Abstract
How endoplasmic reticulum (ER) proteins that are substrates for the ER-associated degradation (ERAD) pathway are recognized for polyubiquitination and proteasomal degradation is largely unresolved. Inositol 1,4,5-trisphosphate receptors (IP3Rs) form tetrameric calcium channels in ER membranes, whose primary role is to control the release of ER calcium stores, but whose levels are also regulated, in an activation-dependent manner, by the ERAD pathway. Here we report that the ER membrane protein SPFH1 and its homolog SPFH2 form a heteromeric ∼2 MDa complex that binds to IP3R tetramers immediately after their activation and is required for their processing. The complex is ring-shaped (diameter ∼250Å), and RNA interference-mediated depletion of SPFH1 and SPFH2 blocks IP3R polyubiquitination and degradation. We propose that this novel SPFH1/2 complex is a recognition factor that targets IP3Rs and perhaps other substrates for ERAD.
The endoplasmic reticulum (ER)2-associated degradation (ERAD) pathway targets aberrant proteins, including irreversibly misfolded proteins and unassembled subunits of multiprotein complexes, for degradation by the ubiquitin-proteasome system (1). Intriguingly, several ER-resident proteins that are stable under normal conditions are also processed by the ERAD pathway. For example, 3-hydroxy-3-methylglutaryl CoA-reductase, the rate-limiting enzyme in sterol synthesis, is targeted for ERAD when sterols are in excess (2), and inositol 1,4,5-trisphosphate (IP3) receptors (IP3Rs), which form tetrameric, IP3- and Ca2+-gated Ca2+ channels in mammalian ER membranes (3), are degraded by ERAD when persistently activated (4).
ERAD substrates appear to be processed via four steps: recognition, retrotranslocation, polyubiquitination, and proteasomal degradation. Recognition can be prompted in various ways, either by generic signals (e.g. surface-exposed hydrophobic patches) or by specific recognition factors (e.g. Insigs, which target mammalian 3-hydroxy-3-methylglutaryl CoA-reductase for ERAD) (2, 5). Following recognition, ERAD substrates are retrotranslocated to the cytosol through an as yet unidentified pore, apparently in concert with the cytosolic AAA ATPase p97, which couples ATP hydrolysis to extraction (6). Once exposed to the cytosol, substrates are polyubiquitinated. E2s (ubiquitin-conjugating enzymes) and E3s (ubiquitin-protein ligases) impart selectivity to substrate ubiquitination, and several are known to be involved in the ERAD pathway, including the E2s Ubc6 and Ubc7, and the E3s yeast Hrd1p and mammalian Hrd1 and Gp78 (7). Finally, polyubiquitinated substrates are delivered to the 26 S proteasome either by shuttle proteins that bind both ubiquitin and the 19 S regulatory cap of the proteasome (8), or by directly interacting with intrinsic subunits of the 19 S cap that contain ubiquitin-binding motifs (9). It appears that some of the aforementioned steps are integrated, because multiprotein complexes that can carry out more than one step are being defined. For example, a complex centered around Hrd1 contains proteins that recognize, polyubiquitinate, and perhaps even retrotranslocate ERAD substrates (10–13).
IP3Rs participate in a wide range of cellular processes, e.g. fertilization, secretion, apoptosis, and development (3). The three mammalian IP3R subtypes (IP3R1, IP3R2, and IP3R3) are each ∼2700 amino acids in length, are tethered to the ER membrane by 6 transmembrane (TM) domains, assemble into homo- and heterotetramers, and are expressed in varying proportions in different tissues (3). They are activated by cell surface receptors that generate IP3, with binding of IP3 and the co-agonist Ca2+ inducing a conformational change that causes channel opening (3). Because persistent activation of IP3Rs leads to their degradation, this conformational change also likely serves as a recognition signal for ERAD (4). This feature makes IP3Rs particularly valuable for studying ERAD, because activation almost instantaneously converts them from stable proteins into ERAD substrates. Thus, we have identified several mediators of IP3R ERAD, notably mammalian Ubc7 (14), the p97-Ufd1-Npl4 complex (15), and most recently, SPFH2 (16).
SPFH2, also known as erlin-2 (17), belongs to a family of ∼100 mammalian proteins (“SPFH proteins”) that contain an “SPFH” domain, an ∼250-amino acid motif named because of minor sequence similarities in the proteins stomatin, prohibitin, flotillin, and HflC/K (18). SPFH proteins tend to have similar properties, including localization to cholesterol-rich, detergent-resistant membranes (DRMs) and assembly into large oligomeric structures (18). However, no universal function has yet been attributed to the SPFH domain, and SPFH proteins have distinct subcellular localizations and roles. For example, stomatin, a plasma membrane protein, binds to and regulates acid-sensing ion channels (19), and prohibitins-1 and -2 are found primarily in the inner mitochondrial membrane, where they carry out a variety of functions (20). Intriguingly, two plasma membrane SPFH proteins, MEC-2 from Caenorhabditis elegans and the mammalian stomatin-like protein, podocin, directly bind cholesterol via their SPFH domains, and it is likely that all SPFH proteins have this property (21). Binding to cholesterol undoubtedly relates to the localization of SPFH proteins in DRMs and suggests that SPFH proteins either recruit cholesterol to membrane microdomains or are restricted by cholesterol to DRMs, where they play other roles. Although the cholesterol content of intracellular membranes is minimal (22), SPFH2 and a closely related protein, SPFH1, also known as erlin-1, localize to putative ER-derived DRMs in a cholesterol-dependent manner (17).
Here, we report that SPFH1 and SPFH2 form an ∼2-MDa ER membrane complex that binds to IP3Rs immediately following their activation and is required for their ERAD. Transmission electron microscopy (TEM) of endogenous SPFH1/2 complexes reveals a double ring-shaped structure with an overall diameter of ∼250 Å. We propose that the SPFH1/2 complex plays a role in the recognition and targeting of IP3Rs and perhaps other substrates for ERAD.
EXPERIMENTAL PROCEDURES
Materials—αT3-1, HeLa, and Rat-1 cells were cultured as described (16). Already available antibodies used were: rabbit polyclonal anti-IP3R1 (23), anti-Hrd1, anti-SPFH2 (16), and anti-α-transaldolase (a kind gift from Dr. A. Perl, SUNY Upstate Medical University, Syracuse, NY); mouse monoclonal anti-ubiquitin clone FK2 (Bio-Mol International), anti-hemagglutinin (HA) epitope clone HA11 (Covance), anti-p97 (Research Diagnostics, Inc.), anti-FLAG epitope clone M5 (Sigma), anti-IP3R3 (BD Transduction Laboratories), and anti-human SPFH1 (a kind gift from Dr. S. M. Robbins, University of Calgary, Calgary, Alberta); rat monoclonal anti-grp94 (Stress-Gen); and horseradish peroxidase- and fluorophore-conjugated secondary antibodies (Sigma). Rabbit polyclonal anti-SPFH1 (which recognizes mouse and rat but not human SPFH1) and anti-FLAG were generated against synthetic peptides corresponding to the rodent SPFH1 C terminus (EPSGESPIQNKENAG) and the FLAG epitope (DYKDDDDK), respectively, and were affinity purified as described (23). Coomassie G-250, digitonin, Fura2-AM, gonadotropin-releasing hormone (GnRH), hygromycin B, N-ethylmaleimide, Polybrene, protease inhibitors, puromycin, Triton X-100, and urea were purchased from Sigma; endothelin 1 (ET1) was from Calbiochem; endoglycosidase H was from New England Biolabs; bisacrylamide, dithiothreitol, Precision Plus™ Protein standards, and SDS were from Bio-Rad; doxycycline was from Clontech; Protein A-Sepharose CL-4B was from Amersham Biosciences; Lipofectamine 2000 and NativeMark Unstained Protein Standard were from Invitrogen; and bortezomib (PS-341) was a kind gift from Millennium Pharmaceuticals.
Cell Lysis, Immunoprecipitation, and SPFH1/2 Complex Immunopurification—Cells grown in monolayer were harvested with lysis buffer (150 mm NaCl, 50 mm Tris-HCl, 1 mm EDTA, 1% Triton X-100 or 1% CHAPS, pH 8.0) supplemented with protease inhibitors (10 μm leupeptin, 10 μm pepstatin, 0.2 mm phenylmethylsulfonyl fluoride, and 0.2 μm soybean trypsin inhibitor) and 1 mm dithiothreitol. When IP3R polyubiquitination was to be measured, cells were harvested with dithiothreitol-free lysis buffer, and 2.5 mm N-ethylmaleimide was added to the lysates for 1 min, followed by 5 mm dithiothreitol. Lysates were incubated on ice for 30 min and clarified by centrifugation at 16,000 × g for 10 min at 4 °C. To immunoprecipitate specific proteins, clarified lysates were incubated with antisera and Protein A-Sepharose CL-4B for 4–16 h at 4 °C, and washed thoroughly with lysis buffer. To elute immunopurified SPFH1/2 complexes for subsequent blue native (BN)-polyacrylamide gel electrophoresis, anti-SPFH1 or anti-SPFH2 immunoprecipitates in 1% Triton X-100-containing lysis buffer were incubated with 100 μg/ml of SPFH1 or SPFH2 peptide antigens used to generate antisera for 48–72 h at 4 °C. To dissociate SPFH1/2 complexes from IP3R1, anti-IP3R1 immunoprecipitates in 1% Triton X-100-containing lysis buffer were incubated for 48 h at 4 °C, which allowed for dissociation of the majority of the co-purifying SPFH1/2 complexes. To elute immunopurified SPFH1/2 complexes for subsequent TEM, anti-SPFH1 immunoprecipitates prepared in 1% Triton X-100-containing lysis buffer were washed and then incubated in either detergent-free lysis buffer or 0.1% Triton X-100-containing lysis buffer with 100 μg/ml SPFH1 peptide for 72 h at 4 °C.
Sample Preparation, PAGE, Immunoblotting, and Mass Spectral Analysis—For SDS-PAGE, samples were boiled in gel loading buffer (24) for 3 min prior to loading. For BN-PAGE, samples prepared in NativePAGE sample buffer (Invitrogen) were supplemented with 0.25% Coomassie G-250 and loaded onto 3–12 or 4–16% gels and run at 4 °C. After electrophoresis, proteins were either stained with Coomassie Blue, or transferred to nitrocellulose (for SDS-PAGE) or polyvinylidene fluoride (for BN-PAGE) membranes and probed. Immunoreactivity was detected using Pierce chemiluminescence substrates and a Genegnome Imager (Syngene Bio Imaging). To identify proteins by matrix-assisted laser desorption ionization time-of-flight, protein bands were excised from SDS-PAGE gels and subjected to in-gel trypsinization and mass spectral analysis at the Molecular Biology Core Facilities at Dana Farber Cancer Institute (Boston, MA). The MS-Fit data base (University of California San Francisco Mass Spectrometry Facility) was used to provide possible identities for the peptides generated from each gel band.
Plasmids and Short Hairpin RNA (shRNA) Constructs—To make deletion and truncation mutants, the sequence encoding mouse SPFH2-HA in pcDNA3 (16) was mutated by inverse PCR and blunt-end ligated. An expressed sequence tag containing mouse SPFH1 cDNA was purchased from ATCC (clone BC011220), and a FLAG or HA epitope sequence was fused to the 3′ end of the open reading frame and inserted into pcDNA3 to generate SPFH1-FLAG or SPFH1-HA. SPFH1-Flag-N108Q was generated using the Stratagene QuikChange PCR mutagenesis kit. cDNAs encoding SPFH1, SPFH2, SPFH1-HA, SPFH2-HA, and SPFH2-HA mutants were also inserted into the pQCXIH vector (BD Biosciences) to generate cDNA-containing retroviruses. Sequences encoding shRNAs were inserted into 6OH1O-pSUPER.retro.puro (16) such that short interfering RNAs were generated in cells. The short interfering RNA-encoding sequences were as follows: Random (actgtcacaagtacctaca), SPFH1si4 (gtaccaggccattgcttct), and SPFH2si6 (gtacaaggctattgcttcc).
Retrovirus-mediated RNA Interference and cDNA Expression in Rat-1 Cells—Retroviruses containing either shRNA-encoding 6OH1O-pSUPER.retro.puro vectors or cDNA-encoding pQCXIH vectors were generated in HEK293T cells, titered, and Rat-1 cells stably expressing tTS were transduced as previously described (16). To deplete either SPFH1 or SPFH2, cells were incubated with SPFH1si4- or SPFH2si6-encoding retrovirus twice, and control cells for these “single knockdown” cells were twice incubated with Random shRNA-encoding virus and named “Random-s.” To deplete both SPFH1 and SPFH2, SPFH2 knockdown cells were incubated twice with SPFH1si4-encoding retrovirus, and control cells for these “double knockdown” cells were generated by incubating Random-s cells with Random shRNA-encoding retrovirus two more times and were named “Random-d.” shRNA expression was induced by treating virally transduced cells with 500 ng/ml doxycycline for 48 h, followed by selection of shRNA-expressing cells with 2.5 μg/ml puromycin for an additional 24–48 h. For stable cDNA expression, Rat-1 cells were transduced with pQCXIH-containing retrovirus for ∼8 h and supplemented with 8 μg/ml Polybrene, followed by selection of transduced cells with 200 μg/ml hygromycin B for 8 days.
TEM—SPFH1/2 complexes eluted in the absence of detergent were used without further processing, whereas complexes eluted in 0.1% Triton X-100 were first subjected to 15–50% glycerol density gradient centrifugation at 180,000 × g for 6 h at 4 °C in lysis buffer containing 0.05% Triton X-100 (SPFH1/2 complexes were typically found in the 35–45% fractions). 5 μl of SPFH1/2 complex-containing samples were applied to glow-discharged, carbon-coated copper grids were incubated for 1 min at room temperature or for >1 h at 4 °C, grids were washed once with water, and were finally stained for 1 min with 1% uranyl acetate. Samples were visualized using a 200 kV transmission electron microscope (Jeol JEM-2100) and images were recorded under minimum dose conditions using a 4096 × 4096 pixel CCD camera (TVIPS F415MP) at a magnification of ×40,000 with an underfocus of ∼1500 nm. The pixel size on the specimen level was calibrated to be 2.82 Å using catalase crystals as a standard. CCD frames were displayed with the Boxer program in the EMAN package (25), and single molecule images were selected manually. Data sets of ∼3800 particles were collected for both the detergent-free and detergent-containing samples. All subsequent image analysis was performed with the Imagic 5 software package (26). The data sets were normalized, band-pass filtered to remove high (>0.14 Å–1) and low (<0.0066 Å–1) spatial frequencies, and a circular mask was applied. The data sets were treated by reference free alignment (27), and the best averages were used as references for multireference alignment as implemented in Imagic 5. At this stage, very similar projection averages were obtained for both data sets. After several rounds of multireference alignment, the data set of the purified sample was sorted into 64 classes and angles were determined for 24 of the best averages using the Imagic command C1 start-up/new projection. The resulting three-dimensional model was forward-projected along 48 directions and the resulting images served as references for a new round of multireference alignment. The projection matching refinement was iterated until no further improvement was observed, using 83 references in the final alignment. At this stage, the two data sets were combined and subjected to two more rounds of refinement by projection matching using 131 references for the final alignment. The resolution of the final model was estimated via the Fourier shell correlation method using 0.5 as cut-off (28). For surface display, the final model was masked using the Imagic command “threed-automatic-masking” using standard parameters and filtered to remove spatial frequencies above 0.05 Å–1. The model was displayed using Chimera (29).
Data Analysis—All experiments were repeated at least once, and representative images of gels or micrographs are shown. Quantitated data are graphed as mean ± S.E. of n independent experiments, with paired Student's t test used to obtain p values.
RESULTS
SPFH1 and SPFH2 Co-purify with Activated IP3Rs—We reported previously that SPFH2 is the most abundant protein that co-immunoprecipitates with activated IP3Rs (16). Subsequent analysis of Coomassie Blue-stained anti-IP3R1 immunoprecipitates from control and GnRH-stimulated αT3-1 cells revealed that in addition to the SPFH2 band at an apparent molecular mass of ∼43 kDa, a fainter band at ∼41 kDa was also visible under stimulated conditions (Fig. 1A, lane 2). Mass spectral analysis identified the ∼41-kDa protein as SPFH1, a close homolog of SPFH2, and we validated the association of SPFH1 and SPFH2 with activated IP3R1 using antisera specific for each protein (Fig. 1A, lanes 3–6). Because SPFH2 was shown to associate with IP3Rs immediately following their activation (16), we tested whether SPFH1 associated in a similar manner. As shown in Fig. 1B, the peak association of SPFH1 and SPFH2 with IP3R1 in GnRH-stimulated αT3-1 cells occurred at 3 min, prior to peak IP3R1 polyubiquitination and the association of p97, which binds to ERAD substrates at least in part via attached polyubiquitin chains (6). Because association of SPFH1 and SPFH2 precedes substantial IP3R1 polyubiquitination, their interaction with activated IP3Rs is not likely dependent on polyubiquitin chain formation. Indeed, in αT3-1 cells preincubated with the proteasome inhibitor bortezomib to deplete intracellular free ubiquitin (30), the rate of GnRH-induced IP3R1 polyubiquitination was slowed, but the association of SPFH2 (Fig. 1C) and SPFH1 (not shown) with activated IP3R1 was unaffected. Thus, SPFH1 and SPFH2 bind to activated IP3R1 prior to and independently of IP3R1 polyubiquitination.
FIGURE 1.
SPFH1 and SPFH2 associate with activated IP3R1 independently of polyubiquitination. A, αT3-1 cells were treated without or with 100 nm GnRH for 3 min, and cell lysates prepared in 1% Triton X-100-containing lysis buffer were incubated with anti-IP3R1 to immunoprecipitate (IP) IP3R1. IPs were subjected to SDS-PAGE and either Coomassie Blue stained (lanes 1–2) or probed in immunoblots (IB) for SPFH1 and/or SPFH2 as indicated (lanes 3–6). Arrows indicate the migration positions of IP3R1, IgG heavy chain, SPFH1, and SPFH2. B, αT3-1 cells were treated with 100 nm GnRH for the times indicated, and anti-IP3R1 IPs and control samples were probed for the indicated proteins. Quantitated data for IP3R1 polyubiquitination and p97, SPFH2, and SPFH1 co-immunoprecipitation are graphed (n = 4). C, αT3-1 cells were incubated with or without 10 μm bortezomib (Bz) for 30 min, followed by treatment with 100 nm GnRH for 0–20 min, and anti-IP3R1 IPs were probed for ubiquitin, SPFH1, and SPFH2. Quantitated data for IP3R1 polyubiquitination and SPFH2 co-immunoprecipitation are graphed (n = 3). Data for SPFH1 were almost identical to that for SPFH2 (not shown).
Molecular Properties of SPFH1 and SPFH2—SPFH1 and SPFH2 are very closely related proteins, sharing ∼70% identity and ∼80% similarity at the amino acid level (supplemental Fig. S1A and Fig. 4A). Their SPFH domains, which account for ∼80% of each protein (residues 27–300 in SPFH1 and 25–298 in SPFH2), share ∼90% identity. In addition to the SPFH domain, SPFH1 and SPFH2 contain several identifiable features: a hydrophobic stretch at their N termini (residues 6–26 in SPFH1 and 4–24 in SPFH2), an N-linked glycosylation site (Asn108 in SPFH1 and Asn106 in SPFH2), and an α-helical stretch predicted to form coiled-coil motifs (residues 179–276 in SPFH1 and 177–274 in SPFH2). The alignment in supplemental Fig. S1A depicts the secondary structure predictions for SPFH1 and SPFH2, emphasizing their similarities.
FIGURE 4.
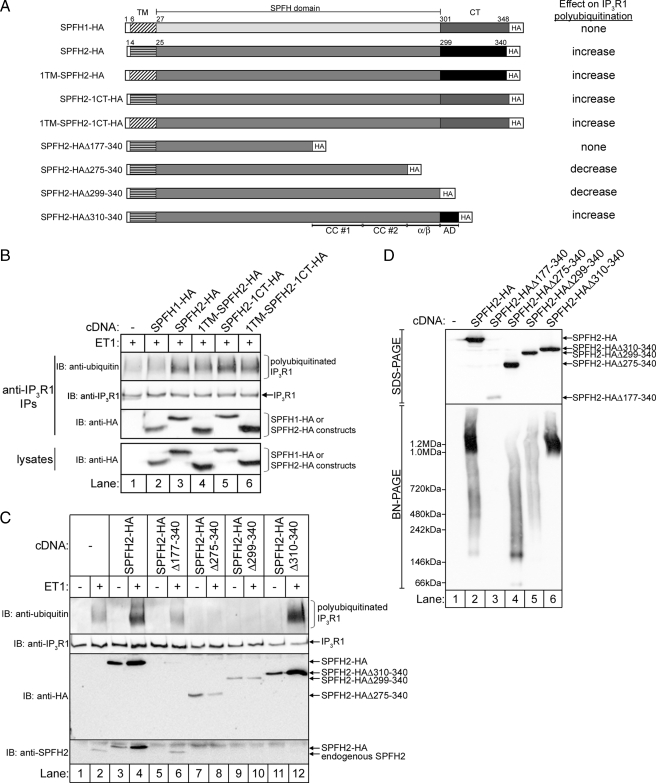
Regions within the C-terminal half of SPFH2 mediate interactions with SPFH1 and complex formation. A, diagram of the SPFH1-FLAG and SPFH2-HA constructs used in B and C, together with averaged estimates of the extent of binding of SPFH2-HA and its mutants to SPFH1-FLAG (n ≥ 5), and the extent to which SPFH2-HA and its mutants assemble into ∼2-MDa complexes. The SPFH domain, CC#1 and CC#2 in both SPFH1-FLAG and SPFH2-HA, and the α/β domain and assembly domain (AD) in SPFH2-HA are indicated. B, HeLa cells were co-transfected with cDNAs encoding wild-type SPFH1-FLAG and either wild-type SPFH2-HA or the indicated SPFH2-HA deletion mutants. Cell lysates prepared in 1% CHAPS-containing lysis buffer were incubated with rabbit polyclonal anti-FLAG, and IPs (top panels) and lysates (bottom panels) were probed with mouse monoclonal anti-FLAG or anti-HA. C, lysates from B were subjected to BN-PAGE and probed with anti-HA. IP, immunoprecipitate; IB, immunoblot.
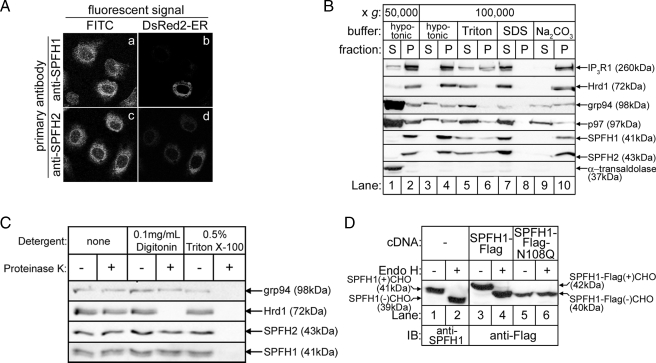
Immunofluorescence microscopy of endogenous SPFH1 and SPFH2 showed that both proteins co-localize with DsRed2-ER (Fig. 2A), suggestive of an ER localization for these proteins (16, 17). SPFH1, like SPFH2 (16), was microsome-associated and, like other integral membrane proteins (SPFH2, IP3R1, and Hrd1), was only solubilized by detergents (Fig. 2B). Interestingly, IP3R1, SPFH1, and SPFH2 were only partially solubilized by Triton X-100 (Fig. 2B, lanes 5 and 6), consistent with the previously reported association of these proteins with DRMs (17, 31). Similar to SPFH2, SPFH1 was resistant to Proteinase K in cells permeabilized with digitonin, but was susceptible in cells treated with Triton X-100 (Fig. 2C), indicating that the majority of SPFH1 mass is located in the ER lumen. Endogenous SPFH1 and exogenous FLAG-tagged SPFH1 (SPFH1-FLAG) were sensitive to endoglycosidase H unless asparagine 108 was mutated to glutamine (Fig. 2D), indicating that SPFH1, like SPFH2, is glycosylated. SPFH1 and SPFH2 are predicted to contain a TM domain at their N termini, and Edman degradation of endogenous SPFH1 and SPFH2 confirmed that these hydrophobic regions are not cleaved signal sequences (not shown). Overall, these data indicate that like SPFH2, SPFH1 is a type II membrane protein anchored to the ER membrane by its N terminus and with the rest of the protein, including an N-linked glycosylation site, protruding into the ER lumen (supplemental Fig. S1B).
FIGURE 2.
SPFH1 and SPFH2 are type II ER membrane glycoproteins. A, Rat-1 cells plated on lysine-coated coverslips were transfected with cDNA encoding DsRed2-ER, a red fluorescent protein targeted to the ER by the calreticulin ER-targeting and KDEL ER-retention sequences. The cells were fixed and stained with anti-SPFH1 (panels a and b) or anti-SPFH2 (panels c and d), followed by fluorescein isothiocyanate (FITC)-conjugated anti-rabbit secondary antibodies as described (16). Images were acquired with a Zeiss LSM510 confocal microscope equipped with a ×63 oil immersion objective. B, essentially as described (16), αT3-1 cells were harvested in hypotonic buffer, sonicated, and fractionated into supernatant (S) and pellet (P) fractions by centrifugation at 50,000 × g for 1 h at 4 °C (lanes 1 and 2). The cytosolic protein α-transaldolase was found in the supernatant, whereas the integral membrane proteins IP3R1, Hrd1, SPFH1, and SPFH2 were found in the pellet. The peripheral membrane protein p97 and the ER luminal protein grp94 were found in both the supernatant and pellet. The 50,000 × g pellets were then resuspended in hypotonic buffer, 1% Triton X-100, 0.5% SDS, or 0.1 m Na2CO3, pH 11.2, and re-centrifuged at 100,000 × g for 1 h at 4 °C (lanes 3–10). Fractions from each centrifugation were then probed for the indicated proteins. Integral membrane proteins were released from the pellet only by detergent (lanes 5–8), whereas peripheral membrane and ER luminal proteins were released from the pellet by detergent or by Na2CO3 treatment (lanes 5–10). C, HeLa cells were treated with the indicated detergents for 10 min, followed by incubation with 1 μg/ml Proteinase K for 30 min. The reactions were quenched with 1 mm phenylmethylsulfonyl fluoride, and samples were probed for the ER luminal protein grp94, the ER membrane protein Hrd1, whose C-terminal epitope is exposed to the cytosol, SPFH1, and SPFH2. D, HeLa cells were transfected with the indicated cDNAs, and cell lysates prepared in 1% Triton X-100-containing lysis buffer were incubated without or with endoglycosidase H for 3 h to cleave N-linked glycans (CHO) and were probed with anti-SPFH1 (lanes 1 and 2) or anti-FLAG (lanes 3–6).
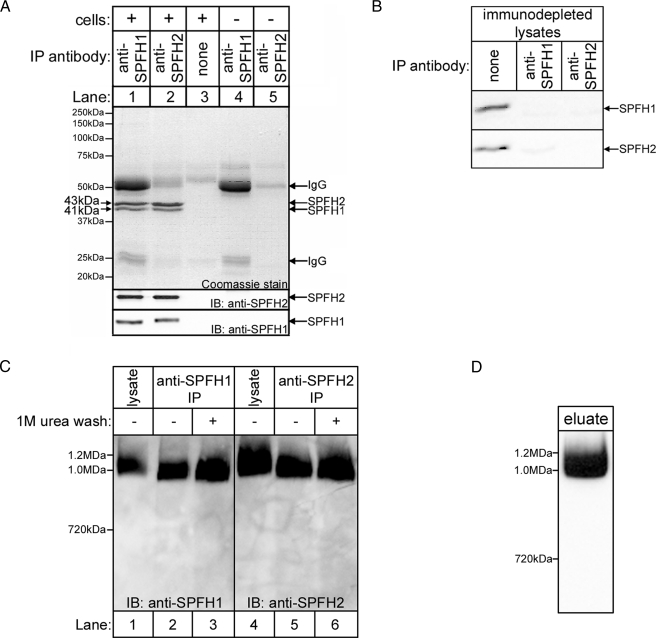
SPFH1 and SPFH2 Form a Complex That Is >1 MDa in Size—Because SPFH1 and SPFH2 co-purify with activated IP3Rs with essentially identical kinetics, we wondered whether they might be physically associated. To test this possibility, we examined Coomassie Blue-stained anti-SPFH1 or anti-SPFH2 immunoprecipitates from αT3-1 cells and observed ∼41- and ∼43-kDa bands, in an ∼1:2 ratio, in both immunoprecipitates (Fig. 3A, lanes 1 and 2). Immunoblots confirmed that these protein bands corresponded to SPFH1 and SPFH2, respectively (Fig. 3A, lower panels), and either immunoprecipitation depleted both proteins from cell lysates (Fig. 3B). Thus, SPFH1 and SPFH2 form a complex. Interestingly, no other protein bands were detectable in the immunoprecipitates (Fig. 3A), indicating that the SPFH1/2 complex is largely devoid of other proteins.
FIGURE 3.
SPFH1 and SPFH2 exist as a complex. A, anti-SPFH1 (lane 1) or anti-SPFH2 (lane 2) IPs fromαT3-1 cell lysates prepared in 1% Triton X-100-containing lysis buffer were subjected to SDS-PAGE and then Coomassie Blue stained (upper panel) or probed for SPFH2 and SPFH1 (lower panels). Controls with either no antibody (lane 3) or no cell lysate (lanes 4 and 5) were also analyzed. B, αT3-1 cell lysates were incubated without or with anti-SPFH1 or anti-SPFH2, and the immunodepleted lysates were subjected to SDS-PAGE and probed for SPFH1 and SPFH2. C, αT3-1 cell lysates (lanes 1 and 4) and immunopurified (IP) SPFH1/2 complex (lanes 2–3 and 5–6) were subjected to BN-PAGE and probed for SPFH1 or SPFH2. The IPs were either not washed (lanes 2 and 5) or were washed (lanes 3 and 6) with 1 m urea prior to elution of SPFH1/2 complexes. D, αT3-1 cells were treated with 100 nm GnRH for 3 min, anti-IP3R1 IPs were prepared, co-purifying SPFH1 and SPFH2 were allowed to dissociate from the IPs, and the eluate was subjected to BN-PAGE and probed for SPFH2. IB, immunoblot.
In αT3-1 cell lysates, the SPFH1/2 complex was ∼1 MDa in size, as estimated by BN-PAGE, which allows for the separation of proteins while still in complexes (Fig. 3C, lanes 1 and 4). Likewise, SPFH1/2 complexes immunopurified with either anti-SPFH1 or anti-SPFH2 migrated at ∼1 MDa (Fig. 3C, lanes 2 and 5), as did urea-washed, immunopurified complexes (Fig. 3C, lanes 3 and 6), indicating that the large size of the SPFH1/2 complex is not due to loosely associated proteins. In addition, SPFH1 and SPFH2 co-purifying with IP3R1 from stimulated cells migrated at ∼1 MDa, indicating that the SPFH1/2 complex, and not the individual proteins, associates with activated IP3Rs (Fig. 3D). Interestingly, size exclusion column chromatography of αT3-1 cell lysates yielded a value of ∼2 MDa for the size of the SPFH1/2 complex (not shown), and a similar discrepancy between BN-PAGE and gel filtration has been noted for the prohibitin 1/2 complex (32, 33), suggesting that BN-PAGE may underestimate the size of high molecular mass complexes. As subsequent TEM yielded a size estimate of ∼2 MDa for the SPFH1/2 complex (Fig. 8), we conclude that the SPFH1/2 complex is ∼2 MDa in size and is composed of a total of ∼50 SPFH1 and SPFH2 molecules in an ∼1:2 ratio.
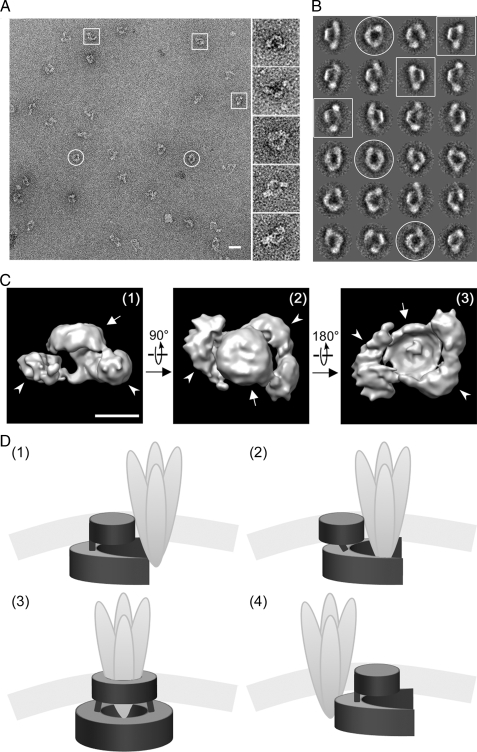
FIGURE 8.
TEM and image analysis of the SPFH1/2 complex. A, a representative micrograph of immunopurified endogenous SPFH1/2 complexes visualized at ×40,000 (scale bar = 400 Å). Putative “top” and “side” views are circled and boxed, respectively. B, 24 class averages, representing ∼50 particles each, resulting from several iterations of multireference alignment and selected for three-dimensional reconstruction are shown, with top and side views indicated as in A. C, final three-dimensional model of the SPFH1/2 complex, determined at a resolution of ∼33 Å and contoured at a volume corresponding to a calculated molecular mass of ∼2 MDa (scale bar = 100 Å). Putative side (1), top (2), and bottom views (3) are shown, with membrane and luminal domains indicated by arrows and arrowheads, respectively. D, four models for how the SPFH1/2 complex (dark gray) might interact with an activated IP3Rs tetramer (light gray). See text for description.
The C-terminal Half of SPFH2 Mediates Its Interactions with SPFH1 and the Formation of ∼2-MDa Complexes—To determine which regions of SPFH1 and SPFH2 mediate their association, we generated constructs encoding wild-type SPFH1-FLAG and either wild-type or mutant SPFH2-HA (Fig. 4A). Initially, four SPFH2-HA internal deletion mutants (SPFH2-HAΔ26–58, Δ59–100, Δ101–176, and Δ177–274) and two C-terminal truncations (SPFH2-HAΔ275–340 and Δ177–340) were generated. Residues 1–25 were preserved in each of these SPFH2-HA constructs, as this region anchors SPFH2 to the ER membrane (17). When SPFH1-FLAG was co-expressed with SPFH2-HA, SPFH2-HAΔ26–58, Δ59–100, or Δ101–176, each construct co-immunoprecipitated strongly with SPFH1-FLAG (Fig. 4B, lanes 3–6). In contrast, SPFH2-HAΔ275–340 bound less well (Fig. 4B, lane 10), and SPFH2-HAΔ177–274 and SPFH2-HAΔ177–340 bound hardly at all (Fig. 4B, lanes 7 and 11). Thus, regions near the C terminus of SPFH2, including the predicted coiled-coil (CC) region from 177–274 and another region C-terminal to 275, mediate binding to SPFH1. The region spanning residues 177–274 in SPFH2 is predicted to form two adjacent CC motifs (supplemental Fig. S1A and Fig. 4A), one from 177–230 (CC#1) and another from 231–274 (CC#2). SPFH2-HAΔ177–230 and Δ231–274 both co-immunoprecipitated with SPFH1-FLAG slightly better than SPFH2-HAΔ177–274, indicating that both CC regions contribute to the interaction with SPFH1-FLAG (Fig. 4B, lanes 8 and 9). To investigate which region C-terminal to residue 275 mediates binding to SPFH1-FLAG, we generated SPFH2-HAΔ299–340 and Δ310–340, both of which co-immunoprecipitated with SPFH1-FLAG nearly as well as wild-type (Fig. 4B, lanes 11 and 12), indicating that the region between residues 275 and 298 also contributes to the interaction between SPFH2-HA and SPFH1-FLAG. Thus, three adjacent regions in SPFH2 govern its interaction with SPFH1: CC#1, CC#2, and a third region at the very C terminus of the SPFH domain, which is predicted to form an α-helix followed by a short β-strand (α/β domain, residues 275–298) (supplemental Fig. S1A and Fig. 4A).
We next investigated the ability of the SPFH2-HA mutants to form high Mr complexes. Although wild-type SPFH2-HA migrated at ∼1 MDa on BN-PAGE (Fig. 4C, lanes 1 and 3), none of the internal deletions or the C-terminal truncations migrated as a ∼1-MDa band except SPFH2-HAΔ310–340 (Fig. 4C, lanes 4–13). Clearly, the difference between SPFH2-HAΔ299–340 and Δ310–340 demonstrates that residues 299–309 are required for the assembly of SPFH2-HA into native-like complexes. Why the internal deletion mutants that interacted well with SPFH1-FLAG (Δ26–58, Δ59–100, and Δ101–176) and contain residues 299–309 do not assemble into native-like complexes is not clear, but may be due to structural perturbations resulting from the deletions. Overall, these data demonstrate that the C-terminal half of SPFH2 is responsible for two distinct interactions: one region (residues 177–298) contains multiple motifs (CC#1, CC#2, and α/β domains) that mediate SPFH1-SPFH2 interactions, whereas the adjacent “assembly domain” (residues 299–309) mediates formation of the ∼50-subunit, native complex (supplemental Fig. S1A and Fig. 4A).
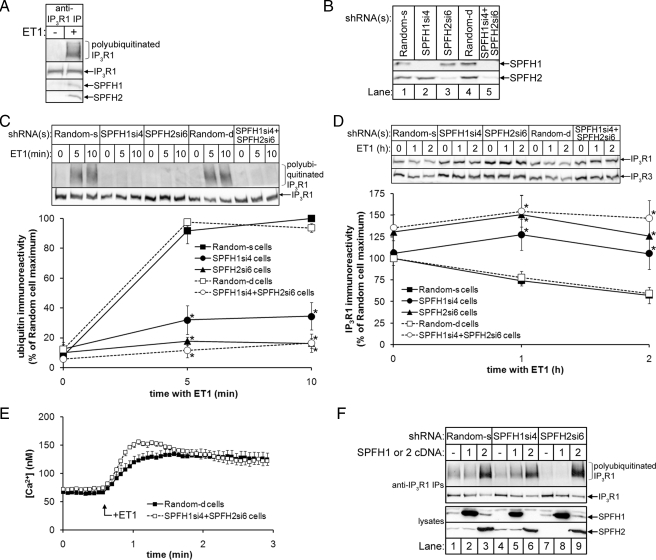
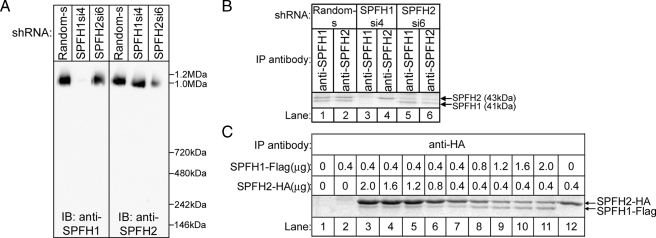
SPFH1 and SPFH2 Are Required for the ERAD of IP3Rs—To assess whether SPFH1 and SPFH2 play a role in IP3R1 processing, we used RNA interference to deplete endogenous SPFH1 or SPFH2, either individually or simultaneously, in Rat-1 fibroblasts, which we have previously used to study IP3R ERAD (15, 16). Both SPFH1 and SPFH2 bind to activated IP3Rs in ET1-stimulated Rat-1 cells (Fig. 5A), and shRNAs targeted against SPFH1 (SPFH1si4) or SPFH2 (SPFH2si6) depleted SPFH1 or SPFH2 by ≥90% as compared with cells expressing control (Random) shRNA (Fig. 5B).
FIGURE 5.
SPFH1 and SPFH2 are required for the polyubiquitination and degradation of activated IP3Rs. A, Rat-1 cells were incubated without or with 10 nm ET1 for 10 min, cell lysates were prepared in 1% Triton X-100-containing lysis buffer, and anti-IP3R1 IPs were probed for ubiquitin, IP3R1, p97, SPFH1, and SPFH2. B, lysates from Rat-1 cells expressing either Random shRNA, SPFH1si4 and/or SPFH2si6 were probed for SPFH1 and SPFH2. C, Rat-1 cells expressing the indicated shRNAs were serum starved, treated with 10 nm ET1 for the indicated times, and anti-IP3R1 IPs were probed for ubiquitin and IP3R1. Quantitated data for IP3R1 polyubiquitination are graphed (n = 6; *, denotes p < 0.001 comparing SPFH1si4, SPFH2si6, or SPFH2si6 + SPFH1si4 cells to Random-s or Random-d cells). D, Rat-1 cells expressing the indicated shRNAs were serum starved and treated with ET1 as in C, and lysates were probed for IP3R1 and IP3R3. Quantitated data for IP3R1 degradation are graphed (n = 8; * denotes p < 0.05 comparing SPFH1si4, SPFH2si6, or SPFH2si6 + SPFH1si4 cells to Random-s or Random-d cells). E, Rat-1 cells expressing the indicated shRNAs and serum starved were loaded with 10 μm Fura2-AM, and 10 nm ET1-induced Ca2+ mobilization was measured as described (n = 6) (15). F, Rat-1 cells expressing the indicated shRNAs and either no cDNA, or mouse SPFH1 or SPFH2 cDNA (which were resistant to the rat-specific shRNAs) were serum starved and treated with 10 nm ET1 for 10 min. Anti-IP3R1 IPs (upper panels) and lysates (lower panels) were probed for the indicated proteins. IP, immunoprecipitate.
Depletion of SPFH1 and/or SPFH2 drastically inhibited ET1-induced IP3R1 polyubiquitination, with SPFH1 “knockdown” leading to an ∼70% decrease, and knockdown of either SPFH2 alone or SPFH1 and SPFH2 together leading to an ∼90% decrease as compared with cells expressing Random shRNA (Fig. 5C). Furthermore, ET1-induced IP3R1 and IP3R3 degradation was significantly inhibited when SPFH1 and/or SPFH2 were depleted (Fig. 5D). These effects did not result from an inhibitory effect of SPFH1 and/or SPFH2 knockdown on IP3R1 activation, because ET1-induced Ca2+ mobilization was not inhibited by knockdown of SPFH1 and SPFH2 (Fig. 5E). Thus, the SPFH1/2 complex mediates an event following IP3R channel opening. Similar inhibition of IP3R1 polyubiquitination and degradation was observed in cells expressing alternate shRNAs directed against SPFH1 and SPFH2 (not shown), ruling out the possibility that these results are due to “off-target” effects of SPFH1si4 and SPFH2si6. Furthermore, reintroduction of SPFH1 rescued the effects of SPFH1 knockdown (Fig. 5F, lanes 4 and 5), but not of SPFH2 knockdown (Fig. 5F, lanes 7 and 8), and reintroduction of SPFH2 rescued the effects of either SPFH1 or SPFH2 knockdown (Fig. 5F, lanes 5–6 and 8–9) and, in fact, markedly enhanced ET1-induced IP3R1 polyubiquitination, even in Random shRNA-expressing cells (Fig. 5F, lane 3). These data and the finding that SPFH1 overexpression does not enhance IP3R1 polyubiquitination, indicate that SPFH2 has functional properties that SPFH1 lacks.
Formation of an ∼2 MDa Complex Is Required for Function—We next exploited the ability of exogenous SPFH2 to enhance IP3R1 polyubiquitination (Fig. 5F, lane 3) to examine the molecular determinants of SPFH2 function. We initially examined SPFH1-SPFH2 chimeras in which the TM domain and/or C-terminal ∼40 amino acids of SPFH2-HA was replaced with that of SPFH1-HA (Fig. 6A) to determine whether these regions of SPFH2, which share the least homology with SPFH1, account for the ability of exogenous SPFH2, but not SPFH1, to enhance IP3R1 polyubiquitination (Fig. 5F). Interestingly, all of these chimeras enhanced IP3R1 polyubiquitination similarly to wild-type SPFH2-HA (Fig. 6B, lanes 3–6), indicating that the functional differences between SPFH1 and SPFH2 must result from the minor sequence differences in their SPFH domains.
FIGURE 6.
Complex assembly is required for IP3R ERAD. A, diagram of the constructs used in B–D, together with their overall effects on IP3R1 polyubiquitination. B, Rat-1 cells expressing either no cDNA, SPFH1-HA, SPFH2-HA, or the indicated HA-tagged SPFH1-SPFH2 chimeras were serum starved, treated with 10 nm ET1 for 10 min, cell lysates were prepared in 1% CHAPS-containing lysis buffer, and anti-IP3R1 IPs and lysates were probed for the indicated proteins. C, Rat-1 cells expressing either no cDNA, or the indicated SPFH2-HA constructs were serum starved, treated with 10 nm ET1 for 10 min, and anti-IP3R1 IPs and lysates were probed for the indicated proteins. D, lysates from non-stimulated cells in C were subjected to SDS-PAGE or BN-PAGE, and were probed with anti-HA. IB, immunoblot.
We also examined the functional properties of several SPFH2-HA C-terminal truncation mutants (Fig. 6A). Although wild-type SPFH2-HA co-immunoprecipitated with activated IP3R1 and enhanced ET1-induced polyubiquitination (Fig. 6C, lanes 1–4), SPFH2-HAΔ275–340 and Δ299–340 inhibited IP3R1 polyubiquitination and interacted only weakly with activated IP3R1 (Fig. 6C, lanes 7–10) and thus, appear to have dominant-negative activity. In contrast, SPFH2-HAΔ310–340 enhanced IP3R1 polyubiquitination and interacted strongly with activated IP3R1 (Fig. 6C, lanes 11 and 12), suggesting that the assembly domain is required for the function of SPFH2-HA. Unfortunately, the activity of SPFH2-HAΔ177–340 could not be meaningfully assessed as its expression was much lower than the other SPFH2-HA constructs (Fig. 6D, lane 3), and the same was true of internal deletions in the region between residues 26 and 275 (not shown). ∼2-MDa complexes were not formed by SPFH2-HAΔ275–340 and Δ299–340, whereas SPFH2-HAΔ310–340 formed complexes similar to wild-type SPFH2-HA (Fig. 6D). Thus, the apparent dominant-negative effect of SPFH2-HAΔ275–340 and Δ299–340 is due to their inability to form ∼2-MDa complexes. Co-immunoprecipitation of endogenous SPFH2 with activated IP3R1 was inhibited when SPFH2-HA, SPFH2-HAΔ275–340, Δ299–340, and Δ310–340 were expressed, but not when SPFH2-HAΔ177–340 was expressed (Fig. 6C). Thus, it appears that any SPFH2-HA construct that is highly expressed can displace endogenous SPFH2 from activated IP3R1, and that only those constructs that assemble into ∼2 MDa complexes (SPFH2-HA and SPFH2-HAΔ310–340) support IP3R polyubiquitination.
Complex Function Is Influenced by the Ratio of SPFH1 to SPFH2—In view of this relationship between ∼2-MDa complex formation and function (Fig. 6), we wondered about the assembly status of the SPFH1 and SPFH2 that remain after RNA interference (Fig. 5) and found that in Rat-1 cells expressing SPFH1si4 or SPFH2si6, the remaining SPFH2 or SPFH1 still migrated at ∼1 MDa on BN-PAGE (Fig. 7A), suggesting that they assemble normally. However, the remaining complexes in cells expressing SPFH1si4 only weakly supported IP3R1 polyubiquitination (to ∼30% of that seen in control cells), and those in cells expressing SPFH2si6 did not support IP3R1 polyubiquitination at all (Fig. 5C), indicating that the remaining near-homomeric assemblies do not function efficiently. However, overexpression of SPFH2 alone did markedly enhance IP3R1 polyubiquitination (Fig. 5F), suggesting that overexpression can compensate for the inefficiency of near-homomeric SPFH2 complexes. Interestingly, the SPFH1:SPFH2 ratio in complexes immunopurified from Rat-1 cells expressing SPFH1si4 or SPFH2si6 differed markedly from that seen in cells expressing Random shRNA; e.g. compare lanes 1 and 5 in Fig. 7B. In addition, when we varied the expression of exogenous SPFH1-FLAG and SPFH2-HA in HeLa cells, the SPFH1-FLAG: SPFH2-HA ratio in complexes changed according to the amount of each protein expressed (Fig. 7C), and both proteins continued to co-migrate at ∼1 MDa on BN-PAGE (not shown). Overall, these results indicate that although essentially any ratio of SPFH1: SPFH2 will allow for assembly of ∼2 MDa complexes, an ∼1:2 ratio of SPFH1:SPFH2 produces optimal activity for the role of the SPFH1/2 complex in IP3R ERAD.
FIGURE 7.
The ratio of SPFH1 to SPFH2 in the SPFH1/2 complex is flexible. A, lysates from Rat-1 cells expressing the indicated shRNAs were prepared in 1% Triton X-100-containing lysis buffer, were subjected to BN-PAGE, and were probed for SPFH1 or SPFH2. B, anti-SPFH1 or anti-SPFH2 IPs from Rat-1 cells expressing the indicated shRNAs were Coomassie Blue stained, and bands corresponding to SPFH1 and SPFH2 are marked. C, HeLa cells were transfected with varying amounts of SPFH1-FLAG and SPFH2-HA cDNA, anti-HA IPs were Coomassie Blue stained, and bands corresponding to SPFH1-FLAG and SPFH2-HA are marked. IP, immunoprecipitate; IB, immunoblot.
The SPFH1/2 Complex Is a Ring-shaped Structure—Negative stain TEM of immunopurified, endogenous SPFH1/2 complexes revealed ring-shaped particles with a diameter of ∼250 Å, containing a central, stain-filled cavity and often two knob-like protrusions suggestive of organized domains (Fig. 8A). Using single-particle image analysis, two-dimensional projections were sorted into classes based on similar orientations (Fig. 8B), which were then used to reconstruct a three-dimensional model (Fig. 8C). This model demonstrates that the SPFH1/2 complex is a double ring-shaped structure, with a larger, apparently incomplete ring (outer diameter ∼250 Å, inner diameter ∼125 Å), joined by weak densities to a smaller disc (diameter ∼125 Å). It is probable that the disc is formed by the TM helices of SPFH1 and SPFH2, which constitute a small portion of the total complex mass and should cluster together. Although it appears as a disc, this domain might also be ring-shaped, as stain-excluding detergent micelles and/or associated lipids could fill a putative central cavity. The larger ring likely represents the luminal domains of SPFH1 and SPFH2 and appears to have pseudo-2-fold symmetry, with two thicker densities joined by a thinner density on one side. The lack of a second thin density that would complete this ring suggests that either the SPFH1/2 complex is constitutively open on one side or that the structure alternates between open and closed states with the open state predominating. It does not appear that the negative stain method prevented us from observing projections showing a complete ring, because very similar projections were observed when SPFH1/2 complexes were prepared for TEM in aurothioglucose (not shown). Interestingly, the estimated mass of the three-dimensional model portrayed in Fig. 8C is ∼2 MDa, which is consistent with the value obtained by size exclusion chromatography, but differs from the ∼1 MDa value derived from BN-PAGE (Fig. 3C).
DISCUSSION
Processing of activated IP3Rs by the ERAD pathway provides a mechanism for regulating the levels of these key mediators of intracellular signaling. In this study, we have identified a high molecular mass (∼2MDa) complex composed of ER membrane proteins SPFH1 and SPFH2 that associates rapidly and strongly with activated IP3Rs and is essential for their ERAD. The complex also mediates, albeit to a lesser extent, the degradation of a subset of model ERAD substrates (16), binds to model ERAD substrates (16), including CFTRΔF508 (34), and also interacts with several known ERAD pathway components (e.g. p97, Hrd1, and Derlin-1) (16). Thus, the SPFH1/2 complex appears to mediate the recognition and targeting of activated IP3Rs and perhaps other substrates for ERAD.
We estimate that the SPFH1/2 complex is ∼2 MDa in size, is composed of ∼50 subunits of SPFH1 and SPFH2 in an ∼1:2 ratio, and assembles via two distinct interactions. The primary interaction between SPFH1 and SPFH2 is mediated by three regions within the SPFH domain (two coiled-coil motifs and the α/β domain), and the secondary interaction requires the assembly domain, located just C-terminal to the SPFH domain, and appears to link together lower order oligomers. These features are consistent with data from another recent analysis of how SPFH1 and SPFH2 interact (35), and are strikingly similar to how other SPFH proteins oligomerize: e.g. coiled-coil regions are essential for oligomerization of prohibitins-1 and -2 and flotillins-1 and -2 (33, 36), and a long, flexible α-helical segment in the solved crystal structure of homotrimeric p-stomatin from Pyrococcus horikoshii is predicted to engage in coiled-coil interactions with adjacent p-stomatin molecules (37). Also, in mammalian stomatin, a short hydrophobic region just C-terminal to the SPFH domain mediates oligomerization and association with DRMs (38). Overall, these findings suggest that all SPFH proteins assemble into high Mr complexes using similar structural motifs.
SPFH1 was stable when SPFH2 was depleted, and vice versa, which contrasts with other SPFH proteins (prohibitins-1 and -2 and flotillins-1 and -2) that are destabilized in the absence of their assembly partners (20, 36). This difference may be because SPFH1 and SPFH2 retain the ability to form native sized (∼2 MDa) complexes when either protein is depleted, whereas these other SPFH proteins may not. Although variation in subunit composition of the SPFH1/2 complex did not affect its assembly, it did influence its function; near homomeric SPFH1 complexes did not function, near homomeric SPFH2 complexes functioned only inefficiently, and the ∼1:2 ratio of SPFH1 to SPFH2 in the native complex produced optimal activity. Assembly to the ∼2 MDa size was, however, absolutely critical for function, because SPFH2 truncation mutants that could not assemble normally exhibited dominant negative activity.
TEM of endogenous SPFH1/2 complex revealed a ring-shaped structure similar to the previously reported two-dimensional projections of the prohibitin1/2 complex (33). These structures are reminiscent of the chaperonin family of molecular chaperones, which interact with exposed hydrophobic regions on nascent proteins and use intrinsic ATPase activity to regulate protein folding (39). Although the SPFH1/2 or prohibitin1/2 complexes have not been reported to hydrolyze ATP, their ring-shaped structures may allow for analogous binding to hydrophobic surfaces, e.g. the SPFH1/2 complex could interact with aberrant-looking, hydrophobic surfaces on IP3Rs that become exposed during IP3R activation. Interestingly, the IP3R channel pore is formed by an intraluminal loop located between the fifth and sixth transmembrane domains and is thought to undergo a conformational change upon IP3R activation (3). This loop is already known to interact with ERp44 (40) and chromogranins A and B (41), and might also provide a docking site for the luminal domain of the SPFH1/2 complex. Models for how the complex might interact with activated IP3Rs are shown in Fig. 8D. In the first model, the apparent opening in the luminal ring “grasps” an activated IP3R tetramer, and in the second model, the luminal domain is more flexible and partially encircles an activated IP3R tetramer. In the third model, the luminal ring alternates between open and closed states, allowing it to fully surround an IP3R tetramer; in this case, the TM domain of the SPFH1/2 complex forms a second ring that parallels opening and closing of the luminal ring. Interestingly, a three-dimensional reconstruction of tetrameric IP3R1 suggests that its membrane and intraluminal region has a diameter of ∼70–126 Å (42), which could easily fit within the ∼125-Å cavity in the putative luminal domain of the SPFH1/2 complex. Finally, in the fourth model, the SPFH1/2 complex associates only peripherally with activated IP3Rs. In each of these models, association of the SPFH1/2 complex targets an IP3R tetramer for ERAD, perhaps by recruiting it to other ERAD pathway components that mediate retrotranslocation and polyubiquitination.
The relationship between the SPFH domain and cholesterol and the tendency of SPFH proteins to form large complexes (18) raises the possibility that SPFH proteins either help organize the lipid content of membrane microdomains by recruiting cholesterol, or are themselves recruited to DRMs by cholesterol. Interestingly, the SPFH1/2 complex appears to reside in DRMs in the ER membrane (17) and IP3Rs redistribute to DRMs upon stimulation of the IP3 signaling pathway (31), suggesting that upon IP3R activation, these proteins might co-localize to regions of the ER membrane enriched in cholesterol. Furthermore, there are alluring hints that ER membrane microdomains play roles in protein processing. For example, immature cellular prion protein is stabilized upon localization to DRMs in the ER, preventing its conversion into the misfolded, pathological scrapie isoform (43). In addition, Shiga-like toxin, a bacterial cytotoxin that exploits the ERAD machinery for entry into the cytosol, co-localizes with certain ERAD machinery components in DRMs (44), and treatment with proteasome inhibitor causes the toxin to accumulate in punctate structures that appear similar to the “quality control compartments” in which some ERAD substrates accumulate upon proteasomal inhibition (45). Overall, these findings suggest that ER membrane microdomains, perhaps enriched in cholesterol and the SPFH1/2 complex, exist as specialized hubs for processing ERAD substrates.
Supplementary Material
Acknowledgments
We thank Yuan Wang for technical assistance, and Dr. James Olzmann, Dr. John Christianson, Dr. Ron Kopito, Dr. Grant Kelley, and Danielle Sliter for many helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK49194 (to R. J. H. W.) and GM58600 and CA100246 (to S. W.). This work was also supported by a predoctoral fellowship from the PhRMA Foundation (to M. M. P. P.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; ERAD, endoplasmic reticulum-associated degradation; IP3, inositol 1,4,5-trisphosphate; IP3R, inositol 1,4,5-trisphosphate receptor; E2, ubiquitin-conjugating enzyme; E3, ubiquitin-protein ligase; TM, transmembrane; DRM, detergent-resistant membrane; TEM, transmission electron microscopy; HA, hemagglutinin; GnRH, gonadotropin-releasing hormone; ET1, endothelin 1; BN, blue native; shRNA, short hairpin RNA; CC, coiled-coil; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
References
- 1.Vembar, S. S., and Brodsky, J. L. (2008) Nat. Rev. Mol. Cell Biol. 9 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeBose-Boyd, R. A. (2008) Cell Res. 18 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foskett, J. K., White, C., Cheung, K. H., and Mak, D. D. (2007) Physiol. Rev. 87 593–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wojcikiewicz, R. J. H. (2004) Trends Pharmacol. Sci. 25 35–41 [DOI] [PubMed] [Google Scholar]
- 5.Ravid, T., and Hochstrasser, M. (2008) Nat. Rev. Mol. Cell Biol. 9 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye, Y., Meyer, H. H., and Rapoport, T. A. (2003) J. Cell Biol. 162 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostova, Z., Tsai, Y. C., and Weissman, A. M. (2007) Semin. Cell Dev. Biol. 18 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raasi, S., and Wolf, D. H. (2007) Semin. Cell Dev. Biol. 18 780–791 [DOI] [PubMed] [Google Scholar]
- 9.Husnjak, K., Elsasser, S., Zhang, N., Chen, X., Randles, L., Shi, Y., Hofmann, K., Walters, K. J., Finley, D., and Dikic, I. (2008) Nature 453 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carvalho, P., Goder, V., and Rapoport, T. A. (2006) Cell 126 361–373 [DOI] [PubMed] [Google Scholar]
- 11.Christianson, J. C., Shaler, T. A., Tyler, R. E., and Kopito, R. R. (2008) Nat. Cell Biol. 10 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lilley, B. N., and Ploegh, H. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye, Y., Shibata, Y., Kikkert, M., van Voorden, S., Wiertz, E., and Rapoport, T. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webster, J. M., Tiwari, S., Weissman, A. M., and Wojcikiewicz, R. J. H. (2003) J. Biol. Chem. 278 38238–38246 [DOI] [PubMed] [Google Scholar]
- 15.Alzayady, K. J., Panning, M. M., Kelley, G. G., and Wojcikiewicz, R. J. H. (2005) J. Biol. Chem. 280 34530–34537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce, M. M. P., Wang, Y., Kelley, G. G., and Wojcikiewicz, R. J. H. (2007) J. Biol. Chem. 282 20104–20115 [DOI] [PubMed] [Google Scholar]
- 17.Browman, D. T., Resek, M. E., Zajchowski, L. D., and Robbins, S. M. (2006) J. Cell Sci. 119 3149–3160 [DOI] [PubMed] [Google Scholar]
- 18.Browman, D. T., Hoegg, M. B., and Robbins, S. M. (2007) Trends Cell Biol. 17 394–402 [DOI] [PubMed] [Google Scholar]
- 19.Price, M. P., Thompson, R. J., Eshcol, J. O., Wemmie, J. A., and Benson, C. J. (2004) J. Biol. Chem. 279 53886–53891 [DOI] [PubMed] [Google Scholar]
- 20.Merkwirth, C., and Langer, T. (2009) Biochim. Biophys. Acta 1793 27–32 [DOI] [PubMed] [Google Scholar]
- 21.Huber, T. B., Schermer, B., Müller, R. U., Höhne, M., Bartram, M., Calixto, A., Hagmann, H., Reinhardt, C., Koos, F., Kunzelmann, K., Shirokova, E., Krautwurst, D., Harteneck, C., Simons, M., Pavenstädt, H., Kerjaschki, D., Thiele, C., Walz, G., Chalfie, M., and Benzing, T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17079–17086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prinz, W. (2002) Semin. Cell Dev. Biol. 13 197–203 [DOI] [PubMed] [Google Scholar]
- 23.Wojcikiewicz, R. J. (1995) J. Biol. Chem. 270 11678–11683 [DOI] [PubMed] [Google Scholar]
- 24.Oberdorf, J., Webster, J. M., Zhu, C. C., Luo, S. G., and Wojcikiewicz, R. J. (1999) Biochem. J. 339 453–461 [PMC free article] [PubMed] [Google Scholar]
- 25.Lüdtke, S. J., Baldwin, P. R., and Chiu, W. (1999) J. Struct. Biol. 128 82–97 [DOI] [PubMed] [Google Scholar]
- 26.van Heel, M., Harauz, G., Orlova, E. V., Schmidt, R., and Schatz, M. (1996) J. Struct. Biol. 116 17–24 [DOI] [PubMed] [Google Scholar]
- 27.Dube, P., Tavares, P., Lurz, R., and van Heel, M. (1993) EMBO J. 12 1303–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Böttcher, B., Wynne, S. A., and Crowther, R. A. (1997) Nature 386 88–91 [DOI] [PubMed] [Google Scholar]
- 29.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004) J. Comput. Chem. 25 1605–1612 [DOI] [PubMed] [Google Scholar]
- 30.Xu, Q., Farah, M., Webster, J. M., and Wojcikiewicz, R. J. H. (2004) Mol. Cancer Ther. 3 1263–1269 [PubMed] [Google Scholar]
- 31.Weerth, S. H., Holtzclaw, L. A., and Russell, J. T. (2007) Cell Calcium 41 155–167 [DOI] [PubMed] [Google Scholar]
- 32.Steglich, G., Neupert, W., and Langer, T. (1999) Mol. Cell. Biol. 19 3435–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatsuta, T., Model, K., and Langer, T. (2005) Mol. Biol. Cell 16 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, X., Venable, J., LaPointe, P., Hutt, D. M., Koulov, A. V., Coppinger, J., Gurkan, C., Kellner, W., Matteson, J., Plutner, H., Riordan, J. R., Kelly, J. W., Yates, J. R., III, and Balch, W. E. (2006) Cell 127 803–815 [DOI] [PubMed] [Google Scholar]
- 35.Hoegg, M. B., Browman, D. T., Resek, M. E., and Robbins, S. M. (2009) J. Biol. Chem. 284 7766–7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solis, G. P., Hoegg, M., Munderloh, C., Schrock, Y., Malaga-Trillo, E., Rivera-Milla, E., and Stuermer, C. A. O. (2007) Biochem. J. 403 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoyama, H., Fujii, S., and Matsui, I. (2008) J. Mol. Biol. 376 868–878 [DOI] [PubMed] [Google Scholar]
- 38.Umlauf, E., Mairhofer, M., and Prohaska, R. (2006) J. Biol. Chem. 281 23349–23356 [DOI] [PubMed] [Google Scholar]
- 39.Horwich, A. L., Fenton, W. A., Chapman, E., and Farr, G. W. (2007) Annu. Rev. Cell Dev. Biol. 23 115–145 [DOI] [PubMed] [Google Scholar]
- 40.Higo, T., Hattori, M., Nakamura, T., Natsume, T., Michikawa, T., and Mikoshiba, K. (2005) Cell 120 85–98 [DOI] [PubMed] [Google Scholar]
- 41.Huh, Y. H., Kim, K. D., and Yoo, S. H. (2007) Biochemistry 46 14032–14043 [DOI] [PubMed] [Google Scholar]
- 42.da Fonseca, P. C. A., Morris, S. A., Nerou, E. P., Taylor, C. W., and Morris, E. P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3936–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarnataro, D., Campana, V., Paladino, S., Stornaiuolo, M., Nitsch, L., and Zurzolo, C. (2004) Mol. Biol. Cell 15 4031–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, D. C., Sillence, D. J., Falguieres, T., Jarvis, R. M., Johannes, L., Lord, J. M., Platt, F. M., and Roberts, L. M. (2006) Mol. Biol. Cell 17 1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamhi-Nesher, S., Shenkman, M., Tolchinsky, S., Fromm, S. V., Ehrlich, R., and Lederkremer, G. Z. (2001) Mol. Biol. Cell 12 1711–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.