-64 at 0 °C limits staining to the plasma membrane. After washing and shifting the temperature to 30 °C, fluorescence disappears from the plasma membrane, appears as distinct punctae, and eventually concentrates in the vacuole membrane (26). The time course of FM4-64 endocytosis is shown in Fig. 5. For wtPDI1, the dye appears rapidly in punctae after raising the temperature. After ∼6–7 min, the FM4-64 fluorescence begins to appear in vacuolar membranes, where it ultimately concentrates. When the isomerase activity of Pdi1p is compromised in the wtPDIa′ truncation mutant, the initial appearance of FM4-64 in the vacuole may be somewhat slower (Table 4). With the tsPDIa′ mutant, the appearance of FM4-64 in the vacuole is ∼3-fold slower than that supported by wild-type Pdi1p (Table 4). Thus, the defect in disulfide isomerization or formation introduced early in the ER by replacing Pdi1p with tsPDIa′ results in a slower trafficking of FM4-64 into the vacuole. FM4-64 internalization may also be correlated with growth (Tables 1 and 4).
FIGURE 5.
Analysis of endocytosis and vesicle trafficking with wild-type Pdi1p, PDIa′, and tsPDIa′ by FM4-64 staining. Cells were treated with FM4-64 as described under “Experimental Procedures” with an assay temperature of 30 °C and imaged with a DeltaVision deconvolution fluorescence microscope with an environment chamber maintained at 30 °C. Excitation was at 622 nm (rhodamine), and emission was observed at 572 nm (TRITC). Images were processed using SoftWoRx or ImageJ software. Individual images displayed are representative single central slices of an optical sectioning image stack. Images were cropped to remove blank space, and the overall brightness and contrast were adjusted uniformly using ImageJ. A series of time lapse images are shown from time points around 0, 2, 7, 20, and 40 min.
TABLE 4.
Initial appearance of FM4-64 in yeast vacuoles for Pdi1p, wtPdi1a′, and tsPdi1a′
The initial time for appearance of FM4-64 at 30 °C into the yeast vacuole was measured as described under “Experimental Procedures.” Pdi1p is wild-type S. cerevisiae protein-disulfide isomerase, PDIa′ is the C-terminal catalytic domain truncation mutant, and tsPDIa′ is a temperature-sensitive mutant of PDIa′.
| PDI species | FM4-64 initial vacuolar appearancea |
|---|---|
| min | |
| Pdi1p | 6.7 (1.2) |
| PDIa′ | 8.2 (2.5) |
| tsPDIa′ | 21.9 (2.5)b |
Results are the averages of at least four independent experiments. Values in parenthesis represent the S.D.
Significantly different from wild type by Student's t test with p < 0.05.
Conclusions—Although the unfolded protein response is essential to compensate for folding defects in the ER, the UPR is not sufficient to compensate for the ER folding defects we introduced. In addition to the expected genes of the stress response, our analysis also revealed clusters of genes involved in the regulation of vesicle trafficking pathways that become essential when disulfide isomerization and formation is compromised in the ER. At this point, it is impossible to exclude the hypothesis that there are no essential proteins that require disulfide isomerization to exit the ER efficiently; however, a significantly slower exit of PDI substrates from the ER might compromise cellular function and viability without mechanisms to compensate. The transcriptional regulation provided by the unfolded protein response is clearly one such mechanism (27); however, the results of synthetic lethality interactions and the observation that net endocytosis is slowed in response to defects in the ER suggest that regulatory networks and signaling pathways may coordinate vesicle trafficking and cell growth to help cells survive transient or permanent defects in ER protein folding. Further research will be required to elucidate the molecular details of these controls and the mechanisms that coordinate ER folding, membrane traffic, and cell growth.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM040379-19.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Fig. 1.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PDI, protein-disulfide isomerase; PDIa′, the C-terminal catalytic domain of S. cerevisiae Pdi1p; tsPDIa′, a temperature-sensitive mutant of PDIa′; wtPDI, wild type PDI; gPDI, genomic PDI; dSLAM, diploid synthetic lethality analysis by microarray; MM, magic medium; DTT, dithiothreitol; TRITC, tetramethylrhodamine isothiocyanate; UPR, unfolded protein response; PM, plasma membrane; GOAT, gene ontology analysis tool.
References
- 1.Schroder, M., and Kaufman, R. J. (2005) Annu. Rev. Biochem. 74 739–789 [DOI] [PubMed] [Google Scholar]
- 2.Gilbert, H. F. (1997) J. Biol. Chem. 272 29399–29402 [DOI] [PubMed] [Google Scholar]
- 3.Edman, J. C., Ellis, L., Blacher, R. W., Roth, R. A., and Rutter, W. J. (1985) Nature 317 267–270 [DOI] [PubMed] [Google Scholar]
- 4.LaMantia, M. L., and Lennarz, W. J. (1993) Cell 74 899–908 [DOI] [PubMed] [Google Scholar]
- 5.Darby, N. J., and Creighton, T. E. (1995) Biochemistry 34 11725–11735 [DOI] [PubMed] [Google Scholar]
- 6.Xiao, R., Solovyov, A., Gilbert, H. F., Holmgren, A., and Lundstrom-Ljung, J. (2001) J. Biol. Chem. 276 27975–27980 [DOI] [PubMed] [Google Scholar]
- 7.Solovyov, A., Xiao, R., and Gilbert, H. F. (2004) J. Biol. Chem. 279 34095–34100 [DOI] [PubMed] [Google Scholar]
- 8.Xiao, R., Wilkinson, B., Solovyov, A., Winther, J. R., Holmgren, A., Lundstrom-Ljung, J., and Gilbert, H. F. (2004) J. Biol. Chem. 279 49780–49786 [DOI] [PubMed] [Google Scholar]
- 9.Anelli, T., and Sitia, R. (2008) EMBO J. 27 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ron, D., and Walter, P. (2007) Nat. Rev. 8 519–529 [DOI] [PubMed] [Google Scholar]
- 11.Gietz, R. D., and Sugino, A. (1988) Gene (Amst.) 74 527–534 [DOI] [PubMed] [Google Scholar]
- 12.Hansen, J., Felding, T., Johannesen, P. F., Piskur, J., Christensen, C. L., and Olesen, K. (2003) FEMS Yeast Res. 4 323–327 [DOI] [PubMed] [Google Scholar]
- 13.Norgaard, P., Westphal, V., Tachibana, C., Alsoe, L., Holst, B., and Winther, J. R. (2001) J. Cell Biol. 152 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amberg, D. C., Burke, D. J., and Strathem, J. N. (2005) Methods in Yeast Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 15.Kim, J. H., Tuziak, T., Hu, L., Wang, Z., Bondaruk, J., Kim, M., Fuller, G., Dinney, C., Grossman, H. B., Baggerly, K., Zhang, W., and Czerniak, B. (2005) Lab. Investig. 85 532–549 [DOI] [PubMed] [Google Scholar]
- 16.Pan, X., Yuan, D. S., Xiang, D., Wang, X., Sookhai-Mahadeo, S., Bader, J. S., Hieter, P., Spencer, F., and Boeke, J. D. (2004) Mol. Cell 16 487–496 [DOI] [PubMed] [Google Scholar]
- 17.Pan, X., Yuan, D. S., Ooi, S. L., Wang, X., Sookhai-Mahadeo, S., Meluh, P., and Boeke, J. D. (2007) Methods 41 206–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein, A. L., and McCusker, J. H. (1999) Yeast 15 1541–1553 [DOI] [PubMed] [Google Scholar]
- 19.Achleitner, G., Gaigg, B., Krasser, A., Kainersdorfer, E., Kohlwein, S. D., Perktold, A., Zellnig, G., and Daum, G. (1999) Eur. J. Biochem. 264 545–553 [DOI] [PubMed] [Google Scholar]
- 20.Travers, K. J., Patil, C. K., Wodicka, L., Lockhart, D. J., Weissman, J. S., and Walter, P. (2000) Cell 101 249–258 [DOI] [PubMed] [Google Scholar]
- 21.Xu, Q., and Shaulsky, G. (2005) Appl. Bioinformatics 4 281–283 [DOI] [PubMed] [Google Scholar]
- 22.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. (2000) Nat. Genet. 25 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rachlin, J., Cohen, D. D., Cantor, C., and Kasif, S. (2006) Mol. Syst. Biol. 2 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuldiner, M., Collins, S. R., Thompson, N. J., Denic, V., Bhamidipati, A., Punna, T., Ihmels, J., Andrews, B., Boone, C., Greenblatt, J. F., Weissman, J. S., and Krogan, N. J. (2005) Cell 123 507–519 [DOI] [PubMed] [Google Scholar]
- 25.Strochlic, T. I., Schmiedekamp, B. C., Lee, J., Katzmann, D. J., and Burd, C. G. (2008) Mol. Biol. Cell 19 4694–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vida, T. A., and Emr, S. D. (1995) J. Cell Biol. 128 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernales, S., Papa, F. R., and Walter, P. (2006) Annu. Rev. Cell Dev. Biol. 22 487–508 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.