Abstract
Peroxisome proliferator-activated receptor (PPARγ) agonists increase insulin sensitivity in humans and are useful for treating human diabetes. Treatment with these agonists leads to increased apoE expression and triglyceride accumulation in adipocytes. The importance of apoE for adipocyte triglyceride accumulation is demonstrated by observations that triglyceride accumulation is impaired in apoE knockout adipocytes treated with PPARγ agonists. The current studies investigate the molecular mechanism for PPARγ stimulation of the adipocyte apoE gene and demonstrate that the liver receptor X (LXR) response element within an apoE gene downstream enhancer is required for the apoE response to PPARγ agonists. The response of the apoE gene to treatment with PPARγ agonists was delayed beyond 12 h suggesting the involvement of an intermediary pathway. The combined addition of PPARγ and LXR agonists did not increase apoE response beyond that observed with addition of either alone. Deletion or mutation of the LXR response element completely eliminated the adipocyte apoE gene response to a PPARγ agonist. Chromatin immunoprecipitation analyses performed using isolated adipocytes, or adipose tissue from mice treated with PPARγ agonists, showed increased LXR binding to the apoE gene after PPARγ agonist treatment. Knockdown of LXR expression completely eliminated the increase in apoE message, protein, and triglyceride in response to PPARγ stimulation. The LXR response element has been previously shown to mediate sterol responsiveness of the apoE gene, and apoE expression plays an important role in adipocyte triglyceride balance. The current observations suggest that the PPARγ-LXR-apoE regulatory cascade could be an important molecular link for cross-talk between adipocyte triglyceride and cholesterol homeostasis.
PPARγ2 is highly expressed in adipocytes where it modulates the expression of a program of genes involved in adipocyte function (1). These include genes involved in TGD and fatty acid metabolism and in the expression of adipokines. Adipocyte lipid metabolism and the secretion of adipokines have a powerful effect on systemic substrate utilization and on overall organismal energy metabolism (2, 3). TZD drugs, currently in clinical use for treatment of human diabetes, are established ligands of adipocyte PPARγ receptors, and we have previously shown that adipocyte apoE expression is responsive to treatment with these drugs (4). Zechner and colleagues first described expression of apoE by adipocytes in 1991 (5), but only recently has additional information been provided regarding regulation and function of adipocyte apoE (6–10). Treating isolated adipocytes or intact humans with PPARγ agonists increases adipocyte and adipose tissue apoE gene expression (4, 6). The pro-inflammatory cytokine, tumor necrosis factor-α, reduces adipocyte apoE expression, an effect mediated by the NF-κB transcriptional complex binding an element in the apoE gene proximal promoter (4, 7). Reactive oxygen species produced by the stromovascular fraction of adipose tissue harvested from obese mice also reduce adipocyte apoE expression via the NF-κB pathway (8). With respect to apoE function, adipocytes from EKO mice are smaller than adipocytes from WT mice, and cultured EKO adipocytes and adipose tissue accumulate less TG compared with WT cells after incubation with apoE-containing lipoproteins (6). This defect in TG accumulation can be corrected by the adenoviral expression of apoE in EKO adipocytes. Furthermore, in EKO adipocytes, TG accumulation in response to treatment with PPARγ agonists is impaired compared with WT adipocytes (6). All of these observations support a role for apoE in adipocyte TG metabolism. Such a role for apoE is different than what has been described for other cell types, for example macrophages, in which apoE has been shown to primarily play a role in cellular sterol metabolism (11, 12). Its role in these cells includes modulating sterol efflux and/or the subcellular distribution of sterol. Consistent with its role in cellular sterol metabolism, the apoE gene is strongly regulated by cellular sterol content acting via the sterol-sensing LXR pathway and LXR response elements present in two duplicated downstream apoE gene enhancers, ME.1 and ME.2 (13).
In previous studies, we established that the response of the apoE gene to PPARγ agonists was not mediated by the apoE proximal promoter but required the presence of at least one of these two apoE downstream gene enhancers (4). Based on the requirement for a downstream enhancer, the presence of an LXRE in the enhancer, the absence of a clearly identifiable PPRE within this enhancer, and the recently established positive effect of PPARγ agonists on LXR gene transcription (14), we hypothesized that the response of the adipocyte apoE gene to PPARγ agonists could be mediated by the LXR pathway (4). Based on similar considerations, a role for the LXR pathway has been posited as mediating the effect of PPARγ agonists on ABCA1 expression in macrophages (14). Because of observations supporting a role for apoE in adipocyte TG metabolism, the involvement of the LXR pathway for mediating the PPARγ response of the adipocyte apoE gene could provide an important nexus of cross-talk between adipocyte TG and sterol metabolism. The studies reported in this manuscript examine the molecular mechanism for PPARγ stimulation of adipocyte apoE gene transcription and establish an important role for the LXR pathway for mediating this response.
EXPERIMENTAL PROCEDURES
Materials—Cell culture media, supplements, and antibiotics were purchased from Invitrogen. Insulin, 1-methyl-3-isobutylxanthine, dexamethasone, GW3965, and 22-OHch were obtained from Sigma. Ciglitazone and rosiglitazone were purchased from Cayman Chemical (Ann Arbor, MI). Pioglitazone was obtained from Takeda Pharmaceutical Co. Antibodies to LXRα/β were obtained from Santa Cruz Biotechnology.
Animals—Eight-week-old C57BL/6J mice were purchased from Jackson Laboratories. Mice were housed in a temperature-controlled room with a 12-h light/dark cycle and were given free access to standard chow and water for 10 days. Pioglitazone (25 mg/kg/day) mixed with chow or the control chow diet was fed for an additional 5 days before mice were sacrificed for isolation of adipose tissue. Intra-abdominal white adipose tissue was collected in ice-cold phosphate-buffered saline. For real-time PCR quantitation of apoE mRNA abundance, total RNA was extracted for synthesis of first-strand cDNA. For the in vivo chromatin immunoprecipitation assay, adipose tissue was prepared according to the EpiQuik™ tissue chromatin immunoprecipitation kit (Epigentek, Brooklyn, NY).
Cell Culture—3T3-L1 cells were obtained from the American Type Culture Collection (Manassas, VA). Cells were maintained for passage in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at <70% confluence. For experiments, differentiation was induced at 2 days post confluence by incubation for 3 days in culture medium containing 0.25 μm dexamethasone, 0.5 mm 3-isobutyl-l-methylxanthine, and 10 μg/ml insulin. This was followed by 10% fetal bovine serum/Dulbecco's modified Eagle's medium containing 5 μg/ml insulin that was renewed every 2 or 3 days. Experiments were performed on post differentiation days 8–10. Triglyceride content of cells was measured using a kit from Wako Chemicals as previously described (6).
Real-time RT-PCR and Immunoblot Analysis—Total RNA was extracted from cells or tissues after the treatments described in figure legends. Each sample of first strand cDNA was synthesized from 1 μg of total RNA using a Thermo-Script™ RT-PCR system according to the manufacturer's instructions (Invitrogen). cDNA (1.5 μl) was amplified by real-time PCR using a TaqSYBR green supermix with ROX (Bio-Rad). Normalization for loading was accomplished by assessing β-actin message abundance. Individual samples were analyzed in triplicate using an Mx3000P quantitative PCR system (Stratagene). Levels for RNA are reported as -fold change compared with control using the comparative quantitation analysis software available on the Mx3000P. Product purity for PCR reactions was confirmed by examination melting curves for the presence of a single peak. Immunoblots on cells or tissues were performed, and the results were quantitated as previously described (7).
Site-directed Mutagenesis—The human apoE promoter fragment –503/+86 alone or with the human apoE ME2 downstream enhancer (7, 13) were cloned into pGL3 luciferase vector as previously described to give rise to pGL503 or pGL503en, respectively. Site-directed deletion or mutation was performed using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). The primer with deleted nucleotides (indicated by the dashed line) was: 5′-GGA CAC CCT CCC CAT CAG CTG CCA————–AAG GCA GCT GGT GGG GAA GGC ATT GG-3′. The primer with mutated nucleotides (as indicated in small cap letters) was: 5′-CCC CAT CAG CTG CCA Gaa TCA CTG GCa aTC AAA GGC AGC TGG TG-3′. These primers and their complementary sequences were used to disrupt the LXR response element in ME2. All deletions and mutations were confirmed by DNA sequencing.
Transient Transfection and Luciferase Activity Assay—Transient transfection of luciferase reporter plasmids was performed using Lipofectamine 2000 (Invitrogen) according to the protocol recommended by the manufacturer in the 12-well plate format. Luciferase assays were performed using the luciferase assay system (Promega, Madison, WI). Following the treatments described in the figure legends, cells were washed twice in cold phosphate-buffered saline and lysed in 120 μl of cell culture lysis reagent provided in the luciferase assay kit. 10 μl of the cell lysate was mixed with 50 μl of luciferase assay solution, and luminescence was determined using a Turner Design Luminometer. All experiments were performed in triplicate and normalized to the activity produced by a cotransfected β-galactosidase expression vector. For RNA interference, differentiated 3T3-L1 cells were trypsinized and plated in fresh growth medium (3 × 105 per ml) and transferred into 12-well plates at 1 ml per well. In each well, 100 pm of a mixture of siRNA targeted to LXRα and LXRβ (L-040649-01-0005, Mouse NR1H3, NM_013839 and L-042839-00-0005, Mouse NR1H2, NM_009473, respectively) complexed with 2 ml of Lipofectamine was added. A non-targeting siRNA provided by the manufacturer was used as a control.
Chromatin Immunoprecipitation—3T3-L1 adipocytes were treated as described in the figure legends. Chromatin immunoprecipitation was carried out using the ChIP-IT Express kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions as previously described (7). Briefly, the cells were fixed, and the isolated nuclear fraction was digested for 13 min at 37 °C by an enzymatic shearing mixture. The digestate was visualized on an agarose gel to ensure shearing efficiency resulted in DNA fragments ranging from 200 to 500 bp. After centrifugation, 10 μl of the supernatant containing sheared chromatin was used as the assay input. The sheared chromatin/ protein was immunoprecipitated using an anti-LXRα/β antibody (sc-13068, Santa Cruz Biotechnology) at 4 °C overnight. After immunoprecipitation, samples were centrifuged, washed, and reverse cross-linked prior to being treated with ribonuclease A and proteinase K. The DNA was purified with DNA mini columns provided with the kit, and real-time PCR was performed with primers (A 5′-AAC TGA CAC GTG GGT TGC AGG GGC AAC A-3′) and the reverse primer (5′-CAA CAT CCT TTC TCT CAG GCT AGA GTC C-3′), designed to amplify a 152-bp portion of the apoE enhancer containing the LXRE. As an internal control, an RNA polymerase II antibody (sc-899, Santa Cruz Biotechnology) was used to immunoprecipitate the mouse GAPDH promoter.
The in vivo ChIP was performed with freshly isolated adipose tissue using the EpiQuik™ tissue ChIP kit according to the manufacturer's instructions. 400 mg of adipose tissue in 2 ml of homogenizing buffer was homogenized using 20 strokes of a Dounce homogenizer. The homogenate was centrifuged at 5000 rpm for 10 min at 4 °C, and the pellet was re-suspended in 1 ml of CP3 buffer (included with the kit) with protease inhibitors. The DNA was sheared by sonication with 10 pulses of 15 s each at 25% Power using a Vibracell 3-mm microtip probe with 40-s incubation on ice between each pulse. The sheared DNA was used for immunoprecipitation as described above.
Statistical Analysis—All the data are presented at means ± S.D. values. Statistical differences were evaluated using analysis of variance (SPSS, Chicago, IL). p < 0.05 was considered significant.
RESULTS
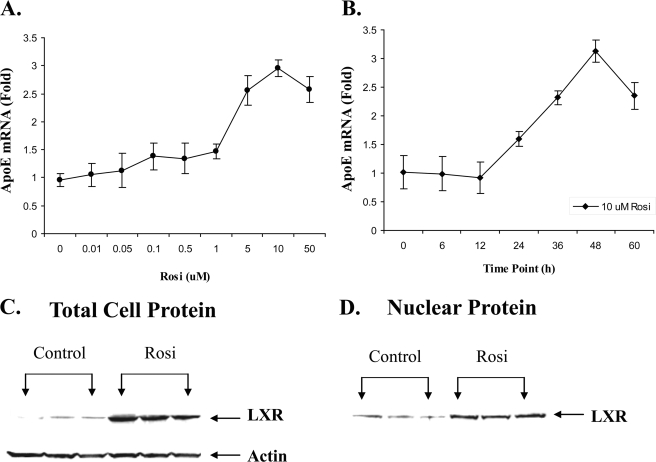
Fig. 1 presents the results of detailed dose response and time course analyses for the response of apoE gene to treatment with rosiglitazone. The dose-related increase with rosiglitazone establishes the apoE response after treatment of adipocytes with three different PPARγ agonists, rosiglitazone (Fig. 1A), ciglitazone, and pioglitazone (4, 6). The time course for the response of the apoE gene to treatment with rosiglitazone (Fig. 1B) demonstrates that the first measurable increase in apoE mRNA levels after addition of rosiglitazone is not detected until after 12 h. This somewhat prolonged lag time for the apoE gene response is consistent with the hypothesis that the apoE gene does not respond directly to PPARγ agonists and that the response requires involvement of an intermediary pathway. Based on considerations presented in the introduction, we focused on a potential intermediary role for the LXR pathway. The LXR gene has been shown to be a target of PPARγ agonists (14, 15), and we examined the response of LXR protein levels in our model system. Fig. 1 (C and D) shows that treatment of 3T3-L1 adipocytes with rosiglitazone increased total cellular (C) and nuclear (D) LXR protein abundance.
FIGURE 1.
Response of adipocyte apoE and LXR expression to rosiglitazone. A, 3T3-L1 adipocytes were treated with the indicated dose of rosiglitazone (Rosi) for 48 h. B, 3T3-L1 adipocytes were treated with 10 μm rosiglitazone or vehicle for the time indicated. The -fold change at each time point is relative to cells treated for the same time with vehicle alone. At the end of the incubation, total RNA was isolated and apoE mRNA was measured as described under “Experimental Procedures.” Values shown are mean ± S.D. C, 3T3-L1 cells were treated with 10 μm rosiglitazone for 48 h. 50 μg of total cell protein was used for immunoblot using an antibody to LXR. D, 30 μg of nuclear protein was used for immunoblot.
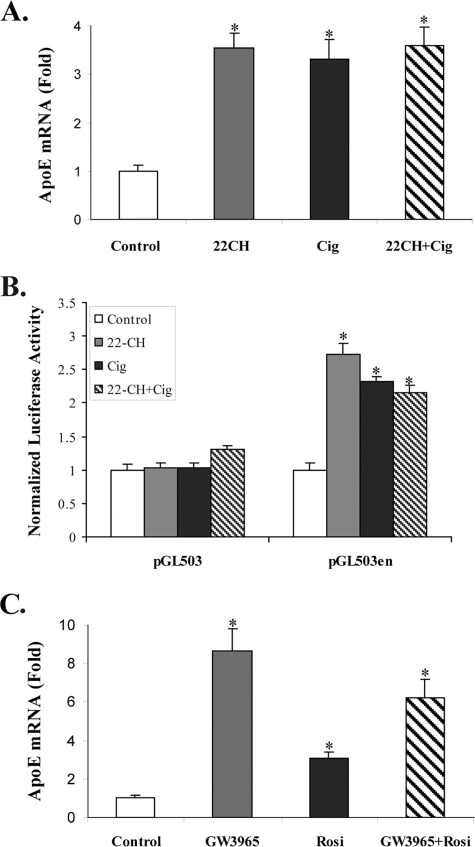
We next measured the response of the adipocyte apoE gene, and of an apoE gene reporter construct, to incubation with an LXR agonist, a PPARγ agonist, or a combination of these (Fig. 2). The addition of 22-OHch (an oxysterol LXR agonist) alone, or ciglitazone alone, led to a 2- to 3-fold increase in the expression of the adipocyte apoE gene and of a reporter construct containing 503 bp of the proximal apoE gene promoter and the apoE gene enhancer ME2 (Fig. 2, A and B). Addition of the agents together, however, did not produce a greater increase in apoE or reporter gene response compared with either agent alone. The reporter construct without ME2 did not respond to either 22-OHch or ciglitazone. An experiment similar to that shown in Fig. 2A, but conducted with the alternate agonists, rosiglitazone and GW3965, gave similar results. Either agent alone significantly stimulated apoE mRNA abundance, but the effect of the agents together was not additive. The absence of an additive response to the combined addition of PPARγ and LXR agonists provided further support for the hypothesis that these agonists act via a common pathway to stimulate adipocyte apoE gene expression.
FIGURE 2.
Activation of the adipocyte apoE gene by PPARγ and LXR ligands is not additive. A, 3T3-L1 cells were treated with ethanol vehicle, 10 μm 22-OHch, 20 μm ciglitazone (Cig), or a combination of these for 48 h before harvest to measure apoE mRNA level. B, 3T3-L1 adipocytes were transfected with the indicated apoE reporter gene constructs and treated as described in A. Luciferase activity was measured as described under “Experimental Procedures.” C, cells were treated with 10 μm rosiglitazone or 1 μm GW3965 for 48 h and harvested as described in A. *, p < 0.005 for comparison to untreated control cells.
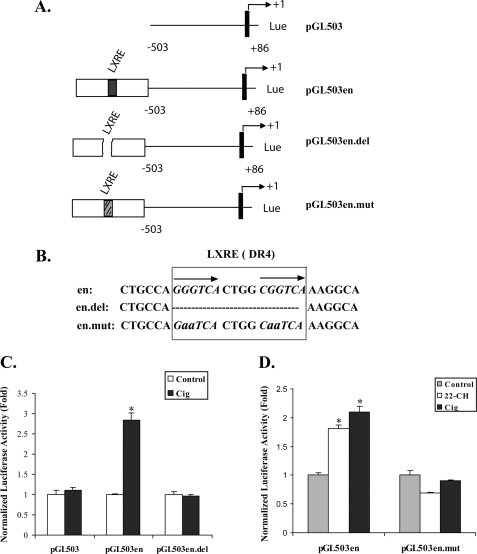
The quantitatively similar response of the endogenous apoE gene and the pGL503en reporter construct to ciglitazone in Fig. 2 indicated that the important gene elements required for increased apoE gene expression in response to PPARγ agonists were contained within this reporter construct. Using the results in Figs. 1 and 2 as a foundation, we designed experiments to more definitively examine the role of the LXR pathway in mediating the PPARγ response of the apoE gene by (a) evaluating the need for the apoE gene LXRE, (b) measuring the recruitment of LXR to the apoE gene in response to PPARγ agonist stimulation in vitro and in vivo, and (c) evaluating the effect of silencing LXR protein expression. Fig. 3 (A and B) provides the details of the four reporter gene constructs used to examine the importance of the apoE gene LXRE. These constructs include a proximal apoE gene promoter construct without the ME2 downstream enhancer, the proximal promoter construct with ME2, the enhancer construct in which the LXR response element is deleted, and the enhancer construct in which the LXR element is mutated. The specific nucleotides deleted or mutated are shown in Fig. 3B. Fig. 3C shows that the promoter construct without ME2 failed to respond to ciglitazone, whereas the promoter construct containing ME2 responded with an approximate 3-fold increase to treatment with ciglitazone. Deletion of the LXR response element within ME2 completely eliminated the response to ciglitazone. Fig. 3D shows that the pGL503en construct responded with 2- to 3-fold increased expression after treatment with 22-OHch or ciglitazone. Mutation of four nucleotides within the LXR element, however, completely eliminated the reporter gene response to both agonists.
FIGURE 3.
Stimulation of an apoE gene reporter construct by ciglitazone (Cig) requires an intact LXRE. A, the reporter constructs used to evaluate apoE gene elements responsive to PPARγ agonists are shown. The numbers on each side of the solid bars indicate the position in base pairs relative to transcription initiation site. The solid bars represent exon-1 of the apoE gene. The open box represents the apoE ME2 enhancer element. The presence of an intact (en), deleted (en. del), or mutated (en. mut) LXRE is indicated. B, sequences of the native apoE gene LXRE, the mutated LXRE, and the LXRE deletion. Lowercase letters indicate mutated nucleotides. Hatch marks represent deleted nucleotides. C, 3T3-L1 cells were transfected with the indicated constructs and incubated with or without 20 μm cig for 48 h. Normalized luciferase activity relative to untreated control is shown. D, 3T3-L1 cells were transfected with the indicated constructs and treated with 20 μm cig or 10 μm 22-OHch for 48 h as shown. Normalized luciferase activity relative to untreated control is shown. *, p < 0.001 for comparison to untreated control. Results shown are mean ± S.D. of three independent experiments each done in triplicate.
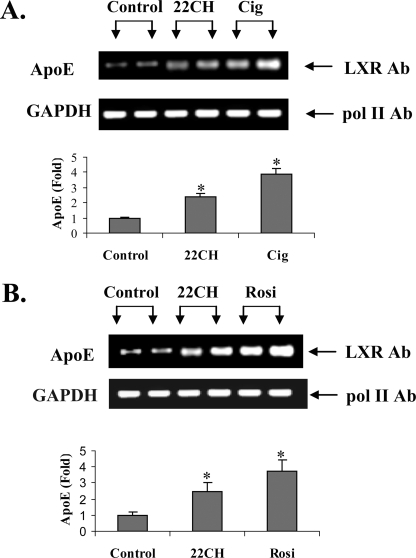
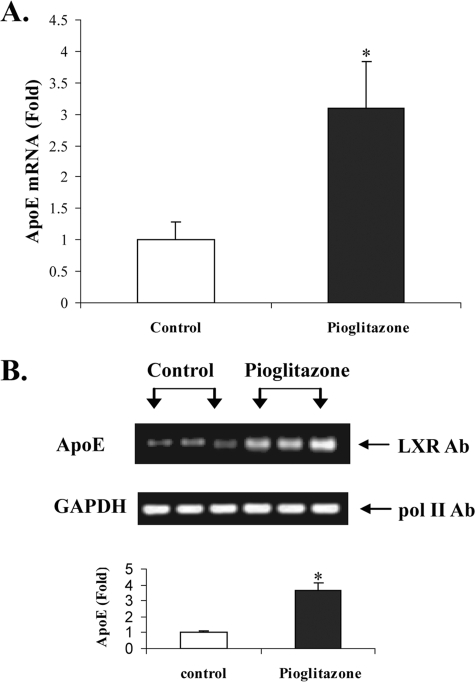
Figs. 4 and 5 show the results of ChIP analyses and confirm that treatment of intact cells (Fig. 4) or mice (Fig. 5) with PPARγ agonists leads to increased LXR binding to the apoE gene. Fig. 4A shows that treatment of intact adipocytes with ciglitazone or 22-OHch increased LXR binding to the apoE gene. A representative agarose gel shows the binding of RNA polymerase II to the GAPDH gene (as an internal control) and the binding of LXR to the apoE gene in response to treatment of adipocytes with 22-OHch or ciglitazone. The bar figure represents PCR quantitation of three separate ChIP analyses, all corrected for the internal GAPDH control, and shows a 4-fold increase in LXR binding to the apoE gene after treatment of adipocytes with ciglitazone. Fig. 4B shows a similar result from an identical experiment using a different PPARγ agonist, rosiglitazone. For Fig. 5, mice were treated for 5 days with pioglitazone at 25 mg/kg/day. At the end of that time, freshly isolated adipose tissue demonstrated a 3-fold increase in apoE mRNA abundance (Fig. 5A). ChIP analysis using freshly isolated adipose tissue demonstrated significantly more LXR protein binding (3.7-fold) at the apoE gene enhancer element.
FIGURE 4.
ChIP analysis of LXR binding to the apoE gene in response to the treatment with PPARγ ligands. 3T3-L1 cells were treated with 10 μg 22-OHch or ciglitazone (Cig)(A) or rosiglitazone (Rosi)(B) for 48 h and then harvested for ChIP analysis as described under “Experimental Procedures.” After immunoprecipitation, a 152-bp amplicon of the apoE enhancer containing the LXRE was amplified by PCR. The amount of LXR bound to the LXR enhancer was assessed using real-time PCR to quantitate immunoisolated DNA. Representative images of agarose gels show RNA polymerase II binding to the GAPDH promoter (as an internal control) and LXR binding to apoE. The real-time PCR results are mean ± S.D. from three different ChIP analyses, each done in triplicate. *, p < 0.05 compared with untreated control.
FIGURE 5.
Treatment with PPARγ ligand in vivo activates LXR binding to the apoE gene. A, mice (three per group) were fed chow or chow plus pioglitazone (Pio, 25 mg/kg per day) for 5 days before harvest of adipose tissue. A, total RNA was extracted, and apoE mRNA was quantitated by RT-PCR as described under “Experimental Procedures.” *, p < 0.005 compared with untreated controls. B, chromatin immunoprecipitation assay was performed as described under “Experimental Procedures” using freshly isolated adipose tissue from pioglitazone-treated and untreated mice. A representative agarose gel shows RNA polymerase II binding to the GAPDH gene (as an internal control) and LXR binding to the apoE gene. Real-time PCR quantitation of immunoisolated DNA after LXR immunoprecipitation is shown as -fold change in treated compared with control mice. *, p < 0.005 compared with untreated mice.
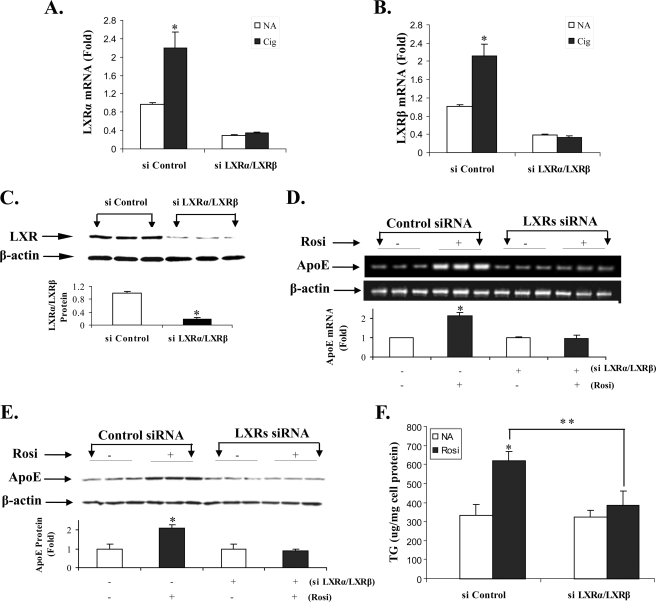
The above results established that treatment with PPARγ agonists increased LXR binding to the apoE gene enhancer in vitro and in vivo, and that the LXR response element in the apoE gene enhancer was required for increased apoE gene expression after PPARγ stimulation. We next performed experiments to further confirm that an intact adipocyte LXR pathway was required for the response of the endogenous adipocyte apoE gene to PPARγ stimulation. Cells were transfected with an siRNA for LXR, or a control siRNA, and after 24 h they were treated with PPARγ agonists as indicated (Fig. 6). The results in Fig. 6 (A and B) show that treatment with a combined LXRα/LXRβ siRNA leads to a 65–80% reduction in LXRα (A) or LXRβ (B) mRNA in unstimulated adipocytes. Further, treatment with the LXR siRNA completely eliminated the response to ciglitazone that is easily observed in cells treated with a control siRNA. For Fig. 6C, cells were harvested for immunoblot analysis using an antibody to LXR that recognizes both LXRα and LXRβ protein. LXR protein abundance is significantly reduced by the LXR siRNA. Fig. 6D shows that treatment of adipocytes with LXR siRNA completely eliminated the increase in apoE mRNA levels in response to rosiglitazone treatment. Silencing LXR also completely prevented the PPARγ-mediated increase in apoE protein (Fig. 6E). We have previously shown that the TG accumulation that follows PPARγ agonist treatment of adipocytes is dependent on the expression of adipocyte apoE (6). This latter observation, along with the results in Fig. 6E, supports the prediction that silencing LXR would prevent the PPARγ-mediated increase in adipocyte TG. The results in Fig. 6F show that treatment with the LXR siRNA completely prevented PPARγ-mediated TG accumulation.
FIGURE 6.
LXR is required for activation of apoE gene expression and TG accumulation by PPARγ agonists. Cells were transfected with 100 pm of combined LXRα/LXRβ siRNA, or a non-targeting control siRNA, as described under “Experimental Procedures.” Twenty-four hours after transfection, cells were washed and incubated in growth medium alone, or with 10 μm rosiglitazone (Rosi) or 20 μm ciglitazone (Cig) as indicated. After an additional 24 h, some cells were harvested for measurement LXRα mRNA abundance (A), LXRβ mRNA abundance (B), or for immunoblot analysis (C) using an antibody that recognizes both LXRα and LXRβ protein. The balance of the cells was allowed to continue incubation for an additional 24 h before being harvested for analysis. D, total RNA was extracted for real-time RT-PCR measurement of apoE mRNA level. The agarose gel shows the result of a single experiment. The bar graph shows -fold change in apoE mRNA level by RT-PCR and represents the mean ± S.D. from three separate experiments each done in triplicate. E, cells were lysed for analysis of apoE and β-actin protein expression by immunoblot analysis as described under “Experimental Procedures.” The -fold change for apoE expression (normalized to β-actin expression) is shown. F, cells were harvested for measurement of TG mass as described under “Experimental Procedures.” *, p < 0.01 comparing PPARγ agonist-treated to untreated samples (A, B, D, E, and F), or comparing samples treated with targeting versus control siRNA (C). **, p < 0.05 for indicated comparison in F.
DISCUSSION
The endogenous expression of apoE in adipocytes has important effects on adipocyte size and on TG content and turnover (6). PPARγ agonists increase adipocyte apoE expression, and the absence of adipocyte apoE expression impairs the lipogenic response to PPARγ agonists. In the current studies, we provide the molecular mechanism for PPARγ stimulation of the adipocyte apoE gene. Based on our previous observations showing that the response of the apoE gene to PPARγ agonists required the presence of a downstream apoE gene enhancer, the presence of an LXRE in this enhancer, and the absence of a clearly identifiable PPRE within this enhancer, we hypothesized that the LXR pathway mediated the apoE gene response to PPARγ agonists (4). Consistent with this hypothesis, we now show that increased expression of an apoE reporter construct to PPARγ agonists requires the presence of an intact LXRE. Either deletion of the LXRE, or the mutation of 4 bp within the LXRE, completely abolishes the apoE gene response to PPARγ agonists, as well as to the LXR agonist 22-OHch. Furthermore, treatment of isolated adipocytes or intact mice with PPARγ agonists increases binding of LXR to the apoE gene LXRE. In addition, and most importantly, silencing LXR expression totally eliminates the response of the endogenous adipocyte apoE gene to PPARγ stimulation. Consistent with the observations of others (14, 15), treatment of adipocytes with PPARγ agonists increases both total cellular and nuclear LXR protein. It is of interest that, despite this increase in LXR protein, the response of the apoE gene to the combined addition of a PPARγ and LXR agonist is not greater than that to either agonist alone. This observation suggests that, in addition to increasing LXR expression, treatment with PPARγ agonists could also stimulate the production of an endogenous cellular ligand for LXR in mature adipocytes, thereby eliminating any incremental stimulation produced by an exogenous LXR ligand. The need to induce an endogenous cellular ligand could also help to explain the lag time of at least 12 h for the response of the apoE gene to PPARγ stimulation. The experiments in this report also demonstrate consistent results with three different PPARγ agonists, decreasing the likelihood that results can be explained by an off-target effect of any one compound.
Numerous observations in the literature suggest that adipocyte cholesterol and TG metabolism are linked (1, 16). Adipocytes contain the largest store of cholesterol in the body, and, in contrast to most other cell types, the bulk of adipocyte cholesterol is maintained in its non-esterified form (17). Large amounts of unesterified cholesterol can be found in the adipocyte plasma membrane and as a component of the TGD (17). There is a strong correlation between adipocyte cell size and cholesterol content and also between the TG and cholesterol content of the adipocyte TGD (17, 18). Adipocyte TG accumulation leads to an increased cellular need for cholesterol as a component of the TGD and to support increasing cell size with the expansion of the plasma membrane. Further, expansion of the TGD may promote the redistribution of cholesterol away from the plasma membrane to the TGD (17, 19). Cholesterol in the adipocyte plasma membrane plays a critical role for maintaining plasma membrane microdomain organization, particularly for the formation and maintenance of plasma membrane caveolae (20). Caveolae are highly abundant in adipocytes and play an important role in vesicular trafficking and signal transduction, including a role in insulin signaling (21). The perturbation of adipocyte plasma membrane cholesterol leads to disruption of caveolar structure, and of plasma membrane receptor distribution and trafficking, and inhibits insulin-stimulated glucose transport and metabolism (20). These observations support a need for coordination between TG and sterol homeostasis in adipocytes to maintain the structural and functional integrity of the adipocyte plasma membrane.
In macrophages, apoE as a target of LXR, participates in modulating cellular sterol balance by influencing cellular cholesterol flux (11, 12). Increased apoE expression facilitates sterol efflux, and the increase in apoE expression in response to sterols can be viewed as a feedback mechanism to prevent macrophage sterol overload (13, 22). However, adipocytes, as noted above, have a large need for cellular cholesterol. This along with a limited capacity to synthesize cholesterol, estimated to be 4% of that in hepatocytes (23), suggests that apoE may play a different role in adipocytes. In line with this notion, others have shown that adipocyte apoE expression does not effectively facilitate sterol efflux and, in fact, participates in a highly effective sterol re-capture mechanism that adds to cell cholesterol stores (24). Sterol efflux from differentiating adipocytes remains very limited despite induction of ABCA1 and apoE expression during adipocyte differentiation (19). Furthermore, treatment of adipocytes with LXR agonists, which stimulate apoE and ABCA1 expression, increases adipocyte lipid accumulation (15). We have shown that adipocyte apoE is required for optimal TG accumulation in adipocytes (6). All of these observations suggest that the LXR-apoE pathway may have a unique role in adipocytes, and may function to link adipocyte TG and sterol metabolism during adipocyte TGD growth.
We have previously demonstrated reciprocal regulation (4) of adipocyte apoE expression by the pro-inflammatory cytokine tumor necrosis factor-α and TZDs. The NFκB transcriptional complex binding at the apoE gene proximal promoter mediates suppression of adipocyte apoE gene transcription by tumor necrosis factor-α. Because TZDs reduce macrophage infiltration and inflammation in adipose tissue (25, 26), the systemic administration of PPARγ agonists could stimulate adipocyte apoE gene expression by two distinct pathways: one involving relief of inflammation-mediated suppression of gene transcription at the proximal promoter and the other working via the LXRE in the downstream enhancer. Adipose tissue is the primary target of the TZD drugs that are used for improving insulin sensitivity in human diabetes. One proposed mechanism for this systemic improvement in insulin sensitivity involves the increased partitioning of TG-rich lipoprotein lipid to adipose tissue (and away from liver and muscle) that is facilitated by PPARγ-mediated increases in the expression of adipocyte lipoprotein lipase, fatty acid transport protein, and aP2 among others (1, 27, 28). We have previously shown that adipocyte apoE is required for adipocyte TG accumulation from TG-rich lipoproteins during PPARγ treatment (6). In this report, we demonstrate that silencing LXR completely eliminates both apoE response and TG accumulation in response to PPARγ agonists.
The linkage between PPARγ and LXR for regulating adipocyte apoE expression and TG accumulation suggests a new pathway by which LXR could influence adipocyte metabolism of TG-rich lipoproteins, and thereby influence systemic insulin sensitivity. An expanding role for LXR in lipid and carbohydrate metabolism is consistent with recent observations demonstrating its role in suppressing hepatocyte sterol synthesis and stimulating adipocyte GLUT4 expression (29, 30). It is not clear that the linkage of the PPARγ and LXR pathways for regulating apoE expression that we have demonstrated in adipocytes can be extended to other cell types. Macrophages, which express high levels of apoE and have functional LXR and PPARγ regulatory pathways, do not respond to PPARγ agonists with increased apoE expression (4). The difference between the cell types could be related to generation of an endogenous LXR ligand after PPARγ stimulation.
In summary, we have identified LXR as a key pathway mediating PPARγ stimulation of the adipocyte apoE gene. LXR is known to mediate sterol responsiveness of the apoE gene (13), and apoE plays a role in adipocyte TG balance (6). The current observations therefore suggest that the PPARγ-LXR-apoE cascade could be an important molecular link mediating cross-talk between adipocyte cholesterol and TG homeostasis.
Acknowledgments
We thank Stephanie Thompson for assistance with manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant DK71711 (to T. M.).
Footnotes
The abbreviations used are: PPAR, peroxisome proliferator-activated receptor; TGD, triglyceride droplet; TZD, thiazolidinedione; TG, triglyceride; LXR, liver receptor X; LXRE, LXR enhancer; 22-OHch, 22-hydroxycholesterol; EKO, apoE knockout; WT, wild type; RT, reverse transcription; siRNA, small interference RNA; ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ME, macrophage enhancer.
References
- 1.Tontonoz, P., and Spiegelman, B. M. (2008) Annu. Rev. Biochem. 77 289–312 [DOI] [PubMed] [Google Scholar]
- 2.Scherer, P. E. (2006) Diabetes 55 1537–1545 [DOI] [PubMed] [Google Scholar]
- 3.Fantuzzi, G., and Mazzone, T. (2007) Arterioscler. Thromb. Vasc. Biol. 27 996–1003 [DOI] [PubMed] [Google Scholar]
- 4.Yue, L., Rassouli, N., Ranganathan, G., Kern, P. A., and Mazzone, T. (2004) J. Biol. Chem. 279 47626–47632 [DOI] [PubMed] [Google Scholar]
- 5.Zechner, R., Moser, R., Newman, T. C., Fried, S. K., and Breslow, J. L. (1991) J. Biol. Chem. 266 10583–10588 [PubMed] [Google Scholar]
- 6.Huang, Z. H., Reardon, C. A., and Mazzone, T. (2006) Diabetes 55 3394–3402 [DOI] [PubMed] [Google Scholar]
- 7.Yue, L., Christman, J. W., and Mazzone, T. (2008) Endocrinology 149 4051–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espiritu, D. J., and Mazzone, T. (2008) Diabetes 57 2992–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, Z. H., Luque, R. M., Kineman, R. D., and Mazzone, T. (2007) Am. J. Physiol. 293 E203–E209 [DOI] [PubMed] [Google Scholar]
- 10.Rao, P., Huang, Z. H., and Mazzone, T. (2007) J. Clin. Endocrinol. Metab. 92 4366–4372 [DOI] [PubMed] [Google Scholar]
- 11.Mazzone, T., and Reardon, C. (1994) J. Lipid Res. 35 1345–1353 [PubMed] [Google Scholar]
- 12.Lin, C.-Y., Duan, H., and Mazzone, T. (1999) J. Lipid Res. 40 1618–1626 [PubMed] [Google Scholar]
- 13.Laffitte, B. A., Repa, J. J., Joseph, S. B., Wilpitz, D. C., Kas, H. R., Mangeldorf, D. J., and Tontonoz, P. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chawla, A., Boisvert, W. A., Lee, C. H., Laffitte, B. A., Barak, Y., Joseph, S. B., Liao, D., Nagy, L., Edwards, P. A., Curtiss, L. K., Evans, R. M., and Tontonoz, P. (2001) Mol. Cell 7 161–171 [DOI] [PubMed] [Google Scholar]
- 15.Juvet, L. K., Andresen, S. M., Schuster, G. U., Dalen, K. T., Tobin, K. A., Hollung, K., Haugen, F., Jacinto, S., Ulven, S. M., Bamberg, K., Gustafsson, J.-A., and Nebb, H. I. (2008) Mol. Endocrinol. 17 172–182 [DOI] [PubMed] [Google Scholar]
- 16.Kalaany, N. Y., Gauthier, K. C., Zavacki, A. M., Mammen, P. P., Kitazume, T., Peterson, J. A., Horton, J. D., Garry, D. J., Bianco, A. C., and Mangelsdorf, D. J. (2005) Cell Metab. 1 231–244 [DOI] [PubMed] [Google Scholar]
- 17.Dugail, I., Le Lay, S., Varret, M., Le Liepvre, X, Dagher, G., and Ferre, P. (2003) Horm. Metab. Res. 35 204–210 [DOI] [PubMed] [Google Scholar]
- 18.Le Lay, S., Krief, S., Farnier, C., Lefrere, I., Liepvre, X., Bazin, R., Ferre, P., and Dugail, E. (2001) J. Biol. Chem. 276 16904–16910 [DOI] [PubMed] [Google Scholar]
- 19.Le, L. S., Robichon, C., Le, L., X, Dagher, G., Ferre, P., and Dugail, I. (2003) J. Lipid Res. 44 1499–1507 [DOI] [PubMed] [Google Scholar]
- 20.Parpal, S., Karlsson, M., Thorn, H., and Stralfors, P. (2001) J. Biol. Chem. 276 9670–9678 [DOI] [PubMed] [Google Scholar]
- 21.Pilch, P. F., Souto, R. P., Liu, L., Jedrychowski, M. P., Berg, E. A., Costello, C. E., and Gygi, S. P. (2007) J. Lipid Res. 48 2103–2111 [DOI] [PubMed] [Google Scholar]
- 22.Mazzone, T., Gump, H., Diller, P., and Getz, G. S. (1987) J. Biol. Chem. 262 11657–11662 [PubMed] [Google Scholar]
- 23.Kovanen, P. T., Nikkila, E. A., and Miettinen, T. A. (1975) J. Lipid Res. 16 211–223 [PubMed] [Google Scholar]
- 24.Vassiliou, G., and McPherson, R. (2004) Arterioscler. Thromb. Vasc. Biol. 24 1669–1675 [DOI] [PubMed] [Google Scholar]
- 25.Bodles, A. M., Varma, V., Yao-Borengasser, A., Phanavanh, B., Peterson, C. A., McGehee, R. E., Jr., Rasouli, N., Wabitsch, M., and Kern, P. A. (2006) J. Lipid Res. 47 2080–2088 [DOI] [PubMed] [Google Scholar]
- 26.Di Gregorio, G. B., Yao-Borengasser, A., Rasouli, N., Varma, V., Lu, T., Miles, L. M., Ranganathan, G., Peterson, C. A., McGehee, R. E., and Kern, P. A. (2005) Diabetes 54 2305–2313 [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. Y., van de Wall E., Laplante, M., Azzara, A., Trujillo, M. E., Hofmann, S. M., Schraw, T., Durand, J. L., Li, H., Li, G., Jelicks, L. A., Mehler, M. F., Hui, D. Y., Deshaies, Y., Shulman, G. I., Schwartz, G. J., and Scherer, P. E. (2007) J. Clin. Invest. 117 2621–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasouli, N., Raue, U., Miles, L. M., Lu, T., Di Gregorio, G. B., Elbein, S. C., and Kern, P. A. (2005) Am. J. Physiol. 288 E930–E934 [DOI] [PubMed] [Google Scholar]
- 29.Laffitte, B. A., Chao, L. C., Li, J., Walczak, R., Hummasti, S., Joseph, S. B., Castrillo, A., Wilpitz, D. C., Mangelsdorf, D. J., Collins, J. L., Saez, E., and Tontonoz, P. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5419–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Y., Rogers, P. M., Su, C., Varga, G., Stayrook, K. R., and Burris, T. P. (2008) J. Biol. Chem. 283 26332–26339 [DOI] [PMC free article] [PubMed] [Google Scholar]