Abstract
Heparan sulfate proteoglycan receptor syndecan-1 interacts with the carboxyl-terminal LG4/5 domain in laminin 332 (α3LG4/5) and participates in cell adhesion and spreading. To dissect the function of syndecan-1 in these processes, we made use of a cell adhesion model in which syndecan-1 exclusively interacts with a recombinantly expressed α3LG4/5 fragment. Plating HT1080 cells on this fragment induces the formation of actin-containing protrusive structures in an integrin-independent manner. Here we show that syndecan-1-mediated formation of membrane protrusions requires dephosphorylation of tyrosine residues in syndecan-1. Accordingly, inhibition of phosphatases with orthovanadate decreases cell adhesion to the α3LG4/5 fragment. We demonstrate that the PDZ-containing protein syntenin-1, known to connect cytoskeletal proteins, binds to syndecan-1 in cells plated on the α3LG4/5 fragment and participates in the formation of membrane protrusions. We further show that syntenin-1 recruitment depends on the dephosphorylation of Tyr-309 located within syndecan-1 PDZ binding domain EFYA. We propose that tyrosine dephosphorylation of syndecan-1 may regulate its association with cytoskeleton components.
Laminins are comprised of three distinct subunits (α, β, and γ) and are multifunctional glycoproteins present in basement membranes, which are essential structures for tissue formation in early development and in adult tissues. A total of 15 laminin isoforms have been identified and are tissue- and developmental stage-specifically expressed. Many studies have highlighted that laminin α chains are required for proper tissue organization and function (1). They also exhibit various biological activities via interaction with specific cellular receptors including integrins, α-dystroglycan, and syndecans (2). Most cell binding domains in laminins are in the G domain, located in the carboxyl-terminal globular domains of the all α chains, consisting of a repeat of 5 laminin G domain-like modules (LG1–5) (3). Although the LG1–3 domain interacts predominantly with integrins, the most extreme carboxyl-terminal pair of LG4/53 modules comprises heparin binding sites and connects with heparan sulfate proteoglycans (4). In laminin 332 (LN332), the α3 chain G1–3 domain binds α3β1, α6β1, and α6β4 integrins to support several biological activities in epithelial basement membranes (5–7), whereas the LG4/5 module was recently shown to bind to transmembrane heparan sulfate proteoglycan receptor syndecans, which mediate interactions with specific ligands via their heparan sulfate chains. Among the four syndecans (syndecan-1–4) identified in mammals, syndecans-1, -2, and -4 have been described as α3LG4/5 receptors (8, 9). An interaction between syndecans-2 and -4 with a recombinantly expressed α3LG4 module was reported (8, 10) and proposed to induce the expression of matrix metalloproteinase-1 through the mitogen-activated protein kinase signaling pathway (11). An interaction of syndecan-1 with either a recombinantly expressed LG4/5 fragment or with the LG4/5 domain in purified precursor LN332 was reported to participate in keratinocytes adhesion and spreading (9, 12). Additional heparin binding activities have been detected within the LG4 or the LG5 subdomain (13–16); however, their interaction through heparan sulfate proteoglycan-type receptors is not established yet. A recent study using recombinantly expressed LN332 reported that syndecan-1 interacts with the domain V in the γ2 chain, which also contains a heparin binding site (17); however, a cell adhesion-promoting activity of this domain was not observed (18).
Syndecans are described either as co-receptors that cooperate with other cell surface receptors or as cell adhesion receptors that independently mediate cell signaling (19, 20). Their short cytoplasmic domains are divided into two conserved regions, C1 and C2, which share common characteristics among all syndecans, and a central variable region, which confers specific properties on each syndecan. The variable domain in syndecan-1 regulates cell spreading and actin cytoskeleton assembly (21) as well as fascin bundling (22). The C1 domain, adjacent to the plasma membrane, is thought to participate in syndecan dimerization and in binding of various intracellular proteins such as ezrin (20). The conserved C2 carboxyl-terminal tetrapeptide sequence present in all syndecans, EFYA, binds some PDZ (Postsynaptic Density-95/Disc large protein/Zonula occludens-1) domain-containing proteins, such as syntenin-1 (23) and CASK (24), which may function as membrane scaffold proteins that recruit signaling and cytoskeletal proteins to the plasma membrane.
We have established a cell adhesion model in which syndecan-1 solely interacts with a recombinantly expressed α3LG4/5 fragment (9). In this model, syndecan-1-mediated cell adhesion to the LG4/5 fragment induces, in an integrin-independent manner, the formation of protrusive adhesion structures through activation of Rac and Cdc42 GTPases (12). However, the syndecan-1-dependent signal transduction pathway leading to these morphological changes is still not well understood. In this paper we made use of this α3LG4/5 adhesion model to dissect syndecan-1-associated intracellular events. We analyzed the level of tyrosine phosphorylation in syndecan-1 after cell adhesion to the LG4/5 fragment and revealed that syndecan-1-mediated formation of protrusions requires dephosphorylation of tyrosine residues in its cytoplasmic tail. We demonstrate that recruitment of the PDZ-containing protein syntenin-1 to syndecan-1 depends on tyrosine dephosphorylation of syndecan-1 PDZ binding domain.
MATERIALS AND METHODS
Cells and Antibodies—Fibrosarcoma cells HT1080 (CCL-212, American Type Culture Collection) and melanoma cells A375 (CRL-1619, American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium containing 2 mm glutamine and 10% fetal calf serum. The HaCaT human keratinocyte cell line (9) was grown in 50% Ham's F-12 and 50% Dulbecco's modified Eagle's medium supplemented with 2 mm glutamine and 10% fetal calf serum. The monoclonal antibody Mi15 against syndecan-1 was from Dako (DakoCytomation, Trappes, France), mAb DL101 and rabbit pAb H-174 against syndecan-1 were from Santa Cruz Biotechnology (Le Perray en Yvelines, France), pAb against syntenin-1 was from Synaptic Systems (Göttingen, Germany), and mAb PY20 antibodies against phosphotyrosine were from Sigma-Aldrich). The pAb anti-glutathione S-transferase (GST) peroxidase conjugate was from Sigma. LN332 and recombinant LG4/5 were prepared and purified as previously described (25, 26).
Plasmids and Transfection—Cloning of full-length human syndecan-1 cDNA in the expression vector pIRES1 neo (Clontech/BD Biosciences) was previously described (12). Mutations of the tyrosine residues (Tyr-276, Tyr-286, Tyr-299, and Tyr-309) to phenylalanine in syndecan-1 cytoplasmic tail and transmembrane domain were obtained by PCR with oligonucleotides containing the expected mutations, cloned in pIRES1 neo, and sequenced (Genome Express, Meylan, France). A375 cells were stably transfected using the FuGENE 6 reagent (Roche Diagnostics). 24 h after transfection cells were distributed in 24-well plates (5 × 104 cells/ml/well) in selective medium containing 600 μg/ml G418 (Invitrogen). Individual colonies were expanded and analyzed by cytofluorometry (fluorescence-activated cell sorter) to quantify syndecan-1 cell surface expression as previously described (9).
To append the cytoplasmic domain of syndecan-1 to the carboxyl terminus of GST, the cDNA encoding this domain (from Arg-277 to Ala-310) was amplified by PCR, subcloned into BamHI and XhoI sites of pGEX-4T1 (Amersham Biosciences), and sequenced (Genome Express). GST fusion proteins were then produced in bacteria and purified on glutathione-Sepharose 4B beads as recommended by the manufacturer's instructions (Amersham Biosciences). Point mutations in the GST-syndecan-1 fusion protein were introduced by PCR using oligonucleotides encoding the respective mutated amino acid residues. A three-letter code referring to the tyrosine residues Tyr-286, Tyr-299, Tyr-309 was hereafter chosen to refer to the native GST-syndecan-1 fusion protein (GST-YYY). The following constructs were obtained: GST-DDD (Tyr-286, Tyr-299, Tyr-309 were changed to Asp), GST-DYY (Tyr-286 was replaced by Asp), GST-YDY (Tyr-299 was changed to Asp), and GST-YYD (Tyr-309 was changed to Asp). For GST fusion proteins with PDZ domains of syntenin-1, the cDNA encoding PDZ1 (from Gly-102 to Pro-194), PDZ2 (from Thr-189 to Ile-274), and PDZ1 and -2 (from Gly-102 to Met-270) domains were amplified by reverse transcription-PCR (Titan one tube RT-PCR kit, Roche Diagnostics) from human keratinocyte total RNA, subcloned into BamHI and XhoI sites of pGEX-4T1 (Amersham Biosciences), and sequenced (Genome Express). Purification from bacteria lysates was as described above.
For human syntenin-1 (accession number NM_005625.3) knock-down experiments, three siRNAs for which the sequences are shared between the five syntenin isoforms (400GCAAGACCUUCCAGUAUAAA, 452UGGAAUUCGUAGAGCAGAA, 903CUCUCAAAUUGCAGACAUA) were synthesized (Sigma-Aldrich). For each individual siRNA, cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen) with 100 nm duplex siRNA according to the manufacturer's recommendations. In all experiments cells were analyzed 48 h after transfection, and viability was assessed using trypan blue. Each siRNA resulted in comparably reduced syntenin expression, and results with only one siRNA were shown. A firefly luciferase GL2 sequence (CGUACGCGGAAUACUUCGA) was used as a negative control (100 nm) as described (27) and resulted in no reduction of syntenin-1 expression.
Cell Adhesion and Inhibition Assays—Multiwell plates (Costar, Dutscher, Brumath, France) were coated with previously determined concentrations of LG4/5 or LN332 substrates (12) by overnight adsorption at 4 °C. Cells were detached with 5 mm EDTA-PBS and rinsed in serum-free medium. After saturation of the wells with 1% BSA, cells were suspended in serum-free medium and seeded (8 × 104 cells/well). After 30 min to 1 h, non-adherent cells were washed with PBS, and the extent of adhesion was determined after fixation of adherent cells followed by staining with 0.1% crystal violet and absorbance measurements at 570 nm as previously described (28). A blank value corresponding to BSA-coated wells was subtracted. Each assay point was derived from triplicate measurements (3 wells per assay point). Adherent cells were photographed using an Axiovert 40 Zeiss microscope coupled to a Coolsnap Fx Camera (Roper Scientific, Evry, France).
Inhibitor Treatments—Sodium orthovanadate, genistein, and staurosporine were from Sigma. Stock solutions of genistein and staurosporine were prepared in dimethyl sulfoxide. Orthovanadate and genistein were incubated with cells for 3 or 16 h at 37 °C in Dulbecco's modified Eagle's medium supplemented with 2 mm glutamine and 2% fetal calf serum. Staurosporine was incubated with cells at 37 °C at the time of assay.
Preparation of Cell Extracts—In the case of pulldown experiments after cell adhesion experiments, 5 × 106 HT1080 cells were plated on BSA- or LG4/5-coated 75-cm2 dishes for the indicated times. The total amount of cells (unadhered and adhered cells) were pooled and extracted with cold radioimmune precipitation assay lysis buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 250 μm phenylmethylsulfonyl fluoride, 1 mm N-ethylmaleimide, 1% Nonidet P-40, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) in order that the same pool of cells are analyzed in the various conditions. Control cultured cells were counted before the extraction. For all protein phosphorylation studies, phosphatase inhibitor mixture set II (Calbiochem) was added to the lysis buffer. All the following procedures were performed at 4 °C. After centrifugation of the extracts, the supernatants were immediately processed for pulldown experiments. In the case of pulldown experiments using exclusively cultured cells, HT1080 or HaCaT cells were extracted with radioimmune precipitation assay lysis buffer, pH 7.4, containing 250 μm phenylmethylsulfonyl fluoride and 1 mm N-ethylmaleimide. After centrifugation, protein concentrations of lysates were determined, and the equivalent amount of proteins was processed for pulldown experiments using beads covered with various ligands.
Pulldown Experiments—For syndecan-1 pulldown assays, the LG4/5 protein was covalently linked to CNBr-activated-Sepharose 4B (Amersham Biosciences). Beads were incubated with extracts for 2 h, washed, and incubated in digestion buffer (20 mm sodium acetate, 5 mm CaCl2, pH 7.0) with 8 milliunits/ml heparitinase I and 50 milliunits/ml chondroitinase ABC (Seikagaku America, Coger, Paris, France) for 2 h at 25 °C. The proteins were resolved either on 8 or 12% SDS-PAGE gels. For syntenin-1 pulldown assays, beads covered with the GST syndecan-1 fusion proteins described above were incubated with cell extracts for 2 h, washed, and resolved on 12% SDS-PAGE gels. Finally, neutravidin-agarose beads (Perbio Science, Bezons, France) were covered with biotinylated synthetic peptides QEEFYA or QEEFpYA (Eurogentec, Anger, France) and incubated with HT1080 extracts for 2 h and treated as above for syntenin-1 pulldown assays. When experiments were performed with GST-fused PDZ domains proteins, beads covered with syndecan-1-biotinylated peptides were incubated with eluted proteins for 1 h in the presence of PBS containing 0.05% Tween, washed, and resolved on 15% SDS-PAGE gels. In all cases gels were transferred to nitrocellulose followed by immunodetection by ECL for either syndecan-1, syntenin-1, or GST.
Immunoprecipitation—Cell lysates were incubated for 4 h at 4 °C with Gamma Bind-G-Sepharose beads (Amersham Biosciences) previously incubated overnight at 4 °C with or without 10 μg of precipitating antibodies (Mi15 mAbs). Beads carrying the immune complexes were washed 3 times with PBS, 0.05% Tween. Immune complexes were dissociated by the addition of denaturing sample buffer, then heated at 95 °C followed by separation by SDS-PAGE on a 8% gel under non-reducing conditions. Gels were transferred to nitrocellulose followed by immunodetection by ECL for syndecan-1 and phosphotyrosine.
Surface Plasmon Resonance Measurements—Biotinylated synthetic peptide RMKKKDEGSYSLEEPKQANGGAYQKPTKQEEFYA and RMKKKDEGSYSLEEPKQANGGAYQKPTKQEEFpYA (Eurogentec) corresponding to the cytoplasmic tail of syndecan-1 either in its unphosphorylated form or comprising a single phosphorylated tyrosine located within the EFYA sequence were immobilized on Biacore sensorchips. For that purpose, flow cells of a CM4 sensorchip were functionalized with 3500 resonance units of streptavidin as described (9), and biotinylated peptides, each at 0.5 mm in HBS-P (10 mm Hepes, 0.15 m NaCl, 0.005% P20, surfactant, pH 7.4) supplemented with phosphatase inhibitor mixture set II, were injected for 5 min across a streptavidin-activated surface. This procedure yields an immobilization level of 600 and 650 resonance units for the unphosphorylated and the phosphorylated peptide, respectively. For binding assays, 100 μl of GST or GST fused to PDZ 1 plus 2 domains were injected at a flow rate of 10 μl/min over the 2 surfaces, after which the formed complexes were washed with HBS-P buffer for 5 min. Phosphatase inhibitor mixture set II was present in all samples, including the Biacore running buffer.
Immunofluorescence Studies—Glass coverslips were coated with LG4/5 (0.3 μm) at 4 °C overnight. After adhesion, cells were fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.1% Triton X-100 for 3 min, rinsed with PBS, and subjected to a treatment with 8 milliunits/ml heparitinase I and/or 50 milliunits/ml chondroitinase ABC for 1 h. After a 1-h incubation with PBS 10% fetal calf serum, antibodies against syndecan-1 and syntenin-1 were successively applied for 1 h. Alexa Fluor 488 and Alexa Fluor 546 anti-mouse or anti-rabbit antibodies were then applied together with Alexa Fluor 647 phalloidin for 30 min. Cells were observed with an Axioplan Zeiss microscope or by laser-scanning confocal microscopy (Zeiss LSM 510). Optical slides of 0.8 μm were selected at the cell-matrix interface.
Molecular Modeling—Two models of the syntenin-1 PDZ2-binding syndecan-1 QEEFYA or QEEFpYA structures were generated using the x-ray-derived coordinate of the syntenin-1 PDZ2-binding syndecan-4 peptide TNEFYA (Protein Data Bank accession number 1OBY) (29) as a template. For each model a geometric optimization of the structure was computed using the sybyl molecular modeling package (SYBYL 7.3, Tripos International, St. Louis, MI). The Tripos forcefield with Gasteiger-Marsilli atomic charge algorithm was applied with a dielectric constant set to 80 with a distance dependent function. To relax the two systems, two short dynamics of 1 ns were computed at constant volume and temperature (300 K). The binding energy of the peptide to the PDZ2 was computed using the ZAP algorithm, which is an enhancement of the Poisson-Boltzmann equation and allows computing the binding energy of two biopolymers. This energy was computed for the two peptides complexes with the Syntenin-1. The binding energy is about -2.92 kcal·mol-1 for the phosphorylated peptide and -18.31 kcal·mol-1 for the unphosphorylated peptide.
Analytical Methods—The following procedures were performed as previously described; that is, SDS-PAGE, followed by the electrophoretic transfer of proteins to nitrocellulose with immunoblot analysis (28).
RESULTS
Tyrosine Phosphorylation of Syndecan-1 after Cell Adhesion to LG4/5—To get an insight into the syndecan-1 signaling pathway, we analyzed the level of tyrosine phosphorylation of syndecan-1 upon cell adhesion to LG4/5. HT1080 were allowed to adhere to LG4/5 for different periods of time, and after cell lysis, syndecan-1 was purified by pulldown experiments using beads covered with LG4/5, as previously characterized (9, 12). After digestion of the glycosaminoglycan chains, the core proteins were immunoblotted with Abs to syndecan-1 and phosphotyrosine (Fig. 1, A and B). As shown in Fig. 1B, freshly detached cells and cultured cells displayed high levels of tyrosine-phosphorylated syndecan-1, leading to the conclusion that some syndecan-1 is phosphorylated in these conditions. After cell adhesion to the LG4/5 fragment, the level of tyrosine-phosphorylated syndecan-1 decreased as time of adhesion to LG4/5 increased. After 5 min of adhesion, tyrosine phosphorylation reduced (Fig. 1, B and C), dropping off completely after 30 min. To verify that the 90-kDa band revealed with the anti-phosphotyrosine antibody was the syndecan-1 core protein, we performed a series of immunoprecipitation experiments using cultured HT1080 cells. Instead of using LG4/5 pulldown assays, syndecan-1 was directly immunoprecipitated from lysates with mAbs to syndecan-1. Upon GAG digestion, syndecan-1 was detected by anti-syndecan-1 immunoblotting. Blotting immunoprecipitated syndecan-1 with an anti-phosphotyrosine mAb revealed a band with the same molecular weight as syndecan-1 (Fig. 1D). When GAG digestion was omitted (Fig. 1D, lanes 2), syndecan-1 was not detected anymore because of high molecular weight GAG chains that prevented efficient electrophoretic transfer to nitrocellulose membranes. Under these conditions, no signal was observed with the anti-phosphotyrosine mAb. These observations strongly suggested that the 90-kDa phosphorylated protein was the syndecan-1 core protein. Altogether, these results demonstrate that some tyrosine residues in the syndecan-1 core protein are phosphorylated and that their dephosphorylation is an early event upon interaction with LG4/5 that might be required for efficient cell adhesion. Preventing dephosphorylation should, therefore, result in the inhibition of adhesion. To test this hypothesis, we performed cell adhesion experiments on LG4/5 in the presence of phosphatase inhibitors (Fig. 1E). Before cell adhesion assays HT1080 cells were treated with orthovanadate, a widely used tyrosine phosphatase inhibitor shown, for instance, to enhance syndecan-1 phosphorylation in cultured cells (30). Integrin-mediated cell adhesion to LN332 was performed concomitantly to establish the specificity of the effect and to ensure the absence of drug toxicity. A concentration of 0.1 mm orthovanadate was sufficient to partially prevent cell adhesion to LG4/5. Integrin-mediated cell adhesion to LN332 was unaffected. We concluded that orthovanadate impedes cell adhesion to LG4/5, although off-target effects of orthovanadate treatment cannot be formally ruled out.
FIGURE 1.
Tyrosine phosphorylation state of syndecan-1 upon HT1080 cell adhesion to α3LG4/5. A and B, 5 × 106 HT1080 cells were allowed to adhere for 0, 5, 30, and 60 min to LG4/5, and lysates corresponding to the total plated cells were incubated with beads covered with LG4/5. A control with cultured HT1080 cells was carried out (lanes C). After washes, bound material was digested with heparitinase I and chondroitinase ABC. Electrophoretic analysis of the bound material was performed on a 8% SDS-polyacrylamide gel under reducing conditions followed by sequential immunoblotting with the mAb PY20 against phosphotyrosine (B) and pAb H-174 against syndecan-1 (A). C, quantification of syndecan-1 tyrosine phosphorylation after adhesion to LG4/5. The data are representative of three independent experiments. D, syndecan-1 was precipitated from HT1080 cell lysates with beads covered with anti-syndecan-1 mAb Mi15. Bound material was either treated (lanes 1) or left untreated (lanes 2) with heparitinase/chondroitinase and analyzed on an 8% SDS-polyacrylamide gel followed by immunoblotting of syndecan-1 and phosphotyrosine. B and D, the migration positions of molecular mass markers are shown on the right. E, inhibition of cell adhesion to the LG4/5 with a phosphatase inhibitor. HT1080 cells were used for cell adhesion to the LG4/5 (0.3 μm, black bars) and LN332 (2 nm, gray bars). HT1080 were seeded at density of 6 × 104 cells/well in serum-free medium after treatment with the indicated concentrations of orthovanadate. For controls, cells were incubated with serum-free medium only. The extent of adhesion was measured as described under “Materials and Methods” and expressed as a percentage of adhesion in the absence of inhibitor. The data are representative of three independent experiments. F and G, effect of genistein and staurosporine treatment on HT1080 cell adhesion toα3LG4/5. Multiwell plates were coated with LG4/5 (0.3 μm), LN332 (2 nm), and 1% BSA. After saturation with 1% BSA, HT1080 cells previously cultured for 16 h (G) in the presence of the indicated concentrations of genistein were seeded in the presence of genistein at the same concentrations. H, HT1080 cells were seeded in the presence of staurosporine at the indicated concentrations. In all experiments HT1080 were seeded at a density of 6 × 104 cells/well in serum-free medium. After 30 min the extent of adhesion was measured and expressed as a percentage of adhesion in the absence of inhibitor.
To further assess the role of syndecan phosphorylation in adhesion, phosphorylation was blocked by the use of specific tyrosine kinase inhibitors (Fig. 1, F and G). In this case constitutive unphosphorylated syndecan-1 should increase cell adhesion. As genistein was shown to inhibit tyrosine phosphorylation of syndecan-1 in adherent B82 fibroblasts (30), HT1080 were treated with genistein for 16 h before cell adhesion assays to LG4/5. Genistein treatments induced a significant and dose-dependent increase in cell adhesion to LG4/5 (Fig. 1F). Adhesion to LG4/5 was specific as no adhesion to BSA nor increased adhesion to LN332 depending on integrins was observed at any genistein concentration used. Staurosporine, another tyrosine kinase inhibitor previously shown to inhibit phosphorylation of syndecan-1 (30), gave identical results (Fig. 1G). No significant change in cell adhesion was observed in integrin-mediated adhesion to LN332 upon staurosporine treatment.
Because tyrosine dephosphorylation was essential for syndecan-1-mediated adhesion to LG4/5, several syndecan-1 mutants were designed to determine critical tyrosine residues within its cytoplasmic domain. We had previously shown that transfection of syndecan-1 in the A375 cell line, which does not express syndecan-1, switched its phenotype from non-adherent to adherent to the LG4/5 fragment with concomitant formation of filopodia (12). We made use of this A375-Syn1 cell model to examine whether any of the three tyrosine residues (Tyr-286, Tyr-299, and Tyr-309) present in the cytoplasmic domain of syndecan-1 (Fig. 2A) is involved in the syndecan-1-mediated adhesion to LG4/5 and subsequent formation of spikes. In a first set of experiments we mutated each individual tyrosine to phenylalanine, a non-phosphorylatable amino acid (Fig. 2A). Transfection of A375 cells with the mutated constructs Syn1-FYY, Syn1-YFY, and Syn1-YYF resulted in expression of syndecan-1 at the cell surface, similar to that of A375-Syn1 cells (Syn1-YYY), as shown by monitoring syndecan-1 expression by flow cytometry (Fig. 2B). To further characterize these syndecan-1 mutants, LG4/5 pulldown experiments were performed (Fig. 2C). Capture of syndecan-1 was carried out by incubating A375, A375-Syn1-YYY, and A375-Syn1-mutants cell lysates with beads covalently covered with LG4/5. Treatment of the LG4/5-bound proteoglycan receptor as described in Fig. 1A revealed a band corresponding to the syndecan-1 core protein in A375-Syn1 mutants cells identical to that isolated from the A375-Syn1 as already reported (12). Cell adhesion experiments to the LG4/5 fragment revealed that all four transfected cell types adhered to the LG4/5 fragment in a dose-dependent manner (Fig. 2D), although adhesion appeared reduced for the A375-Syn1-YYF cell line. Analyzing the morphology of the cells after adhesion to the LG4/5 fragment (Fig. 2E) revealed that only the A375-Syn1-YYF mutant failed to spread and form cellular protrusions otherwise observed with wild-type A375-Syn1-YYY cells (Fig. 2E and Ref. 12). Our results suggest that Tyr-309 in syndecan-1 is critical for spreading and microspike formation in cells adhered to the LG4/5 fragment. To establish that Tyr-309 was phosphorylated in cells, we designed an additional syndecan-1 construct Syn1-FFFY in which Tyr-309 was the only phosphorylatable tyrosine left in the cytoplasmic domain of syndecan-1 (Fig. 2F). In addition, the tyrosine residue (Tyr-276) predicted to be located at the border between the transmembrane domain and the cytoplasmic tail was also mutated to phenylalanine. A control construct in which all tyrosines were mutated Syn1-FFFF was also designed. Constructs were transfected in A375 cells, and A375-Syn1-FFFY and A375-Syn1-FFFF clones were selected for their syndecan-1 expression at the cell membrane by both fluorescence-activated cell sorter and syndecan-1 Western blotting analysis (data not shown). Pulldown experiments were performed by incubating A375-Syn1, A375-Syn1-FFFY, and A375-Syn1-FFFF cell lysates with beads covalently covered with LG4/5. The LG4/5 bound material was GAG-digested and separated by SDS-PAGE followed by Western blotting for syndecan-1 and phosphotyrosine (Fig. 2F). Syndecan-1 was purified from A375-Syn1, A375-Syn1-FFFY, and A375-Syn1-FFFF cell lysates as revealed by anti-syndecan-1 blotting. The anti-phosphotyrosine antibody blotted the syndecan-1 core protein band from the A375-Syn1 and A375-Syn1-FFFY cells but not that of the A375-Syn1-FFFF cells. We, thus, concluded that tyrosine Tyr-309 can be phosphorylated in cells.
FIGURE 2.
Involvement of syndecan-1 tyrosine residues in LG4/5-mediated formation of microspikes. A, schematic diagram of syndecan-1 constructs. The wild-type human syndecan-1 and three mutants of the tyrosine residues (Tyr-286, Tyr-299, and Tyr-309) in the syndecan-1 cytoplasmic tail were cloned in the expression vector pIRES1 neo and stably transfected in A375 cells. B, high expression levels of syndecan-1 transfectants in A375-Syn1-YYY, A375-Syn1-FYY, A375-Syn1-YFY, and A375-Syn1-YYF (bold line) as compared with wild-type A375 cells (thin line), as determined by fluorescence-activated cell sorter analysis with the mAb Mi15 specific for syndecan-1. The dotted line corresponds to negative staining. C, as indicated, syndecan-1 was precipitated from A375, A375-Syn1-YYY, A375-Syn1-FYY, A375-Syn1-YFY, and A375-Syn1-YYF cell lysates with beads covered with LG4/5. Bound material was treated with heparitinase I/chondroitinase ABC and analyzed on an 8% SDS-polyacrylamide gel followed by immunoblotting of syndecan-1. D, dose-dependent cell adhesion of A375-Syn1-YYY, A375-Syn1-FYY, A375-Syn1-YFY, and A375-Syn1-YYF to the LG4/5 fragment. Multiwell plates were coated with increasing concentrations of LG4/5. EDTA-released cells were seeded (8 × 104 cells/well) and incubated for 1 h. E, morphology of A375-Syn1 transfectants after plating for 1 h on surfaces coated with 0.3 μm LG4/5. Bars, 20 μm. F, as indicated, syndecan-1 was precipitated from A375, A375-Syn1, A375-Syn1-FFFY, and A375-FFFF cell lysates with beads covered with LG4/5. Bound material was treated with heparitinase I/chondroitinase ABC and analyzed on an 8% SDS-polyacrylamide gel followed by immunoblotting of syndecan-1 and phosphotyrosine. C and F, molecular markers are indicated on the left.
Syntenin-1 Binds Syndecan-1 upon Cell Adhesion to LG4/5—Tyr-309 belongs to the carboxyl-terminal EFYA motif, which is conserved in all vertebrate syndecans (31) and interacts with proteins containing PDZ domains. Among these, syntenin-1, which contains two PDZ domains, was suggested to serve as an adaptor protein that may couple syndecans to cytoskeletal proteins or cytosolic downstream signal-effectors (23, 32, 33). Because the above results suggested that syndecan-1 Tyr-309 phosphorylation could play a critical role in cell adhesion and formation of cellular protrusions, we next assessed whether phosphorylation of this tyrosine residue could control syntenin-1 recruitment. We first established that syntenin-1 bound tyrosine-unphosphorylated syndecan-1 after cell adhesion to LG4/5 (Fig. 3A). For this purpose HT1080 cells were allowed to adhere to LG4/5 for 60 min, and the amount of syntenin-1 bound to syndecan-1 was determined by pulldown experiments using beads covered with LG4/5 and subsequent Western blotting analysis. As shown in Fig. 3A, cell adhesion to the LG4/5 fragment induced a 45% increase of the amount of syntenin-1 bound to syndecan-1 in the pulldown assay as compared with non adhered cells (p < 0.1). Therefore, syntenin-1 binding to syndecan-1 appears directly correlated with the dephosphorylation level of syndecan-1. Note that the syntenin-1/syndecan-1 interaction was already observed before cell adhesion to LG4/5 as the total pool of syndecan-1 is not phosphorylated under these conditions, and the unphosphorylated syndecan-1 fraction could be prone to syntenin-1 recruitment (see Fig. 1, A and B). To confirm the correlation between syndecan-1 dephosphorylation and syntenin-1 recruitment, HT1080 cells were treated with orthovanadate to prevent syndecan-1 tyrosine dephosphorylation, and syndecan-1 pulldown assays were performed. As shown in Fig. 3B, orthovanadate treatment slightly enhanced the phosphorylation level of tyrosine residues in syndecan-1 and concomitantly decreased the binding of syntenin-1, reinforcing the hypothesis that tyrosine dephosphorylation regulates syntenin-1 recruitment to syndecan-1. To firmly establish the link between the binding of syntenin-1 to unphosphorylated syndecan-1 and the formation of cellular protrusions, we transfected A375 cells with a mutated form of syndecan-1 in which Tyr-309 was changed to aspartic acid. Pulldown experiments using control A375-syn1 and A375-syn1-YYD cell lysates revealed that the Tyr-309-mutated syndecan-1 failed to bind syntenin-1 and inhibited the formation of membrane protrusions after cell adhesion to the LG4/5 fragment (Fig. 3C). Altogether, these results suggest that binding of syntenin-1 to the unphosphorylated Tyr-309 in syndecan-1 allows the formation of protrusions in cells plated on the LG4/5 fragment.
FIGURE 3.
Syntenin-1 is associated with tyrosine unphosphorylated syndecan-1. A, 5 × 106 HT1080 cells were either processed immediately or were allowed to adhere for 60 min to 0.3 μm LG4/5. Lysates corresponding to the total cells were incubated with beads covered with LG4/5. After washes, bound material was digested with heparitinase I and chondroitinase ABC. Electrophoretic analysis of the bound material was performed either on an 8% SDS-polyacrylamide gel followed by sequential immunoblotting with the pAb H-174 against syndecan-1 and the mAb PY20 against phosphotyrosine or 12% SDS-polyacrylamide gel for immunoblotting with the pAb against syntenin-1. The experiment was repeated 4 times, and syntenin-1 contents obtained after cell adhesion to LG4/5 were compared with that obtained with unadhered cells by using Student's t test; p < 0.1 B, syntenin-1 binding to syndecan-1 depending on the level of syndecan-1 tyrosine phosphorylation. HT1080 cells were cultured in the absence or presence of 1 mm orthovanadate for 3 h. Corresponding cell lysates were incubated with beads covered with LG4/5. After washes, bound material was digested with heparitinase I and chondroitinase ABC. Electrophoretic analysis of the bound material was performed either on an 8% SDS-polyacrylamide gel followed by sequential immunoblotting with the pAb H-174 against syndecan-1 and the mAb PY20 against phosphotyrosine or 12% SDS-polyacrylamide gel for immunoblotting with the pAb against syntenin-1. Quantifications of phosphotyrosine in syndecan-1 and of associated syntenin-1 in cells treated with orthovanadate are relative to that of control-untreated cells. C, the mutated form of syndecan-1 bearing an aspartic acid at Tyr-309 was cloned in the expression vector pIRES1 neo and stably transfected in A375 cells. As indicated, syndecan-1 was precipitated from A375-Syn1-YYY and A375-Syn1-YYD cell lysates with beads covered with LG4/5, and bound material was treated with heparitinase I/chondroitinase ABC. Electrophoretic analysis of the bound material was performed either on an 8% SDS-polyacrylamide gel followed by immunoblotting with the pAb H-174 against syndecan-1 or 12% SDS-polyacrylamide gel for immunoblotting with the pAb against syntenin-1. Right panel, morphology of A375-Syn1-YYY and A375-Syn1-YYD cells after plating for 60 min on surfaces coated with 0.3 μm LG4/5. Bars, 20 μm. B and C, the data are representative of three independent experiments.
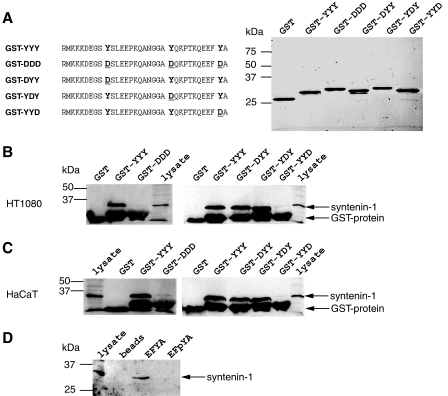
Tyr-309 Phosphorylation in EFYA Prevents Syntenin-1 Binding to Syndecan-1—To further evaluate the effect of syndecan-1 tyrosine phosphorylation on syntenin-1 recruitment and to identify syndecan-1 tyrosine residues implicated in this process, pulldown experiments with fusion proteins between GST and the cytoplasmic domain of syndecan-1 were performed. For this purpose, a native unphosphorylated GST-YYY as well as four mutated forms (Fig. 4A), in which either all or individual tyrosine residues were changed to aspartic acid to mimic phosphorylation (34, 35), were produced in bacteria. The fusion proteins were purified on glutathione-Sepharose beads and first analyzed by SDS-PAGE followed by Coomassie Blue staining to verify the production (Fig. 4A). The beads covered with the various fusion proteins were then incubated with HT1080 cell lysates to assay the binding of syntenin-1. As shown in Fig. 4B, GST-YYY strongly interacted with syntenin-1 as compared with GST alone. In contrast, syntenin-1 failed to interact with the phospho-mimetic mutant GST-DDD. Using the set of individual mutated tyrosine mutants revealed that only the fusion protein GST-YYD, mimicking phosphorylation of Tyr-309, prevented syntenin-1 binding. To verify whether these results were reproducible in the context of epithelial cells, these experiments were repeated with the keratinocyte cell line HaCaT, and identical results were obtained (Fig. 4C). These data suggested that phosphorylation of Tyr-309 might regulate syntenin-1 interaction with syndecan-1. To formally prove this hypothesis, we chose an alternative approach using commercial tyrosine-phosphorylated EFYA peptides corresponding to the last four carboxyl-terminal amino-acids of syndecan-1 including Tyr-309. The minimal sequence EFYA, shown to be required for syntenin-1 binding (23), was coupled to biotin as well as its tyrosine-phosphorylated counterpart EFpYA. Both peptides were captured on neutravidin beads for syntenin-1 pulldown experiments. As shown in Fig. 4D, syntenin-1 bound to EFYA peptides but not to phosphorylated EFpYA peptides. These results demonstrate that phosphorylation of Tyr-309 regulates syntenin-1 binding to the EFYA sequence in syndecan-1 cytoplasmic domain.
FIGURE 4.
Phosphorylation of Tyr-309 in the EFYA sequence of syndecan-1 impedes syntenin-1 binding. A, schematic representation of the cytoplasmic tail of syndecan-1 as well as various tyrosine residue mutants made in the GST fusion proteins used in this study (left panel). 15% SDS-PAGE electrophoresis analysis followed by Coomassie Blue staining of 0.5 μg of each GST fusion protein, including the GST alone (as indicated, right panel). B and C, analysis of syntenin-1 binding to syndecan-1 cytoplasmic tail fusion proteins. Ten μg of GST as well as all GST fusion proteins were coupled to glutathione-Sepharose 4B beads and incubated with HT1080 (B) and HaCaT (C) cell lysates. After washes, bound material was electrophoretically analyzed on a 12% SDS-polyacrylamide gel under reducing conditions followed by immunoblotting with the pAb against syntenin-1. As indicated in each blot, a GST-alone pulldown control experiment was performed in parallel, and 30 μg of HT1080 (B) or HaCaT (C) cell lysate was electrophoretically analyzed to visualize syntenin-1. D, neutravidin-agarose beads were either left uncovered or were covered with 20 μg of biotinylated synthetic peptides EFYA or EFpYA (as indicated) and used for pulldown experiments of syntenin-1 from HT1080 cell lysates as described in B. Ten μg of HT1080 cell lysate was electrophoretically analyzed to visualize syntenin-1. A–D, the migration positions of molecular mass markers are shown on the left.
In light of these observations, at least two different regulatory mechanisms of syntenin-1 binding to syndecan-1 could be proposed. First, phosphorylation of Tyr-309 might directly impede syntenin-1 binding because of a conformation change of the EFYA peptide or to a steric hindrance brought by the phosphate group of Tyr(P)-309. Alternatively, phosphorylated Tyr-309 might recruit another protein preventing syntenin-1 binding to syndecan-1. To discriminate between these two possibilities, we analyzed the impact of Tyr-309 phosphorylation on the direct in vitro interaction between EFYA/EFpYA peptides and recombinantly produced syntenin-1. Syntenin-syndecan-1 interaction relies on the carboxyl-terminal part of syntenin-1, which contains a tandem of PDZ domains (PDZ1 and PDZ2). Both PDZ domains participate in a cooperative manner in the binding to syndecans (32). We, therefore, produced GST fusion proteins including the PDZ1-PDZ2 tandem as well as each individual PDZ domain of syntenin-1. The fusion proteins were purified on glutathione-Sepharose beads and analyzed by SDS-PAGE followed by Coomassie Blue staining (Fig. 5A). These GST-PDZ1, GST-PDZ2, and GST-PDZ1-PDZ2 proteins were eluted from beads and tested for their ability to interact with neutravidin beads covered with either EFYA or EFpYA as described above. As shown in Fig. 5B, both the PDZ1-PDZ2 tandem and the PDZ2 domain alone bound to EFYA peptides, whereas phosphorylation of the tyrosine residue in the EFYA sequence prevented this binding. As expected (32, 29), the PDZ1 domain alone did not interact with EFYA peptides and was used here as a negative control. These results firmly demonstrate that the phosphorylation of Tyr-309 in syndecan directly regulates syntenin-1 recruitment. To further support this view we designed a Biacore-based binding assay in which the binding of GST-PDZ1-PDZ2 tandem to immobilized unphosphorylated syndecan peptide was compared with that of its phosphorylated counterpart. To control the amount of immobilized peptide and overcome the detection sensitivity problems, which would occur with a low molecular mass tetrapeptide, we switched to a longer peptide corresponding to the full-length, 34 residue, cytoplasmic domain of syndecan. As a first step, we repeated the pulldown experiment and confirmed that, using this method, phosphorylation of the Tyr-309 prevented binding of the PDZ1-PDZ2 tandem (Fig. 5C). The two biotinylated peptides were then captured on top of streptavidin-activated sensorchips to similar levels, over which GST or GST fused to the PDZ1-PDZ2 tandem were injected. None of these samples bound to streptavidin (data not shown). GST alone did not interact with the cytoplasmic domain of syndecan (whether it was phosphorylated or not). In contrast, the PDZ1-PDZ2 fusion construct showed a strong interaction with the unphosphorylated peptide, a binding that was fully inhibited by the presence of the phosphate group on the Tyr-309 (Fig. 5D). Altogether, these data demonstrate that the phosphorylation status of the Tyr-309 in syndecan directly regulates syntenin-1 recruitment.
FIGURE 5.
Phosphorylation of Tyr-309 in the EFYA sequence of syndecan-1 impedes syntenin-1 PDZ2 and PDZ1 and -2 domain binding. A, 15% SDS-PAGE electrophoresis analysis of 0.5 μg of GST alone and of GST-fused PDZ1, PDZ2, and PDZ1-PDZ2 domains of syntenin-1, as indicated, followed by Coomassie Blue staining. B, analysis of the GST-PDZ protein binding to the unphosphorylated and phosphorylated EFYA peptide. Neutravidin-agarose beads were either left uncovered or were covered with 20 μg of biotinylated synthetic peptides EFYA or EFpYA (as indicated) and used for pulldown experiments of 0.5 μg of GST alone, GST-PDZ1, GST-PDZ2, and GST-PDZ1-PDZ2, as indicated. 50 ng of each GST-protein was electrophoretically analyzed as a positive control. C, analysis of the GST-PDZ1-PDZ2 protein binding to the 34-residue peptide corresponding to the cytoplasmic tail of syndecan-1 either unphosphorylated or phosphorylated on the tyrosine residue comprised within the PDZ binding domain. Neutravidin-agarose beads were either left uncovered or were covered with 20 μg of biotinylated synthetic peptides (as indicated) and used for pulldown experiments of 0.5 μg of GST-PDZ1-PDZ2, as indicated. 50 ng of GST-PDZ1-PDZ2 was electrophoretically analyzed as a positive control. A–C, the migration positions of molecular mass markers are shown on the left. D, surface plasmon resonance analysis of the interaction of PDZ domains with syndecan-1 cytoplasmic peptides. GST fused to PDZ1-PDZ2 domains of syntenin-1 (125 nm) was injected over chip surfaces activated with either the unphosphorylated syndecan-1 peptide (light gray curve) or its phosphorylated counterpart (medium gray curve) from 0 to 600 s, after which the formed complexes were washed with running buffer. The binding response was recorded in resonance units (RU) as a function of time. The binding curve obtained by injection of GST alone on both surfaces is shown in black.
Syntenin-1 Is Required for LG4/5-induced Microspikes—To determine whether syntenin-1 interaction with syndecan-1 resulted in their cellular colocalization, we next analyzed syntenin-1 distribution in HT1080 and A375-Syn1 cells plated on LG4/5 by confocal microscopy. Although syndecan-1 has a peripheral distribution in freshly spreading cells, it is not present in focal contacts and colocalizes with actin microfilaments when clustered and in long-term adherent cells (36, 37). In cell adhered on the LG4/5 fragment, actin was assembled in typical radial protrusions termed microspikes, along which syndecan-1 staining was found (Fig. 6A). Syntenin, of which staining was very strong all over the cell nuclei, also specifically decorated the cytoplasmic extensions in a manner comparable with that of actin microfilaments and syndecan-1 (Fig. 6A), consistent with the hypothesis that syntenin-1 recruitment in microspikes is mediated by syndecan-1.
FIGURE 6.
Syntenin-1 is localized in LG4/5-induced microspikes. A, EDTA-released HT1080 or A375-syn1 cells were plated on surfaces coated with 0.3 μm LG4/5 for 1 h, fixed, and stained either with the mAb DL101 against syndecan-1 (green), pAb against syntenin-1 (red), and actin (bleu). The merged image of cells plated on LG4/5 shows a juxtaposition of the syndecan-1- and syntenin-1-staining patterns in microspikes. Optical slides of 0.8 μm were selected at the cell-matrix interface. Bars, 10 μm. B and C, behavior of syntenin-1 knock-down cells plated on LG4/5. Low expression levels of syntenin-1 in HT1080 (B) and A375-Syn1 (C) cells transfected with syntenin-specific siRNA as determined by Western blotting are shown. 10 μg of cell lysates from untreated cells (lanes 1) and cells transfected with control-siRNA (lanes 2), syntenin-siRNA (lanes 3), were analyzed on a 12% SDS-polyacrylamide gel followed by immunoblotting of syntenin-1. HT1080 (B) and A375-Syn1 (C) cell adhesion of syntenin-siRNA-transfected cells to the LG4/5 fragment was compared with cells transfected with control siRNA. Bar, 10 μm.
To further assess the direct role of syntenin-1 in microspike formation after cell adhesion to LG4/5, siRNA oligonucleotides specific for human syntenin (syntenin-siRNA) were used to down-regulate its expression. A representative experiment showed that the expression of syntenin-1 was reduced by ∼95% in siRNA-transfected HT1080 and A375-Syn1 cells, as shown by monitoring syntenin-1 expression by Western blotting (Fig. 6, B and C). Cell adhesion experiments to the LG4/5 fragment revealed that syntenin-siRNA-transfected cells adhered to the LG4/5 fragment but failed to spread and form cellular protrusions (only 10% of adhered HT1080 or A375-Syn1 cells developed microspikes versus 60 and 80% in siRNA control-transfected HT1080 and A375-Syn1 cells, respectively, Fig. 6, B and C). These results reinforce the notion that syntenin-1 recruitment to the cytoplasmic tail of syndecan-1 is involved in the formation of cellular protrusions.
DISCUSSION
The transmembrane proteoglycan syndecan-1 participates in cell adhesion to the precursor form of LN332 through an interaction with its carboxyl-terminal LG4/5 domain (9). This interaction promotes cell spreading and contributes to in vitro pre-LN332-induced human keratinocyte migration. Although the biology of this interaction is now well described, the molecular machinery implicated in these processes is still badly understood. To gain insights into theses mechanisms, we have established a cell adhesion model in which syndecan-1 solely interacts with a recombinantly expressed α3LG4/5 fragment without any contribution of integrins to cell adhesion. Plating keratinocytes or HT1080 cells on the LG4/5 fragment does not allow migration but induces the formation of actin-containing protrusive structures through activation of Rac and Cdc42 GTPases (12). To dissect syndecan-1-associated intracellular events, we analyzed the level of tyrosine phosphorylation in syndecan-1 after cell adhesion to the LG4/5 fragment. Here we report for the first time that syndecan-1 is rapidly tyrosine-dephosphorylated upon cell adhesion. The kinetics of this phenomenon is comparable with the rate of attachment to the LG4/5 fragment and can be completed within 5 min, suggesting the involvement of an active tyrosine phosphatase. The link between syndecan-1 phosphorylation and cell adhesion was further strengthened by the use of orthovanadate, which by inhibiting phosphatases dramatically prevented cell adhesion to the LG4/5 fragment. Together these results reinforce already published data showing that syndecan-1 function might be regulated by phospho/dephosphorylation events (30), although this previous work did not provide any explanations for the effect of syndecan-1 dephosphorylation. Interestingly, syndecan-1 phosphorylation results in the cleavage of its ectodomain and its release into cell culture medium (38). Therefore, it is tempting to propose that cell binding to the LG4/5 fragment by inducing syndecan-1 dephosphorylation might protect syndecan-1 from cell surface shedding, thus allowing its involvement in signal transduction induced by pre-LN332. This hypothesis is further supported by our observation that genistein and staurosporine, two potent tyrosine kinase inhibitors previously shown to promote syndecan-1 down-phosphorylation (30), significantly enhanced cell adhesion to LG4/5.
Besides shedding control, tyrosine dephosphorylation of syndecan-1 may promote conformational changes of its cytoplasmic domain and/or provide binding sites for other proteins and regulate syndecan-1 association with cytoskeleton components. Syndecan-1 cytoplasmic domain contains three tyrosine residues that are conserved among all syndecan family members (Tyr-2, Tyr-3, Tyr-4). An additional tyrosine (Tyr-1) is located within the transmembrane domain. Two of the cytoplasmic tyrosine residues are likely targets for phosphorylation based on their surrounding amino acid sequences (31). One of them (Tyr-2) is located within the membrane-proximal conserved region C1 (DEGSY), and the other (Tyr-4) is located within a second conserved sequence (EFYA) at the extreme carboxyl-terminal region C2 (30). The third cytoplasmic tyrosine (Tyr-3) is located within the V region. An in vitro kinase assay has revealed that all tyrosine residues in the cytoplasmic domain of syndecan-3, the closest homologue of syndecan-1, can be phosphorylated (39). However, whether one or several tyrosine residues are targets for phosphorylation on syndecan-1 in vivo is still unknown, and whether these events are associated to functional features has never been assessed. By producing several syndecan-1 mutants in which each individual tyrosine was replaced by phenylalanine and expressing them in cells that do not express syndecan-1, we identified Tyr-4 as the tyrosine residue critical for cell spreading on the LG4/5 fragment and microspikes formation. The observation that Y2F and Y3F mutations did not impair syndecan-1-mediated spreading on the LG4/5 fragment strongly suggests that phosphorylation of these tyrosine residues is not necessary for microspike formation. This conclusion is in agreement with a study reporting that mutation of mouse syndecan-1 Tyr-3 to the unphosphorylatable residue alanine does not abolish the formation of actin-microspikes in cells plated on an immobilized anti-syndecan-1 ectodomain antibody (22). However, in contrast with our observation is an early report by Carey et al. (21) showing by syndecan-1 ectodomain antibody cross-linking experiments that tyrosine 3 of the V region in syndecan-1 is essential for microfilament association. This apparent controversy could reflect that the signaling properties of syndecan-1 might depend on the nature of its ligand. Indeed, at the difference of antibody-mediated cross-linking, cell adhesion to the LG4/5 fragment, a physiological ligand, is mediated by the GAG chains present in the syndecan-1 ectodomain (9), and this may result in distinct syndecan-1 signaling capacities. Interestingly, syndecan-2 is phosphorylated by the EphB2 receptor-tyrosine kinase on Tyr-2 and Tyr-4 residues, leading to dendritic-spine formation in neurons (40). Therefore, it appears that, even if Tyr-4 in the EFYA sequence is conserved among all syndecans, distinct mechanisms are at play for the remodeling of the actin cytoskeleton depending on the syndecan family member and triggering ligands.
As a first step toward the understanding of the mechanisms relying on tyrosine phosphorylation of syndecan-1, we investigated the role of Tyr-4 in cellular functions. Indeed, we showed that mutated Tyr-4 syndecan-1 failed to assemble cellular protrusions. This result strongly suggests that the tyrosine 4 residue is necessary for syndecan-1-mediated spreading on the LG4/5 fragment. This tyrosine residue belongs to the highly conserved C2 region EFYA that binds a number of PDZ domain proteins (for review, see Ref. 20). Among these, syntenin-1, which couples syndecans to cytoskeletal proteins or to downstream signal-effectors in actin microfilament-related adhesion sites (23, 32, 33), was a good candidate to link dephosphorylated syndecan-1 to the induction of microspike formation upon LG4/5 adhesion. Supporting this hypothesis were our pulldown experiments data showing 1) that syntenin-1 is recruited by syndecan-1 in cells plated on the LG4/5 fragment and 2) that syntenin-1 does not bind phosphorylated syndecan-1 in cells treated with orthovanadate. Additional pulldown experiments using several tyrosine phospho-mimetic syndecan-1 fusion proteins, in which tyrosine residues were changed to aspartic acid to mimic phosphorylation (34, 35), suggested that phosphorylation of tyrosine 4 may induce inhibition of syntenin-1 binding to the syndecan-1 cytoplasmic tail. Whether the inhibition of syntenin-1 interaction to the mutated Y4D syndecan-1 is the consequence of its phosphorylated-like state remains, however, questionable, as it has been previously shown that the tyrosine residue Tyr-4 itself is critical for syntenin-1 binding (41). Indeed, mutation of this tyrosine residue to either Ala (23) or Lys (42) results in a significant decrease in syntenin-1 binding without implying phosphorylation events in this process. We definitely solved this ambiguity by showing for the first time that a peptide corresponding to the minimal sequence EFYA known to bind to syntenin-1 (23) failed to interact when the tyrosine residue is phosphorylated. Altogether, our results strongly support a role for the tyrosine 4 dephosphorylation in the regulation of syntenin-1 recruitment by syndecan-1. A similar mechanism was reported for B class ephrins in which phosphorylation of either one of the two successive tyrosine residues within the PDZ binding domain affects the binding of syntenin (43). Other regulatory mechanisms implicating the phosphorylation of a serine residue within the carboxyl terminus of PDZ-binding proteins were reported to either disrupt (44–46) or enhance (47) their interaction with PDZ domains. Involvement of a tyrosine residue has also been described in a study reporting that tyrosine phosphorylation in the PDZ binding domain of the receptor-tyrosine kinase erbB2 abolishes its interaction with the Erbin PDZ domain (48). Recently, studies on syndecan-4 revealed that phosphorylation of Ser-183 (49), a residue in the C1 region, induces a conformational change in the C2 domain even though the phosphorylation site is 20 residues away and impedes the PDZ binding of syntenin-1 (50). Such a mechanism could also occur in syndecan-1 as this Ser residue is conserved through all syndecans in the C1 region. Further experiments will be required to test this hypothesis.
Syntenin contains two PDZ domains (PDZ1 and PDZ2). Each domain consists of six β-strands (β1–β6) and two α-helices (α1 and α2), which fold into a six-stranded β-sandwich (51, 52). Several co-crystallization studies have shown that the carboxyl terminus of the FYA-interacting peptide docks into a cavity formed by the β2 strand, the α2 helix, and a loop connecting β1 and β2 strands (29, 41). The PDZ2 pocket binds syndecan with higher affinity than the PDZ1 pocket, but both pockets are necessary for high affinity binding to oligomeric syndecans, suggesting a cooperative binding mode (32, 33). This was recently confirmed by structural analysis coupling heteronuclear NMR and single x-ray diffraction, reporting that the two PDZ domains participate cooperatively in the binding of syndecan dimers, with PDZ2 being the dominant module (42). Mutational (23, 33) and crystallographic studies (29) have revealed that syntenin-1 mainly recognizes the side chains of the three carboxyl-terminal amino acids in the syndecans, namely FYA, whereas the upstream residues are not involved. Therefore, like all PDZ binding peptides, the terminal carboxylate of the FYA peptide interacts with three hydrogen bonds from three main chain amide groups within the conserved glycine-rich loop preceding the β2 strand in the PDZ domain. Co-structural analysis of a syndecan-4 peptide with individual PDZ domains showed that both Phe and Tyr residues of the carboxyl-terminal syndecan sequence (FYA) play critical roles in the interaction with the PDZ2 domain, each of them being lodged into well defined specificity pockets, respectively, namely S-2 and S-1. The Tyr residue is lodged into the S-1 pocket cushioned by His-208, Ile-212, and Val-222. Most interestingly, the aromatic ring of the tyrosine is additionally involved in an off-center stacking interaction with the critical His-208 residue of the PDZ2 S-1 pocket. Based on this co-structural model (29), we designed a structural model of the syntenin-1 PDZ2 domain together with a syndecan-1 peptide in which the Tyr residue is phosphorylated (Fig. 7). This model reveals that the size of the S-1 pocket is well adapted to the size of the Tyr aromatic ring but is too narrow when the latter contains a phosphate on its hydroxyl function. In addition, the phosphate group would interact with the Asp-204 located within S-1, but this interaction is unfavorable in term of columbic energy of interaction. Finally the π-stacking interaction of the aromatic ring of the tyrosine with His-208 is lost when the tyrosine residue is phosphorylated. Altogether, our data confirm that the tyrosine located within the FYA sequence in syndecan-1 is crucial for its interaction with syntenin-1 and additionally demonstrate that this interaction is regulated by its phosphorylation state.
FIGURE 7.
Co-structural model of syntenin-1 PDZ2-binding syndecan-1 peptide QEEFpYA. Two models of the syntenin-1 PDZ2-binding syndecan-1 QEEFpYA (A) or QEEFYA (B) structures were generated using the x-ray-derived coordinate of the syntenin-1 PDZ2-binding syndecan-4 peptide TNEFYA (Protein Data Bank accession number 1OBY) (29) as a template. The Tyr-1 is in a groove formed by Thr-206, Phe-211, and Ile-212, which orient its aromatic cycle. This groove is well adapted to the size of the Tyr cycle but is too narrow when the latter contains a phosphate on its hydroxyl function. Furthermore, the contact of the phosphate group with the PDZ2 Asp-204 is unfavorable in term of columbic energy of interaction. Finally, the Tyr-1 hydrogen bond with Thr-206 and the π-stacking of its aromatic ring with His-208 are lost when the Tyr-1 is phosphorylated. The binding energy of the peptide to the PDZ2 was computed using the ZAP algorithm, which is an enhancement of the Poisson-Boltzmann equation, allowing computing of the binding energy of two biopolymers. This energy was computed for the two peptides complexes with the syntenin-1. The binding energies are about -2.92 kcal·mol-1 for the phosphorylated Tyr-1 and -18.31 kcal·mol-1 for the unphosphorylated Tyr-1. The color of the surface is mapped with atomic partial charge. Negative charge is colored in blue, neutral charge is in white, and positive charge is in red.
The observation that cell adhesion to the LG4/5 fragment induces tyrosine dephosphorylation in the syndecan-1 cytoplasmic tail, which in turn binds syntenin-1, suggests that syntenin-1 may play a role in the formation of cellular protrusions. This hypothesis is further reinforced by confocal microscopy analysis revealing that syntenin-1 colocalizes with syndecan-1 in actin-rich cellular protrusions formed in cells plated on the LG4/5 fragment. Moreover, we show that knocking-down syntenin-1 expression dramatically reduced the formation of microspikes in these adhered cells. This result corroborates the observation that inhibition of syntenin-1 expression in cultured epithelial cells induces cellular rounding and detachment (53). A role for syntenin-1 in syndecan-1 adhesion sites was previously suggested in a study showing that recombinant enhanced green fluorescent protein-syntenin-1 fusion proteins decorate the plasma membrane and co-cluster with overexpressed syndecans (23). Moreover, cells overexpressing syntenin-1 displayed numerous cellular extensions, suggesting a link between syntenin-1 and cytoskeleton membrane organization (23, 33, 54). Altogether, these data suggest that, as syntenin-1 has no obvious catalytic domain and, therefore, is unlikely to have a signaling function by itself, it could connect syndecan-1 to signaling components and to the actin cytoskeleton.
What could be the physiological relevance of syntenin-1 recruitment by syndecan-1 after cell adhesion to the LG4/5 domain in pre-LN332? Several reports suggest a model in which syntenin-1 recruitment would be essential for cell migration (54, 55); however, a potential link with syndecan was not analyzed. Interestingly syntenin-1 is overexpressed in a number of metastatic breast, gastric, and melanoma cancer cell lines and tumor tissues, in which its expression correlated with the migratory and metastatic potential (54, 56, 57). We have shown that syndecan-1-mediated formation of protrusions in cells plated on pre-LN332 depends on the activation of the Rac and Cdc42 (12), two members of the Rho family GTPase known to regulate actin cytoskeleton and play a pivotal role in cell motility (58). As syntenin-1 was shown to play a role in the formation of actin-dependent cell adhesion structures through regulatory activities of the GTPases Rac, Cdc42, and Rho (33), we propose that syntenin-1 may affect Rho family GTPase activity and, therefore, play an important role in signaling events after syndecan-1-mediated cell adhesion to the LG4/5 domain in pre-LN332.
This work was supported by grants from European Economic Community Contract NMP2-CT-2003-504017, the Association pour la Recherche sur le Cancer (Grants 1128 (to P. R.) and 3985 (to F. L.), the Ligue Nationale contre le Cancer (Loire, Drôme, Savoie), a “Program Cible” Grant from the Rhône-Alpes region, and the Agence Nationale de la Recherche Chemispike program.
Footnotes
The abbreviations used are: LG4/5, recombinant LG4/5 fragment; LN332, laminin 332; GAG, glycosaminoglycan; mAb, monoclonal antibody; pAb, polyclonal antibody; GST, glutathione S-transferase; siRNA, small interfering RNA; PBS, phosphate-buffered saline; BSA, bovine serum albumin.
References
- 1.Miner, J. H., and Yurchenco, P. D. (2004) Annu. Rev. Cell Dev. Biol. 20 255-284 [DOI] [PubMed] [Google Scholar]
- 2.Sasaki, T., Fassler R., and Hohenester, E. (2004) J. Cell Biol. 164 959-963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Timpl, R., Tisi, D., Talts, J. F., Andac, Z., Sasaki, T., and Hohenester, E. (2000) Matrix Biol. 19 309-317 [DOI] [PubMed] [Google Scholar]
- 4.Susuki, N., Yokoyama, F., and Nomizu, M. (2005) Connect. Tissue Res. 46 142-152 [DOI] [PubMed] [Google Scholar]
- 5.Carter, W. G., Ryan, M. C., and Gahr, P. J. (1991) Cell 65 599-610 [DOI] [PubMed] [Google Scholar]
- 6.Sonnenberg, A., de Melker, A. A., Martinez de Velasco, A. M., Janssen, H., Calafat, J., and Niessen, C. M. (1993) J. Cell Sci. 106 1083-1102 [DOI] [PubMed] [Google Scholar]
- 7.Rousselle, P., and Aumailley, M. (1994) J. Cell Biol. 125 205-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Utani, A., Nomizu, M., Matsuura, H., Kato, K., Kobayashi, T., Takeda, U., Aota, S., Nielsen, P. K., and Shinkai, H. (2001) J. Biol. Chem. 276 28779-28788 [DOI] [PubMed] [Google Scholar]
- 9.Okamoto, O., Bachy, S., Odenthal, U., Bernaud, J., Rigal, D., Lortat-Jacob, H., Smyth, N., and Rousselle, P. (2003) J. Biol. Chem. 278 44168-44177 [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama, F., Suzuki, N., Kadoya, Y., Utani, A., Nakatsuka, H., Nishi, N., Haruki, M., Kleinman, H. K., and Nomizu, M. (2005) Biochemistry 44 9581-9589 [DOI] [PubMed] [Google Scholar]
- 11.Utani, A., Momota, Y., Endo, H., Kasuya, Y., Beck, K., Suzuki, N., Nomizu, M., and Shinkai, H. (2003) J. Biol. Chem. 278 34483-34490 [DOI] [PubMed] [Google Scholar]
- 12.Bachy, S., Letourneur, F., and Rousselle, P. (2008) J. Cell. Physiol. 214 238-249 [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, M. P., Engbring, J. A., Nielsen, P. K., Vargas, J., Steinberg, Z., Karmand, A. J., Nomizu, M., Yamada, Y., and Kleinman, H. K. (2001) J. Biol. Chem. 276 22077-22085 [DOI] [PubMed] [Google Scholar]
- 14.Kato, K., Utani, A., Suzuki, N., Mochizuki, M., Yamada, M., Nishi, N., Matsuura, H., Shinkai, H., and Nomizu, M. (2002) Biochemistry 41 10747-10753 [DOI] [PubMed] [Google Scholar]
- 15.Vivès, R., Crublet, E., Andieu, J. P., Gagnon, J., Rousselle, P., and Lortat-Jacob, H. (2004) J. Biol. Chem. 279 54327-54333 [DOI] [PubMed] [Google Scholar]
- 16.Mizushima, H., Takamura, H., Miyagi, Y., Kikkawa, Y., Yamanaka, N., Yasumitsu, H., Misugi, K., and Miyazaki, K. (1997) Cell Growth Differ. 8 979-987 [PubMed] [Google Scholar]
- 17.Sasaki, T., Göhring, W., Mann, K., Brakebusch, C., Yamada, Y., Fässler, R., and Timpl, R. (2001) J. Mol. Biol. 314 751-763 [DOI] [PubMed] [Google Scholar]
- 18.Ogawa, T., Tsubota, Y., Hashimoto, J., Kariya, Y., and Miyazaki, K. (2007) Mol. Biol. Cell 18 1621-1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernfield, M., Gotte, M., Park, P. W., Reizes, O., Fitzgerald, M. L., Lincecum, J., and Zako, M. (1999) Annu. Rev. Biochem. 68 729-777 [DOI] [PubMed] [Google Scholar]
- 20.Couchman, J. R. (2003) Nat. Rev. Mol. Cell Biol. 4 926-937 [DOI] [PubMed] [Google Scholar]
- 21.Carey, D. J., Bendt, K. M., and Stahl, R. C. (1996) J. Biol. Chem. 271 15253-15260 [DOI] [PubMed] [Google Scholar]
- 22.Chakravarti, R., Sapountzi, V., and Adams, J. C. (2005) Mol. Biol. Cell 16 3678-3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grootjans, J. J., Zimmermann, P., Reekmans, G., Smets, A., Degeest, G., Durr, J., and David, G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13683-13688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen, A. R., Woods, D. F., Marfatia, S. M., Walther, Z., Chishti, A. H., Anderson, J. M., and Woods, D. F. (1998) J. Cell Biol. 142 129-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vivinus-Nebot, M., Ticchioni, M., Mary, F., Hofman, P., Quaranta, V., Rousselle, P., and Bernard, A. (1999) J. Cell Biol. 144 563-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belin, V., and Rousselle, P. (2006) Protein Expression Purif. 48 43-48 [DOI] [PubMed] [Google Scholar]
- 27.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001) Nature 411 494-498 [DOI] [PubMed] [Google Scholar]
- 28.Décline, F., Okamoto, O., Mallein-Gerin, F., Helbert, B., Bernaud, J., Rigal, D., and Rousselle, P. (2003) Cell Motil. Cytoskeleton 54 64-80 [DOI] [PubMed] [Google Scholar]
- 29.Kang, B. S., Cooper, D. R., Jelen, F., Devedjiev, Y., Derewenda, U., Dauter, Z., Otlewski, J., and Derewenda, Z. S. (2003b) Structure 11 459-468 [DOI] [PubMed] [Google Scholar]
- 30.Ott, V. L., and Rapraeger, A. C. (1998) J. Biol. Chem. 273 35291-35298 [DOI] [PubMed] [Google Scholar]
- 31.Yoneda, A., and Couchman, J. R. (2003) Matrix Biol. 22 25-33 [DOI] [PubMed] [Google Scholar]
- 32.Grootjans, J. J., Reekmans, G., Ceulemans, H., and David, G. (2000) J. Biol. Chem. 275 19933-19941 [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann, P., Tomatis, D., Rosas, M., Grootjans, J., Leenaerts, I., Degeest, G., Reekmans, G., Coomans, C., and David, G. (2001) Mol. Biol. Cell 12 339-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, W., and Erikson, R. L. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8960-8963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, Y., Spencer, S. D., and Lasky, L. A. (1998) J. Biol. Chem. 273 5765-5770 [DOI] [PubMed] [Google Scholar]
- 36.Rapraeger, A. C., Jalkanen, M., and Bernfield, M. (1986) J. Cell Biol. 103 2683-2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carey, D. J., Stahl, R. C., Cizmeci-Smith, G., and Asundi, V. K. (1994) J. Cell Biol. 124 161-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reiland, J., Ott, V. L., Lebakken, C. S., Yeaman, C., McCarthy, J., and Rapraeger, A. C. (1996) Biochem. J. 319 39-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asundi, V. K., and Carey, D. J. (1997) Biochem. Biophys. Res. Commun. 240 502-506 [DOI] [PubMed] [Google Scholar]
- 40.Ethell, I. M., Irie, F., Kalo, M. S., Couchman, J. R., Pasquale, E. B., and Yamaguchi, Y. (2001) Neuron 31 1001-1013 [DOI] [PubMed] [Google Scholar]
- 41.Kang, B. S., Cooper, D. R., Devedjiev, Y., Derewenda, U., and Derewenda, Z. S. (2003) Structure 11 845-853 [DOI] [PubMed] [Google Scholar]
- 42.Grembecka, J., Cierpicki, T., Devedjiev, Y., Derewenda, U., Kang, B. S., Bushweller, J. H., and Derewenda, Z. S. (2006) Biochemistry 45 3674-3683 [DOI] [PubMed] [Google Scholar]
- 43.Lin, D., Gish, G. D., Songyang, Z., and Pawson, T. (1999) J. Biol. Chem. 274 3726-3733 [DOI] [PubMed] [Google Scholar]
- 44.Cohen, N. A., Brenman, J. E., Snyder, S. H., and Bredt, D. S. (1996) Neuron 17 759-767 [DOI] [PubMed] [Google Scholar]
- 45.Matsuda, S., Mikawa, S., and Hirai, H. (1999) J. Neurochem. 73 1765-1768 [DOI] [PubMed] [Google Scholar]
- 46.Cao, T. T., Deacon, H. W., Reczek, D., Bretscher, A., and von Zastrow, M. (1999) Nature 401 286-290 [DOI] [PubMed] [Google Scholar]
- 47.Hegedüs, T., Sessler, T., Scott, R., Thelin, W., Bakos, E., Váradi, A., Szabó, K., Homolya, L., Milgram, S. L., and Sarkadi, B. (2003) Biochem. Biophys. Res. Commun. 302 454-461 [DOI] [PubMed] [Google Scholar]
- 48.Birrane, G., Chung, J., and Ladias, J. A. (2003) J. Biol. Chem. 278 1399-1402 [DOI] [PubMed] [Google Scholar]
- 49.Horowitz, A., and Simons, M. (1998) J. Biol. Chem. 273 10914-10918 [DOI] [PubMed] [Google Scholar]
- 50.Koo, B. K., Jung, Y. S., Shin, J., Han, I., Mortier, E., Zimmermann, P., Whiteford, J. R., Couchman, J. R., Oh, E. S., and Lee, W. (2006) J. Mol. Biol. 355 651-663 [DOI] [PubMed] [Google Scholar]
- 51.Doyle, D. A., Lee, A., Lewis, J., Kim, E., Sheng, M., and MacKinnon, R. (1996) Cell 85 1067-1076 [DOI] [PubMed] [Google Scholar]
- 52.Morais Cabral, J. H., Petosa, C., Sutcliffe, M. J., Raza, S., Byron, O., Poy, F., Marfatia, S. M., Chishti, A. H., and Liddington, R. C. (1996) Nature 382 649-652 [DOI] [PubMed] [Google Scholar]
- 53.Zimmermann, P., Zhang, Z., Degeest, G., Mortier, E., Leenaerts, I., Coomans, C., Schulz, J., N′Kuli, F., Courtoy, P. J., and David, G. (2005) Dev. Cell 9 377-388 [DOI] [PubMed] [Google Scholar]
- 54.Koo, T. H., Lee, J. J., Kim, E. M., Kim, K. W., Kim, H. D., and Lee, J. H. (2002) Oncogene 21 4080-4088 [DOI] [PubMed] [Google Scholar]
- 55.Meerschaert, K., Bruyneel, E., De Wever, O., Vanloo, B., Boucherie, C., Bracke, M., Vandekerckhove, J., and Gettemans, J. (2007) Exp. Cell Res. 313 1790-1804 [DOI] [PubMed] [Google Scholar]
- 56.Helmke, B. M., Polychronidis, M., Benner, A., Thome, M., Arribas, J., and Deichmann, M. (2004) Oncol. Rep. 12 221-228 [PubMed] [Google Scholar]
- 57.Boukerche, H., Su, Z. Z., Emdad, L., Baril, P., Balme, B., Thomas, L., Randolph, A., Valerie, K., Sarkar, D., and Fisher, P. B. (2005) Cancer Res. 65 10901-10911 [DOI] [PubMed] [Google Scholar]
- 58.Nobes, C. D., and Hall, A. (1995) Biochem. Soc. Trans. 23 456-459 [DOI] [PubMed] [Google Scholar]