Abstract
Anhydrobiotic animals survive virtually complete loss of cellular water. The mechanisms that explain this phenomenon are not fully understood but often include the accumulation of low molecular weight solutes such as trehalose and macromolecules like Late Embryogenesis Abundant (LEA) proteins. Here we report for the first time the occurrence of a mitochondria-targeted LEA gene (Afrlea3m) product in an animal species. The deduced molecular mass of the 307-amino acid polypeptide from the brine shrimp Artemia franciscana is 34 kDa. Bioinformatic analyses reveal features typical of a Group 3 LEA protein, and subcellular localization programs predict targeting of the mature peptide to the mitochondrial matrix, based on an N-terminal, amphipathic presequence. Real-time quantitative PCR shows that Afralea3m mRNA is expressed manyfold higher in desiccation-tolerant embryonic stages when compared with intolerant nauplius larvae. Mitochondrial localization of the protein was confirmed by transfection of human hepatoma cells (HepG2/C3A) with a nucleotide construct encoding the first 70 N-terminal amino acids of AfrLEA3m in-frame with the nucleotide sequence for green fluorescence protein. The chimeric protein was readily incorporated into mitochondria of these cells. Successful targeting of a protein to human mitochondria by use of an arthropod signaling sequence clearly reveals the highly conserved nature of such presequences, as well as of the import machinery. Finally, mitochondria isolated from A. franciscana embryos, which naturally contain AfrLEA3m and trehalose, exhibit resistance to water stress (freezing) as evidenced by an unchanged capacity for oxidative phosphorylation on succinate + rotenone, a resistance that is absent in mammalian mitochondria lacking AfrLEA3m.
Fluctuation in cellular water content is a universal problem confronting a variety of organisms, and any environmental stress that impacts cellular water poses a risk to life (1). Nevertheless, some animals are able to cope with virtually complete loss of cellular water for prolonged times (2, 3), a phenomenon termed anhydrobiosis. Tolerance to water stress (i.e. evaporative water loss, freezing, and osmotic removal of water) is most likely achieved by several different mechanisms designed to protect and repair the structures and functions of cells and tissues. Many desiccation-tolerant organisms respond to water stress by intracellular accumulation of selected sugars, amino acids and derivatives, and methylamines (often occurring with urea) termed “compatible” osmolytes (4–7). Organic solutes such as the non-reducing sugar trehalose can actually stabilize biological structures during severe drying (8–12). Evidence indicates that the presence of small stress proteins and Late Embryogenesis Abundant (LEA)2 proteins is also important for cellular protection during drying in eukaryotic cells (13–16). However, the issue of how subcellular compartments in nucleated cells are protected during water stress has received far less attention (17–20), and this lack of understanding may explain our limited success in stabilizing eukaryotic cells from nontolerant species (21–27). We report here the novel targeting of a LEA protein to the mitochondrial compartment of an animal species and the associated protection against water stress.
Trehalose stabilizes membrane structure through physical intercalation with phospholipids (25, 28) and protects proteins against denaturation incurred during drying (29) and freezing (30, 31). Protective macromolecules are of several types but include Artemia P26, Hsp 21 and Hsp 22, and LEA proteins (13, 32–36). LEA proteins were first identified in land plants (37), and their expression is associated with desiccation tolerance in seeds and anhydrobiotic plants. The expression of LEA proteins is not restricted to plants, having been documented in bacteria, cyanobacteria, fungi, nematodes (38–40), and most recently in desiccation-tolerant embryos of the brine shrimp Artemia franciscana (41) and a chironomid insect larva (14). Among various physiological roles for LEA proteins, stabilization of sugar glasses (vitrified, non-crystalline structure in cells promoted by sugars like trehalose) is often suggested (38), along with protein stabilization via protein-protein interaction (18), ion sequestration (18), and formation of structural networks (39). Such networks have been hypothesized to increase cellular resistance to physical stresses imposed by desiccation (15). Recent studies suggest that a combination of trehalose and protective proteins works synergistically in providing protection during desiccation (16, 43, 44).
It is reasonable to hypothesize that mitochondria are preserved during water stress by basically the same mechanisms that govern protection of the plasma membrane and cytoplasmic proteins. Such stabilization of organelles presumably would require LEA proteins to be targeted to multiple cellular locations. Recently, the occurrence of a plant LEA protein that resides in the mitochondria has been reported for pea seeds (18). Thus far, no evidence exists that animal LEA proteins are accumulated within subcellular structures. Our results document a mitochondria-targeted LEA protein (AfrLEA3m) in desiccation-tolerant embryos of the brine shrimp A. franciscana and underscore the importance of preserving mitochondrial function during water stress in animals.
EXPERIMENTAL PROCEDURES
Animals—Encysted embryos of the brine shrimp, A. franciscana Kellogg (Great Salt Lake population), were either obtained in the dehydrated state (post-diapause) from Sanders Brine Shrimp Co. (Ogden, UT) or collected from the surface of the Great Salt Lake in the hydrated state (diapause) (45). Embryo viability and preservation of the diapause state during storage were evaluated as described previously (46).
Cells—HepG2/C3A cells (ATCC, Manassas, VA), were grown in 75-cm2 cell culture flasks (Corning Inc., Corning, NY) containing OPTI-MEM 1 supplemented with 5% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen). Cells were maintained in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. Medium was renewed every 3–4 days. For subcultures, cells were detached using 0.05% trypsin-EDTA solution (Invitrogen) and replated at 1–3 × 106 cells/plate. The number of cells was determined by counting with a hemocytometer (Hausser and Son, Philadelphia, PA). To ensure that the cell cultures were mycoplasma-free, the MycoAlert mycoplasma detection kit (Cambrex Bio Science Rockland, Inc., Rockland, ME) was used periodically.
Sequencing of Afrlea3m—DNA inserts were sequenced directly from plasmids isolated from bacterial clones picked from a full-length, unidirectional cDNA Library (Lambda Uni-ZAP XR Vector). The library was prepared from poly(A) mRNA purified and pooled from active and diapause stage embryos. Sequencing utilized BigDye terminator chemistry and an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Primer walking was used to ensure that full-length sequence was obtained. Sequences were assembled using Sequencher software (Gene Codes Co., Ann Arbor, MI).
Bioinformatic Analyses—Sequences were compared with the GenBank™/NCBI data base using BLAST software. The Kyte and Doolittle algorithm (47) was used to construct the hydropathy plot (using ExPASy ProtScale). The hydropathy plot was computed with a 9-residue moving average. The SDSC Biology Workbench 3.2 site was used for multiple sequence alignments (CLUSTALW) and evaluation of amino acid abundance (AASTATS). Secondary structure predictions were run with SOPMA (48), Gor 4, and Porter (at University College Dublin). Subcellular localization of proteins was predicted using TargetP, MitoProt, and Predator. De novo repeat detection in the AfrLEA3m protein sequence was performed with the TRUST Server (IBIVU) and RADAR (Rapid Automatic Detection and Alignment of Repeats).
Isolation of Total RNA and Preparation of cDNA for Real-time Quantitative PCR (rtqPCR)—Post-diapause embryos were hydrated and incubated at room temperature in medium equilibrated with air as described previously (45). Diapause embryos were incubated for 4 days as reported previously (46) to allow individuals not in diapause to hatch; larvae and empty shells were removed. To obtain samples of larvae (24 h), swimming nauplii were separated from the shed chorion. After the desired incubation times, RNA extractions were performed with an RNeasy Midi kit (Qiagen, Valencia, CA) as per the manufacturer's instructions for animal tissues. The concentration of RNA in each sample was determined spectrophotometrically (A260). The DyNAmo cDNA synthesis kit (New England Biolabs, Ipswich, MA), which utilizes M-MuLV RNase H+ reverse transcriptase and random hexamer primers, was employed to reverse transcribe total RNA for rtqPCR experiments.
rtqPCR Analysis—Quantification of the reverse-transcribed mRNA from various developmental stages of A. franciscana was performed with an iCycler (Bio-Rad Laboratories) and SYBR Green. PCR reaction mixes were prepared by using a SYBR Green master mix (SABiosciences, Frederick, MD) with a total reaction volume of 25 μl. Cycling parameter were 10 min at 95 °C and then 40 cycles of 95 °C (15 s) and 60 °C (60 s) followed by a melting curve analysis. Every developmental stage was evaluated with 4–6 independent experiments, each with three nested replicates, and with reference dye normalization. The cycle threshold (Ct) value was determined with the manufacturer's software. After determination of the PCR efficiencies (49), all Ct values were normalized to the expression of a bona fide housekeeping gene, α-tubulin (50). Primer sequences for α-tubulin were 5′-CGA CCA TAA AAG CGC AGT CA-3′ and 5′-CTA CCC AGC ACC ACA GGT CTC T-3′, and for Afrlea3m, they were 5′-ATA AAG GTG ACA CTC CGC CAC CAA-3′ and 5′-TGC ACC TTC AGC AAC ATC TTT GGC-3′. Values for the -fold change in expression are given as means ± S.E., and significance was assessed by applying the t test of the mean with Minitab 12 (Minitab Inc., State College, PA).
Transient Expression of GFP Fusion Protein in HepG2/C3A Cells—Using standard techniques, we fused a nucleotide construct encoding for the first 70 N-terminal amino acids of AfrLEA3m in-frame with the nucleotide sequence for a modified GFP protein harbored in a reporter plasmid (pEGFP-N3 vector, Clontech). The Afrlea3m fragment was obtained by PCR that utilized a forward primer containing the Kozak consensus sequence and the XhoI restriction site (5′-AGT TCT CGA GCC ACC ATG GTG TCC AAG CGT TTA ATT AAA AGC-3′), a reversed primer containing the BamHI restriction site (5′-CGC GGA TCC ATA TTC TGG ATC ATA CTC TCC CTT AGC-3′), and the AfrLEA3m cDNA clone from our library as template. PCR was performed with Phusion hot start DNA polymerase (New England Biolabs) as described by the manufacturer's instructions. After agarose gel electrophoreses, the PCR product was cleaned with the Wizard SV gel clean-up system (Promega, Madison, WI). The 210-bp-long PCR product and the destination vector (pEGFP-N3 vector, Clontech) were digested with XhoI and BamHI. After ligation with T4 DNA ligase (New England Biolabs), this vector encoding the AfrLEA3m presequence plus GFP was used to transform XL-1 Blue cells (Stratagene, La Jolla, CA) to amplify the plasmid for final sequence confirmation and transfection of mammalian cells. Finally, FuGENE HD transfection reagent (Roche Diagnostics) was used to transfect HepG2/C3A cells with the expression vector encoding the above chimeric protein. Control cells were transfected with the unmodified pEGFP-N3 vector. Cellular localization of recombinant protein was visualized by confocal imaging 4 days after cell transfection. Images were acquired with a Leica Microsystems TCS SP2 microscope equipped with a Plan Apo 63× N.A. 1.4 oil immersion lens. Three lasers, 488 nm at 15% power, 543 nm at 25% power, and 633 nm at 50% power, were used to sequentially excite GFP, MitoTracker Red and DRAQ 5 (Alexis Biochemicals, San Diego, CA), respectively. Emission passes of 500–535, 558–613, and 655–746 nm were used to detect the signals.
Mitochondrial Respiratory Capacity Pre- and Post-freezing—Mitochondria were isolated from A. franciscana embryos as described by Reynolds and Hand (46), and respiration measurements were performed first on fresh preparations using an Oxygraph-2K (Oroboros Instruments, Innsbruck, Austria; (51)). Briefly, mitochondria were added to a 2.0-ml final volume of respiration medium (240–300 μg/ml mitochondrial protein) that consisted of 0.5 m trehalose, 150 mm KCl, 10 mm KH2PO4, 20 mm HEPES, 1 mm EGTA, 2 mm MgCl2, and 0.5% (w/v) BSA (fatty acid-free, fraction V), titrated to pH 7.5 with KOH (total osmolality, 918 mosm). In some experiments, a mammalian respiration medium (MIRO5, Oroboros Instruments) was used for a comparison that consisted of 110 mm sucrose, 60 mm potassium lactobionate, 20 mm HEPES, 20 mm taurine, 10 mm KH2PO4, 3 mm MgCl2, 0.5 mm EGTA, 0.1% BSA (fatty acid-free, fraction V), titrated to pH 7.1 with KOH (total osmolality, 330 mosm). After temperature equilibration at 25 °C, 5 mm succinate plus 1 μm rotenone was added to the chamber, and state 2 respiration was recorded for several minutes. State 3 respiration was initiated by adding 250 μm ADP. State 4 respiration was recorded for at least 5 min once ADP was depleted. Respiration data were analyzed with DatLab software (Oroboros Instruments). Oxygen flux (Jo2 nmol O2 s-1 mg protein-1) was calculated as the time derivative of oxygen concentration, and the respiratory control ratio (RCR) was calculated as Jo2 (state 3)/Jo2 (state 4). Mitochondria suspended in 500 mm trehalose, 150 mm KCl, 20 mm HEPES, and 1 mm EGTA (pH 7.5) were pipetted in 0.5-ml aliquots into microtubes, wrapped in bubble wrap, and placed in a -80 °C freezer for one month. Samples were thawed, placed on ice, and assayed for respiratory capacity as described above. Mitochondrial protein was assayed using Coomassie Plus™ protein assay reagent and BSA as the standard (Pierce).
RESULTS AND DISCUSSION
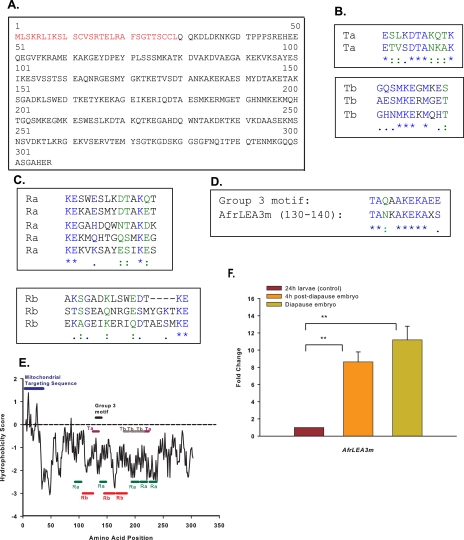
Sequence Analysis—Afrlea3m encodes a polypeptide of 307 amino acids (Fig. 1A) that exhibits high similarity to LEA proteins found in a wide range of organisms including plants (e.g. accession numbers: NP_190872, Arabidopsis thaliana, thale cress, e-value 3e-14; CAF32327, Pisum sativum, pea, 3e-11), a nematode (NP_001024042, Caenorhabditis elegans, 2e-14), and an insect (BAE92617, Polypedilum vanderplanki, 1e-10). High homology is also found to a recently identified LEA protein from A. franciscana (ACA47268, 2e-12) that was reported earlier by us (41). The percentages of identities of amino acid residues between these proteins and AfrLEA3m average about 29%.
FIGURE 1.
Bioinformatic features of AfrLEA3m and its mRNA expression. A, full-length deduced amino acid sequence for AfrLEA3m from embryos of A. franciscana. The 29-amino acid N-terminal leader sequence is highlighted in red. B and C, repetitive amino acid motifs for AfrLEA3m. D, alignment of amino acids 130–140 to a historical 11-mer repeat seen for some group 3 LEA proteins. E, Kyte and Doolittle (47) hydropathy plots for the deduced protein AfrLEA3m indicates strong hydrophilicity (shown by values below zero). A more hydrophobic N-terminal region of about 30 amino acids is indicative of a subcellular targeting sequence. The position of the repetitive motifs were found with the software programs RADAR (Ra, Rb) and TRUST (Ta, Tb), and the group 3 LEA motifs are labeled. F, mRNA expression profile for the gene Afrlea3m from A. franciscana embryos. LEA mRNA is maintained 9–11-fold higher in the two desiccation-tolerant embryonic stages when compared with the desiccation-intolerant nauplius larva. Double asterisks indicate that the paired means are statistically different (t test, p < 0.05).
Based on all three software programs utilized, a mitochondrial localization is projected for AfrLEA3m: 67.6% probability (Target P), 79.4% (MitoProt II), and 54% (Predator). Target P and MitoProt II software predicts AfrLEA3m to be synthesized as a precursor containing a N-terminal presequence of 29 amino acids (Fig. 1A) that is likely to be cleaved after the protein is imported into the mitochondrion. Such N-terminal, amphipathic leader sequences, when present without other internal targeting sequences, direct proteins to the mitochondrial matrix in almost all cases (52–54).
The deduced molecular mass of the AfrLEA3m polypeptide is 34 kDa and would be reduced to 30.9 kDa upon cleavage of the presequence. The primary structure is highly enriched in amino acids with hydrophilic (58.9%) and charged (37%) residues. Cysteine is not found in the mature protein, a common feature of many LEA proteins; however, multiple cysteines are present in the N-terminal targeting sequence of AfrLEA3m. A common characteristic of group 3 LEA proteins is a high α-helix content (55). Secondary structure analysis carried out with several programs predicts a high proportion of α-helices (SOPMA, 84%; Porter, 75%; Gor4, 61%) in AfrLEA3m and reveals the underrepresentation of glycine and arginine. The primary sequence of AfrLEA3m displays several repeated motifs, which is a common feature of low complexity proteins (Fig. 1, B and C). A motif that bears strong resemblance to the historical 11-mer repeat seen for some group 3 LEA proteins (56, 57) is found once in AfrLEA3m (amino acids 130–140) (Fig. 1D), but it does not repeat. A lack of concordance with the historical repeats is not uncommon even among plant LEA proteins (cf. Ref. 18). Finally, a number of important features for AfrLEA3m can be seen in its hydropathy plot (Fig. 1E). With the exception of the amphipathic presequence that is overall more hydrophobic, the remainder of the protein is very hydrophilic. The repetitive motifs in the mature protein mentioned above can be clearly seen in this plot.
Differential Expression of Afrlea3m mRNA in Dehydration-tolerant Stages—There is a strong up-regulation of Afrlea3m mRNA that is tightly associated with the two developmental stages of A. franciscana that are dehydration tolerant: a 9-fold increase in the post-diapause embryo and an 11-fold increase in the diapause embryos, as compared with the dehydration-intolerant nauplius larva (Fig. 1F). These expression profiles are consistent with a functional role for AfrLEA3m in dehydration tolerance.
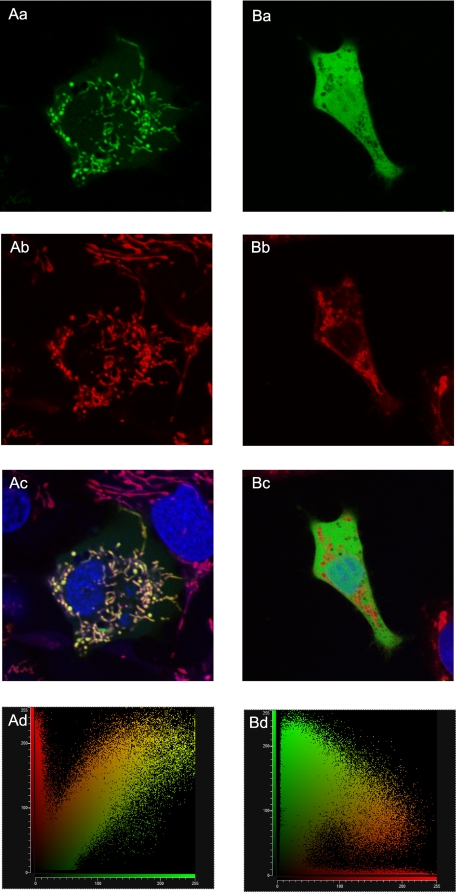
Experimental Evidence for Mitochondrial Targeting—When a nucleotide construct encoding for the first 70 N-terminal amino acids of AfrLEA3m is ligated to the nucleotide sequence for GFP and transfected into human hepatoma cells (HepG2/C3A), the chimeric protein is expressed and then imported into mitochondria (Fig. 2, Aa–Ad). The mitochondrial network of these cells is clearly targeted, as verified by co-localization of Mitotracker red (Fig. 2Ac). In contrast, the GFP lacking the AfrLEA3m leader sequence is not targeted to mitochondria, is dispersed in the cytoplasm (Fig. 2Ba), and does not co-localize with Mitotracker Red (Fig. 2Bc). Quantitative confirmation of the presence and absence of co-localization is provided by the scatter plots presented in Fig. 2 (Ad and Bd). These results demonstrate that a mitochondrial targeting sequence is an intrinsic component of AfrLEA3m and strongly suggest that AfrLEA3m is naturally localized to mitochondria of A. franciscana. The results also indicate the highly conserved nature of the protein import machinery for mitochondria of mammalian and invertebrate cells, and indirectly, of the targeting sequence as well.
FIGURE 2.
Human hepatoma cells (HepG2/C3A) transfected either with a vector encoding a chimeric protein composed of the leader sequence from AfrLEA3m plus GFP (frames Aa–Ad) or with an expression vector encoding only GFP (Ba–Bd). Co-staining with Mitotracker Red highlights the mitochondrial network (Ab and Bb; red color). Green and red fluorescence are co-localized in cells transfected with the chimeric protein (Ac) but not in cells transfected with GFP-only (Bc), where GFP remains in the cytoplasm. Mitotracker Red fluorescence is not found in the nucleus (Ac and Bc; blue indicates the nucleus). Co-localization is quantitatively supported by the scatter plot of red versus green channels in Ad, but substantial separation of green and red florescence is confirmed in Bd.
As noted earlier, a synergistic stabilization of proteins by trehalose and LEA protein has been observed (16), and a similar synergism has been reported during cell desiccation between trehalose and the stress protein p26 (43). Thus, we predict that for optimal mitochondrial stabilization by AfrLEA3m, trehalose must be present inside and outside of mitochondria. Like many sugars, trehalose is not permeable to biological membranes. This is a practical problem because evidence shows that trehalose is most effective as a stabilizer during drying when it is present on both surfaces of a membrane (21, 25, 42, 58, 59). As water is removed, hydroxyl groups of trehalose form hydrogen bonds with phospholipid head groups on each side of the lipid bilayer and prevent lipid phase transitions during rehydration. Thus, in the dried state, direct contact of trehalose is required with all membrane surfaces of cells, organelles, and macromolecules for optimal preservation.
Consistent with the above predictions, our previous studies have shown that mitochondria isolated from rat liver, which contained trehalose artificially loaded into the matrix via transient opening of the mitochondrial permeability transition pore, recovered 86% of their original membrane potential (ΔΨ) upon rehydration following drying to 0.1 g of water/g of solids (trehalose was also present extramitochondrially (17)). By comparison, 30% of ΔΨ was recovered by mitochondria dried with trehalose present only outside and only 20% in controls without any trehalose at all. Oxidative phosphorylation was partially protected by trehalose outside and loaded within the matrix. These data show that stabilizing both the inner and the outer mitochondrial membranes improves mitochondrial performance after drying. Similarly, Yamaguchi et al. (19) showed that when rat liver mitochondria were frozen with trehalose present only extramitochondrially and in the intermembrane space, i.e. absent from the matrix, mitochondrial oxidative phosphorylation was severely compromised. In other words, properties dependent on functionality of the inner membrane were disrupted by freezing, whereas selected properties that only required an intact outer membrane were protected. Importantly, mitochondria isolated from A. franciscana embryos contain trehalose inside the matrix naturally,3 and based on what is reported here, at least one LEA protein (AfrLEA3m) is targeted to the mitochondrial matrix.
Freezing Tolerance of Mitochondria Isolated from A. franciscana Embryos—In light of the above discussion, we chose to freeze mitochondria from A. franciscana embryos in a trehalose solution (with trehalose and AfrLEA3m naturally present internally). Table 1 shows the recovery of remarkably high respiratory control ratios (succinate plus rotenone) for frozen/thawed mitochondria when compared with control, non-frozen mitochondria. These RCR values (ranging from 4.5 to 9.1) are much higher than those reported for mitochondria frozen/thawed without LEA protein but with trehalose present outside (19) or for mitochondria desiccated to 0.1 g of water/g of solids with trehalose present both outside and in the matrix but no LEA protein. Future studies will incorporate the addition of cytoplasmic-targeted LEA proteins on the outside of mitochondria to further improve stability during freezing.
TABLE 1.
Respiration of non-frozen and frozen mitochondria isolated from A. franciscana embryos (succinate + rotenone as substrate) Mitochondria were assayed in medium matching the internal osmotic concentration of A. franciscana mitochondria and in a typical medium used for mammalian mitochondria (MIRO5, see “Experimental Procedures”).
| State 3a | State 4a | RCR | Average RCR | |
|---|---|---|---|---|
| Control (non-frozen) | ||||
| Artemia Resp. medium | 1.42 | 0.215 | 6.60 | 6.5 |
| 1.88 | 0.297 | 6.33 | ||
| MIRO5 | 1.73 | 0.305 | 5.67 | 5.6 |
| 1.74 | 0.310 | 5.61 | ||
| Frozen (–80 °C, 1 monthb) | ||||
| Artemia Resp. medium | 1.06 | 0.250 | 4.24 | 4.5 |
| 1.04 | 0.223 | 4.66 | ||
| MIRO5 | 1.73 | 0.191 | 9.06 | 9.1 |
| 1.74 | 0.191 | 9.11 |
nmol O2 s–1 mg of protein–1
Frozen in 500 mm trelalose, 150 mm KCl, 20 mm Hepes, 1 mm EGTA, pH 7.5
In summary, our results support the presence of a mitochondria-targeted LEA protein for the first time in an animal species. Further, our data indicate a functional importance of AfrLEA3m for stabilization of animal mitochondria during water stress.
Acknowledgments
Appreciation is extended to Matthew Brown of the Socolofsky Microscopy Center at Louisiana State University for assistance with confocal fluorescence microscopy and to Scott Herke of the Genomics Facility for DNA sequencing.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) FJ592175 The amino acid sequence of this protein can be accessed through NCBI Protein Database under NCBI accession number ACM16586.
This work was supported, in whole or in part, by National Institutes of Health Grant 2 RO1 DK046270-14A1.
Footnotes
The abbreviations used are: LEA, Late Embryogenesis Abundant; RtqPCR, real-time quantitative PCR; GFP, green fluorescent protein; BSA, bovine serum albumin; RCR, respiratory control ratio.
J. A. Reynolds, M. A. Menze, G. D. Elliott, M. Toner, and S. C. Hand, unpublished results.
References
- 1.Somero, G. N. (1992) in Water and Life (Osmond, C. B., and Bolis, C. L., eds) pp. 3-18, Springer-Verlag, Berlin
- 2.van Leeuwenhoek, A. (1798, 1807) in The Select Works of Antony van Leeuwenhoek (Hoole, S. ed.), vol. 2, pp. 207-213, G. Sidney, London [Google Scholar]
- 3.Crowe, J. H., and Clegg, J. S. (1973) Anhydrobiosis, Dowden, Hutchinson and Ross, Stroudsburg, PA
- 4.Brown, A. D., and Simpson, J. R. (1972) J. Gen. Microbiol. 72 589-591 [DOI] [PubMed] [Google Scholar]
- 5.Clark, M. E., and Zounes, M. (1977) Biol. Bull. (Woods Hole) 153 468-484 [Google Scholar]
- 6.Somero, G. N. (1986) Am. J. Physiol. 251 R197-R213 [DOI] [PubMed] [Google Scholar]
- 7.Yancey, P. H., Clark, M. E., Hand, S. C., Bowlus, R. D., and Somero, G. N. (1982) Science 217 1214-1222 [DOI] [PubMed] [Google Scholar]
- 8.Xie, G., and Timasheff, S. N. (1997) Biophys. Chem. 64 25-43 [DOI] [PubMed] [Google Scholar]
- 9.Lin, T. Y., and Timasheff, S. N. (1994) Biochemistry 33 12695-12701 [DOI] [PubMed] [Google Scholar]
- 10.Timasheff, S. N. (2002) Biochemistry 41 13473-13482 [DOI] [PubMed] [Google Scholar]
- 11.Bolen, D. W. (2001) Methods Mol. Biol. 168 17-36 [DOI] [PubMed] [Google Scholar]
- 12.Rosgen, J., Pettitt, B. M., and Bolen, D. W. (2007) Protein Sci. 16 733-743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willsie, J. K., and Clegg, J. S. (2001) J. Exp. Biol. 204 2339-2350 [DOI] [PubMed] [Google Scholar]
- 14.Kikawada, T., Nakahara, Y., Kanamori, Y., Iwata, K., Watanabe, M., McGee, B., Tunnacliffe, A., and Okuda, T. (2006) Biochem. Biophys. Res. Commun. 348 56-61 [DOI] [PubMed] [Google Scholar]
- 15.Goyal, K., Tisi, L., Basran, A., Browne, J., Burnell, A., Zurdo, J., and Tunnacliffe, A. (2003) J. Biol. Chem. 278 12977-12984 [DOI] [PubMed] [Google Scholar]
- 16.Goyal, K., Walton, L. J., and Tunnacliffe, A. (2005) Biochem. J. 388 151-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, X. H., Aksan, A., Menze, M. A., Hand, S. C., and Toner, M. (2005) Biochim. Biophys. Acta 1717 21-26 [DOI] [PubMed] [Google Scholar]
- 18.Grelet, J., Benamar, A., Teyssier, E., Avelange-Macherel, M. H., Grunwald, D., and Macherel, D. (2005) Plant Physiol. 137 157-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi, R., Andreyev, A., Murphy, A. N., Perkins, G. A., Ellisman, M. H., and Newmeyer, D. D. (2007) Cell Death Differ. 14 616-624 [DOI] [PubMed] [Google Scholar]
- 20.Hand, S. C., and Hagedorn, M. (2008) in Oceans and Human Health: Risks and Remedies from the Seas (Walsh, P. J., Smith, L. E., Fleming, L. E., Solo-Gabriele, H., and Gerwick, W. H., eds) pp. 611-629, Academic Press, Orlando, FL
- 21.Chen, T., Acker, J. P., Eroglu, A., Cheley, S., Bayley, H., Fowler, A., and Toner, M. (2001) Cryobiology 43 168-181 [DOI] [PubMed] [Google Scholar]
- 22.Gordon, S. L., Oppenheimer, S. R., Mackay, A. M., Brunnabend, J., Puhlev, I., and Levine, F. (2001) Cryobiology 43 182-187 [DOI] [PubMed] [Google Scholar]
- 23.Eroglu, A., Toner, M., and Toth, T. L. (2002) Fertil. Steril. 77 152-158 [DOI] [PubMed] [Google Scholar]
- 24.Acker, J. P., Fowler, B., Lauman, S., Cheley, S., and Toner, M. (2002) Cell Preserv. Tech. 1 129-140 [Google Scholar]
- 25.Crowe, J. H., Crowe, L. M., and Tablin, F. (2005) Integr. Comp. Biol. 44 542-542 [Google Scholar]
- 26.Elliott, G. D., Liu, X. H., Cusick, J. L., Menze, M., Vincent, J., Witt, T., Hand, S., and Toner, M. (2006) Cryobiology 52 114-127 [DOI] [PubMed] [Google Scholar]
- 27.Eroglu, A., Russo, M. J., Bieganski, R., Fowler, A., Cheley, S., Bayley, H., and Toner, M. (2000) Nat. Biotechnol. 18 163-167 [DOI] [PubMed] [Google Scholar]
- 28.Crowe, J. H., Oliver, A. E., and Tablin, F. (2002) Integr. Comp. Biol. 42 497-503 [DOI] [PubMed] [Google Scholar]
- 29.Carpenter, J. F., Martin, B., Crowe, L. M., and Crowe, J. H. (1987) Cryobiology 24 455-464 [DOI] [PubMed] [Google Scholar]
- 30.Carpenter, J. F., Hand, S. C., Crowe, L. M., and Crowe, J. H. (1986) Arch. Biochem. Biophys. 250 505-512 [DOI] [PubMed] [Google Scholar]
- 31.Carpenter, J. F., Crowe, L. M., and Crowe, J. H. (1987) Biochim. Biophys. Acta 923 109-115 [DOI] [PubMed] [Google Scholar]
- 32.Clegg, J. S., Jackson, S. A., and Warner, A. H. (1994) Exp. Cell Res. 212 77-83 [DOI] [PubMed] [Google Scholar]
- 33.Liang, P., Amons, R., Clegg, J. S., and MacRae, T. H. (1997) J. Biol. Chem. 272 19051-19058 [DOI] [PubMed] [Google Scholar]
- 34.Liang, P., Amons, R., Macrae, T. H., and Clegg, J. S. (1997) Eur. J. Biochem. 243 225-232 [DOI] [PubMed] [Google Scholar]
- 35.Qiu, Z., and Macrae, T. H. (2008) FEBS J. 275 3556-3566 [DOI] [PubMed] [Google Scholar]
- 36.Qiu, Z., and Macrae, T. H. (2008) Biochem. J. 411 605-611 [DOI] [PubMed] [Google Scholar]
- 37.Close, T. J., Kortt, A. A., and Chandler, P. M. (1989) Plant Mol. Biol. 13 95-108 [DOI] [PubMed] [Google Scholar]
- 38.Folkert, A., and Hoekstra, F. A. (2005) Integr. Comp. Biol. 45 725-733 [DOI] [PubMed] [Google Scholar]
- 39.Wise, M. J., and Tunnacliffe, A. (2004) Trends Plant Sci. 9 13-17 [DOI] [PubMed] [Google Scholar]
- 40.Close, T. J., and Lammers, P. J. (1993) Plant Physiol. 101 773-779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hand, S. C., Jones, D., Menze, M. A., and Witt, T. L. (2007) J. Exp. Zool. 307A 62-66 [DOI] [PubMed] [Google Scholar]
- 42.Buchanan, S. S., Menze, M. A., Hand, S. C., Pyatt, D. W., and Carpenter, J. F. (2005) Cell Preserv. Tech. 3 212-222 [Google Scholar]
- 43.Ma, X., Jamil, K., Macrae, T. H., Clegg, J. S., Russell, J. M., Villeneuve, T. S., Euloth, M., Sun, Y., Crowe, J. H., Tablin, F., and Oliver, A. E. (2005) Cryobiology 51 15-28 [DOI] [PubMed] [Google Scholar]
- 44.Wolkers, W. F., McCready, S., Brandt, W. F., Lindsey, G. G., and Hoekstra, F. A. (2001) Biochim. Biophys. Acta 1544 196-206 [DOI] [PubMed] [Google Scholar]
- 45.Covi, J. A., and Hand, S. C. (2005) J. Exp. Biol. 208 2783-2798 [DOI] [PubMed] [Google Scholar]
- 46.Reynolds, J. A., and Hand, S. C. (2004) Physiol. Biochem. Zool. 77 366-377 [DOI] [PubMed] [Google Scholar]
- 47.Kyte, J., and Doolittle, R. F. (1982) J. Mol. Biol. 157 105-132 [DOI] [PubMed] [Google Scholar]
- 48.Combet, C., Blanchet, C., Geourjon, C., and Deleage, G. (2000) Trends Biochem. Sci. 25 147-150 [DOI] [PubMed] [Google Scholar]
- 49.Pfaffl, M. W. (2001) Nucleic Acids Res. 29 2001-2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng, Y., Roy, P. J., Liang, P., and MacRae, T. H. (1998) Biochim. Biophys. Acta 1442 419-426 [DOI] [PubMed] [Google Scholar]
- 51.Gnaiger, E., Mendez, G., and Hand, S. C. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 11080-11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiedemann, N., Frazier, A. E., and Pfanner, N. (2004) J. Biol. Chem. 279 14473-14476 [DOI] [PubMed] [Google Scholar]
- 53.Neupert, W., and Herrmann, J. M. (2007) Annu. Rev. Biochem. 76 723-749 [DOI] [PubMed] [Google Scholar]
- 54.Pfanner, N., and Geissler, A. (2001) Nat. Rev. Mol. Cell Biol. 2 339-349 [DOI] [PubMed] [Google Scholar]
- 55.Wise, M. J. (2003) BMC Bioinformatics 4 52-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dure, L., III (1993) Plant J. 3 363-369 [DOI] [PubMed] [Google Scholar]
- 57.Cumming, A. C. (1999) in Seed proteins (Shewry, P., and Casey, R., eds) pp. 753-780, Kluwer Academic Publishers, Boston, MA
- 58.Leslie, S. B., Israeli, E., Lighthart, B., Crowe, J. H., and Crowe, L. M. (1995) Appl Environ. Microbiol. 61 3592-3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun, W. Q., Leopold, A. C., Crowe, L. M., and Crowe, J. H. (1996) Biophys. J. 70 1769-1776 [DOI] [PMC free article] [PubMed] [Google Scholar]