Abstract
Cells are equipped with an efficient quality control system to selectively eliminate abnormally folded and damaged proteins. Initially the cell tries to refold the unfolded proteins with the help of molecular chaperones, and failure to refold leads to their degradation by the ubiquitin proteasome system. But how this proteolytic machinery recognizes the abnormally folded proteins is poorly understood. Here, we report that E6-AP, a HECT domain family ubiquitin ligase implicated in Angelman syndrome, interacts with the substrate binding domain of Hsp70/Hsc70 chaperones and promotes the degradation of chaperone bound substrates. The expression of E6-AP was dramatically induced under a variety of stresses, and overexpression of E6-AP was found to protect against endoplasmic reticulum stress-induced cell death. The inhibition of proteasome function not only increases the expression of E6-AP but also causes its redistribution around microtubule-organizing center, a subcellular structure for the degradation of the cytoplasmic misfolded proteins. E6-AP is also recruited to aggresomes containing the cystic fibrosis transmembrane conductance regulator or expanded polyglutamine proteins. Finally, we demonstrate that E6-AP ubiquitinates misfolded luciferase that is bound by Hsp70. Our results suggest that E6-AP functions as a cellular quality control ubiquitin ligase and, therefore, can be implicated not only in the pathogenesis of Angelman syndrome but also in the biology of neurodegenerative disorders involving protein aggregation.
In the living cell both existing and newly synthesized proteins are at constant risk of misfolding and aggregation. However, cells have a surveillance system that maintains a delicate balance between protecting misfolded proteins with the help of molecular chaperones and promoting rapid and efficient clearance of the misfolded and damaged proteins by ubiquitin proteasome system (UPS)3 (1–4). Any alteration of this homeostatic balance affects normal cellular function and cell viability. Environmental factors such as increased temperature or exposure of various chemical agents can lead to rapid build up of unfolded and damaged proteins inside the cells. Abnormally folded proteins also can be produced in cells resulting from various genetic mutations (5–9). The failure of clearance of misfolded and damaged proteins by UPS can result in the formation of potentially toxic aggregates. Once the aggregation process begins, it further disrupts the function of UPS by overloading its capacity (10).
Degradation of a protein by UPS involves two distinct and successive steps; they are (a) covalent attachment of multiple molecules of ubiquitin to the target protein and (b) degradation of the targeted protein by 26 S proteasome (11). Ubiquitination is a multistep process consisting of activating (E1), conjugating (E2), and ligating (E3) enzymes. The E3 ubiquitin ligase plays a critical role in the substrate selectivity and exists with large diversity (12). However, the molecular mechanisms through which UPS selectively recognizes misfolded proteins are poorly understood. Earlier studies have demonstrated that some chaperones, like Hsp70 or Hsp40, play an important role in the degradation of misfolded proteins in addition to their refolding property (13, 14). But the precise role of chaperones in the clearance of misfolded protein is unclear. Recent identification of E3 ligases that interacts with molecular chaperones raise a strong possibility that the chaperones could ubiquitinate misfolded proteins by directly recruiting E3 ligases (15, 16).
The C terminus of Hsp70-interacting protein (CHIP) is one such E3 ligase. Binding of CHIP to Hsp70 can inhibit the folding of Hsp70-bound substrates and concomitantly facilitates their multiubiquitination (17–19). Initially, CHIP has been demonstrated to regulate the triage decisions of immature cystic fibrosis transmembrane conductance regulator (CFTR) and glucocorticoid receptor in cooperation with Hsp70/Hsp90 (19, 20). Now several lines of evidence suggest that CHIP works as a general cellular quality control E3 ubiquitin ligase (15, 21). CHIP also plays an important role in stress protection and is increasingly implicated in the biology of neurodegenerative disorders involving protein misfolding (22–25).
Cells have an excellent quality control system to deal with the abnormally folded protein. Therefore, it is quite possible that several E3 ligases are involved in the ubiquitination and degradation of the misfolded proteins. E6-AP is a HECT (homologous to E6-AP C terminus) domain family of E3 ubiquitin ligase (26, 27). In addition to its ubiquitination activity, E6-AP also functions as a transcriptional coactivator for steroid hormone receptors (28). E6-AP shows that brain-specific imprinting and loss of function of maternally inherited E6-AP causes Angelman mental retardation syndrome (29–32). Here we demonstrate that E6-AP is involved in the degradation of misfolded protein with the help of molecular chaperones Hsp70/Hsc70 and, hence, can be considered as a quality control E3 ubiquitin ligase.
EXPERIMENTAL PROCEDURES
Materials—MG132, 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT), TRIzol reagent, rabbit polyclonal anti-ubiquitin, anti-luciferase, anti-E6-AP, mouse monoclonal anti-β-tubulin and anti-luciferase, and all cell culture reagents were obtained from Sigma. Lipofectamine® 2000, Opti-MEM, ponasterone A, mouse monoclonal V5 antibody, and the reverse transcription-PCR kit were purchased from Invitrogen. Protein G-agarose beads was purchased from Roche Applied Science. Rabbit polyclonal anti-E6-AP, anti-GAPDH, mouse monoclonal anti-Hsp70, anti-parkin, anti-Myc, and goat polyclonal anti-Hsc70 were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-20 S proteasome was from Calbiochem. Dual luciferase reporter gene assay kit was purchased from Promega. Goat anti-mouse IgG-fluorescein isothiocyanate and IgG-rhodamine, alkaline phosphatase-conjugated anti-mouse, and anti-rabbit IgG were purchased from Vector Laboratories.
Expression Plasmids and Stable Cell Lines—The construction of the full-length and HECT domain-deleted form of E6-AP in pcDNA vector has been described earlier (33). Both constructs were expressed as a fusion of V5 and His tags. The source of plasmids CHIP and parkin were described elsewhere (33, 34). The stable cell lines, Huntington disease (HD) 16Q and HD 150Q, in ecdysone-inducible systems have been described previously (35). The green fluorescence protein (GFP)-CFTR plasmid was a kind gift from Dr. Ron Kopito, Stanford University, and Renilla luciferase expression constructs (PRL-SV40) were obtained from Promega.
Cell Culture, Transfection, Cell Viability Assay, and Purification of Recombinant Proteins—The wild type mouse neuro 2a and Cos-7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics penicillin/streptomycin. The stable cell lines (HD 16Q and HD 150Q) were maintained in the same medium containing 0.4 mg/ml zeocin and 0.4 mg/ml G418. One day before transfection, cells were plated into 6-well tissue cultured plates at a subconfluent density. Cells were transiently transfected with expression vectors using Lipofectamine® 2000 reagent according to the manufacturer's instruction. The transfection for Cos-7 and neuro 2a cells was about 70–80% and 60–70%, respectively. After 24 or 48 h of transfection, cells were used for immunofluorescence staining, coimmunoprecipitation, and immunoblotting. For cell viability assay, neuro 2a cells were transfected with different expression plasmids; 48 h later cells were exposed to variety of stress agents for different time periods. Cell viability was measured by MTT assay as described previously (35). For the purification of full-length and HECT domain-deleted E6-AP, CHIP, and parkin, the Cos-7 cells were transiently transfected with the respective plasmids for 48 h. The collected cells were then subjected to purification of His6-tagged recombinant proteins using nickel-nitrilotriacetic acid purification system according to the manufacturer's instruction.
Coimmunoprecipitation and Immunoblotting Experiment—Cos-7 cells were transiently transfected with full-length and HECT domain-deleted E6-AP plasmids for different time periods. The cells were washed with cold phosphate-buffered saline, collected, pelleted by centrifugation, and lysed on ice for 30 min with Nonidet P-40 lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, Complete protease inhibitor mixture). Cell lysates were briefly sonicated and centrifuged for 10 min at 15,000 × g at 4 °C, and the supernatants (total soluble extract) were used for immunoprecipitation as described earlier (34). For each immunoprecipitation experiment, ∼200 μg of protein in 0.2-ml Nonidet P-40 lysis buffer was incubated with 2.5 μg of primary antibody. The total cell lysate or the immunoprecipitated proteins were separated through SDS-polyacrylamide gel electrophoresis and processed for immunoblotting as described elsewhere (34). All primary antibodies were used in 1:1000 dilutions for immunoblotting except V5, which was used in 1:5000 dilutions.
Immunofluorescence Technique—Cos-7 cells grown in chamber slides were transiently transfected with the appropriate plasmids. In some experiments HD 16Q and HD 150Q cells grown in chamber slides were induced with ponasterone A for 48 h. The cells were then washed twice with phosphate-buffered saline, fixed with 4% paraformaldehyde in phosphate-buffered saline for 20 min, permeabilized with 0.5% Triton X-100 in phosphate-buffered saline for 5 min, washed extensively, and then blocked with 5% nonfat dried milk in Tris-buffered saline-Tween for 1 h. Primary antibody (anti-E6-AP and anti-γ-tubulin, 1:250 dilution, and anti-Hsp70 and anti-20 S proteasome, 1:500 dilution) incubation was carried out overnight at 4 °C. After several washings with Tris-buffered saline-Tween, cells were incubated with rhodamine-conjugated secondary antibody (1:500 dilutions) for 1 h, washed several times, and mounted. Samples were observed using a fluorescence microscope, and digital images were assembled using Adobe Photoshop.
In Vitro Ubiquitination Assay—The in vitro ubiquitylation assay of heat-denatured luciferase was performed as described earlier (21). Briefly, 0.3 μm purified firefly luciferase was incubated with 4 μm Hsp70 (in 50 mm Tris buffer, pH 7.5, containing 4 mm ATP and 2 mm MgCl2) at 43 °C for 10 min and then quickly chilled under ice. One microliter of this reaction mixture was incubated in a reaction volume of 50 μl of 50 mm Tris-HCl, pH 7.5, containing 50 ng of E1, 500 ng of E2 (UBCH7), 2 μg of either E6-AP or HECT-deleted E6-AP, 6 μg of bovine ubiquitin, 1 mm dithiothreitol, 2 mm MgCl2, and 4 mm ATP. The incubation was carried out at 30 °C for 2 h. The reaction was terminated by the addition of SDS sample buffer, boiled, and separated through 6% or 10% SDS-PAGE. Blots were probed with either ubiquitin or luciferase antibody.
Cell Culture Luciferase Assay—Cos-7 cells were transiently transfected with 1 μg of Renilla luciferase expression plasmid along with 2 μg of empty pcDNA3.1 and the full-length and HECT-deleted form of E6-AP plasmids for 24 h. Cells were either maintained in a normal CO2 incubator or heat-stressed at 43 °C for 30 min and then returned to the normal CO2 incubator for 1 h. In some experiments cells were treated with MG132 (10 μm) just before heat stress. Cells were then collected and assayed for luciferase activity according to the manufacturer's protocol.
Statistical Analysis—Statistical differences between groups were determined by one-way analysis of variance. The Student Newman-Keul's test was conducted to compare individual means where analysis of variance indicated statistical differences. The level of significance for all analysis was set at p < 0.05.
RESULTS
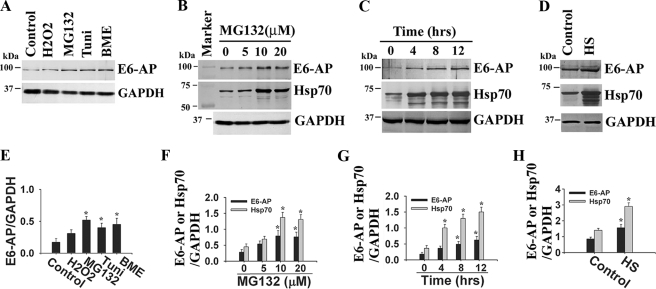
Expression of E6-AP Is Induced under a Variety of Stresses and Protects Stress-induced Cell Death—During our study on the role of E6-AP in degradation of polyglutamine protein, we surprisingly noticed that the expression of E6-AP was significantly increased in cells expressing expanded polyglutamine protein compared with normal repeats (36). The expanded polyglutamine proteins form intracellular aggregates and are known to generate endoplasmic reticula (ER) and oxidative stress. Therefore, we thought that the expression of E6-AP might be regulated by a variety of stresses. To test this hypothesis, we treated mouse neuro 2a cells with oxidative (H2O2), ER (β-mercaptoethanol and tunicamycin), and proteasomal (MG132) stress agents and then checked mRNA and protein levels of E6-AP. We found that the mRNA (supplemental Fig. 1) and protein levels (Fig. 1A) of E6-AP were significantly increased in response to ER and proteasomal stress. The oxidative and heat stress also increased the mRNA and protein levels of E6-AP but comparatively lower ER and proteasomal stress (Fig. 1, A and D). E6-AP protein levels increased dose- and time-dependently in response to proteasome inhibitor, MG132 (Fig. 1, B and C). The induction patterns of E6-AP were very similar to that of Hsp70 in response to proteasomal inhibition or ER stress. However, the basal levels of E6-AP in the neuro 2a cells were comparatively much lower than Hsp70. When we analyzed the 5′-untranslated region of E6-AP gene, we surprisingly found several consensus heat shock factor 1 (HSF1) binding sequences (supplemental Fig. 1I). There are 6 consensus HSF1 binding sequences that have 100% homology with Drosophila and yeast HSF1.
FIGURE 1.
Expression of E6-AP is increased under various stress conditions. A, neuro 2a cells were treated with H2O2 (0.25 mm for 4 h), β-mercaptoethanol (5 mm for 4 h), tunicamycin (5 μg/ml for 6 h), and MG132 (5 μm for 6 h). The cell lysates were then prepared and processed for immunoblot analysis using E6-AP and GAPDH antibodies. B, dose-dependent effects of MG132 on E6-AP and Hsp70 protein levels. MG132 was exposed for 6 h. C, time-dependent effect of MG132 (10 μm) on E6-AP and Hsp70 protein levels. D, effect of heat stress (HS) on E6-AP levels. Cells were exposed to 45 °C for 30 min and then returned to the normal incubator for 2 h. E, F, G, and H, quantitation of the E6-AP band intensities as shown in A, B, C, and D, respectively, collected from three independent experiments using NIH image analysis software. Data were expressed as ratio of E6-AP or Hsp70 to GAPDH. Values are the means ± S.D. of three independent experiments. *, p < 0.05 as compared with respective control. Tuni, tunicamycin; BME, β-mercaptoethanol.
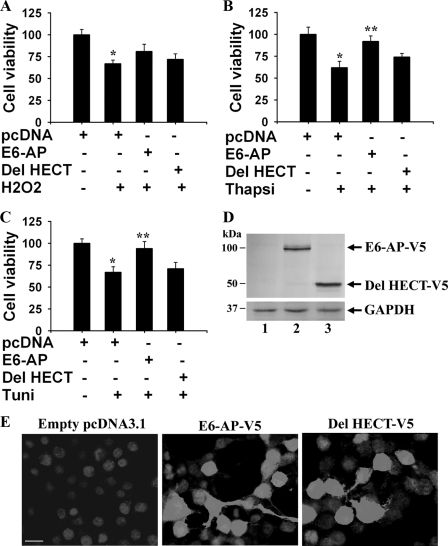
The increased expression of E6-AP in response to a variety of stresses could be a survival response of the cell from the deleterious effects of stress. To confirm this possibility, we transiently transfected full-length and HECT domain-deleted forms of E6-AP into the neuro 2a cells for 48 h and then exposed to a variety of stresses for different time periods. The exposure of cells with oxidative (H2O2) and ER stress (thapsigargin and tunicamycin) agents caused reduction in cell viability (Fig. 2). The overexpression of E6-AP significantly protected the ER stress-induced cell death (Fig. 2). E6-AP also protected oxidative stress-induced cell death to some extent but was not statistically significant. The deletion of HECT domain of E6-AP partially reduced the protective effect, which could be because of the loss of ligase function or decreased stability of E6-AP. Fig. 2, D and E, shows the expression of the full-length and HECT-deleted form of E6-AP after 2 days of transfection.
FIGURE 2.
E6-AP protects various stress-induced cell death. neuro 2a cells were transiently transfected with either empty pcDNA vector or full-length and HECT-deleted (Del) forms of E6-AP (0.5 μg each/well of 24-well tissue culture plate). After 48 h, cells were treated with 0.5 mm H2O2 for 6 h (A), 5 μg/ml thapsigargin for 6 h (B), and 10 μg/ml tunicamycin (C) for 10 h. Cell viability was measured by MTT assay. Values are the means ± S.D. of three independent experiments, each performed in triplicate. *, p < 0.05 as compared with empty pcDNA-transfected experiment. **, p < 0.05 as compared with the tunicamycin- or thapsigargin-treated experiment. D and E, expression of the full-length and HECT-deleted form of E6-AP after 48 h of transfection. V5 antibody was used to detect the overexpressed protein both in immunoblotting (D) and immunofluorescence staining (E). Transfection efficiency was about 60–70%. Lanes 1, 2, and 3 represent empty pcDNA, E6-AP, and HECT domain-deleted construct-transfected samples, respectively. Thapsi, thapsigargin; Tuni, tunicamycin. Scale bar in E, 30 μm.
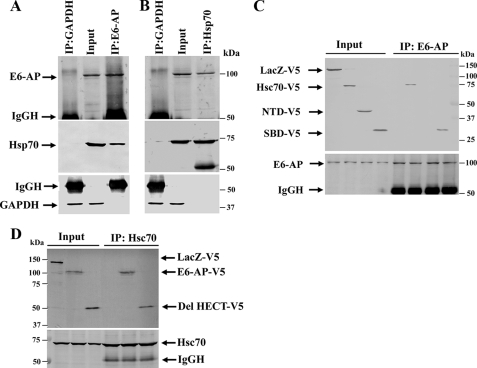
E6-AP Interacts with Hsp70/Hsc70—Because the expression of E6-AP is induced along with Hsp70 in response to a variety of stresses, we presumed that E6-AP might work in cooperation with Hsp70 to protect cell death. Therefore, we studied the interaction of E6-AP with Hsp70. We used Cos-7 cells for this purpose because these cells express very high level of endogenous Hsp70. Cos-7 cell lysates were coimmunoprecipitated with E6-AP and GAPDH antibody, and the blots were probed with E6-AP, Hsp70, and GAPDH antibodies. Fig. 3A showed that the Hsp70 was indeed specifically pulled down by E6-AP antibody. Similarly, E6-AP was also specifically coimmunoprecipitated with Hsp70 antibody (Fig. 3B). The Hsp70/Hsc70 contains N-terminal ATPase domain and C-proximal substrate binding domains. Therefore, we analyzed the domain of Hsc70 that interacts with E6-AP. The full-length and various domains of Hsc70 were transiently transfected into the Cos-7 cells for 24 h, and cell lysates were made and processed for coimmunoprecipitation using E6-AP antibody. Blots were detected with V5 and E6-AP antibodies. As shown in Fig. 3C, the full-length and substrate binding domain of Hsc70 were coimmunoprecipitated along with E6-AP. Our finding suggests that E6-AP interacts with the substrate binding domain of Hsc70. Next, we tried to identify the domain of E6-AP that interacts with the substrate binding domain of Hsc70. The Cos-7 cells were transfected with full-length and HECT-deleted constructs of E6-AP, the cell lysates were coimmunoprecipitated with Hsc70 antibody, and the blots were detected with V5 and Hsc70 antibodies. Fig. 3D showed that full-length and HECT-deleted forms of E6-AP were coimmunoprecipitated by Hsc70. Our results suggest that N-terminal domain of E6-AP interacts with the substrate binding domain of Hsp70/Hsc70.
FIGURE 3.
Interaction of E6-AP with Hsp70. A, Cos-7 cell lysates were coimmunoprecipitated (IP) with E6-AP and GAPDH antibody, and blots were detected with E6-AP, Hsp70, and GAPDH antibodies. B, cell lysates were coimmunoprecipitated with Hsp70 and GAPDH antibody, and blots were probed with Hsp70, E6-AP, and GAPDH antibodies. C, Cos-7 cells were transiently transfected with full-length, ATPase, and substrate binding domain of Hsc70. Twenty-four hours later cells were collected and subjected to coimmunoprecipitation using E6-AP antibody. Blots were probed with V5 and E6-AP antibodies. NTD, N-terminal domain; SBD, substrate binding domain. D, cells were transfected with full-length and HECT domain-deleted constructs of E6-AP for 24 h. Cell lysates were coimmunoprecipitated with Hsc70 antibody, and blots were detected with V5 and Hsc70 antibodies.
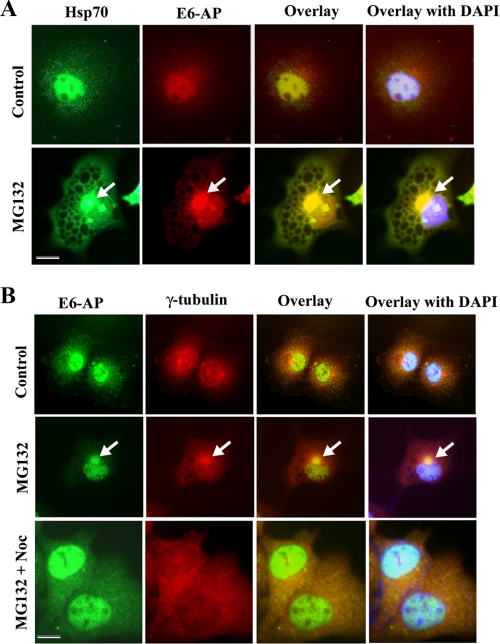
E6-AP Redistributes to Microtubule-organizing Center (MTOC) in Response to Proteasomal Inhibition—Because E6-AP interacts with Hsp70, we further studied the localization of both E6-AP and Hsp70 in Cos-7 cells before and after various stresses. In normal cells E6-AP was localized both in cytoplasmic and nuclear compartment and partially co-localized with Hsp70 (Fig. 4A). Treatment with proteasome inhibitor caused accumulation of E6-AP into a distinct perinuclear region as well as specific nuclear structures (Fig. 4A). A similar pattern of relocalization was also observed in the case of Hsp70. Exposure of heat stress did not show accumulation of Hsp70 or E6-AP into the perinuclear region. However, the E6-AP and Hsp70 levels were increased upon heat stress and were highly concentrated in the nucleus (data not shown). Treatment of ER stress agent, tunicamycin, also did not alter the overall localization pattern of E6-AP (data not shown). To further pinpoint the perinuclear region, we performed an immunofluorescence co-localization study of E6-AP with γ-tubulin. The γ-tubulin is the marker for MTOC, where the majority of cytoplasmic misfolded proteins are degraded. We observed that E6-AP was redistributed around the γ-tubulin-positive MTOC in response to proteasome inhibition (Fig. 4B). The treatment of nocodazole prevented the accumulation of E6-AP around MTOC, suggesting further the involvement of microtubule in this recruitment processes (Fig. 4B).
FIGURE 4.
E6-AP redistributes to aggresomes in response to proteasome inhibition. A, Cos-7 cells were platted onto 2-well chamber slides, and on the following day cells were treated with proteasome inhibitor MG132 (10 μm) for 8 h. The cells were then subjected to double immunofluorescence staining using Hsp70 and E6-AP antibody. Fluorescein isothiocyanate-conjugated secondary antibody was used to label Hsp70, and rhodamine-conjugated secondary antibody was used to stain E6-AP. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The arrow indicates redistribution of E6-AP and Hsp70 around MTOC. B, Cos-7 cells were platted and treated with MG132 in the similar manner as described in A. In some experiments cells were treated with nocodazole (10 μm) along with MG132. Cells were then processed for double immunofluorescence staining with E6-AP and γ-tubulin. Fluorescein isothiocyanate-conjugated secondary antibody was used to label E6-AP and rhodamine-conjugated secondary antibody was used to stain γ-tubulin. Nuclei were stained with 4′,6-diamidino-2-phenylindole. The arrow indicates the localization of E6-AP to aggresome. Scale bar, 20 μm.
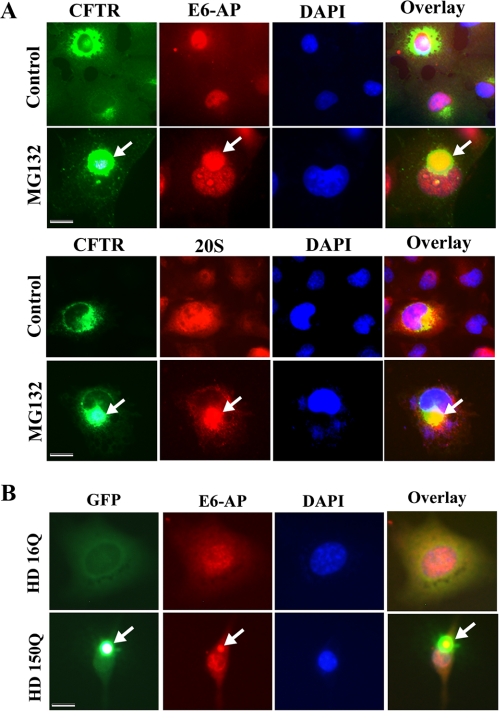
E6-AP Recruits to Aggresomes Formed by CFTR and Expanded Polyglutamine Proteins—Now it is clear that inhibition of cellular proteasome function results in accumulation of E6-AP around MTOC similar to Hsp70. The re-distribution of E6-AP to MTOC suggests that E6-AP might be involved in the degradation of cytoplasmic misfolded proteins. To confirm that, we used a model protein, called CFTR, that is very well known to form aggresomes in response to proteasome inhibition (37). The Cos-7 cells were transiently transfected with GFP-CFTR plasmid, treated with MG132, and then subjected to the immunofluorescence staining of E6-AP. The treatment of MG132 resulted in the formation of distinct perinuclear GFP-CFTR aggresomes that were co-localized with γ-tubulin and Hsp70 (supplemental Fig. 2). E6-AP was also clearly recruited to these aggresomes (Fig. 5A). These GFP-CFTR aggresomes also co-localized with 20 S proteasome components (Fig. 5A). Next, we checked the recruitment of E6-AP to the expanded polyglutamine protein aggregates. The expanded polyglutamine proteins are also known to form aggresomes. The HD 16Q and HD 150Q cells lines were induced with ponasterone A for 48 h and then subjected to immunofluorescence staining of E6-AP. The induction of ponasterone A to HD 16Q cells caused expression of normal repeat-containing tNhtt-16Q-GFP proteins, which was localized into the cytoplasm (Fig. 5B). HD 150Q cells expressed expanded polyglutamine protein, which often formed perinuclear aggregates. The immunofluorescence staining clearly showed the co-localization of E6-AP with the polyglutamine aggregates (Fig. 5B). Recently we showed that E6-AP induced ubiquitination of expanded polyglutamine protein and promotes their degradation by proteasome (36).
FIGURE 5.
E6-AP recruits to aggresomes formed by CFTR and expanded polyglutamine proteins. A, Cos-7 cells were platted onto 2-well chamber slides and transfected with GFP-CFTR plasmid. 36 h post-transfection cells were treated with MG132 (10 μm) for 8 h. The cells were then subjected to immunofluorescence staining using E6-AP and 20 S proteasome antibodies. Rhodamine-conjugated secondary antibody was used to stain E6-AP and 20 S proteasome. The arrow indicates the localization of E6-AP and 20 S proteasome to GFP-CFTR aggresome. B, the HD 16Q and HD 150Q cells were plated into 2-well chamber slide. Cell were induced with 1 μm ponasterone A for 48 h and processed for immunofluorescence staining using E6-AP antibody. Rhodamine-conjugated secondary antibody was used to stain the E6-AP. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The arrow indicates the recruitment of E6-AP to mutant huntingtin aggregates. Scale bar, 20 μm.
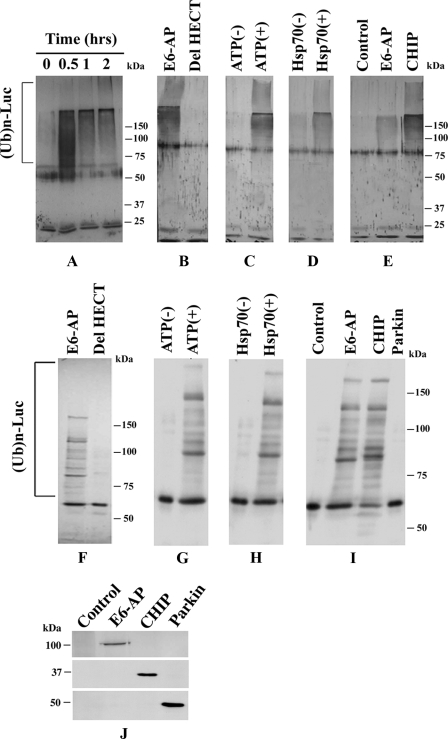
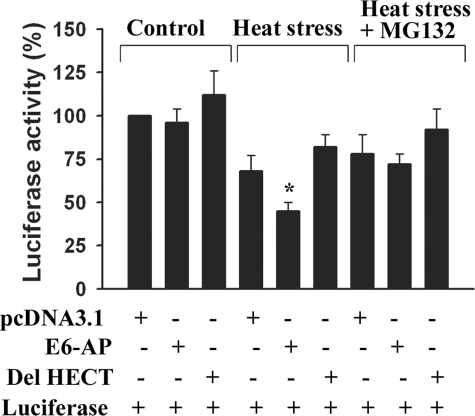
E6-AP Induces Ubiquitination and Degradation of Misfolded Luciferases That Are Bound with Hsp70—The recruitment of E6-AP to aggresomes in response to proteasome impairment indicates that E6-AP is possibly involved in the degradation of misfolded proteins with the help of Hsp70/Hsc70 chaperones. To directly demonstrate the role of E6-AP in the ubiquitination of chaperone bound substrates, we used a fully reconstituted in vitro ubiquitination assay system where thermally denatured luciferase was used as substrate. E6-AP was incubated at 30 °C for 2 h with E1, E2 (UBCH7), ubiquitin, ATP, and firefly luciferase that has been pretreated with Hsp70 at 43 °C for 10 min. As shown in Fig. 6A, E6-AP caused a time-dependent increase in the ubiquitination of the heat denatured luciferase that were captured by Hsp70. The deletion of HECT domain of E6-AP completely blocked the ubiquitination of thermally denatured luciferase (Fig. 6, B and F). The ubiquitination of the denatured luciferase also required ATP and Hsp70 (Fig. 6, C, D, G, and H). Next we compared the effect of various E3 ligases on the ubiquitination of denatured luciferase. CHIP was found to be more efficient than E6-AP in the ubiquitination of misfolded luciferase (Fig. 6, E and I). Parkin had no effect on the ubiquitination of denatured luciferase. We have also studied the degradation of misfolded luciferase in E6-AP-overexpressed cells. The Cos-7 cells were transfected with Renilla luciferase expression construct along with either full-length or HECT domain-deleted plasmids. Twenty-four hours later cells were exposed to heat stress at 43 °C for 30 min and then returned to the incubator for 1 h. In some experiments cells were treated with 10 μm MG132 just before heat stress. Cells were then collected and processed for luciferase activity assay. As shown in the Fig. 7, overexpression of E6-AP led to the rapid degradation of heat-denatured luciferase, which can be prevented upon the addition of MG132. Deletion of HECT domain of E6-AP not only blocked the degradation of denatured luciferase but also probably increased its refolding. The partial protection of cell death by the HECT-deleted form of E6-AP against various stresses demonstrated in Fig. 2 also could be because of the increased refolding of various cellular proteins. How the HECT domain-deleted form of E6-AP increases the refolding of denatured luciferase is not clear at present. The HECT-deleted form of E6-AP could potentially modulate their chaperone activity of Hsp70/Hsc70 because it is capable of interacting with Hsp70/Hsc70 (demonstrated in Fig. 2).
FIGURE 6.
E6-AP ubiquitinates heat-denatured luciferase bound by Hsp70. The firefly luciferase was thermally denatured at 43 °C for 10 min along with Hsp70 and then quickly chilled under ice. The ubiquitination assay of luciferase was then carried out in the presence of E6-AP along with E1 and E2 (UBCH7) at 43 °C for different time periods as described under “Experimental Procedures.” The effect of HECT domain of E6-AP (B and F), ATP (C and G), Hsp70 (D and H), and various ubiquitin ligases (E and I) on ubiquitination of denatured luciferase was carried out at 43 °C for 2 h. A–E, blots were detected with ubiquitin antibody. F–I, blots were detected with luciferase antibody. J, immunoblot analysis of the input of E6-AP, CHIP, and parkin that were used in the ubiquitination reaction in either E or I. E6-AP, CHIP, and parkin were detected by V5, Myc, and parkin antibodies, respectively.
FIGURE 7.
E6-AP promotes degradation of misfolded luciferase. Cos-7 cells were transfected with Renilla luciferase expression construct in the absence or presence of full-length and HECT domain-deleted forms of E6-AP as described under “Experimental Procedures.” Twenty-four hours later cells were exposed to heat stress for 30 min at 43 °C and then returned to the incubator for 1 h. In some experiments cells were treated with 10 μm MG132 just before heat stress. Cells were then collected and processed for luciferase activity assay. Values were normalized as per μg of protein and expressed as percent control. Values are the means ± S.D. of three independent experiments. *, p < 0.05 as compared with heat-stressed empty pcDNA-transfected group.
DISCUSSION
Although we know that cells have a very efficient quality control system, the molecular mechanism through which proteolytic machinery UPS recognizes abnormal proteins is not well understood. Emerging evidence now suggests that some molecular chaperones directly communicate with UPS by recruiting E3 ubiquitin ligases. Considering the efficiency of the cellular quality control system, it is expected that there might be several such E3 ligases, which can be classified as quality control E3 ligases. Here we have demonstrated the identification of one such quality control E3 ligase known as E6-AP. E6-AP is a HECT domain family E3 ligase, and its loss of function causes Angelman mental retardation syndrome (26–31).
First we have shown that the expression of E6-AP is dramatically induced under various stress conditions. The induction pattern of E6-AP looks very similar to Hsp70 in a variety of stressed cells. Exposure of stress would result in a massive buildup of misfolded and damaged proteins, and the cells would try to cope with such a situation. The induction of E6-AP could be an adaptive response similar to Hsp70. The increased levels of E6-AP might try to protect cells by enhancing the degradation of misfolded proteins. In fact, we have observed that overexpression of E6-AP protects various stress-induced cell death. The 5′-untranslated region of E6-AP also consists of several heat shock factor 1 consensus binding sites, which further support our findings. Several other E3 ligases, like parkin and CHIP, also induced under various stresses and protect these stress-induced cell deaths (38, 39).
But how E6-AP could recognize the misfolded proteins? The interaction of E6-AP with Hsp70 might provide the clue. We have observed that the N-terminal region of E6-AP interacts with the C-proximal substrate binding domain of Hsp70/Hsc70, which suggests that E6-AP probably targets Hsp70/Hsc70-bound misfolded substrates for proteasomal degradation. Several co-chaperones and E3 ligases have been shown to interact with the C-proximal substrate binding domain of Hsp70/Hsc70 and regulate their chaperone activity (40). The bindings of E3 ligase CHIP with the C terminus of Hsp70/Hsc70 can stall the folding of Hsp70/Hsc70-bound proteins and at the same time induce the ubiquitination of chaperone-bound proteins for proteasomal degradation (15, 17, 18, 21). However, the functional significance of the interaction of E6-AP with Hsp70 is not clear. It is possible that E6-AP might function in a manner similar to CHIP. The E3 ligases possibly compete with other co-chaperones for the same binding sites in Hsp70/Hsc70, and the critical concentration of all these molecules in a particular conditions might dictate whether a Hsp70/Hsc70-bound misfolded protein will be refolded or ubiquitinated for proteasomal degradation.
To characterize the functional significance of the interaction of E6-AP with the Hsp70/Hsc70, we performed a series of experiments. Surprisingly, we have noticed that E6-AP redistributes to MTOC in response to inhibition of cellular proteasome function. E6-AP also recruits to aggresomes formed by CFTR and expanded polyglutamine proteins. Aggresomes are formed around MTOC by active minus end-directed transport of misfolded proteins through microtubules. The disruption of microtubular network prevents the formation of aggresomes. It has been clearly demonstrated that aggresome formation is a general response of cells to proteasome dysfunction (37, 41, 42). Aggresomes are also highly enriched with molecular chaperones, ubiquitin, and proteasome components (41, 42). The recruitment of E6-AP to MTOC or aggresomes in response to proteasome inhibition could be a desperate attempt of the cells to degrade misfolded and aggregated proteins, and all this clearly suggests that E6-AP is involved in the ubiquitination of misfolded protein for proteasomal degradation. The well known quality control E3 ligase CHIP also has been demonstrated to redistribute and co-localize with aggresomes and degrades variety of misfolded proteins (20, 34, 43). Although the E3 ligase, parkin, has been shown to co-localize with aggresomes, it has not been convincingly shown to degrade misfolded proteins (44, 45). Because overexpressed parkin is very prone to misfold, it can re-locate around MTOC and form its own aggresomes during proteasome inhibition. We have also observed that inhibition of proteasome function sometimes causes recruitment of E6-AP into a discrete nuclear structure. These structures are also highly enriched with Hsp70. We do not know the identity of these structures. These structures might be produced as a part of general cellular response to proteasome inhibition. The recruitment of E6-AP to these structures tempted us to suggest that E6-AP might be involved in the degradation of nuclear misfolded protein. These aspects of E6-AP need further investigation.
Finally we have shown that E6-AP directly ubiquitinates the heat-denatured luciferase that is captured by Hsp70. In Cos-7 cells, overexpression of E6-AP also degrades luciferases that are misfolded upon thermal stress. These finding clearly indicate that the Hsp70-bound misfolded proteins are ubiquitinated by E6-AP for proteasomal degradation. The effect of E6-AP on ubiquitination of misfolded luciferase is comparable with CHIP, which has been demonstrated earlier (21). Interestingly, parkin was unable to ubiquitinate the misfolded luciferase in a similar ubiquitination assay.
Growing evidence suggest that the quality control E3 ligase, CHIP, plays a vital role in the biology of neurodegenerative disorders involving protein misfolding. The CHIP has been shown to reduce the aggregation and cell death mediated by mutant SOD1, α-synuclein, tau, and polyglutamine proteins (24, 34, 43, 46–49). The HD transgenic mice that are deficient in CHIP display an accelerated disease phenotype, whereas a higher level of expression of CHIP in the spinal and bulbar muscular atrophy transgenic mice model ameliorate disease phenotype by enhancing the degradation of polyglutamine-expanded androgen receptor (50, 51). Because E6-AP functions as a quality control ligase, it could be implicated in the biology of neurodegenerative disorders involving protein misfolding and aggregation. In fact, recently we have shown that E6-AP suppresses aggregation and toxicity of polyglutamine disease proteins by promoting their ubiquitination and degradation (36). On the contrary, our findings also indicate that the defect in the cellular quality control system might be involved, at least in part, in the pathogenesis of Angelman syndrome. Altogether, our studies provide evidence that E6-AP promotes ubiquitination of Hsp70/Hsc70 chaperone-bound proteins for proteasomal degradation and, hence, can be regarded as a quality control E3 ligase. The expression of E6-AP is also induced in response to a variety of stressful conditions and protects stress-induced cell death.
Supplementary Material
Acknowledgments
We thank D. Narender and M. Singh for technical assistance.
This work was supported by the Department of Biotechnology, Government of India.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: UPS, ubiquitin proteasome system; CHIP, C terminus of Hsp70-interacting protein; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; HECT, homologous to E6-AP C terminus; ER, endoplasmic reticulum; CFTR, cystic fibrosis transmembrane conductance regulator; MTOC, microtubule-organizing center; HD, Huntington disease; GFP, green fluorescence protein; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Frydman, J. (2001) Annu. Rev. Biochem. 70 603-647 [DOI] [PubMed] [Google Scholar]
- 2.Hartl, F. U., and Hayer-Hartl, M. (2002) Science 295 1852-1858 [DOI] [PubMed] [Google Scholar]
- 3.Bukau, B., Weissman, J., and Horwich, A. (2006) Cell 125 443-451 [DOI] [PubMed] [Google Scholar]
- 4.McClellan, A. J., Tam, S., Kaganovich, D., and Frydman, J. (2005) Nat. Cell Biol. 7 736-741 [DOI] [PubMed] [Google Scholar]
- 5.Hebert, D. N., and Molinari, M. (2007) Physiol. Rev. 87 1377-1408 [DOI] [PubMed] [Google Scholar]
- 6.Scott, M. D., and Frydman, J. (2003) Methods Mol. Biol. 232 67-76 [DOI] [PubMed] [Google Scholar]
- 7.Muchowski, P. J., and Wacker, J. L. (2005) Nat. Rev. Neurosci. 6 11-22 [DOI] [PubMed] [Google Scholar]
- 8.Morimoto, R. I. (2008) Genes Dev. 22 1427-1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lansbury, P. T., and Lashuel, H. A. (2006) Nature 443 774-779 [DOI] [PubMed] [Google Scholar]
- 10.Bence, N. F., Sampat, R. M., and Kopito, R. R. (2001) Science 292 1552-1555 [DOI] [PubMed] [Google Scholar]
- 11.Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373-428 [DOI] [PubMed] [Google Scholar]
- 12.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503-533 [DOI] [PubMed] [Google Scholar]
- 13.Bercovich, B., Stancovski, I., Mayer, A., Blumenfeld, N., Laszlo, A., Schwartz, A.L., and Ciechanover, A. (1997) J. Biol. Chem. 272 9002-9010 [DOI] [PubMed] [Google Scholar]
- 14.Lee, D. H., Sherman, M. Y., and Goldberg, A. L. (1996) Mol. Cell. Biol. 16 4773-4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonough, H., and Patterson, C. (2003) Cell Stress Chaperones 8 303-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai, Y., Soda, M., Hatakeyama, S., Akagi, T., Hashikawa, T., Nakayama, K. I., and Takahashi, R. (2002) Mol. Cell 10 55-67 [DOI] [PubMed] [Google Scholar]
- 17.Ballinger, C. A., Connell, P., Wu, Y., Hu, Z., Thompson, L. J., Yin, L. Y., and Patterson, C. (1999) Mol. Cell. Biol. 19 4535-4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, J., Ballinger, C. A., Wu, Y., Dai, Q., Cyr, D. M., Hohfeld, J., and Patterson, C. (2001) J. Biol. Chem. 276 42938-42944 [DOI] [PubMed] [Google Scholar]
- 19.Connell, P., Ballinger, C.A., Jiang, J., Wu, Y., Thompson, L. J., Hohfeld, J., and Patterson, C. (2001) Nat. Cell Biol. 3 93-96 [DOI] [PubMed] [Google Scholar]
- 20.Meacham, G. C., Patterson, C., Zhang, W., Younger, J. M., and Cyr, D. M. (2001) Nat. Cell Biol. 3 100-105 [DOI] [PubMed] [Google Scholar]
- 21.Murata, S., Minami, Y., Minami, M., Chiba, T., and Tanaka, K. (2001) EMBO Rep. 2 1133-1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai, Q., Zhang, C., Wu, Y., McDonough, H., Whaley, R.A., Godfrey, V., Li, H. H., Madamanchi, N., Xu, W., Neckers, L., Cyr, D., and Patterson, C. (2003) EMBO J. 22 5446-5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian, S. B., McDonough, H., Boellmann, F., Cyr, D. M., and Patterson, C. (2006) Nature 440 551-555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickey, C.A., Patterson, C., Dickson, D., and Petrucelli, L. (2007) Trends Mol. Med. 13 32-38 [DOI] [PubMed] [Google Scholar]
- 25.Min, J. N., Whaley, R. A., Sharpless, N. E., Lockyer, P., Portbury, A. L., and Patterson, C. (2008) Mol. Cell. Biol. 28 4018-4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huibregtse, J. M., Scheffner, M., Beaudenon, S., and Howley, P. M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 2563-2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheffner, M., Nuber, U., and Huibregtse, J. M. (1995) Nature 373 81-83 [DOI] [PubMed] [Google Scholar]
- 28.Ramamoorthy, S., and Nawaz, Z. (2008) Nucl. Recept. Signal. 6 e006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kishino, T., Lalande, M., and Wagstaff, J. (1997) Nat. Genet. 15 70-73 [DOI] [PubMed] [Google Scholar]
- 30.Albrecht, U., Sutcliffe, J. S., Cattanach, B. M., Beechey, C. V., Armstrong, D., Eichele, G., and Beaudet, A. L. (1997) Nat. Genet. 17 75-78 [DOI] [PubMed] [Google Scholar]
- 31.Malzac, P., Webber, H., Moncla, A., Graham, J. M., Kukolich, M., Williams, C., Pagon, R. A., Ramsdell, L. A., Kishino, T., and Wagstaff, J. (1998) Am. J. Hum. Genet. 62 1353-1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamasaki, K., Joh, K., Ohta, T., Masuzaki, H., Ishimaru, T., Mukai, T., Niikawa, N., Ogawa, M., Wagstaff, J., and Kishino, T. (2003) Hum. Mol. Genet. 12 837-847 [DOI] [PubMed] [Google Scholar]
- 33.Mishra, A., and Jana N. R. (2008) Cell. Mol. Life Sci. 65 656-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jana, N. R., Dikshit, P., Goswami, A., Kotliarova, S., Murata, S., Tanaka, K., and Nukina, N. (2005) J. Biol. Chem. 280 11635-11640 [DOI] [PubMed] [Google Scholar]
- 35.Jana, N. R., Zemskov, E. A., Wang, G., and Nukina, N. (2001) Hum. Mol. Genet. 10 1049-1059 [DOI] [PubMed] [Google Scholar]
- 36.Mishra, A., Dikshit, P., Purkayastha, S., Sharma, J., Nukina, N., and Jana, N. R. (2008) J. Biol. Chem. 283 7648-7656 [DOI] [PubMed] [Google Scholar]
- 37.Johnston, J. A., Ward, C. L., and Kopito, R. R. (1998) J. Cell Biol. 143 1883-1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai, Y., Soda, M., and Takahashi, R. (2000) J. Biol. Chem. 275 35661-35664 [DOI] [PubMed] [Google Scholar]
- 39.Dikshit, P., and Jana, N. R. (2007) Biochem. Biophys. Res. Commun. 357 761-765 [DOI] [PubMed] [Google Scholar]
- 40.Fan, C. Y., Lee, S., and Cyr, D. M. (2003) Cell Stress Chaperones 8 309-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopito, R. R. (2000) Trends Cell Biol. 10 524-530 [DOI] [PubMed] [Google Scholar]
- 42.Kopito, R. R., and Sitia, R. (2000) EMBO Rep. 1 225-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin, Y., Klucken, J., Patterson, C., Hyman, B. T., and McLean, P. J. (2005) J. Biol. Chem. 280 23727-23734 [DOI] [PubMed] [Google Scholar]
- 44.Junn, E., Lee, S. S., Suhr, U. T., and Mouradian, M. M. (2002) J. Biol. Chem. 277 47870-47877 [DOI] [PubMed] [Google Scholar]
- 45.Muqit, M. M., Davidson, S. M., Payne Smith, M. D., MacCormac, L. P., Kahns, S., Jensen, P. H., Wood, N. W., and Latchman, D. S. (2004) Hum. Mol. Genet. 13 117-135 [DOI] [PubMed] [Google Scholar]
- 46.Urushitani, M., Kurisu, J., Tateno, M., Hatakeyama, S., Nakayama, K., Kato, S., and Takahashi, R. (2004) J. Neurochem. 90 231-244 [DOI] [PubMed] [Google Scholar]
- 47.Petrucelli, L., Dickson, D., Kehoe, K., Taylor, J., Snyder, H., Grover, A., De Lucia, M., McGowan, E., Lewis, J., Prihar, G., Kim, J., Dillmann, W. H., Browne, S. E., Hall, A., Voellmy, R., Tsuboi, Y., Dawson, T. M., Wolozin, B., Hardy, J., and Hutton, M. (2004) Hum. Mol. Genet. 13 703-714 [DOI] [PubMed] [Google Scholar]
- 48.Sahara, N., Murayama, M., Mizoroki, T., Urushitani, M., Imai, Y., Takahashi, R., Murata, S., Tanaka, K., and Takashima, A. (2005) J. Neurochem. 94 1254-1263 [DOI] [PubMed] [Google Scholar]
- 49.Kumar, P., Ambasta, R. K., Veereshwarayya, V., Rosen, K. M., Kosik, K. S., Band, H., Mestril, R., Patterson, C., and Querfurth, H. W. (2007) Hum. Mol. Genet. 16 848-864 [DOI] [PubMed] [Google Scholar]
- 50.Miller, V. M., Nelson, R. F., Gouvion, C. M., Williams, A., Rodriguez-Lebron, E., Harper, S. Q., Davidson, B. L., Rebagliati, M. R., and Paulson, H. L. (2005) J. Neurosci. 25 9152-9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adachi, H., Waza, M., Tokui, K., Katsuno, M., Minamiyama, M., Tanaka, F., Doyu, M., and Sobue, G. (2007) J. Neurosci. 27 5115-5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.