Abstract
PCSK9 is a secreted protein that degrades low density lipoprotein receptors (LDLRs) in liver by binding to the epidermal growth factor-like repeat A (EGF-A) domain of the LDLR. It is not known whether PCSK9 causes degradation of LDLRs within the secretory pathway or following secretion and reuptake via endocytosis. Here we show that a mutation in the LDLR EGF-A domain associated with familial hypercholesterolemia, H306Y, results in increased sensitivity to exogenous PCSK9-mediated cellular degradation because of enhanced PCSK9 binding affinity. The crystal structure of the PCSK9-EGF-A(H306Y) complex shows that Tyr-306 forms a hydrogen bond with Asp-374 in PCSK9 at neutral pH, which strengthens the interaction with PCSK9. To block secreted PCSK9 activity, LDLR (H306Y) subfragments were added to the medium of HepG2 cells stably overexpressing wild-type PCSK9 or gain-of-function PCSK9 mutants associated with hypercholesterolemia (D374Y or S127R). These subfragments blocked secreted PCSK9 binding to cell surface LDLRs and resulted in the recovery of LDLR levels to those of control cells. We conclude that PCSK9 acts primarily as a secreted factor to cause LDLR degradation. These studies support the concept that pharmacological inhibition of the PCSK9-LDLR interaction extracellularly will increase hepatic LDLR expression and lower plasma low density lipoprotein levels.
Elevated plasma levels of low density lipoprotein cholesterol (LDL-C)6 represent the greatest risk factor for the development of coronary heart disease. Clearance of LDL-C from the plasma occurs primarily by the liver through the action of LDL receptors (LDLRs), which are cell surface glycoproteins that bind to apolipoprotein B100 (apoB100) on LDL particles with high affinity and mediate their endocytic uptake (1). ADH is associated with mutations that reduce plasma LDL clearance and that are found in genes encoding the LDLR (familial hypercholesterolemia (FH)) (2) or apoB100 (familial defective apoB100) (3). Recently, mutations in the gene encoding the serine protease proprotein convertase subtilisin/kexin type 9 (PCSK9) were identified as a third cause of ADH (4). Functional studies have shown that PCSK9 regulates LDL-C levels by mediating LDLR degradation in the liver (5, 6). Mutations in PCSK9 resulting in ADH have been classified as gain-of-function because of their mode of inheritance and their association with elevated LDL-C levels (4, 7, 8). Conversely, loss-of-function mutations in PCSK9 result in lower plasma LDL-C levels, and individuals who carry these mutations are protected from developing coronary heart disease (9–11).
PCSK9 is a member of the mammalian proprotein convertase family of subtilisin-like serine endoproteases. The protein consists of a signal sequence, followed by a prodomain, which serves as both a folding chaperone and an inhibitor of cognate enzymatic activity, a conserved catalytic domain, and a cysteine/histidine-rich C-terminal domain of unknown function (12). Autocatalytic processing of PCSK9 in the endoplasmic reticulum (ER) results in release of the ∼14-kDa prodomain, which remains associated with the ∼60-kDa catalytic/C-terminal domains, occluding the catalytic site (13, 14). PCSK9 is rapidly and efficiently secreted from liver-derived cells in culture and is abundant in human plasma (15, 16). Unlike other proprotein convertases, in which the prodomain is further proteolytically processed to activate the serine protease, the prodomain of secreted PCSK9 remains intact and tightly bound (14, 15, 17).
PCSK9 mRNA is primarily expressed in liver, with lower levels of expression in intestine, kidney, and brain (18). PCSK9 and LDLR expression in liver are co-regulated by SREBP-2 (sterol regulatory element-binding protein 2) (19–21), a transcription factor that is activated when cellular cholesterol levels are depleted (22). Early functional studies demonstrated that PCSK9 promotes LDLR protein degradation within the lysosomal compartment of cells (5, 17); however, it is unclear whether PCSK9-mediated LDLR degradation occurred through the rerouting of nascent LDLRs from the Golgi apparatus to lysosomes (i.e. an intracellular mechanism) or following PCSK9 secretion and reuptake in the endocytic pathway (i.e. an extracellular mechanism).
Secreted PCSK9 binds directly to the first epidermal growth factor-like repeat (EGF-A) within the EGF precursor homology domain of the LDLR, and this binding is required for exogenous PCSK9 to promote LDLR degradation (23). Although autocatalytic activity is obligatory for PCSK9 maturation and secretion, catalytic function is not required for LDLR degradation by exogenous PCSK9 in cultured HepG2 cells (24, 25) or in mouse liver (26). The crystal structure of PCSK9 in complex with the LDLR EGF-A domain reveals that the LDLR-binding site is more than 20 Å away from the PCSK9 catalytic site (27). These findings suggest that PCSK9 plays a chaperone role in diverting the LDLR from the normal endocytic recycling route and toward a degradation pathway.
Here we utilize a soluble recombinant LDLR subfragment consisting of the tandem EGF-like repeats A and B (EGF-AB) to inhibit binding of PCSK9 to cell surface LDLRs to determine the contribution of secreted versus intracellular PCSK9 to overall PCSK9-mediated LDLR degradation in cells. Refinement of this blocking reagent was achieved by introducing a previously uncharacterized FH-associated LDLR mutation in the EGF-A domain, H306Y, that increases PCSK9 binding affinity. Inhibition of secreted PCSK9 action by EGF-AB(H306Y) results in complete recovery of LDLR number and function in HepG2 cells stably overexpressing wild-type PCSK9 or gain-of-function PCSK9 mutants implicated in hypercholesterolemia (D374Y or S127R). These studies demonstrate that PCSK9 acts primarily as a secreted factor in causing LDLR degradation in liver-derived cells.
EXPERIMENTAL PROCEDURES
Purification of Recombinant Proteins—Full-length FLAG-tagged human wild-type PCSK9 and PCSK9(D374Y) were purified from stably expressing HEK-293S cells as described (15). Plasmids containing EGF-AB mutant forms EGF-AB(H306Y) and EGF-AB(L318A) were generated from an EGF-AB:GST fusion cDNA construct (27) using the QuikChange site-directed mutagenesis kit (Stratagene) according to manufacturer's instructions. Δ53-PCSK9 and EGF-AB proteins were expressed and purified as described (27). LDLR extracellular domain (ECD) was expressed and purified as described (28).
Crystallization and Structure Determination—Crystals of Δ53-PCSK9:EGF-AB and Δ53-PCSK9:EGF-AB(H306Y) were formed essentially as described previously (27). Crystals were transferred stepwise into a solution of 0.05 m (NH4)H2PO4, 5 mm CaCl2, and 35% glycerol (final pH 7.4) and flash-frozen in a -160 °C nitrogen stream or directly into liquid propane. Diffraction data were collected at the Advanced Photon Source, beam line 19-ID, and were processed with HKL2000 (29) and the CCP4 suite (30). Refinement was performed with REFMAC (31). Figures were generated with PyMOL (W. L. Delano).
In Vitro Binding Measurements—Infrared dye-labeled LDLR ECD was prepared using the DyLight800 antibody labeling kit (Pierce) as per the manufacturer's instructions. Anti-FLAG M2 monoclonal antibody (mAb) (10 μg) (Sigma) was diluted in TBS-C buffer (50 mm Tris, pH 7.4, 90 mm NaCl, 2 mm CaCl2) and blotted directly onto 0.22-μm pore size nitrocellulose membrane (GE Healthcare) using a BioDot SP slot-blot apparatus (Bio-Rad). Blots were blocked for 30 min in Odyssey blocking buffer (LI-COR Biosciences). All incubations (90 min) and subsequent washes (three for 15 min) were carried out at room temperature with gentle oscillation in TBS-C buffer containing 2.5% nonfat milk. Blots were initially incubated with FLAG-tagged PCSK9 (5 μg/ml) followed by incubation with fluorophore-labeled LDLR-ECD in the absence or presence of unlabeled competitor proteins. Blots were scanned using the LI-COR Odyssey infrared imaging system, and band intensity was quantified using Odyssey version 2.0 software (LI-COR Biosciences). The amount of competitor protein required for 50% inhibition of fluorophore-labeled LDLR-ECD binding (EC50) was determined by fitting data to a sigmoidal dose-response curve using nonlinear regression (GraphPad Software, Inc.).
Tissue Culture Medium—Medium A contained Dulbecco's modified Eagle's medium (Mediatech, Inc.), 1 g/liter glucose, supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin sulfate. Medium B contained Medium A supplemented with 10% fetal calf serum (v/v) (Atlanta Biologicals). Medium C was identical to Medium A but with 4.5 g/liter glucose. Medium D contained Medium C supplemented with 10% fetal calf serum (v/v). Sterol-depleting Medium E contained Medium C supplemented with 5% (v/v) NCLPDS, 10 μm sodium compactin, and 50 μm sodium mevalonate. Sterol-supplemented Medium F contained Medium C with 5% (v/v) NCLPDS, 10 μg/ml cholesterol, and 1 μg/ml 25-hydroxycholesterol.
Transient Transfection of HuH7 Cells—Plasmid pCMV-LDLR(H306Y) was produced by site-directed mutagenesis using the oligonucleotide 5′-cggcggctgttcctacgtctgcaatgac-3′ and pLDLR-17 (32). HuH7 cells were seeded on day 0 at 2.0 × 105 cells in 60-mm dishes and cultured in Medium D at 37 °C and 5% CO2 atmosphere. On day 2, cells were transfected with empty vector (pCMV5), LDLR-WT (pLDLR-17), or LDLR(H306Y) expression plasmids (1 μg) using Lipofectamine 2000 transfection reagent (Invitrogen) as per the manufacturer's instructions. On day 3, cells were refed with sterol-supplemented Medium F. After 18 h, cells were treated with purified FLAG-tagged PCSK9 (see figure legends for specific details) then lysed and subjected to SDS-PAGE and immunoblot analysis as described (6). Mouse anti-human LDLR IgG HL-1 (33), mouse anti-human transferrin receptor (TFR) IgG (Zymed Laboratories Inc.), or mouse anti-FLAG M2 (Sigma) primary antibodies were used to probe immunoblots.
For LDLR degradation assays, cell surface proteins were biotinylated and isolated from whole cell extracts as described (15), and secondary detection was with IRDye800-conjugated secondary donkey anti-mouse antibodies (LI-COR Biosciences). Proteins were visualized and quantified using the LI-COR Odyssey infrared imaging system (LI-COR Biosciences). LDLR protein values were normalized against those of TFR.
Immunofluorescence Staining—Indirect immunofluorescence for PCSK9 was carried out in HuH7 human hepatoma cells. Cells were initially grown to ∼50% confluence on glass coverslips in Medium D. Cells were then incubated for >18 h in sterol-depleting Medium E prior to 30 min of incubation with 5 μg/ml PCSK9 in the absence or presence of 5 μm EGF-AB(H306Y) or EGF-AB(L318A). After incubation, cells were washed with phosphate-buffered saline (PBS), fixed in 4% (w/v) paraformaldehyde in PBS for 15 min, quenched in 1% (w/v) l-glycine in PBS for 10 min, and permeabilized in 0.05% (w/v) Triton X-100 (Pierce) in PBS for 10 min at room temperature. Fixed/permeabilized cells were then blocked 30 min in PBS supplemented with 5% (w/v) BSA, 10% (v/v) goat serum (Sigma), and 0.025% (w/v) Triton X-100. Cells were stained sequentially with a rabbit polyclonal antibody raised against full-length human PCSK9 (228B) (5 μg/ml) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen) (2 μg/ml). Both the 16-h primary incubation (4 °C) and the 1-h secondary incubation (21 °C) were followed by three 20-min washes in PBS with 0.025% Triton X-100. Coverslips were mounted using ProLong Gold with 4′,6-diamidino-2-phenylindole (Invitrogen) and imaged using a Leica TCS SP5 confocal microscope. Fluorescence intensity settings were kept constant for all conditions.
PCSK9 Uptake Inhibition in HuH7 Cells—HuH7 cells were cultured and treated as described above for immunofluorescence staining. Following 30 min of incubation with PCSK9 and EGF-AB peptides (see figure legends for specific details), cells were lysed and subjected to SDS-PAGE and immunoblot analysis as described (6). PCSK9 and actin proteins were detected with mouse anti-FLAG M2 (Sigma) and mouse anti-actin AC40 (Sigma) primary antibodies. Secondary detection was with IRDye800-conjugated secondary donkey anti-mouse antibodies, and proteins were visualized and quantified using the LI-COR Odyssey infrared imaging system (LI-COR Biosciences). LDLR protein values were normalized against those of TFR. PCSK9 quantification was normalized against actin.
PCSK9 Blocking in Stable HepG2 Cells—Human hepatoma cells, HepG2 (ATCC HB-8065) cultured in Medium B at 37 °C and 8.8% CO2 atmosphere were transfected with empty vector or C-terminal FLAG-tagged full-length human wild-type (pCMV-hPCSK9-FLAG), D374Y (pCMV-hPCSK9(D374Y)-FLAG), or S127R (pCMV-hPCSK9(S127R)-FLAG) PCSK9 cDNA using FuGENE 6 (Roche Applied Science) transfection reagent according to the manufacturer's instructions. The cDNAs pCMV-hPCSK9-FLAG and pCMV-hPCSK9(D374Y)-FLAG were described previously (15). The pCMV-hPCSK9(S127R)-FLAG cDNA was created through site-directed mutagenesis of the pCMV-hPCSK9-FLAG vector. Transfected cells were subjected to selection with G418. Surviving colonies were isolated and assessed for PCSK9 expression with mouse anti-PCSK9 IgG-15A6 (15). The stably expressing cell lines were seeded into 60-mm dishes and grown to ∼80% confluence in Medium B. Cells were then washed two times with PBS and incubated for 18 h in sterol-depleting Medium E with no addition, 5 μm EGF-AB(H306Y), or 5 μm EGF-AB(L318A) protein. Following treatments, cells were biotinylated, and whole cell and cell surface protein lysates were isolated as described previously (15). Equal protein fractions were subjected to 8% SDS-PAGE and immunoblot analysis. Mouse anti-human LDLR IgG-HL1 (33) and mouse anti-human TFR IgG (Zymed Laboratories Inc.) primary antibodies were used to probe immunoblots. Secondary detection was with IRDye800-conjugated secondary donkey anti-mouse antibodies. Membranes were scanned and quantified using the LI-COR Odyssey infrared imaging system (LI-COR Biosciences). LDLR protein values were normalized against those of TFR.
DiI-LDL Uptake—LDL was isolated from the plasma of healthy human subjects by ultracentrifugation as described (34). LDL was labeled with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) (Invitrogen). LDL was diluted to 1 mg/ml in PBS with 10 mg/ml NCLPDS, and 150 μg of DiI was added per mg of LDL. The LDL was labeled 18 h at 37 °C. Following labeling, DiI-LDL was re-isolated by ultracentrifugation. The DiI-LDL was then dialyzed for 36 h at 4 °C against four changes of 500 volumes of PBS. DiI-LDL concentration was determined by Modified Lowry Protein Assay kit (Pierce). HepG2 cells stably expressing vector, PCSK9, PCSK9(D374Y), or PCSK9(S127R) were grown to ∼80% confluence in Medium B. Cells were then incubated in Medium E with no addition, 1, 2, or 5 μm EGF-AB(H306Y), or 5 μm EGF-AB(L318A) protein. After 18 h, 100 μg/ml DiI-LDL was added for 2 h. Cells were washed twice in PBS, trypsinized, and transferred to Medium C supplemented with 2.5% NCLPDS. The mean fluorescence intensity of 10,000 live cells, as determined by scatter characteristics, for each sample was determined in the 585 nm range with a FACSCalibur flow cytometer. Replicate samples were assayed for each condition. Experiments were combined by normalization to the average of the vector expressing, no addition condition replicates within each experiment. HuH7 cells were cultured and transfected with empty vector (pCMV5), pLDLR-17, or pCMV-LDLR(H306Y) as described above. The day after transfection, the cells were incubated in Medium E for 18 h. DiI-LDL was added and uptake measured as above. Three replicates were used per sample.
Pulse-Chase Analysis and 125I Uptake—Details of pulsechase analyses of cellular PCSK9 and LDLR processing and 125I-labeled protein cell uptake analyses are in the supplemental material.
RESULTS
Increased Binding Affinity of PCSK9 for LDLR Subfragments Containing the H306Y Mutation—We recently described the co-crystal structure of a complex between PCSK9 and the EGF-A domain of the LDLR at acidic pH (27). The binding site in the catalytic domain of PCSK9 contains the Asp-374 residue that forms a hydrogen bond with EGF-A-H306. The D374Y mutation was previously implicated in ADH (35). The His-306 of the EGF-A is replaced by tyrosine in the FH-associated H306Y LDLR allele (36, 37). The cellular mechanism by which this LDLR mutation causes hypercholesterolemia is not known. Based on the structural model of the PCSK9-EGF-A complex, we predicted that H306Y mutation would result in increased binding affinity for PCSK9 and thus increased susceptibility to PCSK9-mediated degradation (27).
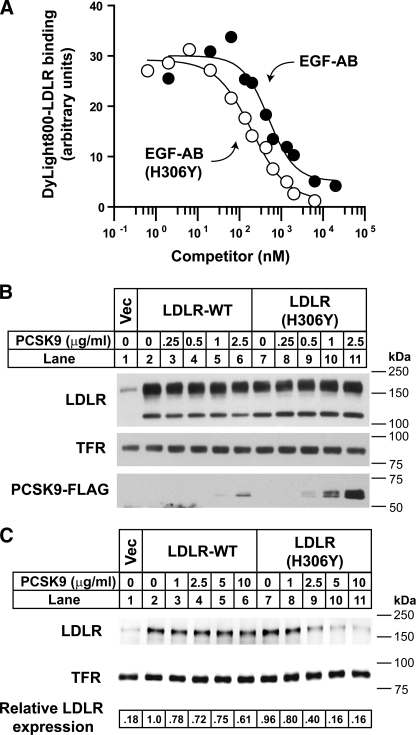
To test this idea, in vitro binding experiments using recombinant LDLR-derived subfragments consisting of the tandem EGF-like repeats A and B (EGF-AB) were performed. The ability of EGF-AB containing the H306Y mutation to bind to PCSK9 was compared with that of wild-type EGF-AB in competition binding assays performed at physiological pH 7.4 (Fig. 1A). EGF-AB(H306Y) bound more avidly to PCSK9 than wild-type EGF-AB, as evidenced by a 2.5-fold enhanced ability to compete for binding with fluorophore-labeled LDLR extracellular domain protein (EC50 of 195.6 nm for EGF-AB(H306Y) versus 497.6 nm for wild-type EGF-AB).
FIGURE 1.
Increased binding and degradation of LDLR(H306Y) by PCSK9. A, competition binding of EGF-AB and EGF-AB(H306Y) to PCSK9. Purified PCSK9 was prebound to slot-blotted anti-FLAG M2 monoclonal antibody (mAb). Blots were incubated for 90 min with 0.1 μg/ml infrared dye (DyLight800)-labeled LDLR-ECD at pH 7.4 in the presence of increasing concentrations of unlabeled EGF-AB (closed circles) or EGF-AB(H306Y) (open circles), and bound fluorophore-labeled LDLR was detected and quantified. B, HuH7 cells transiently expressing LDLR-WT or LDLR(H306Y) were cultured in sterol-supplemented medium (see “Experimental Procedures”) and treated for 1 h with the indicated amount of purified FLAG-tagged PCSK9. Cells were lysed, and cell extracts were subjected to SDS-PAGE and immunoblot analysis as described under “Experimental Procedures.” Internalized PCSK9 was detected with anti-FLAG M2 mAb. LDLR was detected with an anti-LDLR mAb (HL-1). TFR was detected as a control for equal protein loading. C, HuH7 cells transfected as in B were treated with the indicated amount of PCSK9 for 4 h. Following biotinylation, isolated cell surface protein lysates were subjected to SDS-PAGE and immunoblot analysis. LDLR and TFR were detected as in B. Secondary detection used an infrared dye (IRDye800)-labeled antibody. Blots were visualized and quantified using the LI-COR Odyssey infrared imaging system. LDLR levels were normalized to TFR expression. For these and subsequent data, results shown are from representative experiments repeated at least three times with similar results. Vec, vector.
Increased Binding of PCSK9 to Cell Surface LDLRs Containing the H306Y Mutation—As an indirect measure of PCSK9 binding to full-length LDLR, HuH7 human hepatoma cells transiently overexpressing wild-type LDLR (LDLR-WT) or mutant LDLR(H306Y) constructs were incubated for 1 h with low concentrations (0–2.5 μg/ml) of purified FLAG-tagged PCSK9, and cell association of exogenous PCSK9 was quantified by immunoblots of cell extracts using an anti-FLAG antibody. To minimize interference from endogenous LDLR and PCSK9 expression, cells were preincubated in the presence of sterols (25-hydroxycholesterol and cholesterol) to suppress SREBP-2-mediated transcriptional activation of the endogenous PCSK9 and LDLR genes. At these levels of LDLR overexpression, significant LDLR receptor degradation was not observed with the concentrations of PCSK9 used. Nevertheless, cell association of FLAG-tagged PCSK9 was >3-fold higher in cells expressing LDLR(H306Y) compared with LDLR-WT (Fig. 1B). This result supports the direct binding data of Fig. 1A, confirming that LDLR(H306Y) has an increased binding affinity for exogenous PCSK9.
Increased Degradation of LDLR(H306Y) by Exogenous PCSK9—To test the effect of the H306Y mutation on PCSK9-mediated LDLR degradation, transiently transfected HuH7 cells were treated for a longer time period (4 h) and with higher concentrations of purified PCSK9. Following PCSK9 treatment, cell surface proteins were biotinylated, and levels of cell surface LDLRs were quantified by immunoblot analysis. Under basal conditions, cell surface expression of LDLR(H306Y) was nearly identical to that of the LDLR-WT (Fig. 1C, lanes 2 and 7), suggesting that the mutant receptor trafficked normally to the cell surface (see below). As shown in lanes 3–6 and 8–11 of Fig. 1C, LDLR(H306Y) was >2-fold more susceptible to PCSK9-mediated degradation than LDLR-WT.
Structural Basis of Increased Binding to PCSK9 Caused by the H306Y Mutation in the LDLR EGF-A Domain—To elucidate the molecular mechanism responsible for the increased binding of PCSK9 to LDLR(H306Y), the structure of the PCSK9/EGF-A(H306Y) complex was determined. For structural studies, a recombinant PCSK9 protein lacking the N-terminal 21 amino acids of the prodomain region (Δ53-PCSK9) was used, which we have previously shown binds to LDLRs (27). Crystals of Δ53-PCSK9 in complex with either wild-type EGF-AB or EGF-AB(H306Y) were grown at pH 4.8 and transferred to pH 7.4 prior to data collection, and the structures were refined to 2.7 Å (Table 1). The structures of both complexes were nearly identical with the previously determined structure of Δ53-PCSK9: EGF-AB at pH 4.8 (27) with a root mean square deviation across all C-α atoms of 0.3 Å (EGF-AB wild-type at neutral pH) and 0.4 Å (EGF-AB H306Y) when compared with the acidic pH structure. Similar to the previous structure, electron density was not visible for the EGF-B domain.
TABLE 1.
Data collection and refinement statistics
| EGF-AB | Wild-type, pH 7.4 | H306Y, pH 7.4 |
|---|---|---|
| Space group | P4(1)2(1)2 | P4(1)2(1)2 |
| a, b | 116.047 Å | 115.661 Å |
| c | 133.629 Å | 133.401 Å |
| Resolution (Å) (final shell) | 40.0 to 2.70 | 40.0 to 2.70 |
| (2.77 to 2.70) | (2.77 to 2.70) | |
| Reflections | ||
| Total | 110,489 | 117,756 |
| Unique | 24,921 | 25,367 |
| Completeness | 96.6% (81.9%) | 99.3% (92.8%) |
| Rsym | 8.4% | 8.1% |
| Rcryst | 22.6% (34.7%) | 23.4% (33.2%) |
| Rfree | 26.1% (46.5%) | 27.8% (41.2%) |
| Root mean square deviation | ||
| Bond length | 0.006 Å | 0.006 Å |
| Bond angle | 0.973° | 0.971° |
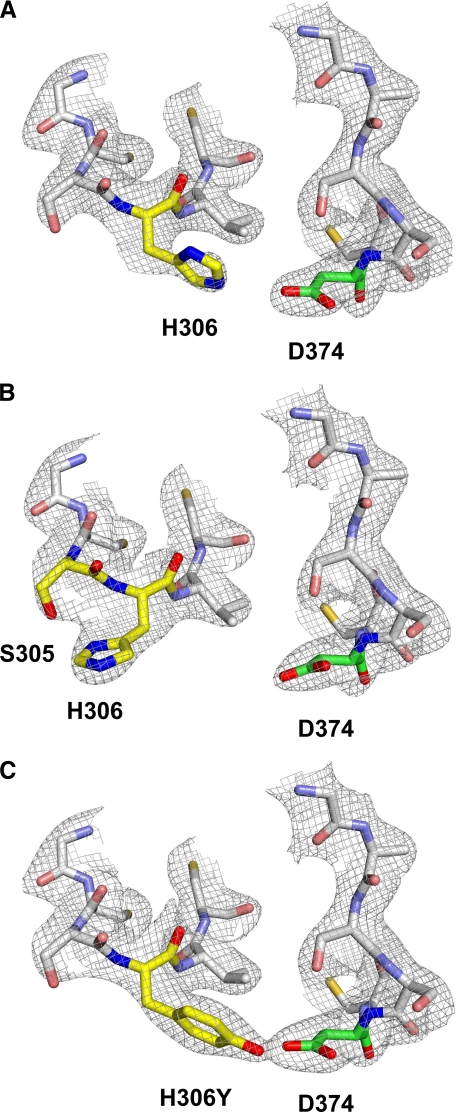
At acidic pH, His-306 of EGF-A is 4 Å from Asp-374 of PCSK9, forming an intermolecular salt bridge (Fig. 2A); however, at neutral pH, His-306 of EGF-A is rotated away from Asp-374 of PCSK9 and is 3.1 Å from Ser-305 of EGF-A forming an intramolecular hydrogen bond (Fig. 2B). At neutral pH, H306Y is in a conformation similar to that seen for His-306 at acidic pH, with the hydroxyl group of H306Y 3.0 Å from Asp-374 forming an intermolecular hydrogen bond (Fig. 2C). These data provide a structural mechanism for increased binding between PCSK9 and LDLR harboring the FH-associated H306Y mutation.
FIGURE 2.
Structure of the PCSK9-EGF-A complex. The sigmaA weighted 2Fo - Fc electron density map contoured at 1σ shows the conformational change that occurs upon protonation of His-306. EGF-A and PCSK9 are represented as a stick model. Residues involved in the pH-dependent conformational change are colored according to element type as follows: nitrogen, blue; oxygen, red; EGF-A carbon, yellow; PCSK9 carbon, green. All other residues are colored gray. A, at acidic pH, His-306 of EGF-A forms a salt bridge with Asp-374 of PCSK9. B, at neutral pH, His-306 of EGF-A forms an intramolecular hydrogen bond with Ser-305. C, FH mutation H306Y of EGF-A is able to form a hydrogen bond with Asp-374 of PCSK9 at neutral pH.
We next confirmed that the H306Y mutation had no effect on other well characterized aspects of LDLR function in HuH7 cells. Pulse-chase radiolabeling analysis showed that newly synthesized LDLR(H306Y) and LDLR-WT underwent similar rates of processing in the secretory pathway, indicating that the H306Y mutation did not affect the kinetics of LDLR folding in the ER or O-linked glycosylation in the Golgi apparatus (supplemental Fig. 1A). The biotinylation experiments of Fig. 1C demonstrated that LDLR(H306Y) was expressed at normal levels on the cell surface. Cells expressing LDLR(H306Y) were able also to internalize fluorescent LDL (DiI-LDL) at normal levels (supplemental Fig. 1B). Together, these data indicate that the H306Y mutation does not affect cellular LDLR function but that the mutation results in FH as a result of increased binding to PCSK9.
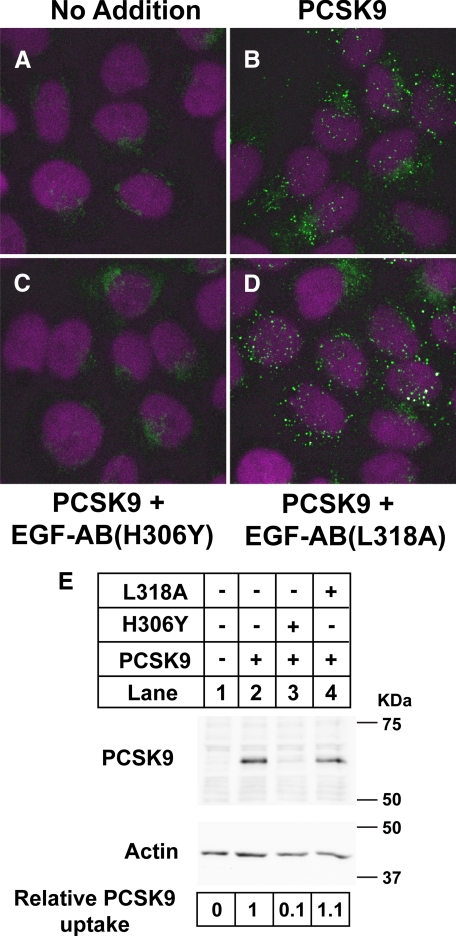
LDLR Subfragments Containing the H306Y Mutation Potently Inhibit PCSK9 Binding to Cell Surface LDLRs—To determine the cellular site of action of PCSK9, and hence the mechanism for increased degradation of LDLR(H306Y) by PCSK9, a recombinant EGF-AB(H306Y) peptide was used to inhibit the interaction of secreted PCSK9 and the LDLR on the cell surface. First, indirect immunofluorescence was performed in HuH7 cells to test whether EGF-AB(H306Y) blocked the cellular uptake of recombinant PCSK9 added to the medium of cells. In the absence of exogenously added PCSK9, weak diffuse immunostaining of endogenous PCSK9 was detected, with a localized concentration near the perinuclear region in some cells (Fig. 3A). Addition of recombinant PCSK9 to the culture medium resulted in robust cellular uptake of PCSK9 and immunostaining of punctate vesicular structures throughout the cytoplasmic space, which were indicative of early/late endosomes (Fig. 3B). In contrast, only weak fluorescence corresponding to endogenous PCSK9 immunostaining was evident upon simultaneous addition of EGF-AB(H306Y) at 5 μm (Fig. 3C). A control EGF-AB peptide with a mutation (L318A) in a residue known to be required for binding of PCSK9 to full-length LDLRs (23) had no effect on PCSK9 uptake (Fig. 3D), indicating that the EGF-AB(H306Y) peptide prevented PCSK9 endocytosis through a direct interaction.
FIGURE 3.
EGF-AB(H306Y) blocks uptake of PCSK9 in HuH7 cells. HuH7 cells were treated 30 min with 5 μg/ml purified full-length FLAG-tagged PCSK9 alone or in combination with 5 μm EGF-AB(H306Y) blocking peptide or EGF-AB(L318A) negative-control peptide. A–D, cells were immunostained for PCSK9 (green) and nuclear stained with 4′,6-diamidino-2-phenylindole (magenta) as described under “Experimental Procedures.” E, cells were lysed and subjected to SDS-PAGE and immunoblotting for FLAG-tagged PCSK9 as described in the legend to Fig. 1B. Actin was detected as a control for equal protein loading. PCSK9 and actin levels were visualized and quantified using an IRDye800-labeled secondary antibody and the LI-COR Odyssey infrared imaging system. PCSK9 levels were normalized to actin expression.
To quantify the blocking efficiency of EGF-AB(H306Y) observed in the immunofluorescence experiments, extracts from cells treated under the same conditions were subjected to SDS-PAGE, and immunoblotting analyses were carried out using an antibody to detect FLAG-tagged PCSK9. Fig. 3E shows that PCSK9 efficiently associated with HuH7 cells (lane 2) and that cell association was inhibited by 90% in the presence of 5 μm EGF-AB(H306Y), whereas 5 μm EGF-AB(L318A) had no effect (lanes 3 and 4). Thus, low micromolar concentrations of EGF-AB(H306Y) are effective at inhibiting cell surface binding of exogenously added recombinant PCSK9.
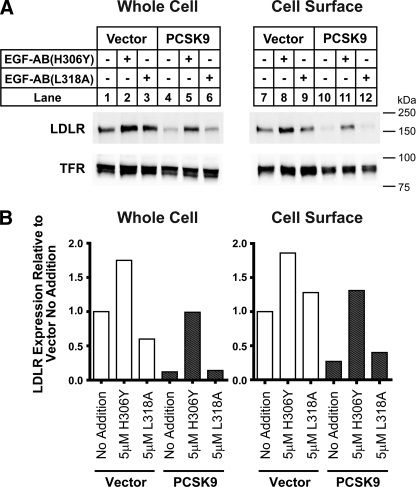
PCSK9 Functions as a Secreted Factor in Mediating LDLR Degradation in HepG2 Cells—We next utilized the EGF-AB(H306Y) peptide as an antagonist to selectively inhibit endogenously synthesized and secreted PCSK9 action. HepG2 cell lines stably expressing C-terminal FLAG-tagged wild-type PCSK9 were produced, and a representative clone was selected for further studies (HepG2-PCSK9-WT). These cells had ∼7-fold higher PCSK9 mRNA levels compared with control cells transfected with empty vector (HepG2-Vec) (data not shown).
For all experiments, cell monolayers were washed extensively prior to culture in sterol-depleting medium (see “Experimental Procedures”) to induce LDLR gene transcription and protein expression. Secreted PCSK9 concentrations in the medium were ∼10-fold higher for PCSK9 overexpressing cells than control cells (∼2 μg/ml for HepG2-PCSK9-WT cells versus ∼0.2 μg/ml for HepG2-Vec cells) as measured by ELISA. We have previously used this ELISA to determine the PCSK9 concentrations in human plasma (50–600 ng/ml) (15); thus the medium PCSK9 concentrations for HepG2-Vec cells were within the physiological range found in human plasma, whereas concentrations for PCSK9 overexpressing cells were moderately elevated.
Under basal conditions, LDLR protein levels of HepG2-PCSK9-WT cells in cell lysates and at the cell surface were reduced by >70% compared with those of control cells (Fig. 4). PCSK9 overexpression did not affect transferrin receptor (TFR) levels. The addition of 5 μm EGF-AB(H306Y) to the culture medium for 18 h increased LDLR protein levels 3–4-fold in both cell lysates and on the cell surface of HepG2-PCSK9-WT cells. This level of LDLR protein expression approximated that of untreated control cells. A >50% increase in LDLR protein levels in HepG2-Vec control cells was also measured following EGF-AB(H306Y) treatment. Addition of the PCSK9-binding defective EGF-AB(L318A) peptide had no appreciable effect on LDLR protein levels in either cell line (Fig. 4, A and B). The above results were confirmed using two independent HepG2 cell lines overexpressing PCSK9 (data not shown).
FIGURE 4.
EGF-AB(H306Y) treatment restores LDLR numbers in PCSK9 overexpressing cells. HepG2 cells stably overexpressing empty vector or wild-type PCSK9 (WT) were cultured in sterol-depleting medium (see “Experimental Procedures”) and treated for 18 h with 5 μm EGF-AB(H306Y) or negative-control EGF-AB(L318A) peptides. A, cell surface proteins were isolated following biotinylation, and whole cell and cell surface protein extracts were subjected to SDS-PAGE and immunoblot analysis for LDLR. LDLR and TFR levels were visualized as described in the legend to Fig. 1C. B, LDLR bands in A were quantified using the LI-COR Odyssey infrared imaging system. Values were normalized to TFR and graphed relative to untreated vector-transfected control cells.
The results in Fig. 4 suggested that the EGF-AB(H306Y) peptide blocked binding of secreted PCSK9 to cell surface LDLRs, thus antagonizing PCSK9-mediated LDLR degradation. Alternatively, secreted PCSK9 may reenter cells and act upon nascent LDLRs in the secretory pathway. To test this possibility, HepG2 cells were treated with cycloheximide (CHX) to inhibit protein synthesis for a 2-h preincubation and also during a 4-h incubation with purified PCSK9 (5 μg/ml). CHX effectively inhibited protein synthesis but did not appreciably affect LDLR degradation caused by PCSK9 (69% reduction in LDLR protein levels in absence of CHX versus 60% reduction in the presence of CHX) (supplemental Fig. S2, A and B). This result indicates that exogenous PCSK9 mediates degradation of mature LDLRs.
To further rule out the possibility that PCSK9 was acting intracellularly, we measured the cell association, uptake, and lysosomal degradation of 125I-labeled EGF-AB(H306Y) and PCSK9 proteins in HepG2 cells using a pulse-chase protocol. Following a 2-h chase, ∼2 ng of 125I-PCSK9 per mg of protein was cell-associated, and ∼4 ng was recovered as mono-125I-tyrosine because of lysosomal degradation and subsequent excretion into the culture medium (38) (supplemental Fig. S2C). In contrast, negligible amounts of 125I-labeled EGF-AB(H306Y) were cell-associated or recovered as mono-125I-tyrosine in the culture medium, indicating absent or very low cellular uptake of this peptide (supplemental Fig. S3B). Thus the ability of the EGF-AB(H306Y) peptide to antagonize PCSK9-mediated LDLR cellular degradation is manifest at the cell surface.
Gain-of-Function S127R and D374Y PCSK9 Mutants Also Function as Secreted Factors in Promoting Cellular LDLR Degradation—We next investigated the effect of EGF-AB(H306Y) on LDLR protein levels in HepG2 cells stably transfected with the mutant forms of PCSK9 associated with hypercholesterolemia. Two PCSK9 gain-of-function mutations (S127R and D374Y) were studied because of the disease severity associated with these mutations (4, 35). HepG2-PCSK9(S127R) and HepG2-PCSK9(D374Y) cells had 12- and 7-fold higher PCSK9 mRNA levels than HepG2-Vec cells, respectively (data not shown).
To assess PCSK9 autocatalytic processing and secretion in these HepG2 cell lines, pulse-chase radiolabeling analysis was performed using an anti-FLAG monoclonal antibody to immunoprecipitate radiolabeled FLAG-tagged PCSK9 proteins. In agreement with a previous report (17), PCSK9(S127R) was processed from a proprotein to a cleaved form at a slower rate than wild-type PCSK9 or PCSK9(D374Y) (supplemental Fig. S3). Nevertheless, the concentration of PCSK9 in the culture medium as measured by ELISA was increased 6-fold in HepG2-PCSK9(S127R) cells and 5-fold in HepG2-PCSK9(D374Y) cells compared with HepG2-Vec control cells after an 18-h culture period.
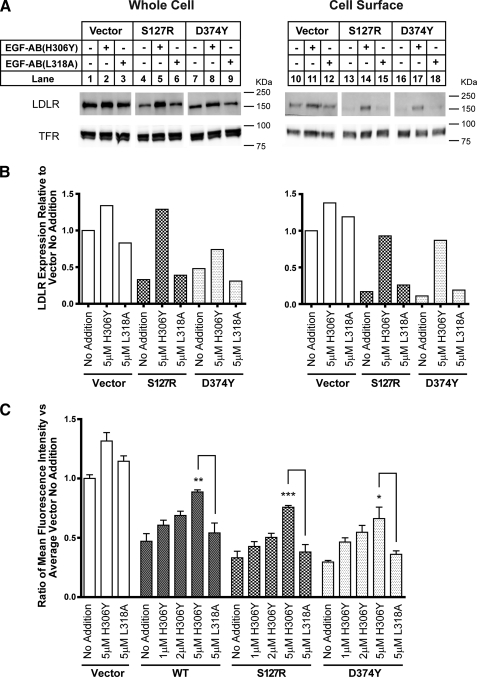
LDLR protein levels were markedly reduced in cell extracts and at the cell surface for both mutant PCSK9 expressing cell lines compared with control cells (Fig. 5, A and B). For consistency with previous experiments, the EGF-AB(H306Y) peptide was used for blocking experiments with HepG2-PCSK9(D374Y) cells as described above, although the binding affinity of this peptide to purified PCSK9(D374Y) is the same as wild-type EGF-AB (data not shown). Incubation of HepG2-PCSK9(S127R) and HepG2-PCSK9(D374Y) cells with 5 μm EGF-AB(H306Y) for 18 h resulted in the recovery of whole cell and cell surface LDLR protein levels to those approximating HepG2-Vec control cells cultured under basal conditions, whereas incubation with 5 μm of the PCSK9-binding defective peptide EGF-AB(L318A) had no effect on LDLR protein levels (Fig. 5, A and B).
FIGURE 5.
EGF-AB(H306Y) treatment restores LDLR number and function in gain-of-function PCSK9 overexpressing cell lines. A and B, HepG2 cells stably overexpressing empty vector, PCSK9(S127R), or PCSK9(D374Y) were cultured in sterol-depleting medium (see “Experimental Procedures”) and treated for 18 h with 5 μm EGF-AB(H306Y) or negative-control EGF-AB(L318A) peptides. LDLR and TFR levels in cell surface and whole cell protein extracts were visualized and quantified as described in the legend to Fig. 4. C, empty vector, PCSK9-WT, PCSK9(S127R), or PCSK9(D374Y) overexpressing cells were treated for 18 h with the indicated concentration of EGF-AB(H306Y) or EGF-AB(L318A) and then incubated for 2 h with 100 μg/ml DiI-LDL. Cells were trypsinized, and the mean fluorescence intensity of each sample was measured by flow cytometry. Six replicate samples were used per condition. * indicates a statistical difference between columns with significance, p < 0.05, by one-way analysis of variance and post-test pairwise comparison using Tukey's method; **, p < 0.005; ***, p < 0.001.
As a measure of LDLR function, cellular uptake of DiI-LDL was assessed by fluorescence-activated cell sorter analysis. Under basal conditions, DiI-LDL uptake was decreased >50% in HepG2-PCSK9-WT cells compared with HepG2-Vec controls (Fig. 5C). LDLR function was more severely affected in cells overexpressing PCSK9 gain-of-function mutants, with a >70% reduction in DiI-LDL uptake in both HepG2-PCSK9(S127R) and HepG2-PCSK9(D374Y) cells. Addition of 1–5 μm EGF-AB(H306Y) for 18 h prior to DiI-LDL incubation resulted in a dose-dependent increase in DiI-LDL uptake in all PCSK9 overexpressing cells. In HepG2-PCSK9-WT cells, a dose of 5 μm resulted in DiI-LDL uptake levels approximating those of untreated control cells. This same level of peptide caused a >2-fold increase in DiI-LDL uptake in HepG2-PCSK9(S127R) and HepG2-PCSK9(D374Y) cells; however, overall levels were still suppressed ∼20–30% compared with untreated control cells. Incubation with 5 μm EGF-AB(L318A) peptide had no appreciable effect on DiI-LDL uptake in any cell line (Fig. 5C). The increase in DiI-LDL uptake following EGF-AB(H306Y) incubation was statistically significant for all three PCSK9 overexpressing cell lines compared with negative control values. Interestingly, 5 μm EGF-AB(H306Y) treatment resulted in a moderate but consistently detectable ∼30% increase in DiI-LDL uptake in HepG2-Vec cells.
Combined, these results support the conclusion that wild-type PCSK9 as well as the S127R and D374Y gain-of-function mutant forms of PCSK9 largely act to degrade cellular LDLRs following their secretion.
DISCUSSION
In this study, we show that a previously uncharacterized FH-associated LDLR allele (H306Y) exhibits enhanced binding to PCSK9, and we provide the structural basis for this increased affinity. An LDLR EGF-AB fragment containing the H306Y mutation blocks the binding of secreted PCSK9 to LDLRs, which results in the restoration of LDLR number and activity in HepG2 cells that overexpress wild-type PCSK9 or that express two gain-of-function PCSK9 mutants, D374Y and S127R. Our results provide evidence that PCSK9 acts extracellularly to cause subsequent degradation of LDLRs in liver cells.
The structure of PCSK9 bound to EGF-AB identifies the regions of PCSK9 that interact with the EGF-A domain of the LDLR and provides a possible structural explanation for the underlying mechanism by which a previously uncharacterized mutation in the LDLR (H306Y) causes FH (27). FH-associated loss-of-function mutations in the LDLR represent one of the most common inborn errors of metabolism. More than 150 loss-of-function FH-associated LDLR alleles have been characterized at the molecular and cellular level. These mutations are divided into five classes based on the mechanism underlying the defect: class 1, null alleles; class 2, transport-defective alleles, which partially or completely fail to reach the cell surface; class 3, ligand binding-defective alleles; class 4, internalization-defective alleles, which fail to cluster in coated pits; and class 5, recycling-defective alleles (39).
Our studies provide the first description of an FH-associated LDLR allele (H306Y) that results in a molecular gain-of-function, i.e. enhanced LDLR binding to PCSK9. This increased affinity of PCSK9 to the LDLR would in turn lead to enhanced LDLR destruction, decreased plasma LDL-C clearance, and hypercholesterolemia. Accordingly, we propose that the H306Y LDLR allele be considered as a class 6 FH-associated LDLR allele. Patients with this novel type of LDLR mutation will be especially important to identify, as they would particularly benefit from anti-PCSK9 therapies.
Unlike other biological ligands of the LDLR, such as apoB and apoE that are released from the receptor in the acidic environment of the endosome, the binding affinity for PCSK9 increases substantially at acidic pH, which presumably contributes to the ability of PCSK9 to promote LDLR degradation in the lysosome (14, 23, 40). The structural data of Fig. 2 reveal that a conformational change occurs upon protonation of His-306 in the EGF-A repeat, allowing for the formation of a salt bridge with Asp-374 of PCSK9 that likely contributes to the increased affinity of PCSK9 for the LDLR at acidic pH. At neutral pH, His-306 is not protonated and is thus unable to form a hydrogen bond with Asp-374 of PCSK9, instead forming an intramolecular hydrogen bond with Ser-305. Similarly, H306Y in the EGF-A repeat of the LDLR adopts the acidic pH conformation, even at neutral pH, providing the molecular basis of the enhanced PCSK9 binding affinity caused by the H306Y mutation that ultimately results in increased LDLR degradation and hypercholesterolemia.
We took advantage of the increased binding affinity of the EGF-A H306Y mutation to address an unresolved issue in PCSK9 biology, whether PCSK9 functions primarily intracellularly to degrade LDLRs or as a secreted protein that acts on LDLRs at the cell surface. Support for an intracellular mechanism is derived from a study in which deletion of the autosomal recessive hypercholesterolemia (LDLRAP) gene, which encodes an adaptor protein required for LDLR endocytosis in hepatocytes, did not alter the ability of PCSK9 to degrade hepatic LDLRs when overexpressed in vivo (6). Maxwell et al. (41) also demonstrated that PCSK9 enhanced degradation of mature LDLR, and to a lesser extent a precursor form of the LDLR, in a post-ER compartment of HepG2 cells. Finally, Nassoury et al. (42) showed that the ER-localized proform of PCSK9 bound to the LDLR in the early secretory pathway when both proteins were overexpressed in cultured cells and that this interaction directed subsequent cellular trafficking of PCSK9. Support for an extracellular mechanism is derived from studies in which exogenous PCSK9, either present in conditioned medium or added in purified recombinant form, mediated LDLR degradation in the endosomal/lysosomal compartment of cells (15, 23, 40, 43). Studies using a parabiosis model (15) or direct PCSK9 infusion (26) show that PCSK9 in plasma is also capable of lowering liver LDLR levels in mice.
The ability of the recombinant EGF-AB(H306Y) peptide fragment to function as an antagonist that rescues expression of LDLRs in HepG2 cells stably overexpressing PCSK9 suggests that the binding of secreted PCSK9 to the LDLR EGF-A domain at the cell surface is the initiating event leading to eventual LDLR degradation in lysosomes (Fig. 3). Although the soluble full-length LDLR extracellular domain protein (44) or EGF-A containing peptides (45, 46) have been used to inhibit the ability of exogenously added PCSK9 to mediate LDLR degradation, these studies did not address whether the majority of PCSK9 activity was intracellular or extracellular.
EGF-AB(H306Y) also normalized LDLR protein levels and function in HepG2 cells overexpressing two gain-of-function PCSK9 mutants, D374Y or S127R, which are associated with hypercholesterolemia (Fig. 5). Co-crystallization studies showed that Asp-374 in PCSK9 forms a critical salt bridge with His-306 in the LDLR EGF-A domain (27). Based on this structural model, it is predicted that D374Y in PCSK9 would result in a more favorable hydrogen bond distance for this interaction, at least in part explaining an increase in LDLR binding affinity associated with the mutation (14, 15, 40). Despite the increase in affinity, the EGF-AB(H306Y) peptide still inhibited the ability of intracellularly expressed PCSK9(D374Y) to mediate LDLR degradation.
The mechanism by which the PCSK9(S127R) protein results in hypercholesterolemia has been suggested to be intracellular (47), because this amino acid substitution delays autocatalytic cleavage and secretion (17), and there is only a modest increase in affinity for the LDLR (14, 40). The inhibition studies shown in Fig. 5 indicate that PCSK9(S127R) primarily functions extracellularly to degrade the PCSK9-LDLR complex.
The current data provide initial evidence that PCSK9 functions predominantly as a secreted factor in mediating cellular LDLR degradation. Although our studies cannot completely exclude the existence of an intracellular pathway for PCSK9-mediated LDLR degradation, they do suggest that if such a pathway exists, it plays only a minor role in LDLR regulation. Consequently, the molecular and cellular characterization of the FH-associated H306Y LDLR allele not only provides further evidence of the central role of PCSK9 in controlling plasma LDL-C levels in humans, but it also supports that this role is mainly carried out by secreted PCSK9. This conclusion implies that inhibition of the extracellular interaction between PCSK9 and the LDLR with agents such as antibodies or peptides represents a valid therapeutic approach for the treatment of hypercholesterolemia.
While this manuscript was in preparation, Bottomley et al. (46) published studies reporting the co-crystal structure of the EGF-AB(H306Y) and PCSK9 and reached similar interpretations and conclusions.
Supplementary Material
Acknowledgments
We thank Maya Palnitkar, Lisa Henry, Y. K. Ho, Linda Donnelly, Norma Anderson, Tuyet Dang, Lisa Beatty, Shomanike Head, and Ijeoma Onwuneme for technical assistance; Hans Deisenhofer for advice and support; and David W. Russell for critical reading of the manuscript. Results shown in this study are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne National Laboratory is operated by University of Chicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02–06CH11357.
The atomic coordinates and structure factors (codes 3GCW and 3GCX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grants HL-38049 and HL-20948 (to J. D. H.). This work was also supported by The Perot Family Foundation.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Figs. S1–S3, and additional references.
Footnotes
The abbreviations used are: LDL-C, low density lipoprotein cholesterol; ER, endoplasmic reticulum; LDLR, low density lipoprotein receptor; PCSK9, proprotein convertase subtilisin/kexin type 9; EGF-A, epidermal growth factor-like repeat A; TFR, transferrin receptor; mAb, monoclonal antibody; ADH, autosomal dominant hypercholesterolemia; FH, familial hypercholesterolemia; ECD, extracellular domain; PBS, phosphate-buffered saline; DiI, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; LDL, low density lipoprotein; ELISA, enzyme-linked immunosorbent assay; CHX, cycloheximide; NCLPDS, newborn calf lipoprotein-deficient serum.
References
- 1.Goldstein, J. L., Brown, M. S., Anderson, R. G., Russell, D. W., and Schneider, W. J. (1985) Annu. Rev. Cell Biol. 1 1-39 [DOI] [PubMed] [Google Scholar]
- 2.Hobbs, H. H., Russell, D. W., Brown, M. S., and Goldstein, J. L. (1990) Annu. Rev. Genet. 24 133-170 [DOI] [PubMed] [Google Scholar]
- 3.Innerarity, T. L., Mahley, R. W., Weisgraber, K. H., Bersot, T. P., Krauss, R. M., Vega, G. L., Grundy, S. M., Friedl, W., Davignon, J., and McCarthy, B. J. (1990) J. Lipid Res. 31 1337-1349 [PubMed] [Google Scholar]
- 4.Abifadel, M., Varret, M., Rabes, J. P., Allard, D., Ouguerram, K., Devillers, M., Cruaud, C., Benjannet, S., Wickham, L., Erlich, D., Derre, A., Villeger, L., Farnier, M., Beucler, I., Bruckert, E., Chambaz, J., Chanu, B., Lecerf, J. M., Luc, G., Moulin, P., Weissenbach, J., Prat, A., Krempf, M., Junien, C., Seidah, N. G., and Boileau, C. (2003) Nat. Genet. 34 154-156 [DOI] [PubMed] [Google Scholar]
- 5.Maxwell, K. N., and Breslow, J. L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7100-7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park, S. W., Moon, Y.-A., and Horton, J. D. (2004) J. Biol. Chem. 279 50630-50638 [DOI] [PubMed] [Google Scholar]
- 7.Leren, T. P. (2004) Clin. Genet. 65 419-422 [DOI] [PubMed] [Google Scholar]
- 8.Timms, K. M., Wagner, S., Samuels, M. E., Forbey, K., Goldfine, H., Jammulapati, S., Skolnick, M. H., Hopkins, P. N., Hunt, S. C., and Shattuck, D. M. (2004) Hum. Genet. 114 349-353 [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J., Pertsemlidis, A., Kotowski, I. K., Graham, R., Garcia, C. K., and Hobbs, H. H. (2005) Nat. Genet. 37 161-165 [DOI] [PubMed] [Google Scholar]
- 10.Cohen, J. C., Boerwinkle, E., Mosley, T. H., and Hobbs, H. H. (2006) N. Engl. J. Med. 354 1264-1272 [DOI] [PubMed] [Google Scholar]
- 11.Zhao, Z., Tuakli-Wosornu, Y., Lagace, T. A., Kinch, L., Grishin, N. V., Horton, J. D., Cohen, J. C., and Hobbs, H. H. (2006) Am. J. Hum. Genet. 79 514-523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton, J. D., Cohen, J. C., and Hobbs, H. H. (2007) Trends Biochem. Sci. 2 71-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naureckiene, S., Ma, L., Sreekumar, K., Purandare, U., Lo, C. F., Huang, Y., Chiang, L. W., Grenier, J. M., Ozenberger, B. A., Jacobsen, J. S., Kennedy, J. D., DiStefano, P. S., Wood, A., and Bingham, B. (2003) Arch. Biochem. Biophys. 420 55-67 [DOI] [PubMed] [Google Scholar]
- 14.Cunningham, D., Danley, D. E., Geoghegan, K. F., Griffor, M. C., Hawkins, J. L., Subashi, T. A., Varghese, A. H., Ammirati, M. J., Culp, J. S., Hoth, L. R., Mansour, M. N., McGrath, K. M., Seddon, A. P., Shenolikar, S., Stutzman-Engwall, K. J., Warren, L. C., Xia, D., and Qiu, X. (2007) Nat. Struct. Mol. Biol. 14 413-419 [DOI] [PubMed] [Google Scholar]
- 15.Lagace, T. A., Curtis, D. E., Garuti, R., McNutt, M. C., Park, S. W., Prather, H. B., Anderson, N. N., Ho, Y. K., Hammer, R. E., and Horton, J. D. (2006) J. Clin. Investig. 116 2995-3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert, G., Ancellin, N., Charlton, F., Comas, D., Pilot, J., Keech, A., Patel, S., Sullivan, D. R., Cohn, J. S., Rye, K.-A., and Barter, P. J. (2008) Clin. Chem. 54 1038-1045 [DOI] [PubMed] [Google Scholar]
- 17.Benjannet, S., Rhainds, D., Essalmani, R., Mayne, J., Wickham, L., Jin, W., Asselin, M. C., Hamelin, J., Varret, M., Allard, D., Trillard, M., Abifadel, M., Tebon, A., Attie, A. D., Rader, D. J., Boileau, C., Brissette, L., Chretien, M., Prat, A., and Seidah, N. G. (2004) J. Biol. Chem. 279 48865-48875 [DOI] [PubMed] [Google Scholar]
- 18.Seidah, N. G., Benjannet, S., Wickham, L., Marcinkiewicz, J., Jasmin, S. B., Stifani, S., Basak, A., Prat, A., and Chretien, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 928-933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubuc, G., Chamberland, A., Wassef, H., Davignon, J., Seidah, N. G., Bernier, L., and Prat, A. (2004) Arterioscler. Thromb. Vasc. Biol. 24 1454-1459 [DOI] [PubMed] [Google Scholar]
- 20.Horton, J. D., Shah, N. A., Warrington, J. A., Anderson, N. N., Park, S. W., Brown, M. S., and Goldstein, J. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12027-12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong Jeong, H., Lee, H.-S., Kim, K.-S., Kim, Y.-K., Yoon, D., and Wook Park, S. (2008) J. Lipid Res. 49 399-409 [DOI] [PubMed] [Google Scholar]
- 22.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) J. Clin. Investig. 109 1125-1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, D.-W., Lagace, T. A., Garuti, R., Zhao, Z., McDonald, M., Horton, J. D., Cohen, J. C., and Hobbs, H. H. (2007) J. Biol. Chem. 282 18602-18612 [DOI] [PubMed] [Google Scholar]
- 24.McNutt, M. C., Lagace, T. A., and Horton, J. D. (2007) J. Biol. Chem. 282 20799-20803 [DOI] [PubMed] [Google Scholar]
- 25.Li, J., Tumanut, C., Gavigan, J.-A., Huang, W.-J., Hampton, E. N., Tumanut, R., Suen, K. F., Trauger, J. W., Spraggon, G., Lesley, S. A., Liau, G., Yowe, D., and Harris, J. L. (2007) Biochem. J. 406 203-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grefhorst, A., McNutt, M. C., Lagace, T. A., and Horton, J. D. (2008) J. Lipid Res. 49 1303-1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon, H. J., Lagace, T. A., McNutt, M. C., Horton, J. D., and Deisenhofer, J. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1820-1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudenko, G., Henry, L., Henderson, K., Ichtchenko, K., Brown, M. S., Goldstein, J. L., and Deisenhofer, J. (2002) Science 298 2353-2358 [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50, 760-76315299374 [Google Scholar]
- 31.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53, 240-255 [DOI] [PubMed] [Google Scholar]
- 32.Russell, D. W., Brown, M. S., and Goldstein, J. L. (1989) J. Biol. Chem. 264 21682-21688 [PubMed] [Google Scholar]
- 33.van Driel, I. R., Goldstein, J. L., Sudhof, T. C., and Brown, M. S. (1987) J. Biol. Chem. 262 17443-17449 [PubMed] [Google Scholar]
- 34.Brown, M. S., Dana, S. E., and Goldstein, J. L. (1974) J. Biol. Chem. 249 789-796 [PubMed] [Google Scholar]
- 35.Naoumova, R. P., Tosi, I., Patel, D., Neuwirth, C., Horswell, S. D., Marais, A. D., van Heyningen, C., and Soutar, A. K. (2005) Arterioscler. Thromb. Vasc. Biol. 25 2654-2660 [DOI] [PubMed] [Google Scholar]
- 36.Day, I. N., Whittall, R. A., O'Dell, S. D., Haddad, L., Bolla, M. K., Gudnason, V., and Humphries, S. E. (1997) Hum. Mutat. 10 116-127 [DOI] [PubMed] [Google Scholar]
- 37.Fouchier, S. W., Defesche, J. C., Umans-Eckenhausen, M. W., and Kastelein, J. P. (2001) Hum. Genet. 109 602-615 [DOI] [PubMed] [Google Scholar]
- 38.Goldstein, J. L., Basu, S. K., and Brown, M. S. (1983) Methods Enzymol. 98 241-260 [DOI] [PubMed] [Google Scholar]
- 39.Hobbs, H. H., Brown, M. S., and Goldstein, J. L. (1992) Hum. Mutat. 1 445-466 [DOI] [PubMed] [Google Scholar]
- 40.Fisher, T. S., Surdo, P. L., Pandit, S., Mattu, M., Santoro, J. C., Wisniewski, D., Cummings, R. T., Calzetta, A., Cubbon, R. M., Fischer, P. A., Tarachandani, A., De Francesco, R., Wright, S. D., Sparrow, C. P., Carfi, A., and Sitlani, A. (2007) J. Biol. Chem. 282 20502-20512 [DOI] [PubMed] [Google Scholar]
- 41.Maxwell, K. N., Fisher, E. A., and Breslow, J. L. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2069-2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nassoury, N., Blasiole, D. A., Tebon Oler, A., Benjannet, S., Hamelin, J., Poupon, V., McPherson, P. S., Attie, A. D., Prat, A., and Seidah, N. G. (2007) Traffic 8 718-732 [DOI] [PubMed] [Google Scholar]
- 43.Cameron, J., Holla, O. L., Ranheim, T., Kulseth, M. A., Berge, K. E., and Leren, T. P. (2006) Hum. Mol. Genet. 15 1551-1558 [DOI] [PubMed] [Google Scholar]
- 44.Qian, Y.-W., Schmidt, R. J., Zhang, Y., Chu, S., Lin, A., Wang, H., Wang, X., Beyer, T. P., Bensch, W. R., Li, W., Ehsani, M. E., Lu, D., Konrad, R. J., Eacho, P. I., Moller, D. E., Karathanasis, S. K., and Cao, G. (2007) J. Lipid Res. 48 1488-1498 [DOI] [PubMed] [Google Scholar]
- 45.Shan, L., Pang, L., Zhang, R., Murgolo, N. J., Lan, H., and Hedrick, J. A. (2008) Biochem. Biophys Res. Commun. 375 69-73 [DOI] [PubMed] [Google Scholar]
- 46.Bottomley, M. J., Cirillo, A., Orsatti, L., Ruggeri, L., Fisher, T. S., Santoro, J. C., Cummings, R. T., Cubbon, R. M., Lo Surdo, P., Calzetta, A., Noto, A., Baysarowich, J., Mattu, M., Talamo, F., De Francesco, R., Sparrow, C. P., Sitlani, A., and Carfi, A. (2008) J. Biol. Chem. 284 1313-1323 [DOI] [PubMed] [Google Scholar]
- 47.Homer, V. M., Marais, A. D., Charlton, F., Laurie, A. D., Hurndell, N., Scott, R., Mangili, F., Sullivan, D. R., Barter, P. J., Rye, K. A., George, P. M., and Lambert, G. (2008) Atherosclerosis 196 659-666 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.