Abstract
ARFRP1 and ARL1, which are both ARF-like small GTPases, are mammalian orthologs of yeast Arl3p and Arl1p, respectively. In yeast, Arl3p targeted to trans-Golgi network (TGN) membranes activates Arl1p, and the activated Arl1p in turn recruits a GRIP domain-containing protein; this complex regulates retrograde transport to the TGN and anterograde transport from the TGN. In the present study, using RNA interference-mediated knockdown of ARFRP1 and ARL1, we have examined whether the orthologs of Arl3p-Arl1p-GRIP story serve similar functions in mammalian cells. However, we have unexpectedly found differential roles of ARL1 and ARFRP1. Specifically, ARL1 and ARFRP1 regulate retrograde transport of Shiga toxin to the TGN and anterograde transport of VSVG from the TGN, respectively. Furthermore, we have obtained evidence suggesting that a SNARE complex containing Vti1a, syntaxin 6, and syntaxin 16 is involved in Shiga toxin transport downstream of ARL1.
The ARF/ARL3 family of small GTPases play crucial roles in membrane trafficking and in other cellular processes by interacting with various effector proteins (1, 2). The functions of ARF proteins in membrane traffic have been well established, whereas much less is known about the functions of ARL proteins. Accumulating lines of evidence in yeast, however, have suggested a cascade model in which N-acetylated Arl3p is targeted to the trans-Golgi network (TGN) membranes where it causes activation of Arl1p; activated Arl1p in turn recruits a GRIP (golgin-97/RanBP2/Imh1p/p230) domain-containing protein, Imh1p, and regulates retrograde transport from endosomes to the TGN via tethering of endosome-derived vesicles to the TGN and anterograde transport from the TGN to the plasma membrane (3–7).
It has been generally accepted that the Arl3p-Arl1p-GRIP cascade also occurs in mammalian cells. The mammalian orthologs of yeast Arl3p and Arl1p are ARFRP1 and ARL1, respectively. ARL1 has been shown to recruit GRIP domain-containing golgins, golgin-97 and golgin-245/p230/tGolgin-1, onto TGN membranes (8–11); it also participates in retrograde transport from endosomes to the TGN (10). Our laboratory and other groups previously showed, by expressing a dominant-negative mutant of ARFRP1, that the small GTPase functions upstream of ARL1 and the golgins and regulates anterograde traffic from and retrograde traffic to the TGN (11, 13). However, our subsequent experiments using the dominant-negative mutant of both ARL1 and ARFRP1 have suggested that these mutants might nonspecifically affect the recruitment of TGN-localizing proteins, and ARL1 might not necessarily act downstream of ARFRP1.
In the present study, we have therefore re-evaluated the roles of ARL1 and ARFRP1 by RNAi-mediated knockdown of these small GTPases. We have found that ARL1 predominantly regulates retrograde transport of Shiga toxin B fragment (StxB) to the TGN, whereas ARFRP1 regulates anterograde transport of vesicular stomatitis virus G protein (VSVG) from the TGN. Furthermore, our data have suggested that Q-SNAREs (syntaxin 6, syntaxin 16, and Vti1a) participate in the retrograde transport process by functioning downstream of ARL1.
EXPERIMENTAL PROCEDURES
Antibodies, Reagents, and Plasmids—Preparation and affinity purification of anti-ARL1 and anti-ARFRP1 antibodies were described previously (11). Polyclonal rabbit anti-golgin-97 antibody (14) was kindly provided by Nobuhiro Nakamura (Kanazawa University). Monoclonal mouse antibodies to golgin-245, GM130, GGA3, syntaxin 6, Vti1a, Vti1b, and CD4 were purchased from BD Biosciences. Polyclonal rabbit antibody to green fluorescent protein (GFP) was from Molecular Probes. Monoclonal mouse antibody to cation-independent mannose 6-phosphate receptor (CI-MPR) and polyclonal rabbit anti-β-COP antibody were from Affinity Bioreagents. Monoclonal rat anti-β′-COP antibody was from StressGen. Monoclonal rat anti-HA antibody was from Roche Applied Science. Polyclonal rabbit antibody to syntaxin 16 was from Synaptic Systems. AlexaFluor-conjugated secondary antibodies were from Molecular Probes. Recombinant StxB was purified as described previously (15) and conjugated with AlexaFluor488 using an AlexaFluor488 protein labeling kit (Molecular Probes).
An expression vector for untagged ARL1 or C-terminally HA-tagged ARL1 was constructed by subcloning the entire coding sequence of human ARL1 into the pCAG vector. An expression vector for N-terminally EGFP-tagged VSVG tsO45 (pCIpreEGFP-VSVG) was constructed by exchanging the mature VSVG tsO45 segment from pcDNA3-VSVG-EGFP (16), originally provided by Jennifer Lippincott-Schwartz (National Institutes of Health) (17), for the CI-MPR segment of pCIpreEGFP-CI-MPR tail (a kind gift from Satoshi Waguri, Fukushima Medical University) (18). Construction of an expression vector for C-terminally HA-tagged ARFRP1 was described previously (11). Construction of an expression vector for a CD4-furin fusion protein containing the exoplasmic domain of CD4 and the transmembrane and cytoplasmic domains of furin was described previously (19).
Cell Culture, RNAi Suppression, VSVG Transport Experiments, Antibody Uptake Experiments, and Immunofluorescence Analysis—Culture of HeLa cells and transfection of expression plasmids were performed as described previously (11, 20). A HeLa cell line stably expressing CD4-furin was established as described previously (21). Knockdown of ARFRP1, ARL1, Vti1a, Vti1b, or syntaxin 6 was performed as previously described (22). Briefly, pools of siRNAs directed for the mRNA regions covering nucleotide residues 1–546, 1–606, 1–540, 1–699, and 1–470, respectively (when the A residue of the initiation Met codon is assigned as residue 1) were prepared using a BLOCK-iT RNAi TOPO transcription kit and a BLOCK-iT Dicer RNAi kit (Invitrogen). Single siRNA oligonucleotides were synthesized at Dharmacon: GUGGAUGGUGAAGUGUGUC for ARFRP1 and AAGAAGAGCUGAGAAAAGCCA for ARL1. Control siRNA was purchased from Dharmacon. The cells were transfected with the siRNAs using Lipofectamine 2000 (Invitrogen) and incubated overnight. The transfected cells were then transferred to a culture dish containing coverslips, further incubated for 48 h, and processed for immunofluorescence and immunoblot analyses.
StxB uptake experiments were performed as described previously (11, 20, 21). Briefly, HeLa cells were mock treated (a pool of siRNAs for LacZ) or treated with siRNAs for ARFRP1, ARL1, Vti1a, Vti1b, or syntaxin 6 for 72 h and incubated with AlexaFluor488-conjugated StxB at 19 °C for 50 min. After medium change, the cells were incubated for 30 min at 37 °C and processed for immunofluorescence analysis. Uptake of anti-CD4 antibody by CD4-furin-expressing HeLa cells was analyzed as described previously (19, 20). The fluorescence intensity of StxB or CD4-furin that reached the Golgi region was quantified using IP-Lab4.0 software (Solution Systems, Japan). The Golgi structures containing GM130 or β′-COP were defined as regions of interest in more than 50 cells, and the pixel intensity of StxB or CD4-furin in the GM130 or β′-COP regions of interest was estimated. Transport of EGFP-VSVG from the Golgi to the cell surface was examined as described previously (11). HeLa cells transfected with pCIpreEGFP-VSVG were incubated at 40 °C overnight and then at 19 °C for 2 h to arrest the protein at the Golgi and finally incubated at 32 °C for up to 30 min. The cells were stained with anti-GFP antibody without permeabilization.
RESULTS
Dominant-negative Mutants of ARL1 Might Disorganize the TGN—We previously showed that exogenous expression of an HA-tagged dominant-negative ARFRP1 mutant, ARFRP1-(T31N)-HA, in HeLa cells causes redistribution of ARL1 and golgin-245 from the TGN (Fig. 1, A and A′, and supplemental Fig. S1) and blocks both exit of VSVG from the TGN (see supplemental Fig. S2) and retrograde transport of StxB from endosomes to the TGN (see supplemental Fig. S3). Furthermore, exogenous expression of dominant-negative ARL1, ARL1(T31N), redistributed golgin-245 (Fig. 1B and supplemental Fig. S1) and blocked transport of VSVG and StxB (see supplemental Figs. S2 and S3); note that we used an untagged version of the ARL1 mutant, because the expression efficiency of the mutant with any epitope tag sequence examined (HA, Myc, or FLAG) was extremely low for an unknown reason. Taken together with a previously proposed model in yeast (3–7), these observations suggested that ARFRP1 functioned upstream of ARL1 and GRIP domain-containing golgins and participated in both anterograde transport of VSVG and retrograde transport of StxB.
FIGURE 1.
Effects of overexpression of ARFRP1(T31N)-HA or ARL1(T31N) on the localization of Golgi proteins. HeLa cells were transfected with an expression vector for C-terminally HA-tagged ARFRP1(T31N) (A) or untagged ARL1(T31N) (B–D) and doubly stained with antibodies to HA and ARL1 (A and A′), golgin-245 and ARFRP1 (B and B′), ARL1 and GM130 (C and C′), or ARL1 and CI-MPR (D and D′). Cells overexpressing ARFRP1(T31N)-HA or ARL1(T31N) are indicated by asterisks; nontransfected cells are indicated by crosses. In B and B′, cells overexpressing ARL1(T31N) were indirectly identified by redistribution of golgin-245, because of antibody restriction.
However, we also made an observation suggesting that ARFRP1 may not function upstream of ARL1. As shown in Fig. 1 (B and B′), the ARL1(T31N) expression resulted in redistribution of not only golgin-245 but also ARFRP1 (cells indicated by asterisks); note that the cells expressing exogenous ARL1(T31N) were indirectly identified by redistribution of golgin-245, because double-staining for ARL1 and ARFRP1 was technically impossible because of antibody restriction. This observation was incompatible with the notion that ARFRP1 functions entirely upstream of ARL1. ARL1(T31N) expression also caused redistribution of CI-MPR (Fig. 1, D and D′), TGN46, and GGA3 (data not shown), all of which are markers of TGN, but not that of GM130, a cis-Golgi marker (Fig. 1, C and C′). It is therefore possible that the ARL1 mutant causes redistribution of ARFRP1 (Fig. 1B′) by disorganizing the structure of the TGN.
Depletion of ARL1 Results in Redistribution of Specific TGN Proteins—To prevent possible misinterpretation caused by overexpression of dominant-negative mutants, we next took an RNAi approach. As shown in Fig. 2A, immunoblot analysis demonstrated that treatment of HeLa cells with a pool of siRNAs for ARFRP1, which were prepared using a Dicer RNAi system (see “Experimental Procedures”), resulted in the significant depletion of ARFRP1 but did not affect the level of ARL1, golgin-97, or SNAREs, and a pool of siRNAs for ARL1 specifically knocked down ARL1. When examined by immunofluorescence analysis, the ARFRP1 siRNA treatment abolished Golgi-like staining for ARFRP1 (Fig. 2B). Inconsistent with the data obtained by overexpression of the dominant-negative ARFRP1 mutant (Fig. 1A and supplemental Fig. S1), however, the ARFRP1 knockdown did not change the Golgi localization of ARL1, golgin-245 (Fig. 2, C and D), or golgin-97 (data not shown). Treating cells with the ARL1 siRNAs abolished not only the ARL1 staining but also the staining for its direct effectors, golgin-245 (Fig. 2, F and G) and golgin-97 (data not shown), in the Golgi region. The localization of golgin-245 was restored by overexpression of wild-type ARL1 (not siRNA-resistant version) (Fig. 3A, panels a and a′, asterisks) but not ARFRP1 (Fig. 3A, panels b and b′, crosses), supporting the specificity of the RNAi. In contrast to the data obtained by the ARL1(T31N) overexpression (Fig. 1B′), however, the ARL1 knockdown did not cause redistribution of ARFRP1 from the Golgi region (Fig. 2E). A simple explanation for these observations is that ARL1 determines the TGN association of the GRIP domain-containing golgins, whereas ARFRP1 is not directly involved in the association of ARL1 or the golgins with TGN membranes.
FIGURE 2.
Specific knockdown of ARFRP1 and ARL1 by RNAi and its effect on localization of GRIP domain-containing golgins. HeLa cells were treated with a pool of siRNAs for ARFRP1 (A–D) or ARL1 (A and E–G) or control siRNAs (LacZ; A and H–J) as described under “Experimental Procedures.” A, immunoblot analysis of lysates of the cells with antibody to ARFRP1, ARL1, golgin-97, Vti1a, or syntaxin 6. B–J, immunofluorescence analysis of the cells with antibody to ARFRP1 (B, E, H, and inset of D), ARL1 (C, F, I, and inset of G), or golgin-245 (D, G, and J). The insets are zoom-out images of the cells stained with antibody to ARFRP1 (D) or ARL1 (G). Asterisks indicate knockdown cells.
FIGURE 3.
Recovery of localization of TGN proteins by exogenous ARL1-HA expression. HeLa cells treated with a pool of siRNAs for ARL1 were transfected with an expression vector for ARL1(WT)-HA (A and B) or ARFRP1(WT)-HA (A), and doubly stained with antibody to HA (A, panels a and b, and B, panels a–c) and golgin-245 (A, panels a′ and b′), Vti1a (B, panel a′), syntaxin 6 (B, panel b′), or syntaxin 16 (B, panel c′). Mislocalization of SNAREs and golgin-245 was rescued by expression of ARL1 (*) but not that of ARFRP1 (+) (A, panels b and b′).
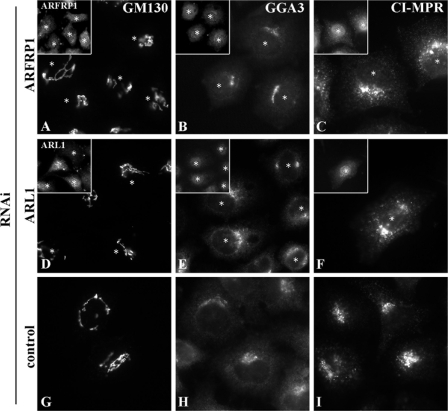
We next examined whether knocking down ARFRP1 or ARL1 affects the localization of other Golgi proteins. As shown in Fig. 4, the depletion of ARFRP1 or ARL1 did not significantly affect the localization of GM130 (cis-Golgi proteins) or GGA3 or CI-MPR (localized to the TGN and endosomes).
FIGURE 4.
Golgi proteins whose localization were not affected by knockdown of ARFRP1 or ARL1. HeLa cells treated with a pool of siRNAs for ARFRP1 (A–C) or ARL1 (D–F), or control siRNAs (G–I) were stained with antibody to GM130 (A, D, and G), GGA3 (B, E, and H) or CI-MPR (C, F, and I). The insets are zoom-out images of the cells stained with antibody to ARFRP1 (A–C) or ARL1 (E and F). The asterisks indicate knockdown cells.
However, the depletion of ARFRP1 and ARL1 differentially affected the localization of SNAREs that are involved in transport from endosomes to the TGN. Vti1a is a Q-SNARE that is mainly localized to the TGN and involved in retrograde transport of Stx from early/recycling endosomes to the TGN (23, 24), reviewed in (25, 26). As shown in Fig. 5, knocking down ARL1 (E) but not ARFRP1 (A) abolished the Vti1a staining in the TGN region. By contrast, the ARL1 knockdown did not significantly affect the localization of Vti1b (F), which is a close homolog of Vti1a and mainly associated with late endosomes and lysosomes (25, 26).
FIGURE 5.
Effect of knockdown of ARFRP1 and ARL1 on the localization of TGN SNAREs. HeLa cells treated with a pool of siRNAs for ARFRP1 (A–D) or ARL1 (E–H), or control siRNAs (I–L) were stained with antibody to Vti1a (A, E, and I), Vti1b (B, F, and J), syntaxin 6 (C, G, and K), or syntaxin 16 (D, H, and L). The insets are zoom-out images of the cells stained with antibody to ARFRP1 (A–C), ARL1 (E–G), or golgin-245 (H). The asterisks indicate knockdown cells.
Because Vti1a is known to form a SNARE complex together with syntaxin 6, syntaxin 16, and VAMP3/4, and to regulate retrograde transport from early/recycling endosomes (24), we also examined effects of the ARL1 knockdown on the localization of syntaxin 6 and syntaxin 16. As shown in Fig. 5G, the syntaxin 6 staining disappeared in the Golgi region and was dispersed more peripherally in the ARL1-depleted cells than in the control (K) and ARFRP1-depleted (C) cells. The syntaxin 16 staining also disappeared in the Golgi region in the ARL1-depleted cells (Fig. 5H); note that the ARL1-depleted cells were indirectly identified by redistribution of golgin-245, because double staining for ARL1 and syntaxin 16 was technically impossible because of antibody restriction. The ARFRP1-depleted cells cannot be identified for the same reason (Fig. 5D). The syntaxin 16 localization is almost intact (more than 80%) in the ARFRP1-depleted cells. Mislocalization of Vti1a, syntaxin 6, and syntaxin 16 can be rescued by overexpression of wild-type ARL1 (Fig. 3B). Thus, ARL1 depletion appears to affect the localization of a specific set of SNAREs involved in retrograde transport from endosomes to the TGN.
Differential Effects of Depletion of ARFRP1 and ARL1 on Anterograde and Retrograde Transport—Because overexpression of the dominant-negative mutant of either ARFRP1 or ARL1 blocks exit of VSVG from the TGN (supplemental Fig. S2) and retrograde transport of StxB from endosomes to the TGN (supplemental Fig. S3), we next examined the effects of knockdown of either ARFRP1 or ARL1 on these transport processes. As shown in Fig. 6, in the control (C–C″) and ARFRP1-depleted (A–A″) cells, significant fractions of StxB were transported from endosomes to the Golgi region by 30 min of incubation at 37 °C. However, the knock-down of ARL1 significantly inhibited transport of StxB, most of which was arrested at punctate endosomal structures (B–B″).
FIGURE 6.
Inhibition of StxB transport from endosomes to the Golgi by ARL1 depletion. HeLa cells treated with siRNAs for ARFRP1 (A), ARL1 (B), or control siRNAs (C) were incubated with AlexaFluor488-conjugated StxB at 19 °C for 50 min to accumulate StxB at early/recycling endosomes and then at 37 °C for 30 min to allow its transport toward the Golgi. The cells were then stained with anti-GM130 antibody to identify the Golgi region (A′–C′). Merged images of StxB and GM130 are shown in A″–C″. D, pixel intensity of AlexaFluor488-StxB in GM130-positive regions was estimated as described under “Experimental Procedures.” The values are the means of more than 50 cells from two independent experiments.
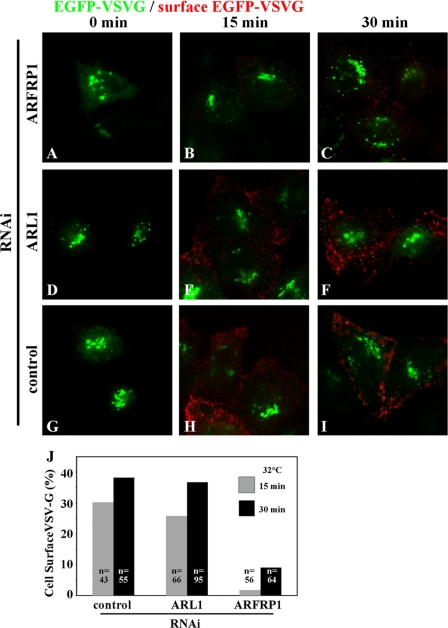
In contrast, VSVG transport from the TGN to the cell surface was significantly retarded in cells depleted of ARFRP1 but not in those depleted of ARL1 (Fig. 7); EGFP-VSVG molecules that reached the cell surface were detected by incubating fixed cells with anti-GFP antibody under nonpermeabilized conditions (see “Experimental Procedures”).
FIGURE 7.
Inhibition of VSVG exit from the TGN by ARFRP1 depletion. HeLa cells transfected with an expression vector for N-terminally EGFP-tagged VSVG tsO45 were treated with siRNAs for ARFRP1 (A–C), ARL1 (D–F), or control siRNAs (G–I) and incubated at 40 °C overnight and then at 19 °C for 2 h to accumulate EGFP-VSVG at the Golgi (0 min; A, D, and G). The temperature was then shifted to 32 °C for 15 min (B, E, and H) or 30 min (C, F, and I) to allow EGFP-VSVG transport toward the plasma membrane. To detect cell surface expression of EGFP-VSVG, the cells were stained with anti-GFP antibody under nonpermeabilized conditions (red). The cells expressing EGFP-VSVG were identified by the epifluorescence of EGFP (green). The images are representative of three independent experiments. J, percentages of cells with cell surface expression of EGFP-VSVG in total EGFP-VSVG-expressing cells were estimated after temperature shift to 32 °C for 15 or 30 min. n, numbers of cells counted.
The results presented so far were obtained using cells treated with pools of siRNAs that were prepared using a Dicer RNAi system. To corroborate that the observed block in transport of StxB and VSVG indeed resulted from specific depletion of ARL1 and ARFRP1, respectively, but not from off-target effects, we also examined the effects of single siRNA oligonucleotide for ARL1 and that for ARFRP1 on transport of StxB and VSVG, respectively. As shown in supplemental Fig. S4, treatment of cells with the ARL1 siRNA caused specific reduction in the ARL1 protein and a block in retrograde transport of StxB to the Golgi. Treating cells with the ARFRP1 siRNA specifically retarded the VSVG transport to the cell surface (supplemental Fig. S4).
StxB is transported from the plasma membrane to the Golgi via early and recycling endosomes (24, 27). We next examined whether ARL1 depletion affected another retrograde transport process, furin transport via late endosomes to the TGN (28, 29). To this end, we made use of a HeLa cell line that stably expresses CD4-furin (21), which is a chimeric protein with the exoplasmic domain of CD4 and the transmembrane and cytoplasmic domains of furin (19), and examined its retrograde transport by following extracellularly added anti-CD4 antibody as described previously (19, 21, 28, 29). As shown in supplemental Fig. S5, however, the retrograde transport of the furin construct from endosomes to the TGN was not significantly altered by the ARL1 depletion. The internalized CD4-furin colocalizes well with a Golgi marker, β′-COP, after 90 min of chase in control as well as in ARL1 knockdown cells. As we showed previously, the knockdown of BIG1 and BIG2 inhibited the CD4-furin transport (21). Therefore, depletion of ARL1 inhibits the retrograde transport of StxB from early/recycling endosomes but not that of CD4-furin from late endosomes to the Golgi.
Depletion of Vti1a or Syntaxin 6 Blocks Retrograde Transport of StxB—Taking into account our observations that knocking down ARL1 redistributed Vti1a and syntaxin 6 (Fig. 5) and blocked the retrograde transport of StxB (Fig. 6), we next asked whether knockdown of these Q-SNAREs also affect StxB transport. For this purpose, we first examined knockdown efficiency of these SNARE proteins. As shown in Fig. 8A, immunoblot analysis confirmed specific depletion of Vti1a and syntaxin 6 by treatment of HeLa cells with corresponding siRNAs. When examined by immunofluorescence analysis, RNAi for syntaxin 6 specifically abolished its Golgi-like staining; the staining for Vti1a, Vti1b, golgin-245, ARFRP1, or ARL1 was not significantly affected (Fig. 8, H–M). When the cells were treated with siRNAs for Vti1a, the Golgi-like staining for not only Vti1a (Fig. 8B) but also syntaxin 6 (Fig. 8C) disappeared. In this context, it is noteworthy that syntaxin 6 localization has been reported to be also perturbed in syntaxin 16 knockdown cells (30, 31). These observations together suggest that the majority of syntaxin 6 is complexed with Vti1a and syntaxin 16 and thereby stabilized on the membrane. Knocking down Vti1b did not affect the localization of any proteins examined other than Vti1b itself (N-S).
FIGURE 8.
Knockdown of Vti1a, syntaxin 6, and Vti1b by RNAi. HeLa cells were treated with a pool of siRNAs for Vti1a (A–G), syntaxin 6 (A and H–M), or Vti1b (A and N–S), or control siRNAs (A and T–Y), as described under “Experimental Procedures.” A, immunoblot analysis of lysates of the cells with antibody to Vti1a, Vti1b, syntaxin 6, or golgin-97. B–Y, immunofluorescence of the cells using antibody to Vti1a (B, H, N, and T), syntaxin 6 (C, I, O, and U), Vti1b (D, J, P, and V), golgin-245 (E, K, Q, and W), ARFRP1 (F, L, R, and X), or ARL1 (G, M, S, and Y).
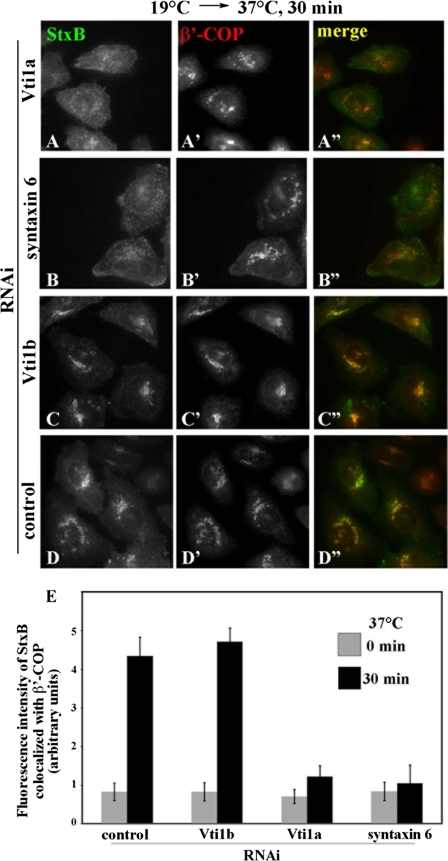
We then examined the effects of knocking down these SNAREs on the StxB transport (Fig. 9). As expected, retrograde transport of StxB from endosomes to the Golgi was significantly inhibited in cells depleted of Vti1a (Fig. 9A) or syntaxin 6 (Fig. 9B) but not affected in the control cells (Fig. 9D) or in those depleted of Vti1b (Fig. 9C). The block in the retrograde StxB transport is unlikely to be an indirect consequence of a block in anterograde Golgi trafficking, because VSVG transport was unaffected by syntaxin 6 knockdown (data not shown), like previously reported for syntaxin 16 knockdown (30). Taken together with the data from the ARL1 knockdown, these observations make it likely that ARL1 participates in StxB retrograde transport from early/recycling endosomes to the TGN by regulating the localization or function of Vti1a and syntaxin 6. It is also possible, however, that ARL1 and the SNAREs are independently involved in the transport process.
FIGURE 9.
Inhibition of StxB transport from endosomes to the Golgi by depletion of Vti1a or syntaxin 6. HeLa cells treated with siRNAs for Vti1a (A), syntaxin 6 (B), Vti1b (C), or control siRNAs (D) were examined for transport of AlexaFluor488-StxB from early/recycling endosomes to the Golgi as described in the legend for Fig. 6. The cells were then stained with anti-β′-COP antibody to identify the Golgi region (A′–D′). The images are representative of three independent experiments. The merged images of StxB and β′-COP are shown in A″–D″. E, pixel intensity of AlexaFluor488-StxB in the β′-COP-positive regions was estimated as described under “Experimental Procedures.” The values are the means of more than 50 cells from two independent experiments.
DISCUSSION
In the present study, we have revealed two important points regarding the roles of ARL1 and ARFRP1 in mammalian cells. First, ARL1 and ARFRP1 have distinct roles in membrane trafficking; ARL1 preferentially regulates the retrograde transport of StxB from early/recycling endosomes to the TGN, whereas ARFRP1 participates predominantly in the transport of VSVG from the TGN. Second, ARL1 regulates the localization and/or recycling of Vti1a, syntaxin 6, and syntaxin 16, and these Q-SNAREs participate in the StxB retrograde transport.
In previous studies by our laboratory and other groups, a dominant-negative ARFRP1 mutant, ARFRP1(T31N), was overexpressed. The results of these studies suggested that ARFRP1 serves as an upstream regulator of ARL1, indirectly determines TGN association of GRIP domain-containing golgins, and regulates the anterograde transport of VSVG from the TGN as well as the retrograde transport of Shiga toxin from endosomes to the TGN (5, 11, 13). Because overexpression of a dominant-negative ARL1 mutant results in essentially the same phenotypes as those of the ARFRP1(T31N) overexpression (9, 10), we previously concluded that yeast and mammalian cells use the same Arl3p/ARFRP1-Arl1p/ARL1-GRIP cascade to regulate both retrograde transport to and anterograde transport from the TGN. However, RNAi-mediated knockdown of ARFRP1 and ARL1 in the current study has revealed that the ARFRP1 depletion does not affect the TGN association of ARL1 or GRIP domain-containing golgins and that the ARL1 depletion blocks StxB retrograde transport but not anterograde VSVG transport, whereas ARFRP1 depletion inhibits anterograde but not retrograde transport. A possible explanation for the apparent inconsistency between the data obtained by the overexpression and knockdown studies is that the overexpression of these dominant-negative mutants could indirectly affect transport through the TGN by sequestering their binding partners or inducing an alteration in TGN organization. Another possibility is that other ARF family GTPase(s) could function as an upstream regulator of ARL1, thereby compensating for the ARFRP1 deficiency in the knockdown cells. We think this is unlikely, however, because ARL3, which is the mammalian ARF GTPase most similar to ARFRP1 and shares ∼42% of its amino acids with ARFRP1 (32), associates mainly with centrosomes (33). In support of our data on nonoverlapping functions of ARFRP1 and ARL1, Schürmann and co-workers (34) recently reported by knockdown experiments that ARFRP1, but not ARL1, regulates E-cadherin transport from the Golgi to the plasma membrane.
Schürmann and co-workers (13) previously reported that depletion of ARFRP1 using shRNA prevented the Golgi association of ARL1. We have taken essentially the same approach using the same ARFRP1 shRNA construct but failed to obtain similar results. We have found that, after depletion of ARFRP1 using the shRNA, the cell viability decreased dramatically, and even in a few surviving cells the Golgi structure itself was disrupted. Therefore, it is difficult to determine whether the depletion of ARFRP1 inhibited membrane recruitment of ARL1 and golgins under the experimental conditions employed by Schürmann and co-workers (35). The discrepancy between the siRNA- and shRNA-treated cells in the viability might come from the knockdown efficiency. The almost complete depletion of ARFRP1 by the shRNA probably affect the cell viability, which reminds us that the knock-out of ARFRP1 was embryonic lethal; the embryonic ectoderm underwent apoptotic cell death during gastrulation. In contrast, the ARFRP1 depletion using siRNAs may be less efficient than that using the shRNA but enough to inhibit VSVG transport. Although the lack of apparent effects of the ARFRP1 siRNA treatment on Golgi association of ARL1 (Fig. 2C) or on StxB transport (Fig. 6 and supplemental Fig. S4) might be attributable to incomplete depletion of ARFRP1 (Fig. 2A and supplemental Fig. S4), treating cells with the ARFRP1 siRNAs did retard VSVG transport from the TGN to the cell surface (Fig. 7 and supplemental Fig. S4). Therefore, ARFRP1 seems to have a preferential role in the VSVG transport. While the present study was in progress, Schürmann and co-workers (34) reported that the absence of ARFRP1 induces the disorganization of trans-Golgi and suggested that the redistribution of ARL1 and golgins from the TGN region in ARFRP1 knock-out cells is attributable to the disorganization of the TGN itself, supporting our view.
In the present study, we have shown that depleting ARL1 leads to delocalization of Q-SNAREs, Vti1a, syntaxin 6, and syntaxin 16, which (together with the VAMP3/4 R-SNARE) are able to form a SNARE complex (Ref. 24; reviewed in Refs. 25 and 26). A previous study showed that addition of antibody against either of these Q-SNAREs to a permeabilized cell system or overexpression of the cytoplasmic domain of either of these Q-SNAREs in intact cells specifically inhibited StxB transport (24). More recently, RNAi-mediated knockdown of syntaxin 16 was shown to inhibit StxB transport (30, 36). Consistent with these previous results, we have shown that depletion of Vti1a or syntaxin 6 also inhibits StxB retrograde transport. Although recent work showed that depletion of syntaxin 6 had no apparent affect on StxB retrograde transport (37), our finding that depletion of syntaxin 6 inhibits the StxB retrograde transport is consistent with the previous result of reconstituted transport assay (24).
What connects ARL1 and the Q-SNAREs? It is most likely that GRIP domain-containing golgins mediate between them; this model is supported by the following observations: (i) Lu et al. reported that a variety of conditions (addition of anti-golgin-97 antibody in a permeabilized cell system, overexpression of the golgin-97 GRIP domain, RNAi-mediated golgin-97 knockdown, or microinjection of anti-golgin-97 antibody) all inhibited StxB retrograde transport (10); (ii) Yoshino et al. (38) reported that RNAi-mediated knockdown of golgin-245/p230/t-Golgin-1 inhibited StxB transport; and (iii) Gleeson and co-workers (37, 39) reported that knocking down GCC185, but not GCC88, impaired StxB transport, although GCC185 and GCC88 were reported to bind poorly to ARL1 (40). Thus, the impairment of each golgin blocks StxB transport, suggesting that these golgins are unable to compensate for one another even though they are all under the regulation of ARL1. It is therefore a critical issue to examine whether knockdown of these golgins affects the localization of Vti1a, syntaxin 6 and/or syntaxin 16. Thus far, however, our attempts have been unsuccessful because impairing golgins results in fragmentation of the Golgi structures as reported previously (10, 38); therefore, we have not unequivocally determined the localization of these Q-SNAREs.
In the context of golgin-SNARE interactions, it is worth mentioning recent reports of Pfeffer and co-workers (31, 41). Their data suggested that syntaxin 10 (the SNARE most similar to syntaxin 6), in complex with syntaxin 16 and Vti1a, mediates transport of CI-MPR from late endosomes. In contrast, a complex containing syntaxin 6, syntaxin 16, and Vti1a mediates transport from early endosomes of the cholera toxin B subunit, which takes the same transport route as StxB; GCC185 participates in the former transport (12, 31). Furthermore, they showed that ARL1 recruits GCC185 to Golgi membranes in concert with Rab6, although ARL1 alone binds poorly to GCC185, and that GCC185 binds to syntaxin 16 (41).
The next challenge will be to determine whether other golgins are able to interact with SNAREs that are important for the transport from early/recycling endosomes to the Golgi. Future experiments will seek to elucidate the exact mechanism by which two different retrograde transport pathways (one from early/recycling endosomes, the other is from late endosomes) are regulated by coordination of tethering molecules and SNARE complexes.
Supplementary Material
Acknowledgments
We thank Nobuhiro Nakamura, Jennifer Lippincott-Schwartz, and Satoshi Waguri for kindly providing materials.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Japan Society for Promotion of Science, the Protein 3000 Project, the Targeted Proteins Research Program, the International Young Scientists Career Development Organization of Kyoto University, the Uehara Memorial Foundation, and the NOVARTIS Foundation (Japan) for the Promotion of Science, and the Hayashi Memorial Foundation for Female Natural Scientists.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: ARF, ADP-ribosylation factor; ARL, ARF-like; ARFRP, ARF-related protein; CI-MPR, cation-independent mannose 6-phosphate receptor; GFP, green fluorescent protein; RNAi, RNA interference; siRNA, small interfering RNA; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; StxB, Shiga toxin B fragment; TGN, trans-Golgi network; VSVG, vesicular stomatitis virus G protein; HA, hemagglutinin; shRNA, small hairpin RNA.
References
- 1.Burd, C. G., Strochlic, T. I., and Setty, S. R. G. (2004) Trends Cell Biol. 14 687-694 [DOI] [PubMed] [Google Scholar]
- 2.D'Souza-Schorey, C., and Chavrier, P. (2006) Nat. Rev. Mol. Cell Biol. 7 347-358 [DOI] [PubMed] [Google Scholar]
- 3.Panic, B., Whyte, J. R. C., and Munro, S. (2003) Curr. Biol. 13 405-410 [DOI] [PubMed] [Google Scholar]
- 4.Setty, S. R. G., Strochlic, T. I., Tong, A. H. Y., Boone, C., and Burd, C. G. (2004) Nat. Cell Biol. 6 414-419 [DOI] [PubMed] [Google Scholar]
- 5.Behnia, R., Panic, B., Whyte, J. R. C., and Munro, S. (2004) Nat. Cell Biol. 6 405-413 [DOI] [PubMed] [Google Scholar]
- 6.Setty, S. R. G., Shin, M. E., Yoshino, A., Marks, M. S., and Burd, C. G. (2003) Curr. Biol. 13 401-404 [DOI] [PubMed] [Google Scholar]
- 7.Liu, Y.-W., Lee, S.-W., and Lee, F.-J. S. (2006) J. Cell Sci. 119 3845-3855 [DOI] [PubMed] [Google Scholar]
- 8.Van Valkenburgh, H., Shern, J. F., Sharer, J. D., Zhu, X., and Kahn, R. A. (2001) J. Biol. Chem. 276 22826-22837 [DOI] [PubMed] [Google Scholar]
- 9.Lu, L., and Hong, W. (2003) Mol. Biol. Cell 14 3767-3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu, L., Tai, G., and Hong, W. (2004) Mol. Biol. Cell 15 4426-4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin, H.-W., Kobayashi, H., Kitamura, M., Waguri, S., Suganuma, T., Uchiyama, Y., and Nakayama, K. (2005) J. Cell Sci. 118 4039-4048 [DOI] [PubMed] [Google Scholar]
- 12.Reddy, J. V., Schweizer-Burguete, A., Sridevi, K., Ganley, I. G., Nottingham, R. M., and Pfeffer, S. R. (2006) Mol. Biol. Cell 17 4353-4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zahn, C., Hommel, A., Lu, L., Hong, W., Walther, K., Florian, S., Joost, H.-G., and Schürmann, A. (2006) Mol. Membr. Biol. 23 475-485 [DOI] [PubMed] [Google Scholar]
- 14.Yoshimura, S., Yamamoto, A., Misumi, Y., Sohda, M., Barr, F. A., Fujii, G., Shakoori, A., Ohno, H., Mihara, K., and Nakamura, N. (2004) J. Biochem. (Tokyo) 135 201-216 [DOI] [PubMed] [Google Scholar]
- 15.Johannes, L., Tenza, D., Antony, C., and Goud, B. (1997) J. Biol. Chem. 272 19554-19561 [DOI] [PubMed] [Google Scholar]
- 16.Kasai, K., Shin, H.-W., Shinotsuka, C., Murakami, K., and Nakayama, K. (1999) J. Biochem. (Tokyo) 125 780-789 [DOI] [PubMed] [Google Scholar]
- 17.Presley, J. F., Cole, N. B., Schroer, T. A., Hirschberg, K., Zaal, K. J. M., and Lippincott-Schwartz, J. (1997) Nature 389 81-85 [DOI] [PubMed] [Google Scholar]
- 18.Waguri, S., Dewitte, F., Le Borgne, R., Rouillé, Y., Uchiyama, Y., Dubremetz, J.-F., and Hoflack, B. (2003) Mol. Biol. Cell 14 142-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi, S., Nakagawa, T., Banno, T., Watanabe, T., Murakami, K., and Nakayama, K. (1995) J. Biol. Chem. 270 28397-28401 [DOI] [PubMed] [Google Scholar]
- 20.Shin, H.-W., Morinaga, N., Noda, M., and Nakayama, K. (2004) Mol. Biol. Cell 15 5283-5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishizaki, R., Shin, H.-W., Mitsuhashi, H., and Nakayama, K. (2008) Mol. Biol. Cell 19 2650-2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishizaki, R., Shin, H.-W., Iguchi-Ariga, S. M. M., Ariga, H., and Nakayama, K. (2006) Genes Cells 11 949-959 [DOI] [PubMed] [Google Scholar]
- 23.Kreykenbohm, V., Wenzel, D., Antonin, W., Atlachkine, V., and Fischer von Mollard, F. (2002) Eur. J. Cell Biol. 81 273-280 [DOI] [PubMed] [Google Scholar]
- 24.Mallard, F., Tang, B. L., Galli, T., Tenza, D., Saint-Pol, A., Yue, X., Antony, C., Hong, W., Goud, B., and Johannes, L. (2002) J. Cell Biol. 156 653-664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, W. (2005) Biochim. Biophys. Acta 1744 120-144 [DOI] [PubMed] [Google Scholar]
- 26.Jahn, R., and Scheller, R. H. (2006) Nat. Rev. Mol. Cell Biol. 7 631-643 [DOI] [PubMed] [Google Scholar]
- 27.Mallard, F., Antony, C., Tenza, D., Salamero, J., Goud, B., and Johannes, L. (1998) J. Cell Biol. 143 973-990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallet, W. G., and Maxfield, F. R. (1999) J. Cell Biol. 146 345-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schapiro, F. B., Soe, T. T., Mallet, W. G., and Maxfield, F. R. (2004) Mol. Biol. Cell 15 2884-2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Y., Tai, G., Johannes, L., Hong, W., and Tang, B. L. (2005) Mol. Membr. Biol. 22 313-325 [DOI] [PubMed] [Google Scholar]
- 31.Ganley, I. G., Espinosa, E., and Pfeffer, S. R. (2008) J. Cell Biol. 180 159-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kahn, R. A., Cherfils, J., Elias, M., Lovering, R. C., Munro, S., and Schurmann, A. (2006) J. Cell Biol. 172 645-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou, C., Cunningham, L., Marcus, A. I., Li, Y., and Kahn, R. A. (2006) Mol. Biol. Cell 17 2476-2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahn, C., Jaschke, A., Weiske, J., Hommel, A., Hesse, D., Augustin, R., Lu, L., Hong, W., Florian, S., Scheepers, A., Joost, H.-G., Huber, O., and Schürmann, A. (2008) J. Biol. Chem. 283 27179-27188 [DOI] [PubMed] [Google Scholar]
- 35.Mueller, A. G., Moser, M., Kluge, R., Leder, S., Blum, M., Büttner, R., Joost, H.-G., and Schürmann, A. (2002) Mol. Cell Biol. 22 1488-1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amessou, M., Fradagrade, A., Falguières, T., Lord, J. M., Smith, D. C., Roberts, L. M., Lamaze, C., and Johannes, L. (2007) J. Cell Sci. 120 1457-1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieu, Z. Z., Derby, M. C., Teasdale, R. D., Hart, C., Gunn, P., and Gleeson, P. A. (2007) Mol. Biol. Cell 18 4979-4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshino, A., Setty, S. R. G., Poynton, C., Whiteman, E. L., Saint-Pol, A., Burd, C. G., Johannes, L., Holzbaur, E. L., Koval, M., McCaffery, J. M., and Marks, M. S. (2005) J. Cell Sci. 118 2279-2293 [DOI] [PubMed] [Google Scholar]
- 39.Derby, M. C., Lieu, Z. Z., Brown, D., Stow, J. L., Goud, B., and Gleeson, P. A. (2007) Traffic 8 758-773 [DOI] [PubMed] [Google Scholar]
- 40.Derby, M. C., van Vliet, C., Brown, D., Luke, M. R., Lu, L., Hong, W., Stow, J. L., and Gleeson, P. A. (2004) J. Cell Sci. 117 5865-5874 [DOI] [PubMed] [Google Scholar]
- 41.Schweizer Burguete, A., Fenn, T. D., Brunger, A. T., and Pfeffer, S. R. (2008) Cell 132 286-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.