Abstract
Amplification of the complement cascade through the alternative pathway can lead to excessive inflammation. Targeting C3b, a component central to the alternative pathway of complement, provides a powerful approach to inhibit complement-mediated immune responses and tissue injury. In the present study, phage display technology was employed to generate an antibody that selectively recognizes C3b but not the non-activated molecule C3. The crystal structure of C3b in complex with a Fab fragment of this antibody (S77) illustrates the structural basis for this selectivity. Cleavage of C3 to C3b results in a plethora of structural changes within C3, including the rearrangement of macroglobulin domain 6 enabling binding of S77 to the adjacent macroglobulin domain 7 domain. S77 blocks binding of factor B to C3b inhibiting the first step in the formation of the alternative pathway C3 convertase. In addition, S77 inhibits C5 binding to C3b. This results in significantly reduced formations of anaphylatoxins and membrane-attack complexes. This study for the first time demonstrates the structural basis for complement inhibition by a C3b-selective antibody and provides insights into the molecular mechanisms of alternative pathway complement activation.
Next to its importance in immune surveillance, the alternative pathway (AP)3 of complement activation is of central importance in immune responses (1) where it can account for up to 80% of total complement activation (2). A first step in the AP of complement activation is the cleavage of C3 into C3a and C3b. C3b then binds pro-enzyme factor B (fB) and properdin (P) to form the C3 convertase responsible for subsequent cleavage of the substrates C3 and C5 (3). In addition to acting as the noncatalytic subunit of the protease fB, C3b serves as the binding partner for several complement regulators and receptors, including complement factor H (fH) and complement receptor 1 (CR1).
Our understanding of complement activation at the structural levels has been greatly advanced with solving the structures of the central complement component C3, the first cleavage product C3b and second product C3c (4–6). The cleavage of C3 to C3b, the first step in activation of the alternative complement pathway, is accompanied by large conformational changes. These changes result in the exposure of new surfaces required for convertase assembly and regulation (7). Most importantly, C3b serves as the binding partner for fB that, upon cleavage by factor D (fD), forms the catalytic subunit of the AP convertase.
Because levels and turnover of C3 in serum are high (8) reagents that target systemic C3 require high dosing to establish therapeutically relevant concentrations in the circulation. In the present study, phage-display technology was employed to generate S77, an antibody that selectively binds C3 activation products but not native C3, targeting only a small portion (0.5%) of total C3 proteins present in serum. To further define the structural basis for this selectivity, we solved the crystal structure of S77 in complex with C3b at 3.1-Å resolution. Because epitopes exposed after the first C3 cleavage step also serve as binding sites for complement receptors and modulators, we further determined if S77 inhibits binding of complement regulators fH and CR1 to C3b and whether S77 interferes with the formation of the complement convertase. Together, we demonstrate the utility of phage display technology to generate a C3b-specific antibody and further elucidate the molecular basis by which this antibody interferes with AP complement activation.
EXPERIMENTAL PROCEDURES
All complement proteins were obtained from CompTech except C3 and its fragments. C3 was generated as described elsewhere (9). C3b was derived from C3 by cleavage with a C3 convertase and purified as described elsewhere (10). Briefly, C3 was incubated with CVF, fB, and fD (CompTech) in a 10:10:1 molar ratio at 37 °C for 1 h in the presence of 10 mm MgCl2. The C3b fragments were subsequently isolated by a strong anion exchanger Mono Q 5/50 and Superdex S-200 10/300 GL gel filtration column (Amersham Biosciences) for a purity of >95% by Coomassie Blue-stained gel. iC3b and C3c were generated by incubating C3b, fI, and fH in 1:10:10 molar ratios at 37 °C for 1 h. The reaction was diluted with a 0.05× volume of 20 mm Tris, pH 8.0, containing 5 mm EDTA and loaded onto a Mono Q (Amersham Biosciences) column equilibrated in the same buffer. Protein was eluted with a 0.0 to 0.5 m NaCl gradient. The Identity of C3c and iC3b fragments was confirmed by Edman degradation. Generation of iC3 (hydrolyzed C3) and recombinant soluble CR1 (sCR1) consisting of long homologous repeats A and C is described in the supplemental materials. The generation of CRIg-Fc has been described elsewhere (10). S77 was expressed in Escherichia coli as described (10). Anti-neuropilin Fab (11) was used as a control antibody.
Selection of Phage Antibodies That Specifically Bind to C3b—The human synthetic phage antibody libraries displayed bivalent Fab fragments on M13 phage, and the diversity was generated by use of oligonucleotide-directed mutagenesis in selected positions of three heavy chain CDRs (12). Nunc 96-well Maxi-Sorp immunoplates (Nunc) were coated overnight at 4 °C with C3b (10 μg/ml) and blocked for 1 h with PT buffer (phosphate-buffered saline, 0.05% Tween 20) supplemented with 1% bovine serum albumin. The antibody phage libraries were added and incubated overnight. The plates were washed with PT buffer, and bound phage were eluted with 50 mm HCl and 500 mm NaCl for 30 min and neutralized with an equal volume of 1 m Tris base. Recovered phages were amplified in E. coli XL-1 blue cells. During subsequent selection rounds, the amplified antibody phage were preincubated with 500 nm C3 before being added to antigen-coated plates to counter-select against phage antibodies that could also bind human C3. The binding time of antibody-antigen was reduced to 2–3 h, and the stringency of plate washing was gradually increased (11). The phage antibodies that specifically bound to C3b but not C3 were reformatted to full-length IgGs by cloning VL and VH regions of individual clones into LPG3 and LPG4 vectors, respectively (13). Antibodies were transiently expressed in mammalian cells and purified with protein A columns. Methods for affinity maturation of the candidate antibody, measuring binding specificity by an enzyme-linked immunosorbent assay (ELISA) and affinity determination of the antibody by surface plasmon resonance are described in the supplemental materials.
Crystallization, Data Collection, Structure Solution, and Refinement—Crystals of the C3b-S77 complex were obtained within a week at 19 °C from 4-μl hanging drops, consisting of a 1:1 ratio of protein solution (10 mg/ml protein in 50 mm NaCl, 25 mm Tris, pH 7.8) to mother liquor (10% polyethylene glycol 4000, 0.2 m MgCl2, 100 mm Na-HEPES, pH 7.0), suspended over mother liquor. Crystals were harvested, briefly placed in a solution containing 20% glycerol, 10% polyethylene glycol 4000, 0.2 m MgCl2, and 0.1 m Na-HEPES, pH 7.0, and then flash frozen in liquid N2. Diffraction data were collected at Advanced Light Source beamline 5.0.2 and processed using HKL2000 (14). Crystals of the C3b-S77 complex diffracted to 3.1-Å resolution, belonged to space group C2 with cell parameters of a = 216.4 Å, b = 180.4 Å, c = 154.6 Å, and β = 115.73°. Crystals contained two complexes each composed of one C3b molecule bound to one S77 fragment in the asymmetric unit. The structure was solved by molecular replacement with the coordinates of C3b from the C3b-CRIg complex (pdb code 2ICE) using the program Phaser. Refinement using Refmac (15) and manual adjustments with program O (16) resulted in a model with an Rcryst of 21.7% and Rfree of 28.3% (supplemental Table S1). With the exception of the CUB and the C345C domains, all domains were well defined in the electron density.
Methods for detecting fB, fH, and CR1 binding to C3b, decay acceleration assays, fH and CR1 co-factor activity for fI-mediated proteolytic cleavage of C3b, convertase assays, and the C5 binding assay are described in the supplemental materials.
C5 Convertase Assay—C5 convertase was assembled on zymosan particles (CompTech) as described previously (17) with modification described in the supplemental materials.
RESULTS
Generation of a Phage Antibody Selective for Cleavage Products of C3—To identify antibodies that distinguish C3 and C3b, a protocol was designed that employed subtractive phage panning to counter-select phage antibodies that bind to C3. Phage that bound C3b were identified, the Fab sequences were grafted onto the human IgG1 framework of herceptin (18), and the sequences were expressed in mammalian cells. Antibody YW144.2.45 IgG was selected out of a panel of 15 clones based on its binding affinity (KD = 160 nm measured by surface plasmon resonance; results not shown) and its selectivity for C3b but not to C3.
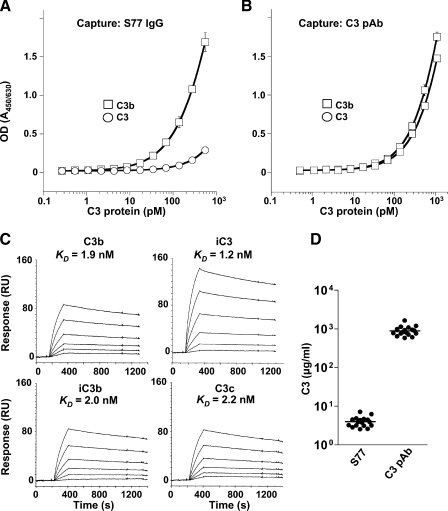
To further improve its affinity, YW144.2.45 was affinity-matured using three different complementarity determining region (CDR) combinatorial libraries. Selected amino acids in the CDRs 1, 2, and 3 of both the light chain and the heavy chain (see supplemental Fig. S1) were randomized, and a high stringency selection process was applied to improve the on- and off-rate of the antibody. After six rounds of sorting, phage antibodies that bound specifically to C3b and that displayed higher affinity compared with parental clones were reformatted to full-length IgGs. Out of eight antibodies, YW144.2.45.S77 (hereafter referred to as S77 IgG), had the highest affinity as measured by surface plasmon resonance (results not shown) and was selected for further analysis. S77 IgG contained seven amino acid changes (supplemental Fig. S1) in the CDRs of the heavy chain and two changes in CDR L3 of the light chain and had a 10-fold higher affinity when compared with the parent antibody. Furthermore, S77 IgG bound C3b but not C3 as shown by ELISA (Fig. 1, A and B). Residual binding to C3 at higher concentrations of S77 IgG most likely resulted from partial hydrolysis of C3 when exposed to water. Surface plasmon resonance studies using S77 (the Fab fragment of the antibody) further confirmed binding of S77 to C3b, hydrolyzed C3 (iC3), iC3b, and C3c with affinities ranging from 1.2 to 2.2 nm (Fig. 1C). S77 did not bind to C3dg (results not shown). In line with the selective recognition of C3b, iC3b, and C3c but not C3 by S77, the antibody detected only a small fraction (0.5%) of the total C3 proteins present in human serum (Fig. 1D).
FIGURE 1.
Generation of a phage antibody that selectively binds to C3b, but not native C3. Binding of S77 IgG (A) or a polyclonal anti-C3 antibody (C3 pAb, B) to C3b or C3 protein. Binding is expressed as the ratio of optical densities of the reaction mixtures measured at 450 and 630 nm. C, Biacore analysis of binding affinity of S77 to C3b, iC3, iC3b, and C3c. S77 was captured on a CM5 sensor chip. Increasing concentrations of S77 (3.1, 6.25, 12.5, 25, 50, and 100 nm) were injected for 180 s. The KD is calculated from binding curves showing response at equilibrium plotted against the concentration. D, the plasma concentration of C3 protein recognized by S77 IgG is significantly lower than the plasma concentration of total C3 proteins. Data in A and B represent average ± S.D. of triplicate measurements.
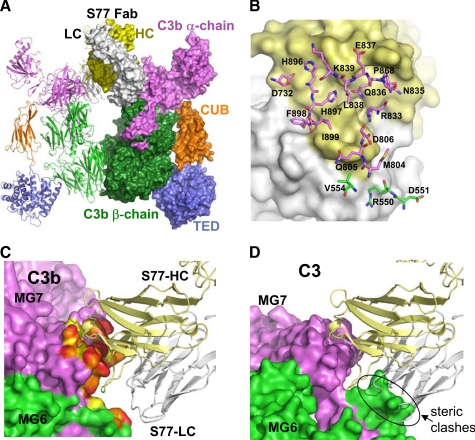
Crystal Structure of S77 in Complex with C3b Indicates Why S77 Recognizes C3b and Not C3—To determine the structural basis for S77's selective recognition of C3b and not C3, the structure of S77 in complex with C3b was solved at 3.1-Å resolution. Crystals contained two complexes, each composed of one C3b molecule and one Fab fragment, in the asymmetric unit (Fig. 2A). Both complexes superimposed well onto each other with minor conformational changes in the C1r/C1s, Uegf, and bone morphogenetic protein-1 (CUB), thioester bond-containing domain (TED), and the C-terminal C345C domain. Similarly, C3b in complex with S77 also superimposed well onto the C3b-CRIg complex (pdb 2ICF) with 574 Cα positions of the β-chain superimposing with <0.8 Å r.m.s.d. and 824 Cα positions of the α-chain superimposing with <3 Å r.m.s.d. This indicates that C3b, with the exception of the C345C domain, and to a lesser extent the CUB and TED domains are very rigid. The interface between C3b and the Fab buries a total of ∼1930 Å2 of solvent-accessible surface. The Fab contacts residues from the α′NT (∼50 Å2), the MG6 domain (∼140 Å2), and the MG7 domain (∼830 Å2) of C3b (Fig. 2). The bulk of these interactions is mediated by the heavy chain CDR-H1 (∼210Å2), CDR-H2 (∼180 Å2), and CDR-H3 (360 Å2) with additional contributions from the light-chain CDRs (∼150 Å2). In addition to a number of hydrophilic interactions, a few hydrophobic interactions exist; most notably Phe-898 that protrudes from the surface of C3b binding into a hydrophobic pocket formed by residues from CDR-H3 and CDR-L2 of the Fab (Fig. 2B). Comparison of C3b-Fab complex with structures of C3b in its unbound form (pdb code: 2I07) or bound to CRIg (pdb code: 2ICF) reveals that minimal conformational changes are required in C3b to allow binding to the Fab, with the largest difference occurring at the position of the Phe-898 side chain. S77 recognizes an epitope that is present on the primary sequence level in C3, the un-activated molecule, as well as in C3b or C3c, yet it selectively binds C3b and C3c. Comparison of C3 and C3b structures reveal the underlying basis for this selectivity: whereas the overall domain structure of both the MG6 and the MG7 domains is very similar in C3b and C3 (r.m.s.d. of 0.5 Å for 97 Cα positions in MG6 and r.m.s.d. 0.65 for 101 Cα positions in MG6), the orientation of these domains with respect to each other is quite different in these molecules. The rotation of MG6 with respect to MG7 of ∼15° (4) occurring during C3 activation, allows the Fab to recognize an epitope formed by both domains. Superimposition of the MG7 domain of the C3b-Fab complex onto the MG7 domain of C3 shows that numerous steric clashes of the Fab and the MG6 domain within C3 prevent the Fab from binding to C3 (Fig. 2, C and D). In this respect binding of S77 to C3b or C3c is analogous to binding of CRIg, which also recognizes a binding epitope that is formed only after conformational changes occur during C3 activation (i.e. conversion to either iC3 or C3b).
FIGURE 2.
Crystal structure of S77 in complex with C3b. A, structure of the C3b-S77 dimer in the asymmetric unit. One C3b-S77 complex is shown as a ribbon diagram; the other is in surface representation. The β-chains of C3b are depicted in green, the α-chains shown in violet, light blue (TED domain), and orange (CUB domain). S77 is shown with the light chains colored white, and the heavy chains colored yellow. B, the Fab surface is shown with the light chain atoms in white and heavy chain in yellow. All amino acids that have at least one atom closer than 4.5 Å to the S77 are shown in stick representation and are labeled with carbon atoms of residues from the β-chain and α-chain colored green and violet, respectively. C, the S77 binding site of C3b. Atoms of C3b that are closer than 4.5, 4, and 3.5 Å are colored yellow, orange, and red, respectively. D, the surface of C3 is shown after superimposing the MG7 domain of C3 onto the MG7 domain of the C3b-S77 complex. Although the MG7 domains of C3 and C3b superimpose very well (r.m.s.d. 0.5 Å for 97 common Cα positions), the different relative orientation of MG7 in respect to MG6 leads to steric clashes between the light chain of the S77 and the MG6 domain in a potential complex, thus preventing S77 binding to C3.
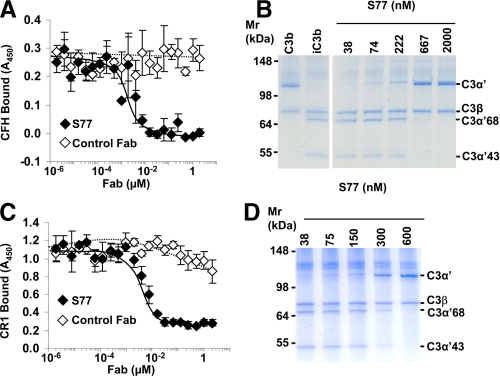
S77 Inhibits fH and CR1 Binding to C3b and Factor I-mediated Cofactor Activity—After cleavage, C3 undergoes large conformational changes that result in the exposure of new surfaces that form binding sites for various complement receptors and regulators. Binding sites for fH and CR1 (19) are located within residues 727–768 of the α′NT and MG6 domain of C3b, a region that also harbors binding sites for fB (20, 21). S77 inhibited fH binding to C3b by ELISA (Fig. 3A) and fH cofactor activity for factor I (fI)-mediated cleavage of C3b to iC3b in a fluid-phase cofactor assay (Fig. 3B). S77 also inhibited CR1 binding to C3b (Fig. 3C) and sCR1-mediated fI cofactor activity (Fig. 3D). Because S77 binding sites on C3b do not overlap with the binding sites of CR1 and fH on C3b (supplemental Fig. S2, A and B), the Fab fragment likely prevents access of fH, CR1, and factor B to these residues via steric clashes. Thus, S77 binds to a site on C3b that is important for fH and CR1 functional activity.
FIGURE 3.
S77 inhibits fH and sCR1 binding to C3 and inhibits fH and sCR1 co-factor activity. A, microtiter plates were coated with C3b, and fH was added in the presence of increasing concentrations of S77 or control Fab. Binding of fH to C3b was determined using an anti-fH antibody and a secondary HRPO-conjugated antibody. Absorbance of the reaction mixture was measured at 450 nm. B, cofactor activity for fI-mediated cleavage of C3b was measured by incubating C3b and fI with fH or increasing concentrations of S77 or control Fab. The mixture was incubated at 37 °C, and the samples were analyzed by gel-electrophoresis and Simply Blue staining. C, S77 inhibits CR1 binding to C3b. Microtiter plates coated with C3b were incubated with sCR1 and increasing concentrations of S77 or control Fab. Binding of sCR1 to C3b was detected with an anti-CR1 primary antibody and an HRPO-conjugated secondary antibody. D, sCR1 cofactor activity was determined as described under B except that fH was replaced by sCR1. Data in A and C are expressed as mean ± S.D. of four repeats.
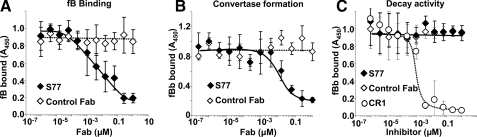
Formation, but Not the Stability, of the Alternative Pathway C3 Convertase Is Inhibited by S77—Because C3b is a central subunit of both the C3 and C5 convertases of the alternative pathway we further determined whether S77 binding to C3b could inhibit the activity of both convertases. The first step in the generation of C3 convertase is binding of factor B to iC3 or C3b, after which exposure of a scissile bond allows factor D-mediated cleavage of factor B to form the proteolytic active serine protease C3bBb (22). S77 inhibited binding of factor B to C3b (Fig. 4A) and consequently prevented the formation of C3 convertase (Fig. 4B). Unlike CR1, S77 did not decay the C3 convertase (Fig. 4C) nor did S77 interfere with P binding (supplemental Fig. S3). Thus, S77 inhibits the formation, but not the stability, of the alternative pathway convertase.
FIGURE 4.
S77 affects the formation, but not stability, of the alternative pathway C3 convertase. A, S77 inhibits binding of factor B to C3b. C3b was coated on microtiter plates. S77 or control Fab was added, followed by addition of factor B. Binding of factor B to C3b was determined with anti-factor B antibody and secondary antibodies conjugated to HRPO. B, S77 Fab inhibits formation of C3 convertase. C3b was coated on microtiter plates. S77 or control Fab was added, followed by addition of fB and factor D. Binding of fBb to C3b was determined with anti-fB antibody and secondary antibody conjugated to HRPO. C, S77 does not decay the C3 convertase. Microtiter plates were coated with C3b, incubated with factor B and factor D followed by the addition of S77, sCR1 or control Fab. Factor Bb was detected with goat anti-human factor B and donkey anti-goat antibody conjugated to HRPO. Absorbance of the reaction product was read at 450 nm. Data are expressed as mean ± S.D. of four repeats.
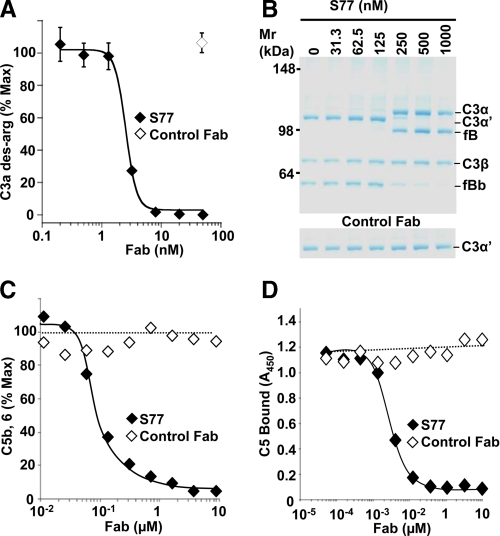
Blockade of the Alternative Pathway C3 and C5 Convertases by S77—Next, the effect of S77 on activity of a fluid phase C3 convertase was determined. Generation of C3a des-Arg (Fig. 5A) and C3b (Fig. 5B) was inhibited by S77, confirming that S77 prevents formation of the active C3-convertase. Because S77 also recognizes iC3, S77 may additionally inhibit factor B binding to iC3, preventing formation of the initial fluid-phase C3 convertase of the AP (23).
FIGURE 5.
S77 inhibits the alternative pathway C3 and C5 convertase. A, C3 and factors B and D were incubated in the presence of increasing concentrations of S77 or control Fab. The concentration of C3a des-Arg reaction product was determined by ELISA. Data are expressed as mean ± S.D. of triplicate measurements. B, in a separate experiment, the effect of S77 or control Fab on C3 convertase activity was determined by gel electrophoresis. Experiments were repeated three times with similar results. C, S77 inhibits C5 convertase activity. C5 convertase was assembled on the surface of zymosan particles. A fixed concentration of C5 and convertase was mixed with increasing concentrations of S77 or control Fab. Convertase activity was determined using a chicken erythrocyte hemolytic assay and expressed as percentage of hemolysis in the absence of inhibitor. D, C3b was coated on microtiter plates and incubated with a mixture of C5 and increasing concentrations of S77 or control Fab. C5 bound to plate-coated C3b was detected with a polyclonal antibody against C5 and a HRPO-conjugated secondary antibody.
At the next level of complement activation, C3b dimers constitute the noncatalytic subunit of the C5 convertase with two potential binding sites for S77. To determine if S77 could block C5 convertase in addition to C3 convertase activity, a C5 convertase was generated by assembling C3b subunits on the surface of zymosan particles. The presence of S77 prevented cleavage of the substrate C5 into C5a and C5b. Together, these results indicate that S77 inhibited the alternative pathway at the level of both the C3 and C5 convertases (Fig. 5C). To be cleaved by the complement convertases, C5 needs to bind to the C3b subunit of the C5 convertase. S77 inhibited binding of C5 to C3b multimers in an ELISA-based assay (Fig. 5D). Thus, in addition to interfering with the binding of fB to C3b, S77 inhibited C5 binding to C3b, a step required for C5 proteolytic activation and formation of C5a and the membrane attack complex.
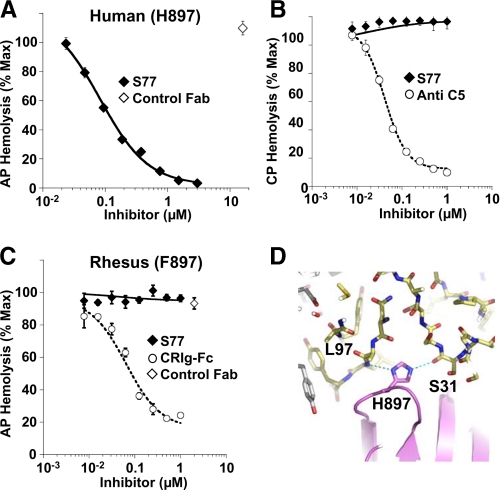
S77 Inhibits the Alternative, but Not Classical Pathway of Complement Activation in Human Serum—Both, the AP and classical pathway (CP) C5 convertases contain at least one C3b molecule that could potentially serve as a binding partner for S77. Therefore, we next determined if S77 could block terminal C5 cleavage and membrane attack complex formation mediated by the AP or CP convertases. S77 inhibited hemolytic activity in C1q-depleted serum (Fig. 6A) but did not inhibit the CP of complement in fB-depleted human serum (Fig. 6B) indicating that S77 is a selective inhibitor of the AP convertases.
FIGURE 6.
S77 inhibits the alternative, but not classical, pathway C5 convertase in human serum. A, S77 inhibits AP-mediated hemolysis. Rabbit erythrocytes were incubated with C1q-depleted human serum in the presence of increasing concentrations of S77 or control Fab. Hemolysis was determined by absorbance of the supernatant at 412 nm. Convertase activation was expressed as percentage of hemolysis in the absence of inhibitor. B, S77 does not inhibit CP convertase activation. IgM-coated sheep erythrocytes were incubated with factor B-depleted human serum and increasing concentrations of S77 or anti-C5 antibody. C, S77 lacks complement inhibitory activity in rhesus serum where His-897 is substituted by Phe-897. CRIg-Fc was used as a positive control. D, His-897 of C3b is within hydrogen bonding distance to the main-chain atoms of the heavy chain CDR1 and CDR3 of S77. The exchange of His-897 against phenylalanine, the respective amino acid in the C3 sequence of rhesus, leads to the loss of these hydrophilic interactions. Data in A–C are expressed as mean ± S.D. of triplicate measurements.
While testing species cross-reactivity of the Fab, we observed the absence of blocking potential in serum from baboon, cynomologus, and rhesus monkey (results not shown). To further elucidate the basis for lack of species cross-reactivity, we sequenced the stretch of rhesus C3b that encompasses the S77 binding site. Importantly, His-897 in human C3b is replaced by the more hydrophobic Phe in C3b from rhesus (supplemental Fig. S2A). This specificity of the Fab toward human C3b has a clear structural rationale. His-897, located in the center of the interface between C3b and the Fab, is ideally positioned to form two hydrogen bonds to the main-chain carbonyl of Ser-31 of CDR-H1 and the main-chain nitrogen of Leu-97 in CDR-H3 (Fig. 6D). The exchange of His-897 against Phe introduces a much more hydrophobic residue into the hydrophilic environment in this part of the interface abolishing the ability of the Fab to recognize rhesus C3b and hence to inhibit alternative pathway complement activation in rhesus serum (Fig. 6C). Thus, we provide the structural basis for the selectivity of S77 for human and not rhesus C3b and provide evidence that complement inhibition by S77 requires binding of the Fab to the C3b molecule.
DISCUSSION
Complement activation has been associated with an increasing number of disease indications, including ischemia-reperfusion injury, membranoproliferative glomerulonephritis, cardiopulmonary bypass, asthma, autoimmune disease and age-related macular degeneration (1, 8, 24). Various inhibitors have been generated that target complement activation (see for review Ref. 2). These consist of natural regulators of complement activation (25), monoclonal antibodies that block enzymatic activity of the serine proteases factor D and factor B (26, 27), or a cyclic peptide that blocks enzyme-substrate interaction (28). Systemic dosing of therapeutics that target complement components in the circulation may result in unfavorable pharmacokinetics. Antibodies that recognize C3b directly and thus target the active C3- and C5-convertase therefore have an advantage over antibodies that target both the pro-molecule and the active variant. Phage display offers the advantage of altering the binding conditions such that selection against specific epitopes can be achieved using fluid-phase competitors. Using this approach, an antibody was generated that selectively blocked the alternative pathway of complement by inhibiting binding of fB to the C3b subunit of C3 convertase, an essential step in the activation of the AP. In addition, S77 inhibited C5 binding to the C5 convertase. This mechanism of action confers the ability of S77 to inhibit both the formation and the activity of already formed convertase, explaining its potent inhibition of AP complement activation in human serum.
S77 interfered with both fluid-phase and solid-phase AP convertase activity. Similar to CRIg, S77 did not interfere with CP complement activation, which is surprising given that C3b is part of the heterodimeric C3bC4b subunit of the CP C5 convertase. A possible explanation for this is that residual binding of C5 to the C4b subunit of the CP convertase, which is not recognized by S77, precludes inhibition of binding activity. Similar to S77, a previously described monoclonal antibody that binds C3b and iC3b was shown to selectively inhibit the AP of complement (29). Selectivity for a single pathway is of much importance from a safety point of view, because it permits residual complement activation through one of the other pathways to mount an appropriate host response to pathogens.
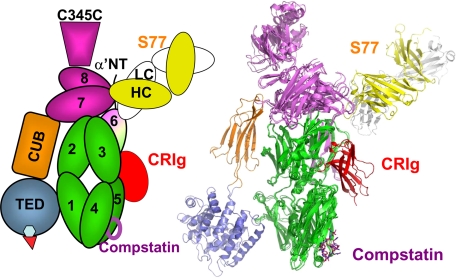
Comparing the crystal structure of C3b and C3 shows that cleavage of the anaphylatoxin domain results in large conformational change in TED/CUB domains and resulting exposure of hydrophobic surface amenable to interaction (Janssen, et al. (4) and Wiesmann, et al. (6)). The reorientation of the MG6 and other domains in C3b creates binding sites that account for the selective interaction of a variety of molecules to C3b. S77 binds selectively to the MG6 and MG7 domains in the domain arrangement present in C3b and C3c and sterically blocks the interaction of C3b with complement fH, CR1, fB, and C5. Although both CRIg and S77 share binding affinity for C3b, iC3b, and C3c, they recognize different sites of C3b. CRIg binds primarily to the residues of the C3b β-chain (6), whereas S77 binds predominantly to residues from the α-chain (Fig. 7). Interestingly, the binding sites for CRIg as well as for S77 are located on the same face of C3b: on the opposite site of the CUB and TED domain, the latter mediating the opsonization of particles by C3b. In addition, compstatin also binds to this face (28) (Fig. 7). As all three molecules, soluble CRIg, S77, and compstatin, are able to prevent binding of substrate to the complement convertases, it appears likely that the molecular surface of the face of C3b, opposite of the TED domain, contains the binding surface for the substrates C3 and C5.
FIGURE 7.
Structural basis for complement inhibition by S77, CRIg, and compstatin. The superposition of the C3b-S77 complex, the C3c-CRIg complex, and the C3c-compstatin complex shows that all three inhibitors recognize the same “face” of the complement protein; their binding sites are located on the opposite site of the TED position in C3b. C3b-C3c and S77 are colored as described in Fig. 2, CRIg is shown in red, and compstatin is depicted as purple sticks.
Mapping approaches employing blocking antibodies and synthetic peptides have implicated the 727–767 segment at the N terminus of C3b α′-chain as contributing to the interactions with fB, fH, and CR1 (19–21). One monoclonal anti-C3c antibody, clone C3-9, shares some properties with S77. C3-9 binds to a neoepitope present in C3b (30) and inhibits fH, CR1, and fB binding to EC3b cells (20). Moreover, negative-stain electron microscopy combined with image classification identified binding of a Fab fragment of this antibody to a region between the C345C knob and the MG1–6 ring, identified as the MG7/MG8 region of C3b (31). Because there is no high resolution structure of this antibody in complex with C3b, we cannot confirm if the binding sites of C3-9 overlap with those of S77.
S77 does not recognize native C3 but recognizes C3 proteins formed following C3 hydrolysis or cleavage by C3 convertase, which account for 0.5% of total C3 proteins present in serum (Fig. 1D). This selectivity provides therapeutic advantage over targeting C3, which is highly abundant in human serum. Moreover, S77 selectively inhibits the alternative, but not classic, pathway of complement activation, avoiding complete shutdown of the complement-mediated host response. The antibody is fully humanized, avoiding unwanted neutralizing immune responses by the host. Thus, although further studies are required to determine the therapeutic potential of the S77, this study provides a unique approach for generating a selective complement inhibitor and further provides structural insight into the function of the complement convertases central to various chronic inflammatory diseases.
Supplementary Material
Acknowledgments
We thank the staff at the Advanced Light Source for help with data collection.
The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Dept. of Energy under contract DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, Figs. S1–S3, and Table S1.
The atomic coordinates and structure factors (code 3G6J) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: AP, alternative pathway; fB, factor B; P, properdin; fH, factor H; fD, factor D; CR1, complement receptor 1; ELISA, enzyme-linked immunosorbent assay; CDR, complementarity determining region; CUB, C1r/C1s, Uegf, and Bone morphogenetic protein-1; TED, thioester bond-containing domain; CP, classical pathway; r.m.s.d., root mean square deviation.
References
- 1.Thurman, J. M., and Holers, V. M. (2006) J. Immunol. 176 1305–1310 [DOI] [PubMed] [Google Scholar]
- 2.Ricklin, D., and Lambris, J. D. (2007) Nat. Biotechnol. 25 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller-Eberhard, H. J., and Gotze, O. (1972) J. Exp. Med. 135 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen, B. J., Christodoulidou, A., McCarthy, A., Lambris, J. D., and Gros, P. (2006) Nature 444 213–216 [DOI] [PubMed] [Google Scholar]
- 5.Janssen, B. J., Huizinga, E. G., Raaijmakers, H. C., Roos, A., Daha, M. R., Nilsson-Ekdahl, K., Nilsson, B., and Gros, P. (2005) Nature 437 505–511 [DOI] [PubMed] [Google Scholar]
- 6.Wiesmann, C., Katschke, K. J., Yin, J., Helmy, K. Y., Steffek, M., Fairbrother, W. J., McCallum, S. A., Embuscado, L., DeForge, L., Hass, P. E., and van Lookeren Campagne, M. (2006) Nature 444 217–220 [DOI] [PubMed] [Google Scholar]
- 7.Gros, P., Milder, F. J., and Janssen, B. J. (2008) Nat. Rev. Immunol. 8 48–58 [DOI] [PubMed] [Google Scholar]
- 8.Walport, M. J. (2001) N. Engl. J. Med. 344 1058–1066 [DOI] [PubMed] [Google Scholar]
- 9.Hammer, C. H., Wirtz, G. H., Renfer, L., Gresham, H. D., and Tack, B. F. (1981) J. Biol. Chem. 256 3995–4006 [PubMed] [Google Scholar]
- 10.Helmy, K. Y., Katschke, K. J., Jr., Gorgani, N. N., Kljavin, N. M., Elliott, J. M., Diehl, L., Scales, S. J., Ghilardi, N., and van Lookeren Campagne, M. (2006) Cell 124 915–927 [DOI] [PubMed] [Google Scholar]
- 11.Liang, W. C., Dennis, M. S., Stawicki, S., Chanthery, Y., Pan, Q., Chen, Y., Eigenbrot, C., Yin, J., Koch, A. W., Wu, X., Ferrara, N., Bagri, A., Tessier-Lavigne, M., Watts, R. J., and Wu, Y. (2007) J. Mol. Biol. 366 815–829 [DOI] [PubMed] [Google Scholar]
- 12.Lee, C. V., Liang, W. C., Dennis, M. S., Eigenbrot, C., Sidhu, S. S., and Fuh, G. (2004) J. Mol. Biol. 340 1073–1093 [DOI] [PubMed] [Google Scholar]
- 13.Carter, P., Presta, L., Gorman, C. M., Ridgway, J. B., Henner, D., Wong, W. L., Rowland, A. M., Kotts, C., Carver, M. E., and Shepard, H. M. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 4285–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski, Z. and Minor, W. (1997) Methods Enzymol. 276 307–326 [DOI] [PubMed] [Google Scholar]
- 15.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 16.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard. (1991) Acta Crystallogr. Sect. A 47 110–119 [DOI] [PubMed] [Google Scholar]
- 17.Rawal, N., and Pangburn, M. K. (1998) J. Biol. Chem. 273 16828–16835 [DOI] [PubMed] [Google Scholar]
- 18.Wu, Y., Eigenbrot, C., Liang, W. C., Stawicki, S., Shia, S., Fan, B., Ganesan, R., Lipari, M. T., and Kirchhofer, D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 19784–19789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oran, A. E., and Isenman, D. E. (1999) J. Biol. Chem. 274 5120–5130 [DOI] [PubMed] [Google Scholar]
- 20.Becherer, J. D., Alsenz, J., Esparza, I., Hack, C. E., and Lambris, J. D. (1992) Biochemistry 31 1787–1794 [DOI] [PubMed] [Google Scholar]
- 21.Lambris, J. D., Lao, Z., Oglesby, T. J., Atkinson, J. P., Hack, C. E., and Becherer, J. D. (1996) J. Immunol. 156 4821–4832 [PubMed] [Google Scholar]
- 22.Milder, F. J., Gomes, L., Schouten, A., Janssen, B. J., Huizinga, E. G., Romijn, R. A., Hemrika, W., Roos, A., Daha, M. R., and Gros, P. (2007) Nat. Struct. Mol. Biol. 14 224–228 [DOI] [PubMed] [Google Scholar]
- 23.Pangburn, M. K., Schreiber, R. D., and Muller-Eberhard, H. J. (1981) J. Exp. Med. 154 856–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickering, M. C., Cook, H. T., Warren, J., Bygrave, A. E., Moss, J., Walport, M. J., and Botto, M. (2002) Nat. Genet. 31 424–428 [DOI] [PubMed] [Google Scholar]
- 25.Weisman, H. F., Bartow, T., Leppo, M. K., Marsh, H. C., Jr., Carson, G. R., Concino, M. F., Boyle, M. P., Roux, K. H., Weisfeldt, M. L., and Fearon, D. T. (1990) Science 249 146–151 [DOI] [PubMed] [Google Scholar]
- 26.Leinhase, I., Rozanski, M., Harhausen, D., Thurman, J. M., Schmidt, O. I., Hossini, A. M., Taha, M. E., Rittirsch, D., Ward, P. A., Holers, V. M., Ertel, W., and Stahel, P. F. (2007) J. Neuroinflam. 4 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Undar, A., Eichstaedt, H. C., Clubb, F. J., Jr., Fung, M., Lu, M., Bigley, J. E., Vaughn, W. K., and Fraser, C. D., Jr. (2002) Ann. Thorac. Surg. 74 355–362; discussion 362 [DOI] [PubMed] [Google Scholar]
- 28.Janssen, B. J., Halff, E. F., Lambris, J. D., and Gros, P. (2007) J. Biol. Chem. 282 29241–29247 [DOI] [PubMed] [Google Scholar]
- 29.DiLillo, D. J., Pawluczkowycz, A. W., Peng, W., Kennedy, A. D., Beum, P. V., Lindorfer, M. A., and Taylor, R. P. (2006) Mol. Immunol. 43 1010–1019 [DOI] [PubMed] [Google Scholar]
- 30.Hack, C. E., Paardekooper, J., Smeenk, R. J., Abbink, J., Eerenberg, A. J., and Nuijens, J. H. (1988) J. Immunol. 141 1602–1609 [PubMed] [Google Scholar]
- 31.Nishida, N., Walz, T., and Springer, T. A. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 19737–19742 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.