Abstract
The processive Serratia marcescens chitinases A (ChiA) and B (ChiB) are thought to degrade chitin in the opposite directions. A recent study of ChiB suggested that processivity is governed by aromatic residues in the +1 and +2 (aglycon) subsites close to the catalytic center. To further investigate the roles of aromatic residues in processivity and to gain insight into the structural basis of directionality, we have mutated Trp167, Trp275, and Phe396 in the -3, +1, and +2 subsites of ChiA, respectively, and characterized the hydrolytic activities of the mutants toward β-chitin and the soluble chitin-derivative chitosan. Although the W275A and F396A mutants showed only modest reductions in processivity, it was almost abolished by the W167A mutation. Thus, although aglycon subsites seem to steer processivity in ChiB, a glycon (-3) subsite seems to be adapted to do so in ChiA, in line with the notion that the two enzymes have different directionalities. Remarkably, whereas all three single mutants and the W167A/W275A double mutant showed reduced efficiency toward chitin, they showed up to 20-fold higher activities toward chitosan. These results show that the processive mechanism is essential for an efficient conversion of crystalline substrates but comes at a large cost in terms of intrinsic enzyme speed. This needs to be taken into account when devising enzymatic strategies for biomass turnover.
Enzymatic degradation of recalcitrant polysaccharides in biomass, such as cellulose (β-1,4-linked glucoses) and the cellulose derivative chitin (β-1,4-linked N-acetylglucosamines), is of great biological and economical importance. The annual production of these biopolymers is around one trillion and 100 billion tons, respectively (1, 2), which makes them an almost unlimited source of raw material for environmentally friendly and biocompatible products, as well as biofuels. The development of sufficiently effective enzymes for the conversion of insoluble recalcitrant polysaccharides is one of the key issues in current research on biomass conversion, in particular in relation to production technology for cellulosic ethanol (3, 4). Chitin is the main component in the exoskeletons of invertebrates, the cell walls of fungi, and the digestive tracts of insects. Degradation of chitin and its soluble derivative chitosan could, if efficient enzyme technology were available, give bioactive chito-oligoosaccharides, with a number of potential applications in agriculture and medicine (5–8).
Because of the crystalline and inaccessible nature of cellulose and chitin, enzymes have developed special tactics to ensure efficient degradation. In addition to their catalytic domains, cellulases and chitinases often have one or multiple carbohydrate-binding modules that are beneficial for enzyme efficiency because they adhere to and sometimes disrupt the substrate (9–14). Furthermore, chitin-degrading organisms produce accessory proteins that disrupt the crystalline substrate, thus increasing the efficiency of chitinases (15, 16). Similar proteins may also exist for degradation of cellulose (17).
Another common feature of enzymes acting on crystalline substrates is the ability to remain attached to their substrates in between subsequent hydrolytic reactions, called processive action (18, 19). Processive degradation is thought to improve catalytic efficiency because single polymer chains are prevented from reassociating with the insoluble material in between catalytic cycles (20, 21), thus reducing the number of times the enzyme has to carry out the energetically unfavorable process of gaining access to a single chain. Processive enzymes often have long and deep substrate-binding clefts as illustrated by the first structures of processive cellulases (19, 22). There are, however, exceptions, such as the processive Cel9A cellulase from Thermobifida fusca, which has a relatively open and shallow active site cleft (23, 24). The substrate-binding sites in processive chitinases and cellulases are lined with aromatic residues, in particular tryptophan residues. These residues are thought to facilitate processivity by functioning as a flexible and hydrophobic sheath along which the polymer chain can slide during the processive mode of action (see Figs. 1 and 2 and Refs. 25 and 26). The hydrophobic interactions are nonspecific and concern large interaction surfaces, thus providing strong but “fluid” binding while avoiding tight binding at any specific sites that could hamper the sliding movement (27, 28). There is little experimental data in support of the idea that processivity is important for enzyme efficiency toward cellulose, which may be due to the fact that it is difficult to address this phenomenon experimentally when working with crystalline substrates (29). Recently it was shown that processivity of family 18 chitinases can be accurately assessed using the water-soluble polymeric chitin derivative chitosan, which is produced by partial deacetylation of chitin (29–31).
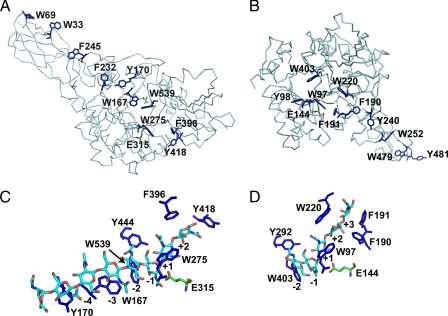
FIGURE 1.
Position of aromatic residues and enzyme-substrate interactions in ChiA and B from S. marcescens. A and B show the α-carbon chain of the complete ChiA (A; Protein Data Bank code 1EDQ) and ChiB (B; Protein Data Bank code 1E15) enzyme as well as the aromatic side chains that line the substrate-binding cleft of the catalytic domain and the surface of the chitin-binding domain (which points to the upper left in A and to the lower right in B). The catalytic domains are oriented in approximately the same way. C and D show the details of the crystal structure of ChiA-E315Q in complex with (GlcNAc)8 (C; Protein Data Bank code 1EHN) and ChiB-E144Q in complex with (GlcNAc)5 (D; Protein Data Bank code 1E6N), with aromatic amino acids in dark blue and the GlcNAc oligomer in light blue (carbon), dark blue (nitrogen), and red (oxygen). In all of the panels, the side chain of the catalytic acid (Glu315 in ChiA and Glu144 in ChiB) is shown in green (in C and D, the catalytic acid is shown for illustration purposes). The numbers indicate the subsite to which the sugar above is bound. In ChiA (47, 62), the chitin-binding domain (comprising residues 24–137) extends the substrate-binding cleft toward the nonreducing end of the substrate; dimeric products are thought to be released from subsites +1 and +2. In ChiB (37, 48), the chitin-binding domain (comprising residues 451–499) extends the substrate-binding cleft toward the reducing end of the substrate; dimeric products are thought to be released from subsites -1 and -2.
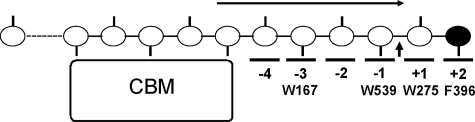
FIGURE 2.
Schematic picture of ChiA from S. marcescens in complex with a single chitin chain. The catalytic domain of the enzyme has six subsites, numbered from -4 to +2. CBM indicates a FnIII-type carbohydrate-binding module thought to be involved in chitin-binding (12). The reducing end sugar is colored black. Approximate positions of important aromatic residues that have hydrophobic interactions with sugar residues are shown. The glycosidic bond between the sugar residues in subsite +1 and -1 is enzymatically cleaved (illustrated with an arrow). A correctly positioned N-acetyl group (shown as sticks) in the -1 subsite is essential for the substrate-assisted catalysis (35–37). The scheme shows the situation during processive action when only dimers are produced. The arrow indicates the supposed direction of the sliding of the substrate through the active site cleft (48, 49). In the case of chitosan, which consists of both N-acetylated glucosamines (GlcNAc or A units) and glucosamines (GlcN or D units), complexes formed during processive action may be nonproductive because the sugar bound in the -1 subsite may lack the catalytically crucial N-acetyl group. This leads to the production of longer even-numbered oligomers, which is diagnostic for processivity (30, 31, 38).
In chitin (and cellulose), the sugar units are rotated 180° relative to their neighboring residues, so that the smallest structural unit and the product of processive enzymatic action is a disaccharide (see Fig. 2). Family 18 chitinases, such as the processive chitinases A (ChiA)2 and B (ChiB) from Serratia marcescens, use a substrate-assisted reaction mechanism, meaning that productive binding only occurs when the sugar positioned in the -1 subsite (32, 33) of the enzyme has a correctly positioned N-acetyl group (see Figs. 1 and 2 and Refs. 34–37). Thus, during processive degradation of partially deacetylated chitin (chitosan), nonproductive complexes may emerge. It has been shown for both ChiA and ChiB that this does not necessarily lead to dissociation of the polymer but that the processive movement continues. This leads to the production of longer even-numbered oligomers, which is diagnostic for processivity (30, 31, 38).
The role of aromatic residues in the substrate-binding clefts of family 18 chitinases and on the (putative) binding surfaces of their carbohydrate-binding modules has been addressed in several studies (39–45). These studies were mainly focused on residues remote from the catalytic center and did not address processivity nor chitosan as a substrate. The main conclusion from these studies is that aromatic residues are important for interactions with the substrate and that mutation of these residues tends to lead to modest reductions of chitin hydrolyzing activity. A recent study by Horn et al. (21) showed that mutations of tryptophans very close to the catalytic center of ChiB had dramatic effects on enzyme properties. The Trp97 → Ala mutation in the +1 subsite of ChiB was shown to almost completely abolish processivity. Most importantly, although this mutation led to a reduction in chitin hydrolyzing activity, it also led to a large increase in the activity toward water-soluble oligomeric and polymeric substrates (e.g. 29-fold toward chitosan). This result indicated that processivity is beneficial for hydrolyzing insoluble substrates but may drastically reduce the efficiency of the enzyme toward more accessible substrates.
In addition to ChiB, S. marcescens produces another processive enzyme, ChiA (43, 46). Even though ChiA and ChiB have similar catalytic centers with a highly conserved -1 subsite where the catalytically active acetamide group of the substrate binds, they are believed to be fundamentally different in that they degrade chitin chains in opposite directions. ChiB is thought to move toward the reducing end (releasing dimeric products from its -1 and -2 subsites), and ChiA is thought to move toward the nonreducing end (releasing products from its +1 and +2 subsites) (see Fig. 1 and Refs. 37 and 47–49). The availability of these two enzymes and the analytical possibilities provided by chitosan provide an unique possibility to gain further insight into the contribution of processivity to biomass conversion. Therefore, we have now studied the catalytic properties of ChiA and three variants with mutations in the -3 (W167A), +1 (W275A), or +2 (F396A) subsite for both insoluble and soluble polymeric substrates. Size exclusion chromatography was used to separate chito-oligomers after partial and complete degradation of chitosan, and the composition of the oligomer fractions was used for comparison of the degrees of processivity. Hydrolytic activities were determined using crystalline β-chitin and chitosan as substrates. The results show how aromatic residues contribute to processivity and its directionality. Furthermore, the results show that the benefit of processivity strongly depends on the accessibility of the polymeric substrate, a finding that has important implications for future development of enzymes for biomass conversion.
EXPERIMENTAL PROCEDURES
Chemicals—Squid pen β-chitin (180 μm) was purchased from France Chitin (Marseille, France). Chitosan, with a degree of N-acetylation of 63% (FA = 0.63), an intrinsic viscosity, [η]of 730 ml/g, and with a number average degree of polymerization (DPn) of 800, was prepared by homogeneous N-deacetylation of milled (1.0-mm sieve) shrimp shell chitin (50) and was converted to the chitosan hydrochloride salt (51). This procedure results in a chitosan with a random distribution of N-acetylated and de-N-acetylated units (52). Mutagenic primers were purchased from Operon Biotechnologies (Huntsville, AL). All other chemicals were purchased from Sigma.
Enzymes—The chitinase gene chia from S. marcescens strain BJL200 was expressed in Escherichia coli TOP10 (Invitrogen) under control of its own promoter (53). Mutagenesis was performed using the QuikChange™ site-directed mutagenesis kit from Stratagene (La Jolla, CA) essentially as described by the manufacturer. The primers used for the mutagenesis are listed in Table 1. To confirm that the chia gene contained the desired mutations and to check for the occurrence of nondesirable mutations, the mutated genes were sequenced using the ABI PRISM™ dye terminator cycle sequencing ready reaction kit and an ABI PRISM 377 DNA sequencer (PerkinElmer Life Sciences). Wild-type and mutant chitinases were produced by growing cells for 16–18 h at 37 °C in Luria-Bertani medium containing 50 μg/ml ampicillin. Periplasmatic extracts were produced as described by Brurberg et al. (54). After being filtered trough 20-μm filters, the extracts were loaded on a column packed with chitin beads (New England, Biolabs) equilibrated in 100 mm sodium phosphate buffer, pH 8.0. After washing the column with this same buffer, the enzymes were eluted with 20 mm acetic acid and dialyzed against 20 mm Tris, 4% v/v isopropanol for 20 h at 4 °C. Enzyme purity was verified by SDS-PAGE and estimated to be >95% in all cases. Protein concentrations were determined by using the Quant-It™ protein assay kit and Qubit™ fluorometer from Invitrogen.
TABLE 1.
Primers used for site-directed mutagenesis
| Mutant | DNA template | Primer | |
|---|---|---|---|
| F396A | ChiA wild type | Forward | 5′-GACTTCTATGGCGCCGCCGATCTGAAGAAC-3′ |
| Reverse | 5′-GTTCTTCAGATCGGCGGCGCCATAGAAGTC-3′ | ||
| W275A | ChiA wild type | Forward | 5′-GATCGGCGGCGCGACGCTGTCCG-3′ |
| Reverse | 5′-CGGACAGCGTCGCGCCGCCGATC-3′ | ||
| W167A | ChiA wild type | Forward | 5′-CGTCGAGGCGGGCGTTTAC-3′ |
| Reverse | 5′-GTAAACGCCCGCCTCGACG-3′ | ||
| W167A/W275A | W275A mutant | Forward | 5′-CGTCGAGGCGGGCGTTTAC-3′ |
| Reverse | 5′-GTAAACGCCCGCCTCGACG-3′ |
Degradation of Chitosan—A 20 mg/ml chitosan solution (in doubly distilled H2O) was mixed with equal volumes of buffer (0.08 m NaAc, 0.2 m NaCl, pH 5.5, and 0.1 mg/ml bovine serum albumin (used to prevent unspecific enzyme binding to the test tube)), giving a final chitosan concentration of 10 mg/ml. Hydrolysis was carried out at 37.0 °C in a shaking incubator using an enzyme concentration of 5 μg/ml. Degradation was allowed to proceed for increasing time intervals. The reactions were stopped by lowering the pH to 2.5 by the addition of 1.0 m HCl and immersing the samples in boiling water for 2 min.
Size Exclusion Chromatography of Chitosan Oligomers—Oligomers produced by enzymatic depolymerization of chitosan were separated on three XK 26 columns packed with Superdex™ 30 (Pharmacia Biotech) with an overall dimension of 2.60 × 180 cm. Before analysis, the reaction samples were filtrated through a 10,000 molecular weight cut-off Vivaspin ultrafiltration filter (Sartorius Stedim Biotech, France) to remove the bulk of the remaining polymeric material. For analysis of samples taken during the initial and halfway stages of the chitosan degradation, a sample volume corresponding to 20 mg of starting chitosan was injected onto the column. The relative amounts of oligomers were monitored with an Gilson (model 133) on-line refractive index detector at the highest detector sensitivity (RI = 1), and the data were logged with Adam 4561/4017+ basic data acquisition modules and software (Advantech). For reaction end points, the injected samples corresponded to 10 mg of starting chitosan, and the detector sensitivity was 4-fold lower. The mobile phase was 0.15 m ammonium acetate, pH 4.5, and the flow rate was 0.8 ml/min. Raw data chromatographic spectra were base line-corrected using the statistical software R, version 2.6.1 (The R Foundation for Statistical Computing). It has been shown that this method allows the separation of mixtures of partially N-acetylated oligomers according the degree of polymerization (DP), regardless of chemical composition in the separation range between a DP of 4 and a DP of ∼20. Studies with standard samples have shown that there is a linear relationship between peak areas and the amount (mass) of injected oligomer, irrespective of DP and the degree of N-acetylation (31).
1H NMR Spectroscopy—1H NMR spectroscopy was used to calculate the number average degree of polymerization (DPn) in the chitosan reaction mixtures as described previously (31). The NMR spectra were recorded on a Varian Gemini 300 instrument in D2O at 300 MHz. The extent of chitosan degradation is given as the degree of scission, α (= 1/DPn), which represents the fraction of glycosidic linkages that has been cleaved. Complete conversion of the polymer to dimers (DP = 2) would yield an α of 0.50.
Degradation of β-Chitin—Hydrolysis of β-chitin (0.05 mg/ml) was carried out in 50 mm sodium acetate with 0.1 mg/ml bovine serum albumin at pH 6.3. The reaction tubes were incubated at 37.0 °C in a reciprocal shaker to avoid settling of the β-chitin particles. The enzyme concentrations were 0.4 μm. Reaction samples of 50 μl were withdrawn at regular time intervals, and the enzyme was inactivated by adding 50 μl of acetonitrile. All of the reactions were run in triplicate, and all of the samples were stored at -20 °C until further analysis. Concentrations of (GlcNAc)2 were determined using HPLC, as described by Horn and co-workers (46).
RESULTS
The Enzymes—Fig. 1C shows aromatic residues in the -6 to +2 subsites of ChiA. We focused on residues close to the catalytic center, meaning that Tyr170 and Tyr418 were not considered (Fig. 1C). Tyr444, located in the -1 subsite, was also not considered because it is positioned such that its aromatic ring only has minor interactions with the -1 sugar (Fig. 1C). Of the remaining residues Trp275 in the +1 subsite corresponds to Trp97 in ChiB, whereas Phe396 in the +2 subsite corresponds to Trp220 in ChiB (Fig. 1D). The most prominent residue on the glycon side is Trp539. This residue, however, is fully conserved in family 18 chitinases and is followed by a conserved cis-peptide bond, indicating a major structural role in the catalytically crucial -1 subsite. We were not able to produce variants of ChiA (or ChiB) in which Trp539 (Trp403 in ChiB) was mutated to a nonaromatic residue. The closest aromatic residue on the glycon side that could be mutated was Trp167, which dominates enzyme-substrate interactions in the -3 subsite (Fig. 1C).
Effect of the Mutations on Processivity—A highly acetylated chitosan of high molecular weight can be considered as a water-soluble chitin analogue, which is well suited for studying processivity, because it can bind both productively and nonproductively to chitinases. As explained above and in the legend to Fig. 2, processive enzymes will predominantly produce even-numbered fragments of varying lengths during the initial phase of degrading chitosan. This predominance will be stronger in the more processive enzymes. However, even highly processive enzymes will eventually produce odd-numbered products, because of rebinding of previously released even numbered products (31). It has previously been shown that ChiA initially binds to chitosan primarily in an endo-mode and then acts processively (30).
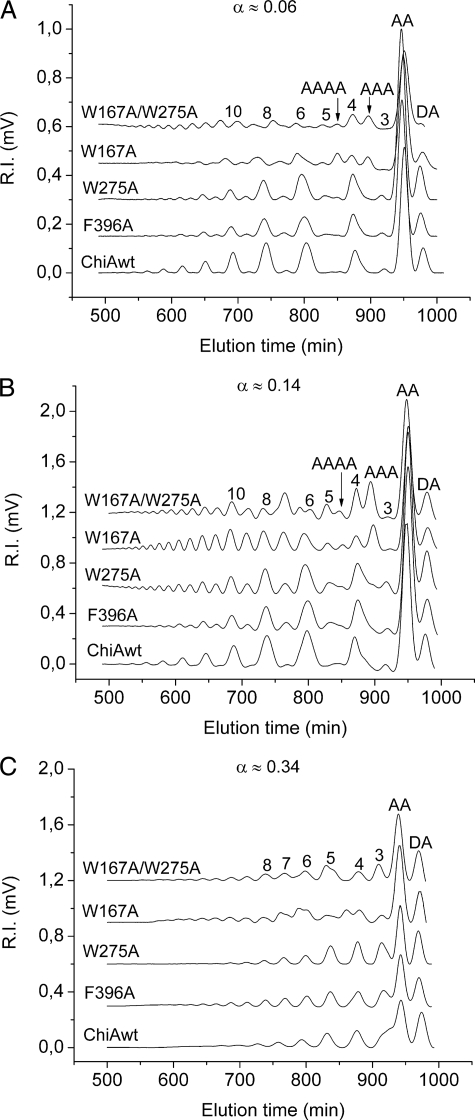
The effects of the W167A, W275A, and F396A mutations on ChiA processivity were studied using a chitosan substrate with an acetylation degree of 63% (FA = 0.63). The reactions were stopped at different stages (as determined by the α value, i.e. the fraction of cleaved glycosidic bonds), and the oligomer fractions were analyzed using size exclusion chromatography. Fig. 3 shows chromatograms of the oligomer fractions at α ≈ 0.06 and 0.14 and, after completion of the reaction, at α ≈ 0.34. As observed before (30, 38), wild-type ChiA yielded primarily even-numbered oligomers during the early stages of hydrolysis (α ≈ 0.06 and 0.14; Fig. 3 A and B), whereas odd-numbered products were almost absent. All of the mutants showed reduced processivity compared with the wild-type enzyme but to varying extents. The data obtained at α ≈ 0.06 (Fig. 3A) showed a clear reduction in processivity for W167A and the W167A/W275A mutants, as shown by the almost even distribution of odd- and even-numbered fragments. Product profiles for the two other mutants showed a clear dominance of even-numbered fragments, but the relative amounts of odd-numbered fragments were slightly higher compared with wild-type ChiA (Fig. 3A). At α ≈ 0.14 (Fig. 3B), the reduced processivity of the mutants became more apparent, in particular for the W167A mutant (Fig. 3B). The oligomer fraction from the W167A degradation had an almost uniform distribution of even- and odd-numbered fragments. At α ≈ 0.14 reduced processivity was also clearly visible for the W275A and F396A mutants (shown by the clearly visible peaks for odd-numbered products), but the observed effects were not as dramatic as for the W167A mutant. The product spectrum observed for the W167A/W275A mutant differed somewhat from the other product profiles but still clearly showed a lack of processivity (Fig. 3B). At the near end point of hydrolysis (α ≈ 0.34; Fig. 3C), all of the enzyme variants produced similar oligomeric fractions.
FIGURE 3.
Size exclusion chromatography of oligomer products obtained after the degradation of chitosan (FA = 0. 63, DPn = 800) with wild-type and mutant ChiAs. The peaks are annotated according to their content, either by a sequence (A, acetylated unit; D, deacetylated unit) or by the length of the oligomer (the latter peaks contain oligomers of equal lengths but different compositions and sequences). The annotation of the peaks is based on the use of standard samples and previous sequencing using NMR (31, 38). The chromatograms for W167A and W167A/W275A contain a few peaks that are not annotated (see text). For the reaction samples at α ≈ 0.06 (A) and α ≈ 0.14 (B), a volume corresponding to 20 mg of starting chitosan was used for the analysis. For the near end point samples (α ≈ 0.34, C), a volume corresponding to 10 mg of starting chitosan was used, and detector sensitivity was 4-fold lower. Thus, for quantitative comparison of the panels, the signal in C must be multiplied by a factor eight. α indicates the fraction of cleaved glycosidic bonds, e.g. cleavage of 6% the glycosidic bonds corresponds to α = 0.06. Complete conversion of the substrate to dimers would yield α = 0.50, whereas complete conversion of an FA = 0.63 chitosan is expected to yield a maximum α value in the order of 0.35 (38). See “Experimental Procedures” for a more thorough description.
The chromatograms for the W167A and W167A/W275A mutants showed some atypical features such as the production of relatively large amounts of fully acetylated trimers, A3, and tetramers, A4, in the early stages of the reaction (marked in Fig. 3, A and B), as well as heterogeneity in the pentamer and hexamer peaks (visible in Fig. 3, B and C). The presence of unusual products (in addition to abundant standard end products such as AA and DA dimers) was confirmed by matrix-assisted laser desorption ionization time-of-flight analyses, which showed the presence of several oligomer species not observed for the wild-type enzyme, including fully acetylated pentamers (A5) and hexamers (A6) (results not shown).
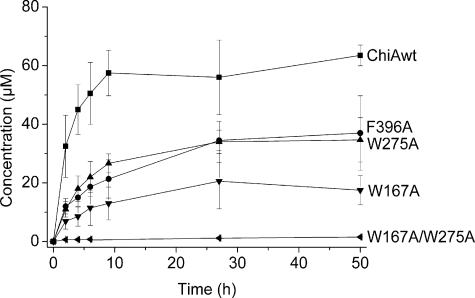
Hydrolyzing Activity toward β-Chitin and Chitosan—Degradation experiments with insoluble crystalline β-chitin showed that all the mutants were considerably less efficient than wild-type ChiA, both in terms of rate and the yield after 50 h of degradation (Fig. 4). The W275A and F396A mutants showed similar (reduced) initial rates and ∼50% of the wild-type yield. The W167A mutant showed an even lower initial rate, and the yield was only around 30% of the wild type. The double mutant W167A/W275A showed the lowest efficiency toward β-chitin and had practically no activity toward this insoluble substrate.
FIGURE 4.
Hydrolysis of β-chitin with wild-type and mutant ChiAs. 0.05 mg/ml β-chitin in 50mm sodium acetate, pH 6.1, and 0.1 mg/ml bovine serum albumin was incubated with 0.4 μm enzyme at 37 °C. Chitin degradation was approximated by analyzing the amount of liberated (GlcNAc)2, the absolutely dominant product, by HPLC (see “Experimental Procedures”). The error bars represent the standard deviation of three separate analytical runs.
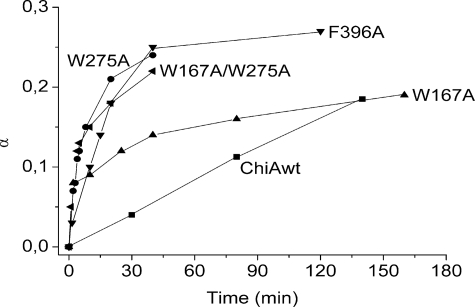
Fig. 5 shows that all mutants had considerably elevated activities toward the soluble chitosan polymer. The initial specific activity of wild-type ChiA was calculated to be 12 s-1, and the rate was constant up to α ≈ 0.18. The W275A and F396A mutants had specific initial rates that were calculated to be 219 and 80 s-1, respectively. The -3 subsite mutant, W167A showed a similar initial rate as the W275A mutant, but already in the early phase of reaction, at α ≈ 0.08, the rate decreased markedly. In contrast, W275A and F396A kept their high initial activities up to α ≈ 0.13 and α ≈ 0.18, respectively. The reaction kinetics of the double mutant, W167A/W275A (initial specific activity of 231 s-1) resembled that of W275A.
FIGURE 5.
Degradation of chitosan with wild-type and mutant ChiAs. A10 mg/ml chitosan solution in 40 μm sodium acetate, 100 μm NaCl, pH 5.5, and 0.1 mg/ml bovine serum albumin was incubated with enzyme (5 μg of enzyme/10 mg of chitosan). Degradation was allowed to proceed for increasing time intervals. The reactions were stopped by lowering the pH to 2.5 by the addition of 1.0 m HCl and immersing the samples in boiling water for 2 min. The fraction of cleaved glycosidic bonds (α) at different time points during the reaction was determined by NMR, as described under “Experimental Procedures.”
DISCUSSION
In a previous study (21), it was shown that mutation of Trp97 in the +1 subsite of ChiB (Fig. 1D) reduces processivity and enzyme efficiency toward crystalline chitin while increasing specific activity toward chitosan a spectacular 29-fold. A similar but less spectacular effect was seen for the W220A mutation in the +2 subsite of ChiB. The present study provides deeper insight into the unique roles of aromatic residues close to the catalytic centers of processive enzymes acting on crystalline substrates. The very special role of these residues is perhaps best illustrated by the W167A/W275A double mutant of ChiA, which has lost most of its processivity and activity toward crystalline chitin while being almost 20-fold more active toward the water-soluble polymeric substrate chitosan.
All three individual mutations affected processivity, albeit to varying extents (Fig. 3). The largest effect was observed for the W167A mutation in the -3 subsite, whereas the effects of the W275A and F396A mutations, located in the +1 and +2 subsites were clearly smaller. These observations support previous suggestions from both structural analysis (48) and substrate binding studies (43, 49) that ChiA moves toward the nonreducing end of the chitin chain. This would imply that Trp167 binds to the longer (polymeric) part of the substrate and would thus be more important for maintaining the enzyme-substrate association during processive action than Trp275 and Phe396, which interact with the dimeric product (Figs. 1 and 2). In ChiB, the situation would be opposite, explaining why the W97A mutation had a much larger effect on processivity of this enzyme than the analogous W275A mutation had on processivity in ChiA. Unfortunately, there are no options for mutating aromatic residues in the glycon subsites of ChiB (see legend to Fig. 1).
As expected, the less processive ChiA variants displayed reduced activities toward chitin, the largest reduction being observed for the least processive variants (W167A and W167A/W275A) (Fig. 4). The double mutant showed practically no hydrolytic activity toward β-chitin. It is not possible to conclude from the data in Fig. 3 whether the additive effect of the W275A and W167A mutations of chitin degradation efficiency is correlated with an accumulation of processivity effects. Considering the general importance of aromatic residues for chitin binding and chitin degradation efficiency (43, 44), it is conceivable that other effects play a role, such as the mere ability of extracting and binding a single chitin chain from the crystalline substrate. The large differences in degradation yields for chitin even after prolonged incubation (Fig. 4) show that, indeed, the mutant enzymes have varying abilities to access the substrate. The dramatic decrease in activity in the double mutant could also be a result of a structural effect of the double mutation. This, however, is not very likely considering the fact that the double mutant is highly active toward chitosan and thus must have its structural integrity preserved.
Although the mutants showed decreased catalytic efficiencies toward crystalline β-chitin, they all showed a substantial increase in activity toward the water-soluble chitin derivative chitosan (Fig. 5). Of the single mutants, W275A showed the largest increase in initial rate, 18-fold compared with the wild-type enzyme. The double mutant, with almost no activity toward chitin, showed a similar increase. This provides a dramatic illustration of the fact that the rate-limiting steps for the degradation of insoluble and soluble substrates are different, as previously suggested for both chitinous (21) and cellulosic (55, 56) substrates. For insoluble substrates, enzyme-substrate associations that end with the enzyme gaining access to single polymer chains as well as maintenance of the association in between hydrolytic steps (processivity) are essential for efficient hydrolysis and likely to comprise the rate-limiting step. For soluble, single chain substrates, the catalytic reaction itself, in particular product release, may become the rate-limiting step. The present results as well as the previous results for ChiB clearly show that aromatic residues play major and unique roles in both these rate-limiting processes.
The aromatic residues lining the polymer-binding sites form a flexible hydrophobic sheath, which results in “fluid-like” binding of the polymeric substrate (26). Kinetic experiments on two processive enzymes (the cellobiohydrolase CBHII and ChiB) using oligomeric substrates have shown that the “sticky” character of the binding gives an off rate, koff, that is much lower than kcat (Ref. 57 and Norberg et al.,3 respectively). In terms of processivity, low off rates may be translated into a higher probability for sliding rather than dissociation of the substrate. Although essential for efficient degradation of insoluble substrates, processivity and low off rates clearly decrease the catalytic efficiency toward more soluble substrates with higher diffusion rates. This can explain why the almost nonprocessive and less “sticky” mutants W167A in ChiA and W97A in ChiB (21) showed a large increase in activity toward chitosan.
The effect of the W275A and F396A mutations on processivity was less extreme than for the W167A mutant, but, nevertheless, these mutants also showed high activities toward chitosan. This may be due to faster release of the dimeric product in these mutants. Fig. 1C illustrates that the Trp275 and Phe396 residues interact strongly with the product in the +1 and +2 subsites. It is well known that dimeric products may bind strongly to glycoside hydrolases to the extent that in some cases significant product inhibition is observed (58). Strong binding of such dimeric products to the actual product release sites seems counterintuitive but may be explained by the fact that binding to the -1 subsite has an unfavorable energetic effect because of sugar distortion; this needs to be compensated by tight binding interactions in the neighboring subsites, as for example shown in classical work on lysozyme (59, 60).
Apart from affecting product affinities and processivity, the mutations made in this study are likely to affect substrate binding specificities (e.g. the preferences/tolerance for acetylated (A) or deacetylated (D) sugars), the general ability to bind and extract single chitin chains from insoluble material, and, as shown by Aronson et al. (40), transglycosylation abilities. Thus, one would expect to see additional mutational effects on product profiles and on noninitial conversion rates, i.e. the conversion rates that apply when the substrate is becoming more and more heterogeneous because of the emergence of intermediate products. Indeed, Fig. 3 shows that the W167A mutation leads to accumulation of some previously unseen products, which may result from transglycosylation and/or the fact that W167A has lost the ability to bind productively to some of the intermediate oligomeric products that would have been hydrolyzed further by the wild-type enzyme. Furthermore, Figs. 4 and 5 not only show differences in initial rates but also that the mutants differ with respect to the rate change during the reaction and reaction yields. Such rate changes are due to certain sequences in chitosan or certain subfractions of the chitin being cleaved slower than others or perhaps not being cleaved at all. This leads to bi- or mutiphasic behavior in the enzyme reactions and has been observed and discussed previously (15, 31).
Although the negative effects of tight binding of dimeric products on the efficiency of glycoside hydrolases have been recognized previously, the present results show that the slow release of the polymeric products necessary for processivity is another major rate-limiting factor. Although such slow release is beneficial when degrading crystalline substrates, it makes the enzymes intrinsically slow, as shown by the mutational effects on water-soluble chitosan hydrolyzing activity in this study.
The present study also shows that ChiA and ChiB are finely tuned for opposite directionalities. These are not only due to the opposite locations of the substrate-binding domains (Fig. 1) but also to the positioning of aromatic side chains close to the catalytic center. The presence of Trp167 in ChiA adapts this protein to “fluid” binding of polymeric products in the glycon subsites. Although Trp97/275 in the +1 subsite is conserved, the other aglycon subsites in ChiB seem more adapted to such “fluid” binding than in ChiA. In subsite +2 Phe396 in ChiA is replaced by Trp220 in ChiB. Furthermore, ChiB has two phenylalanines in the +3 subsite that may contribute to polymer binding in this enzyme and that both are lacking in ChiA.
The negative correlation between processivity and enzyme efficiency toward soluble substrates can explain several previous studies on cellulases and chitinases, which have shown that mutation of aromatic residues in polymer-binding sites reduces activity toward crystalline substrates but yields unaltered or even slightly increased activity toward soluble and more accessible substrates (41, 44, 55). Recently Li et al. (24) and Zhou et al. (61) introduced several point mutations in the processive endoglucanase, Cel9A-68 from T. fusca and tested the mutants for the degree of processivity (by comparing the ratio of soluble versus insoluble reducing ends) and activity toward different substrates. Some of the mutations near the catalytic center (including mutation of two tryptophan residues) led to slightly reduced processivity, decreased activity toward crystalline cellulose, and slightly increased activity toward the more accessible carboxymethylcellulose. These results suggest that the apparent link between processivity and enzyme efficiency observed for chitinases is a more general phenomenon, which may be valid for many enzyme-substrate systems. In the case of chitinases, this phenomenon can be observed very clearly because of the combination of a substrate-assisted reaction mechanism in family 18 enzymes with the availability of partially deacetylated soluble polymeric chitin that is natural in the sense that it does not pose unnatural sterical hindrance when sliding through the active site cleft (as opposed to carboxymethylcellulose).
The new knowledge on the role of processivity suggests new avenues for the development of enzymatic tools for biomass conversion. It may be wise to focus to a larger extent on making the substrate more accessible, for example by using pretreatment steps and/or optimized accessory proteins, enabling the use of enzymes with a lower degree of processivity.
Acknowledgments
We thank Pawel Sikorski for helpful discussions.
This work was supported by Grants 164653 and 174972 from the Norwegian Research Council.
Footnotes
The abbreviations used are: ChiA, chitinase A from S. marcescens; ChiB, chitinase B from Serratia marcescens; A, N-acetylated glucosamine; D, glucosamine; HPLC, high pressure liquid chromatography.
A. L. Norberg, V. Karlsen, I. Alne Hoell, I. Bakke, K. M. Vårum, V. G. H. Eijsink, and M. Sørlie, manuscript submitted.
References
- 1.Kim, J., Yun, S., and Ounaies, Z. (2006) Macromolecules 39 4202-4206 [Google Scholar]
- 2.Tharanathan, R. N., and Kittur, F. S. (2003) Crit. Rev. Food. Sci. Nutr. 43 61-87 [DOI] [PubMed] [Google Scholar]
- 3.Ragauskas, A. J., Williams, C. K., Davison, B. H., Britovsek, G., Cairney, J., Eckert, C. A., Frederick, Jr., W. J., Hallett, J. P., Leak, D. J., Liotta, C. L., Mielenz, J. R., Murphy, R., Templer, R., and Tschaplinski, T. (2006) Science 311 484-489 [DOI] [PubMed] [Google Scholar]
- 4.Merino, S., and Cherry, J. (2007) Adv. Biochem. Eng. Biotechnol. 108 95-120 [DOI] [PubMed] [Google Scholar]
- 5.Ephraim, C. (1993) Arch. Insect. Biochem. Physiol. 22 245-261 [DOI] [PubMed] [Google Scholar]
- 6.Oliveira, E., El Gueddari, N., Moerschbacher, B., Peter, M., and Franco, T. (2008) Mycopathologia 166 163-174 [DOI] [PubMed] [Google Scholar]
- 7.Sakuda, S., Isogai, A., Matsumoto, S., and Suzuki, A. (1987) J. Antibiot. 40 296-300 [DOI] [PubMed] [Google Scholar]
- 8.Vinetz, J. M., Valenzuela, J. G., Specht, C. A., Aravind, L., Langer, R. C., Ribeiro, J. M. C., and Kaslow, D. C. (2000) J. Biol. Chem. 275 10331-10341 [DOI] [PubMed] [Google Scholar]
- 9.McCartney, L., Gilbert, H. J., Bolam, D. N., Boraston, A. B., and Knox, J. P. (2004) Anal. Biochem. 326 49-54 [DOI] [PubMed] [Google Scholar]
- 10.Lehtio, J., Sugiyama, J., Gustavsson, M., Fransson, L., Linder, M., and Teeri, T. T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 484-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrard, G., Koivula, A., Soderlund, H., and Beguin, P. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10342-10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe, T., Ito, Y., Yamada, T., Hashimoto, M., Sekine, S., and Tanaka, H. (1994) J. Bacteriol. 176 4465-4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Din, N., Damude, H. G., Gilkes, N. R., Miller, R. C., Jr., Warren, R. A. J., and Kilburn, D. G. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 11383-11387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boraston, A. B., Bolam, D. N., Gilbert, H. J., and Davies, G. J. (2004) Biochem. J. 382 769-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaaje-Kolstad, G., Horn, S. J., van Aalten, D. M. F., Synstad, B., and Eijsink, V. G. H. (2005) J. Biol. Chem. 280 28492-28497 [DOI] [PubMed] [Google Scholar]
- 16.Vaaje-Kolstad, G., Houston, D. R., Riemen, A. H. K., Eijsink, V. G. H., and van Aalten, D. M. F. (2005) J. Biol. Chem. 280 11313-11319 [DOI] [PubMed] [Google Scholar]
- 17.Karkehabadi, S., Hansson, H., Kim, S., Piens, K., Mitchinson, C., and Sandgren, M. (2008) J. Mol. Biol. 383 144-154 [DOI] [PubMed] [Google Scholar]
- 18.Davies, G., and Henrissat, B. (1995) Structure 3 853-859 [DOI] [PubMed] [Google Scholar]
- 19.Rouvinen, J., Bergfors, T., Teeri, T., Knowles, J. K., and Jones, T. A. (1990) Science 249 380-386 [DOI] [PubMed] [Google Scholar]
- 20.Teeri, T. T. (1997) Trends Biotechnol. 15 160-167 [Google Scholar]
- 21.Horn, S. J., Sikorski, P., Cedekvist, J. B., Vaaje-Kolstad, G., Sørlie, M., Synstad, B., Vårum, K. M., and Eijsink, V. G. H. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 18089-18094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Divne, C., Ståhlberg, J., Reinikainen, T., Ruohonen, L., Pettersson, G., Knowles, J. K., Teeri, T. T., and Jones, T. A. (1994) Science 265 524-528 [DOI] [PubMed] [Google Scholar]
- 23.Sakon, J., Irwin, D., Wilson, D. B., and Karplus, P. A. (1997) Nat. Struct. Mol. Biol. 4 810-818 [DOI] [PubMed] [Google Scholar]
- 24.Li, Y., Irwin, D. C., and Wilson, D. B. (2007) Appl. Environ. Microbiol. 73 3165-3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Divne, C., Ståhlberg, J., Teeri, T. T., and Jones, T. A. (1998) J. Mol. Biol. 275 309-325 [DOI] [PubMed] [Google Scholar]
- 26.Varrot, A., Frandsen, T. P., von Ossowski, I., Boyer, V., Cottaz, S., Driguez, H., Schulein, M., and Davies, G. J. (2003) Structure 11 855-864 [DOI] [PubMed] [Google Scholar]
- 27.Breyer, W. A., and Mattthews, B. W. (2001) Protein Sci. 10 1699-1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer, J. E. W., and Schulz, G. E. (1997) Protein Sci. 6 1084-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eijsink, V. G. H., Vaaje-Kolstad, G., Vårum, K. M., and Horn, S. J. (2008) Trends Biotechnol. 26 228-235 [DOI] [PubMed] [Google Scholar]
- 30.Sikorski, P., Sørbotten, A., Horn, S. J., Eijsink, V. G. H., and Vårum, K. M. (2006) Biochemistry 45 9566-9574 [DOI] [PubMed] [Google Scholar]
- 31.Sørbotten, A., Horn, S. J., Eijsink, V. G. H., and Vårum, K. M. (2005) FEBS J. 272 538-549 [DOI] [PubMed] [Google Scholar]
- 32.Biely, P., Kratky, Z., and Vrsanska, M. (1981) Eur. J. Biochem. 119 559-564 [DOI] [PubMed] [Google Scholar]
- 33.Davies, G. J., Wilson, K. S., and Henrissat, B. (1997) Biochem. J. 321 557-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Synstad, B., Gåseidnes, S., Van Aalten, D. M. F., Vriend, G., Nielsen, J. E., and Eijsink, V. G. H. (2004) FEBS J. 271 253-262 [DOI] [PubMed] [Google Scholar]
- 35.Terwisscha van Scheltinga, A. C., Armand, S., Kalk, K. H., Isogai, A., Henrissat, B., and Dijkstra, B. W. (1995) Biochemistry 34 15619-15623 [DOI] [PubMed] [Google Scholar]
- 36.Tews, I., Terwisscha van Scheltinga, A. C., Perrakis, A., Wilson, K. S., and Dijkstra, B. W. (1997) J. Am. Chem. Soc. 119 7954-7959 [Google Scholar]
- 37.van Aalten, D. M. F., Komander, D., Synstad, B., Gaseidnes, S., Peter, M. G., and Eijsink, V. G. H. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8979-8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horn, S. J., Sørbotten, A., Synstad, B., Sikorski, P., Sørlie, M., Vårum, K. M., and Eijsink, V. G. H. (2006) FEBS J. 273 491-503 [DOI] [PubMed] [Google Scholar]
- 39.Aronson, N. N., Jr., Halloran, B. A., Alexeyev, M. F., Amable, L., Madura, J. D., Pasupulati, L., Worth, C., and Van Roey, P. (2003) Biochem. J. 376 87-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aronson, N. N., Jr., Halloran, B. A., Alexeyev, M. F., Zhou, X., Wang, Y., Meehan, E. J., and Chen, L. (2006) Biosci. Biotechnol. Biochem. 70 243-251 [DOI] [PubMed] [Google Scholar]
- 41.Katouno, F., Taguchi, M., Sakurai, K., Uchiyama, T., Nikiaidou, N., Nonaka, T., Sugiyama, J., and Watanabe, T. (2004) J. Biochem. (Tokyo) 136 163-168 [DOI] [PubMed] [Google Scholar]
- 42.Suginta, W., Songsiririttigul, C., Kobdaj, A., R., O., and Svasti, J. (2007) Biochim. Biophys. Acta 1770 1151-1160 [DOI] [PubMed] [Google Scholar]
- 43.Uchiyama, T., Katouno, F., Nikaidou, N., Nonaka, T., Sugiyama, J., and Watanabe, T. (2001) J. Biol. Chem. 276 41343-41349 [DOI] [PubMed] [Google Scholar]
- 44.Watanabe, T., Ariga, Y., Sato, U., Toratani, T., Hashimoto, M., Nikiaidou, N., Kezuka, Y., Nonaka, T., and Sugiyama, J. (2003) Biochem. J. 376 237-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pantoom, S., Songsiriritthigul, C., and Suginta, W. (2008) BMC Biochem. 9 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sikorski, P., Stokke, B. T., Sørbotten, A., Vårum, K. M., Horn, S. J., and Eijsink, V. G. H. (2005) Biopolymers. 77 273-285 [DOI] [PubMed] [Google Scholar]
- 47.Perrakis, A., Tews, I., Dauter, Z., Oppenheim, A. B., Chet, I., Wilson, K. S., and Vorgias, C. E. (1994) Structure 2 1169-1180 [DOI] [PubMed] [Google Scholar]
- 48.van Aalten, D. M. F., Synstad, B., Brurberg, M. B., Hough, E., Riise, B. W., Eijsink, V. G. H., and Wierenga, R. K. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5842-5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hult, E., Katouno, F., Uchiyama, T., T., W., and Sugiyama, J. (2005) Biochem. J. 388 851-856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sannan, T., Kurita, K., Ogura, K., and Iwakura, Y. (1978) Polymer 19 458-459 [Google Scholar]
- 51.Draget, K. I., Vårum, K. M., Moen, E., Gynnild, H., and Smidsrød, O. (1992) Biomaterials 13 635-638 [DOI] [PubMed] [Google Scholar]
- 52.Vårum, K. M., Anthonsen, M. W., Grasdalen, H., and Smidsrød, O. (1991) Carbohydr. Res. 211 17-23 [DOI] [PubMed] [Google Scholar]
- 53.Brurberg, M. B., Eijsink, V. G. H., and Nes, I. F. (1994) FEMS Microbiol. Lett. 124 399-404 [DOI] [PubMed] [Google Scholar]
- 54.Brurberg, M. B., Nes, I. F., and Eijsink, V. G. H. (1996) Microbiology 142 1581-1589 [DOI] [PubMed] [Google Scholar]
- 55.Koivula, A., Kinnari, T., Harjunpaa, V., Ruohonen, L., Teleman, A., Drakenberg, T., Rouvinen, J., Jones, T. A., and Teeri, T. T. (1998) FEBS Lett. 429 341-346 [DOI] [PubMed] [Google Scholar]
- 56.Zhang, S., and Wilson, D. B. (1997) J. Biotechnol. 57 101-113 [DOI] [PubMed] [Google Scholar]
- 57.Harjunpää, V., Teleman, A., Koivula, A., Ruohonen, L., Teeri, T. T., Teleman, O., and Drakenberg, T. (1996) Eur. J. Biochem. 240 584-591 [DOI] [PubMed] [Google Scholar]
- 58.Claeyssens, M., Vantilbeurgh, H., Tomme, P., Wood, T. M., and McRae, S. I. (1989) Biochem. J. 261 819-825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schindler, M., Mirelman, D., and Sharon, N. (1977) Biochim. Biophys. Acta 482 386-392 [DOI] [PubMed] [Google Scholar]
- 60.Chipman, D. M., and Sharon, N. (1969) Science 165 454-465 [DOI] [PubMed] [Google Scholar]
- 61.Zhou, W., Irwin, D. C., Escovar-Kousen, J., and Wilson, D. B. (2004) Biochemistry 43 9655-9663 [DOI] [PubMed] [Google Scholar]
- 62.Papanikolau, Y., Prag, G., Tavlas, G., Vorgias, C. E., Oppenheim, A. B., and Petratos, K. (2001) Biochemistry 40 11338-11343 [DOI] [PubMed] [Google Scholar]