Abstract
The innate immune system provides an initial defense system against microbial infections and contributes to the development of adaptive immune response. Type I interferons play a pivotal role for the first line of defense against virus infections, and dendritic cells (DCs) are important sensors of pathogens responsible for priming of adaptive immune responses in lymphoid organs. Here we have investigated the role and mechanisms of activation of the MAPK pathway in innate immune responses induced by Sendai virus, a negative sense single-stranded RNA virus. Both p38 and JNK were activated in fibroblasts and DCs after infection with Sendai virus in a manner dependent on virus replication and RIG-I. Virus replication was also required for stimulation of interferon production in both cell types and interleukin-12 production in DCs. Blocking of p38 MAPK activation by the specific inhibitor SB202190 abolished the expression of these cytokines. p38 MAPK exerted its function independent of the MAPK-activated protein kinases MK2, MNK, and MSK1/2. We also observed that TRAF2 and TAK1 were essential for RIG-I-mediated activation of p38 MAPK. Interestingly, the kinase activity of p38 MAPK was required for its own phosphorylation, which was kinetically associated with TAB1 interaction. By contrast, the canonical p38 upstream kinase MKK3 was not involved in the p38-dependent response. Thus, activation of p38 MAPK by RIG-I proceeds via a TRAF2-TAK1-dependent pathway, where the enzymatic activity of the kinase plays an essential role. The p38 MAPK in turn stimulates important processes in the innate antiviral response.
Activation of immune responses against microbial pathogens depends on the recognition of pathogen-associated molecular patterns (PAMPs)5 by pattern recognition receptors (PRRs), which trigger a first line of defense against the infection and also promote development of antigen-specific adaptive immune response. The type I interferon (IFN) system forms the major component of innate immune responses against viral infections (1-3), primarily by directly inhibiting viral replication and activating cellular antiviral activities (4). Dendritic cells (DCs) are sentinel cells of the immune system having the capacity to initiate both innate and adaptive immune responses (5). Upon pathogen recognition, DCs become activated and up-regulate expression of co-stimulatory molecules (e.g. CD40 and CD86), secrete cytokines (such as interleukin (IL)-12), and migrate to secondary lymphoid organs to initiate maturation of the T cell response.
Cellular recognition of viruses and other microbial pathogens is mediated by germ line-encoded PRRs, which recognize PAMPs and activate intracellular signal transduction pathways that regulate the activation of host immune responses (4). The Toll-like receptors represent a class of membrane-bound PRRs, which recognize extracellular and intracellular PAMPs (4). Toll-like receptors are expressed in a cell type-specific manner and play an important role in DC maturation (6). Another class of PRRs involved in activation of antiviral response is the DEXD/H-box helicase family receptors, which includes three members: RIG-I (retinoid acid-inducible gene I), MDA5 (melanoma differentiation-associated gene 5), and Lgp2 (Laboratory of Genetics and Physiology 2) (4, 7, 8). RIG-I recognizes double-stranded RNA and uncapped 5′-triphosphate RNA molecules, which are found in the viral genomes of many RNA viruses or produced during viral replication (9-12); MDA5 recognizes double-stranded RNA (13, 14); and Lgp2 has been shown to have both a negative and a positive role in RIG-I/MDA5 signaling (15-17). RIG-I and MDA5 are expressed in many cell types, including fibroblasts and conventional CD11c+ DCs (14, 18), and they play an essential role in activation of the innate immune response against RNA viruses, including the production of IFN-α/β and maturation of DCs (14, 18).
PAMP-PRR interaction triggers several intracellular signaling pathways leading to activation of host cell cytokine gene expression and enhancement of other cellular functions. The pathway leading to activation of nuclear factor (NF)-κB is activated by most if not all PRRs. NF-κB plays a central role in driving the expression of proinflammatory mediators (4, 19). In addition, a subset of PRRs activate members of the IFN regulatory factor (IRF) family (4), which are essential for triggering the type I and type III IFN responses and providing efficient antiviral response (20, 21). A third important PRR-activated signaling pathway is the mitogen-activated protein kinase (MAPK) pathway, which have multiple functions, including cell cycle control, cell survival, and regulation of transcription, mRNA stability, and translation (22). These functions are mediated via the activation of MAPKs (p38, JNK, and ERK), which act by phosphorylating specific target proteins directly or via the activation of downstream MAPK-activated protein kinases (MKs) such as MAPK-interacting kinases (MNKs), mitogen- and stress-activated protein kinases (MSKs), MK2, and MK3 (22-25).
The RIG-I signaling pathway is dependent on the adaptor protein MAVS/IPS-1/CARDIF/VISA (26-29), which is located in the outer mitochondrial membrane (26), where the intracellular signaling is initiated (28). To induce type I IFN response, MAVS signals to IRF-3, through a pathway regulated by TRAF3 (30, 31). RIG-I also stimulates the NF-κB pathway, and recent work has shown that RIG-I signaling shares significant similarity with the signaling pathway downstream of the TNF receptor. For instance, both receptors rely on TRADD, RIP1, and FADD to activate NF-κB (27, 32-34). However, many of the details in the regulation of signaling by TNF-α and RIG-I to NF-κB need to be clarified. Similarly, the mechanism of how RIG-I activates the p38 MAPK is not well characterized.
In the present work, we show that the activation of the RIG-I pathway leads to activation of p38 MAPK, via a process that is dependent on TRAF2, TAK1, and p38 kinase activity. The activity of p38 in turn is essential for viral induction of type I IFNs as well as activation of conventional DCs.
EXPERIMENTAL PROCEDURES
Cells and Virus—Mouse embryonic fibroblasts (MEFs) derived from C57BL/6, Rig-I+/-, Rig-I-/- (S. Akira, Osaka Japan), Traf2-/-, Traf6+/-, Traf6-/- (T. Mak, Toronto, Canada), and Tak1-/- mice (14, 18, 35, 36) were maintained in Dulbecco's modified Eagle's medium supplemented with 15% fetal calf serum, 200 IU/ml penicillin, and 100 μg/ml streptomycin. For experiments, cells were plated in 96-well tissue culture plates in a concentration of 0.8 × 105 cells/well. The plates were left overnight in 5% CO2 at 37 °C to allow the cells to attach before further experiments were carried out. For isolation of total cellular RNA and Luminex experiments, cells were plated in 6-well tissue culture plates in a concentration of 5 × 106 cells/well in 2 ml of medium.
The murine dendritic cell line BC-1 was maintained in Iscove's modified Dulbecco's medium containing 10% fetal calf serum, 200 IU/ml penicillin, 100 μg/ml streptomycin, 600 mg/ml l-glutamine, 50 μm 2-mercaptoethanol, 30% NIH/3T3 conditioned medium, and 10 ng/ml recombinant murine granulocyte-macrophage colony-stimulating factor (R&D Systems). For experiments, cells were plated in 96-well tissue culture plates in a concentration of 1 × 105 cells/well. The plates were left overnight in 5% CO2 at 37 °C to allow the cells to attach before the experiments. For isolation of total cellular RNA and Luminex experiments, cells were plated in 6-well tissue culture plates in a concentration of 3 × 106 cells/well in 2 ml of medium.
Bone marrow (BM)-derived DCs were prepared as described elsewhere (37). Briefly, C57BL/6, MK2-/-, MSK1/2-/-, and MKK3-/- mice were sacrificed by cervical dislocation, and femur and tibia were removed. Bone marrow cells were flushed from the bone shafts with PBS, pipetted to break up cell aggregates, centrifuged, washed, counted, and put into culture in Petri dishes (Falcon; BD Biosciences). The cells were cultured at 2 × 105 cells/ml in culture medium containing 40 ng/ml recombinant murine granulocyte-macrophage colony-stimulating factor (R&D systems) in 5% CO2 at 37 °C. Fresh medium containing 40 ng/ml granulocyte-macrophage colony-stimulating factor was added after 3 days, and on day 5, half of the medium was replaced with fresh medium. After 7 days of culture, non-adherent cells were harvested for flow cytometry or infection with infectious or UV-inactivated Sendai virus (SeV). The cells were 80% CD11c-positive, as determined by flow cytometry (data not shown). For isolation of cell culture supernatant, cells were seeded at a density of 1 × 105 cells/well in a 96-well tissue culture plate and left overnight in an atmosphere of 5% CO2 at 37 °C to allow the cells to settle, after which they were infected with virus and/or chemical inhibitors/activators as indicated. For total RNA isolation and Luminex experiments, cells were plated on 6-well tissue culture plates in a concentration of 3 × 106 cells/well. The plates were left for 2 h in 37 °C in a humidified atmosphere with 5% CO2 before further treatment.
SeV (strain Cantell) was grown in 11-day-old embryonated hen eggs, as previously described (38). The infectivity titer of the virus in DCs was 4 × 109 plaque-forming units/ml (39). UV inactivation of the virus was performed by exposing the virus to UV light for 5 min unless otherwise indicated. The uninfected hen egg allantoic fluid did not stimulate proinflammatory cytokine expression in DCs, and the virus preparation did not contain lipopolysaccharide (data not shown).
Reagents—For inhibition of specific molecules in the MAPK pathways, the following compounds were used: SB202190 (p38 inhibitor) (Sigma), PD169316 (p38 inhibitor) (Sigma), BIRB 796 (p38 inhibitor) (Axon Medchem), Sp600125 (JNK inhibitor) (Sigma), and U0126 (MEK-1 inhibitor) (Sigma). CGP057380 (MNK inhibitor) was kindly provided by Dr. Hermann Gram (Novartis Pharma AG, Basel, Switzerland).
Flow Cytometry—Cells were counted and resuspended in RPMI 1640 supplemented with 10% fetal calf serum at a concentration of 1 × 107 cells/ml. Cells were incubated in a 96-well plate on ice in the dark for 40 min with combinations of the fluorescein isothiocyanate- and PE-conjugated antibodies at a concentration of 0.5 mg/ml. Antibodies used (all from BD Pharmingen) were as follows: fluorescein isothiocyanate-conjugated hamster anti-mouse monoclonal CD11c, fluorescein isothiocyanate-conjugated CD11b, PE-conjugated anti-mouse monoclonal CD8α (Ly-2), PE-conjugated anti-mouse CD86 and CD40, PE-conjugated anti-mouse I-A (major histocompatibility complex I), and PE-conjugated anti-mouse H-2K (major histocompatibility complex II). As isotype control antibodies, we used fluorescein isothiocyanate-conjugated hamster IgG1, λ1, PE-conjugated monoclonal rat IgG2a, and PE-conjugated monoclonal mouse IgG2a. The labeled cells were fixed in 1% paraformaldehyde diluted in PBS for 15 min, and fixed cells were kept in PBS at 4 °C until they were analyzed. Acquisition and analysis were performed with a flow cytometer (Coulter FS500). The data were stored in list mode files. A total of 30,000 cells were analyzed in each experiment by using a single laser system with a wavelength of 488 nm. Compensation was determined before the acquisition of data.
Cytokine Determinations—To measure cytokine production, culture supernatants were harvested after 12-24 h of infection. Cytokine levels in the supernatants were determined by ELISA. 96-well tissue culture plates were coated overnight at 4 °C with 100 μl of anti-IL-12 p40 (6 μg/ml; Pharmingen) capture antibodies diluted in coating buffer (15 mm NaHCO3, 35 mm Na2CO3, 0.2% sodium azide, pH 9.6). The cells were washed three times with washing buffer (PBS plus 0.05% Tween), blocked for 3 h with blocking buffer (PBS, 5% sucrose, 0.05% sodium azide, 1% bovine serum albumin), after which 100 μl of serial dilutions of IL-12 (rmIL12-p40; Pharmingen) standard or cell supernatants were added in duplicates or triplicates. The plates were left overnight at 4 °C. The plates were washed three times with washing buffer, and 100 μl of anti-IL-12 p40/p70 (Pharmingen) detection antibodies diluted in a reagent diluent (TBS, 0.05% Tween, 0.1% bovine serum albumin) was added and left to incubate for 2-3 h at room temperature. The plates were washed three times, incubated for 20 min with Streptavidin-horseradish peroxidase (R&D Systems) at room temperature, washed again three times, and developed with 100 μl of substrate reagent (Substrate Reagent Pack; R&D Systems) at room temperature for 2-5 min followed by the addition of 2 n H2SO4 to stop the reaction. Cytokine concentration was measured by an automated ELISA reader.
For measurement of IL-12p70 and CXCL10, we used Luminex kits purchased from Bio-Rad and BIOSOURCE, respectively. Briefly, the filter plate was washed with assay buffer, and 50 μl of freshly vortexed antibody-conjugated beads was added to each well. The plate was washed with assay buffer, and samples and standards were added. After a brief shake (30 s at 1,100 rpm), the plate was incubated at room temperature in the dark for 2 h with light shaking (300 rpm). After one wash step, 25 μl of the detection antibody was added to each well, and the plate was shaken and incubated as above. Subsequently, the plate was washed and incubated for 30 min with 50 μl of a streptavidin-PE solution with shaking (30 s at 1,100 rpm, 10 min at 300 rpm). Finally, the plate was washed, and 125 μl of assay buffer was added to each well, and the plate was shaken for 10 s at 1,100 rpm. Measurement was carried out immediately on the Bio-Plex reader.
IFN-α/β Bioassay—IFN-α/β bioactivity was measured by an L929 cell-based bioassay. L929 cells (2 × 104 cells/well in 100 μl) grown in minimum Eagle's medium supplemented with 5% fetal calf serum and antibiotics were incubated overnight at 37 °C in successive 2-fold dilutions of samples or murine IFN-α/β as a standard. Subsequently, vesicular stomatitis virus (VSV/V10) was added to the wells, and the cells were incubated for 2-3 days. The dilution giving 50% protection was defined as 1 unit/ml of IFN-α/β. To ensure that the bioassay was useful for measurement of IFN activity in samples containing p38 MAPK inhibitors, we performed control experiments, where samples spiked with IFN in the presence or absence of the inhibitors were subjected to IFN measurement by bioassay. We found that the concentrations of p38 MAPK inhibitor used in this study did not affect the result of the bioassay (data not shown).
Quantitative Reverse Transcription-PCR—Total RNA was extracted with the High Pure RNA isolation kit (Roche Applied Science) according to the recommendations of the manufacturer. For cDNA generation, RNA was subjected to reverse transcription with oligo(dT) as primer and Expand reverse transcriptase (both from Roche Applied Science). The cDNA was amplified by PCR using the following primers: IFN-β, 5′-CACTGGGTGGAATGAGACTAT-3′ (forward) and 5′-GACATCTCCCACGTCAATC-3′ (reverse); ISG56, 5′-GCCAGGAGGTTGTGCATC-3′ (forward) and 5′-ACCATGGGAGAGAATGCTGAT-3′ (reverse); IL12-p40, 5′-ACGTGAACCGTCCGGAGTAA-3′ (forward) and 5′-CCACAAAGGAGGCGAGACTCT-3′ (reverse); β-actin, 5′-TAGCACCATGAAGATCAAGAT-3′ (forward) and 5′-CCGATCCACACAGAGTACTT-3′ (reverse). The products were measured by the use of SYBR Green I (from Qiagen). All primers were obtained from DNA Technology (Aarhus, Denmark).
Detection of Phosphoproteins—For detection of the phosphorylation status of IκBα, p38, and JNK, we used Luminex technology. Briefly, the filter plate was washed with assay buffer, and 50 μl of freshly vortexed antibody-conjugated beads were added to each well. The plate was washed with assay buffer, and samples and standards were added. After a brief shake (30 s at 1,100 rpm), the plate was incubated at 4 °C overnight in the dark with light shaking (300 rpm). After one wash step, 25 μl of the detection antibody was added to each well, and the plate was shaken and incubated as above. Subsequently, the plate was washed and incubated for 30 min with 50 μl of a streptavidin-PE solution with shaking (30 s at 1,100 rpm, 10 min at 300 rpm). Finally, the plate was washed, 125 μl of assay buffer was added to each well, and the plate was shaken for 10 s at 1,100 rpm and read immediately on the Bio-Plex machine.
Immunoprecipitation and Western Blotting—For co-immunoprecipitations, cells were washed twice in phosphate-buffered saline and lysed in 850 μl of lysis buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol, 0.5% (v/v) Nonidet P-40 containing 1 mm phenylmethylsulfonyl fluoride, 0.01% (v/v) aprotinin, and 1 mm sodium orthovanadate). For immunoprecipitation, rabbit polyclonal anti-p38α (Abcam, Cambridge, UK) was precoupled to protein A-Sepharose beads overnight at 4 °C. The beads were then washed twice in lysis buffer and incubated with 1.5 mg of cell lysate/sample overnight at 4 °C. The immune complexes were washed three times in lysis buffer, boiled, and analyzed by standard SDS-PAGE and Western blotting techniques using rabbit polyclonal anti-p38α (as above) and rabbit polyclonal anti-TAB1 antibodies (Cell Signaling Technology, Danvers, MA) for blotting.
RESULTS
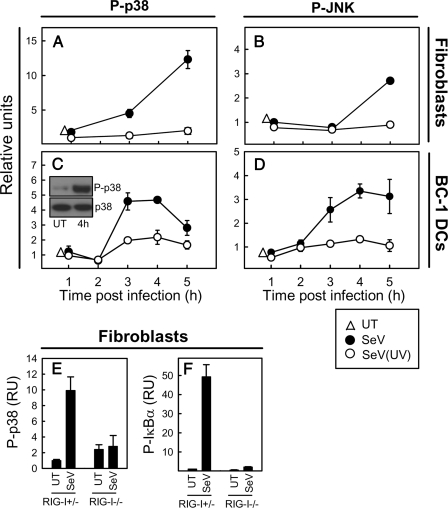
SeV Replication Stimulates Activation of MAPKs in a RIG-I-dependent Manner—To examine the ability of SeV to stimulate activation of MAPKs, we first studied MEFs, in which we observed that the virus strongly activated p38 and to a lesser extent also JNK (Fig. 1, A and B). For both kinases, the activation was dependent on the ability of the virus to replicate, since UV-irradiated virus was not able to activate these kinases. The MAPKs ERK1/2 were not activated by SeV (data not shown).
FIGURE 1.
SeV-mediated activation of p38 MAPK is dependent on viral replication and RIG-I. MEFs (A and B) or BC-1 DCs (C and D) were infected with live or UV-inactivated SeV at an MOI of 1. At the indicated time points after infection, cells were collected, cell extracts were prepared, and the phosphorylation levels of p38 and JNK were measured using Luminex technology. For comparison, p38 samples were also analyzed by Western blotting (C, inset). E and F, Rig-I+/- and Rig-I-/- MEFs were infected with SeV for 5 h, at which point cell extracts were prepared. Phosphorylation levels of p38 and IκBα were measured using Luminex technology. The data are shown as means of duplicate cultures ± S.D. UT, untreated cells.
To study the mechanism of DC activation during viral infection, we used a murine DC cell line, BC-1 (40), which was grown in parallel with BM-DCs. BC-1 cells resemble myeloid DCs, as assessed by their high expression of CD11b and CD11c and low expression of CD8α (Fig. S1). The cells express low levels of CD40 and major histocompatibility complex class I and II and intermediary levels of CD86, thus displaying a semimature phenotype. When looking for MAPK activation in response to SeV infection in BC-1 cells, we observed the same pattern of activation as in MEFs, although the kinetics of activation was somewhat faster in DCs (Fig. 1, C and D).
To study whether virus-induced activation of MAPKs was dependent on RIG-I, we compared the ability of Rig-I+/- and Rig-I-/- MEFs to activate p38 MAPK during SeV infection. As shown in Fig. 1E, SeV-mediated activation of p38 was fully dependent on RIG-I, hence resembling the activation of the NF-κB pathway (Fig. 1F). In BC-1 cells, the cellular response to SeV infection was not inhibited by chloroquine, which was capable of inhibiting TLR9 (Toll-like receptor 9) signaling (Fig. S2), thus showing that the response was independent of endosomal Toll-like receptors and hence indicating that the RIG-I pathway was responsible for activation of this cell type in response to SeV infection.
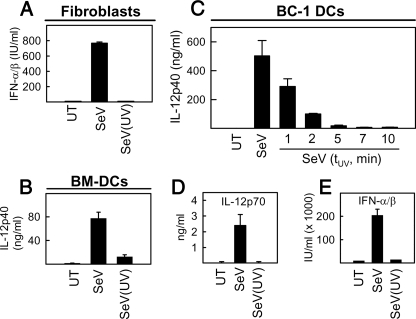
SeV Replication and p38 MAPK Activation Are Essential for Stimulation of Type I IFN Production and Activation of DCs—To examine whether the secretion of cytokines in response to virus infection required viral replication and was dependent on MAPKs, we stimulated MEFs, BC-1, and BM-DCs with infectious or UV-inactivated virus for subsequent examination of cytokine production. As shown in Fig. 2A, MEFs responded to SeV infection with a robust type I IFN response, which was fully dependent on viral replication. In DCs (BM-DCs or BC-1 cells), the production of IL-12p40 was also observed to be dependent on the replication of the virus (Fig. 2, B and C). Similar findings were made when examining expression of the bioactive IL-12 p70 heterodimer (Fig. 2D). In BC-1 cells, the type I IFN response was also abolished in cells stimulated with UV-inactivated virus (Fig. 2E).
FIGURE 2.
Viral replication is required for induction of type I IFN production in fibroblasts and full activation of dendritic cells. MEFs (A), BM-DCs (B), and BC-1 DCs (C-E) were infected with live or UV-inactivated SeV at MOI 1. Cell culture supernatants were collected 18 h after infection, and cytokine levels were determined. In C, the cells received infectious virus (MOI 1) or a similar dose of virus that had been UV-treated for the indicated times (1-7 min). The data are shown as means of triplicates ± S.D. UT, untreated cells.
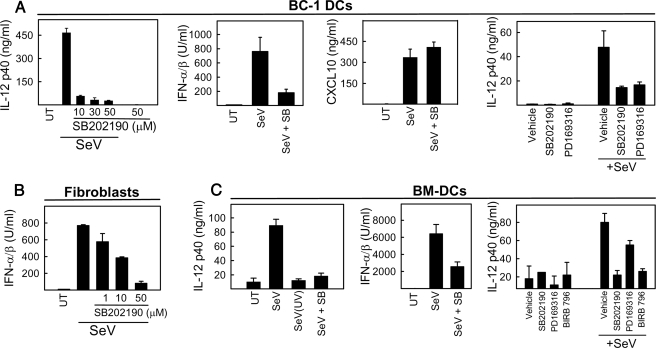
To investigate the involvement of the different MAPK pathways in induction of IFN production and maturation of DCs, we treated the cells with SB202190, SP600125, or U0126, which inhibit p38, JNK, and the ERK activators MEK1/2, respectively, and subsequently infected them with SeV. Supernatants were harvested for measurement of relevant cytokines. As seen in Fig. 3, A-C, treatment of MEF, BC-1 DC, and BM-DC cells with SB202190 inhibited virus-induced production of IFN-α/β and IL-12. By contrast, inhibition of the JNK and ERK pathways did not affect SeV-induced IL-12 production in BC-1 cells (Fig. S3). Importantly, SeV-induced expression of CXCL10, which is regulated through an IFN-stimulated response element and two NF-κB sites (41), was not affected by treatment with SB202190 (Fig. 3A). Given the well known nonspecific effects of all inhibitors, we wanted to corroborate the above findings by using other p38 inhibitors working through different mechanisms. We found that both PD169316 and BIRB 796 were able to inhibit SeV-induced IL-12 production in the BC-1 DCs and BM-DCs in a manner comparable with SB202190 (Fig. 3, A and C). Thus, RIG-I-mediated activation of p38 MAPK in response to SeV infection is important for stimulation of both type I IFN response and activation of DCs.
FIGURE 3.
MAPK p38 activity is essential for viral induction of type I IFNs in fibroblasts and full activation of dendritic cells. BC-1 DCs (A), MEFs (B), and BM-DCs (C) were pretreated with p38 MAPK inhibitors 30 min prior to infection with SeV (MOI 1). Cell culture supernatants were collected 12 h after infection, and cytokine levels were determined. Unless indicated otherwise, the concentrations used were as follows: SB202190, 30 μm; PD169316, 10 μm; BIRB 796, 10 μm. The data are shown as means of triplicates ± S.D. UT, untreated cells.
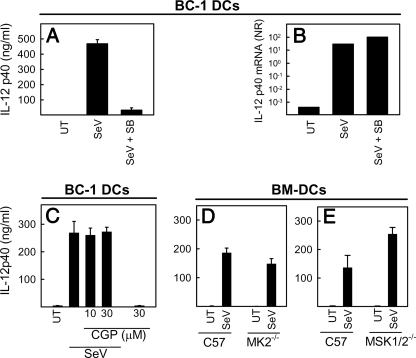
p38 MAPK Activates Innate Antiviral Immunity at the Posttranscriptional Level Independent of MNK1, MK2, and MSK1/2—In order to examine the level at which p38 MAPK was affecting SeV-induced gene expression, we compared the ability of SB202190 to inhibit IL-12p40 accumulation at the protein versus mRNA levels. Interestingly, the potent inhibition of IL-12p40 at the protein level (Fig. 4A), was not observed at the level of IL-12 p40 mRNA (Fig. 4B), suggesting that p38 MAPK acts through a mechanism affecting cytokine expression at the posttranscriptional level.
FIGURE 4.
SeV-induced DC activation through p38 is exerted at the posttranscriptional level and is independent of MNK1, MK2, and MSK1/2. A and B, BC-1 DCs were pretreated with SB202190 (30 μm) 30 min prior to infection with SeV (MOI 1). Supernatants and total RNA were harvested 12 and 5 h, after infection, respectively, and analyzed for the presence of IL-12p40 protein (A) and IL-12p40 mRNA (B). C, BC-1 DCs were seeded and pretreated with the MNK1 inhibitor CGP57380 30 min before infection with SeV at MOI 1. Cell culture supernatants were collected 12 h after infection, and IL-12p40 levels were determined by ELISA. D and E, BM-DCs were generated from C57BL/6, MK2-/-, and MSK1/2-/- mice and plated for 2 h in tissue culture plates before treatment as described above. At 12 h after SeV infection, cell culture supernatants were collected, and IL-12p40 protein production was determined by ELISA. The data are shown as means of triplicates ± S.D. UT, untreated cells.
The p38 MAPK exerts its activities through phosphorylation of downstream target molecules, including protein kinases, transcription factors, and other regulatory proteins (22). To examine the role of MKs in virus-stimulated innate immune response, we first treated BC-1 cells with the MNK1 inhibitor CGP57380, infected the cells with SeV, and analyzed the expression of IL-12p40. As shown in Fig. 4C, CGP57380 treatment had no affect on the expression of IL12p40, although it did inhibit the production of CXCL8 in response to anisomycin in HeLa cells (Fig. S4). Next we examined whether BM-DCs from MK2-/- and MSK1/2-/- mice showed impaired cytokine production in response to infection with live SeV. We found that MK2-/- and MSK1/2-/- DCs displayed unimpaired ability to produce IL-12p40 in response to SeV infection (Fig. 4, D and E). Similar findings were obtained when type I IFN production was measured (data not shown). Thus, p38 MAPK stimulates virus-specific innate immune responses through a posttranscriptional mechanism, which is independent of MNK1, MK2, and MSK1/2.
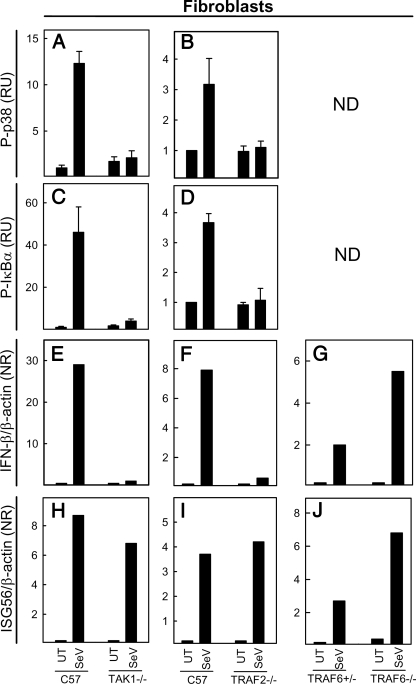
RIG-I-mediated Activation of p38 and NF-κB Is Dependent on TAK1 and TRAF2—The signaling pathway downstream of RIG-I has been reported to share similarities with TNF-α signaling (32), and we were interested in studying the potential role of TRAF2 or TAK1 in RIG-I signaling, both of which are essential for TNF-α-induced NF-κB activation (35, 36). MEFs from C57BL/6, TRAF2-/-, and TAK1-/- mice were infected with SeV, and cell lysates were harvested at 5 h postchallenge, followed by an examination of the phosphorylation status of p38 and IκBα. Although the virus potently stimulated phosphorylation of both p38 and IκBα in wild type cells, lack of either TAK1 or TRAF2 abolished the activation of MAPK and NF-κB pathways as well as the downstream IFN-β response (Fig. 5, A-F). The impaired responsiveness of the TRAF2-/- and TAK1-/- MEFs to SeV was not due to a general defect in the cells, since they responded to TNF-α as reported in the literature (35, 36, 42) and mounted a normal STAT2 response after stimulation with IFN-α (Fig. S5). We also found that TRAF2 and TAK1 were not involved in viral activation of IRF-3 and expression of ISG56 (data not shown) (Fig. 5, H and I). Finally, we observed that TRAF6-/- cells were fully capable of stimulating expression of IFN-β and ISG56 (Fig. 5, G and J). Thus, in our system, TRAF2 and TAK1 are selectively involved in RIG-I-induced NF-κB and MAPK activation.
FIGURE 5.
RIG-I-mediated activation of the MAPK and NF-κB pathways is dependent on TRAF2 and TAK1, which are essential for virus-induced type I IFN expression. Wild type, Tak1-/-, Traf2-/-, Traf6+/-, and Traf6-/- MEFs were seeded in triplicates and infected with SeV at MOI 1. 5 (A-D) and 6 (E-J) h postchallenge, cell extracts and total RNA, respectively, were harvested for measurement of the phosphorylation status of p38 (A and B) and IκBα (C and D) as well as mRNA expression of IFN-β (E-G) and ISG56 (H-J). The data are shown as means of duplicate cultures ± S.D. ND, not done. UT, untreated cells.
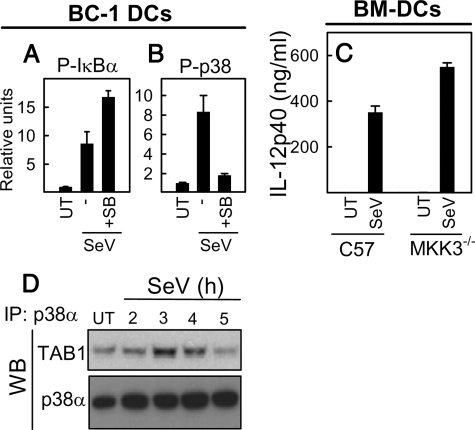
SeV-induced p38 Activation Is Dependent on p38 Kinase Activity and Correlated with p38-TAB1 Interaction—It has been reported that p38α can be activated via autophosphorylation by a mechanism that is dependent on association of p38 with TAB1 (44), and this mechanism has been suggested to be involved in the cellular response to intracellular pathogens (45). Therefore, we examined the effect of inhibition of p38 MAPK on its own phosphorylation. Interestingly, the kinase activity of p38 was required for phosphorylation of p38, whereas phosphorylation of IκBα was still taking place when p38 was inhibited by SB202190 (Fig. 6, A and B). Activation of p38 through autophosphorylation is independent of the canonical p38 upstream kinase MKK3 (45). To examine this phenomenon in more detail, we stimulated wild type and MKK3-/- BM-DCs with SeV and analyzed for virus-induced expression of IL-12p40, which is dependent on p38 MAPK (Fig. 3, B and C). Virus infection induced a strong production of IL-12p40 from wild type cells, and this response was even weakly enhanced in MKK3-/- cells (Fig. 6C).
FIGURE 6.
Viral activation of p38 MAPK requires its own kinase activity and leads to association of p38 MAPK with TAB1. A and B, BC1 DCs were pretreated with 30 μm SB202190 30 min before infection with SeV at MOI 1. 4 h later, cells were collected, cell extracts were prepared, and the amounts of phosphorylated p38 and IκBα were determined by Luminex. C, BM-DCs were generated from C57BL/6 and MKK3-/- mice and plated for 2 h in tissue culture plates before treatment as described above. At 12 h after SeV infection, cell culture supernatants were collected, and IL-12p40 protein production was determined by ELISA. D, BC-1 DCs were infected with SeV (MOI 1) for the indicated time intervals, and cell extracts were prepared and subjected to immunoprecipitation with anti-p38α antibodies. The immunoprecipitates (IP) were separated on SDS-PAGE and immunoblotted (WB) with anti-TAB1 antibodies. UT, untreated cells.
To investigate the potential interaction between p38α MAPK and TAB1, we infected BC-1 cells for the indicated times, collected cells, and prepared whole cell extracts. The p38α MAPK was immunoprecipitated, and possible association with TAB1 was detected by Western blotting. We observed that SeV infection lead to an increased interaction between p38α and TAB1 at the 3 and 4 h time points (Fig. 6D), thus coinciding with the enhanced phosphorylation of p38 (Fig. 6A). In summary, activation of p38 MAPK in response to SeV infection in DCs is dependent on the kinase activity of p38 and correlates with the kinetics of p38-TAB1 association.
DISCUSSION
The innate immune response against microbial pathogens is activated through host recognition of PAMPs through PRRs, which in turn trigger intracellular signaling pathways leading to stimulation of gene expression (4, 6). PRR-stimulated gene expression induces both immediate host defense and also initiates development of adaptive immunity, mainly through activation of DCs (6). During viral infections, IFNs are expressed and constitute perhaps the most important line of innate antiviral defense (1-3). In the present work, we demonstrate that innate recognition of a negative strand RNA paramyxovirus SeV, which is mediated by the cytoplasmic PRR RIG-I, leads to activation of the p38 MAPK pathway, through a pathway dependent on TRAF2, TAK1, and the kinase activity of p38 MAPK. We further show that the activation of p38 MAPK is essential for virus-induced expression of type I IFNs in fibroblasts and IL-12 in DCs. Thus, the activation of the p38 MAPK pathway is important both for triggering the first line of antiviral defense and to achieve full activation of DCs.
RIG-I-mediated signaling proceeds through the adaptor MAVS (26-29), which is located in the outer mitochondrial membrane (28). The signaling to IRF-3 has been reported to proceed through TRADD, TRAF3, TANK, NAP1, NEMO, TBK1, and IKKε (30-32, 46-48), and the pathway to NF-κB involves TRADD, TRAF2/6, RIP1, FADD, caspase-8/10, NEMO, and IKKα/β (27, 29, 32-34, 48). However, the signaling pathway from RIG-I to the MAPK pathway is not well described. We observed that SeV stimulated the activation of p38 MAPK in a manner that was dependent on RIG-I and viral replication and that this process involved TRAF2, TAK1, and the kinase activity of p38. The potential activation of p38 MAPK by autophosphorylation is discussed in detail below. For comparison, we also investigated the involvement of TRAF2, TRAF6, and TAK1 in RIG-I-mediated activation of IRF-3 and NF-κB and found that stimulation of the NF-κB pathway was dependent on TRAF2 and TAK1. However, IRF-3 was found to be activated independently of these signaling proteins. Thus, the RIG-I-activated signalosome leading to IRF-3 activation is clearly distinct from the signalosome activating NF-κB, which seems to include most of the components of the signaling pathway leading to the activation of MAPKs. Our data suggest that RIG-I activates NF-κB and p38 MAPK through a common upstream pathway, which diverges only downstream of TAK1.
MAVS/IPS-1 contains consensus TRAF2 and TRAF6 binding motifs and is reported to interact with both proteins (29). In our system, only TRAF2 was essential for phosphorylation of IκBα and p38 and expression of IFN-stimulated genes. This is in contrast to a recent report by Yoshida et al. (49) showing that TRAF6 and MEKK1 play important roles in RLR signaling to NF-κB and MAPKs. In the same paper, it was also concluded that TAK1 is not involved in the RLR antiviral pathway. At the present stage, it is difficult to explain the different findings of the two studies, since MEFs and conventional DCs were used in both studies. In our work, we used SeV as activator of RIG-I, whereas Yoshida et al. (49) used VSV. It is possible that the viruses used differentially interfere with TRAF2/6 signaling, thus revealing stimulus-dependent roles for TRAF2/6 in RLR signaling. Alternatively, the requirement for TRAF2 versus 6 may be partly cell type-dependent. This idea is supported by the fact that Yoshida et al. (49) demonstrated a complete loss of responsiveness to VSV in TRAF6-/- conventional DCs but only a minor reduction in the response in TRAF6-/- MEFs. Therefore, it is possible that both a TRAF2-TAK1 and a TRAF6-MEKK1 signaling cassette can be activated downstream of MAVS/IPS-1, and the relative importance of these signaling systems in activation of NF-κB and MAPKs by RLRs depends on the cell type and the activating stimuli.
SeV-induced p38 MAPK activation peaked at 3-4 h postinfection and was dependent on viral replication. The signaling pathway to p38 MAPK is well described and proceeds through stimulus-specific activation of MAPK kinase kinases, which activate the p38 MAPK phosphorylating MAPK kinases, MKK3 and MKK6 (50). However, on some occasions, p38 MAPK can be activated via an alternative mechanism where p38α MAPK associates with TAB1 and undergoes autophosphorylation (44). We observed that p38 MAPK associated with TAB1 at early times of infection, and this association preceded or occurred at the same time as p38 MAPK phosphorylation. In addition, we found that the phosphorylation of p38 MAPK was strongly reduced after inhibiting the kinase activity of p38 MAPK by SB202190. Finally, our data showed that DCs from MKK3-/- mice displayed unimpaired activation after viral infection. Collectively, the data suggest that autophosphorylation of p38 MAPK is involved in p38 MAPK activation downstream of RIG-I, although it also remains possible that the physical association between the inhibitor compound and p38 may block the access of other kinases. It has recently been shown that p38 MAPK autophosphorylation drives IL-12 production during infection with the intracellular protozoan Toxoplasma gondii (45). This finding, together with our observations in the present work, support the concept that intracellular pathogens activate a common pathway leading to the activation of p38 MAPK in select cell types through the mechanism of autophosphorylation and that this is a key event in the regulation of cytokine gene expression.
Concerning the mechanism through which p38 MAPK exerts its activity on RIG-I-dependent gene expression, we found that this was mainly occurring through a posttranscriptional mechanism, since inhibition of p38 MAPK reduced virus-induced expression of IL-12p40 at the protein but not mRNA level. The activities of p38 MAPK are mediated through direct phosphorylation of effector molecules, including transcription factors, or through activation of downstream signaling molecules (51). MKs represent an important class of p38 downstream target molecules affecting diverse functions, including transcription, mRNA stability, and translation (22-25). However, we did not observe any effect of MK2 deficiency or MNK1 inhibition on virus-induced IL-12 expression. Similarly, MSK1/2 deficiency did not lead to reduced cytokine production; rather, we observed an increase in SeV-induced IL-12 response in cells with this genotype. This is in line with a recent publication identifying MSK1/2 as a negative regulator of the MAPK pathway (43). Therefore, p38 MAPK stimulates RIG-I-dependent gene expression through a posttranscriptional mechanism independent of MNK1, MK2, and MSK1/2.
In conclusion, we show that SeV infection of fibroblasts and myeloid DCs stimulates RIG-I-mediated activation of p38 MAPK and that this proceeds through a pathway dependent on TRAF2, TAK1, and the kinase activity of p38 MAPK. The activation of p38 MAPK in turn promotes virus-induced expression of type I IFNs and IL-12. Thus, p38 MAPK is important for mounting the first line of defense against viral infections and also takes part in initiation of the development of adaptive immunity.
Supplementary Material
Acknowledgments
The technical assistance of Kirsten Stadel Petersen and Birthe Søby is greatly acknowledged.
This work was supported by Danish Medical Research Council Grant 271-06-0438), Danish Natural Science Research Council Grant 272-05-0222, Lundbeck Foundation Grants 116/06 and R17-A1526, the Kathrine og Vigo Skovgards Fond, the Aarhus University Research Foundation, and the Research Program in Molecular Medicine, Faculty of Health Science, Aarhus University.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S5.
Footnotes
The abbreviations used are: PAMP, pathogen-associated molecular patterns; PRR, pattern recognition receptor; IFN, interferon; DC, dendritic cell; IL, interleukin; NF, nuclear factor; IRF, IFN regulatory factor; MAPK, mitogen-activated protein kinase; MK, MAPK-activated protein kinase; MNK, MAPK-interacting kinases; MSK, mitogen- and stress-activated protein kinase; MEF, mouse embryo fibroblast; BM, bone marrow; SeV, Sendai virus; PE, phycoerythrin; ELISA, enzyme-linked immunosorbent assay; JNK, c-Jun N-terminal kinase; ERK, extracellular signal-regulated kinase; RLR, RIG-like receptor.
References
- 1.Dupuis, S., Jouanguy, E., Al Hajjar, S., Fieschi, C., Al Mohsen, I. Z., Al Jumaah, S., Yang, K., Chapgier, A., Eidenschenk, C., Eid, P., Al Ghonaium, A., Tufenkeji, H., Frayha, H., Al Gazlan, S., Al Rayes, H., Schreiber, R. D., Gresser, I., and Casanova, J. L. (2003) Nat. Genet. 33 388-391 [DOI] [PubMed] [Google Scholar]
- 2.Leib, D. A., Harrison, T. E., Laslo, K. M., Machalek, M. A., Moorman, N. J., and Virgin, H. W. (1999) J. Exp. Med. 189 663-672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ank, N., Iversen, M. B., Bartholdy, C., Staeheli, P., Hartmann, R., Jensen, U. B., Dagnaes-Hansen, F., Thomsen, A. R., Chen, Z., Haugen, H., Klucher, K., and Paludan, S. R. (2008) J. Immunol. 180 2474-2485 [DOI] [PubMed] [Google Scholar]
- 4.Mogensen, T. H., and Paludan, S. R. (2005) J. Mol. Med. 83 180-192 [DOI] [PubMed] [Google Scholar]
- 5.Reis e Sousa, C. (2004) Curr. Opin. Immunol. 16 21-25 [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki, A., and Medzhitov, R. (2004) Nat. Immunol. 5 987-995 [DOI] [PubMed] [Google Scholar]
- 7.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immunol. 5 730-737 [DOI] [PubMed] [Google Scholar]
- 8.Kawai, T., and Akira, S. (2006) Nat. Immunol. 7 131-137 [DOI] [PubMed] [Google Scholar]
- 9.Weber, F., Wagner, V., Rasmussen, S. B., Hartmann, R., and Paludan, S. R. (2006) J. Virol. 80 5059-5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahasi, K., Yoneyama, M., Nishihori, T., Hirai, R., Kumeta, H., Narita, R., Gale, M., Jr., Inagaki, F., and Fujita, T. (2008) Mol. Cell 29 428-440 [DOI] [PubMed] [Google Scholar]
- 11.Pichlmair, A., Schulz, O., Tan, C. P., Naslund, T. I., Liljestrom, P., Weber, F., and Reis e Sousa, C. (2006) Science 314 997-1001 [DOI] [PubMed] [Google Scholar]
- 12.Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K. K., Schlee, M., Endres, S., and Hartmann, G. (2006) Science 314 994-997 [DOI] [PubMed] [Google Scholar]
- 13.Gitlin, L., Barchet, W., Gilfillan, S., Cella, M., Beutler, B., Flavell, R. A., Diamond, M. S., and Colonna, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8459-8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K. J., Yamaguchi, O., Otsu, K., Tsujimura, T., Koh, C. S., Reis e Sousa, C., Matsuura, Y., Fujita, T., and Akira, S. (2006) Nature 441 101-105 [DOI] [PubMed] [Google Scholar]
- 15.Venkataraman, T., Valdes, M., Elsby, R., Kakuta, S., Caceres, G., Saijo, S., Iwakura, Y., and Barber, G. N. (2007) J. Immunol. 178 6444-6455 [DOI] [PubMed] [Google Scholar]
- 16.Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., Foy, E., Loo, Y. M., Gale, M., Jr., Akira, S., Yonehara, S., Kato, A., and Fujita, T. (2005) J. Immunol. 175 2851-2858 [DOI] [PubMed] [Google Scholar]
- 17.Rothenfusser, S., Goutagny, N., Diperna, G., Gong, M., Monks, B. G., Schoenemeyer, A., Yamamoto, M., Akira, S., and Fitzgerald, K. A. (2005) J. Immunol. 175 5260-5268 [DOI] [PubMed] [Google Scholar]
- 18.Kato, H., Sato, S., Yoneyama, M., Yamamoto, M., Uematsu, S., Matsui, K., Tsujimura, T., Takeda, K., Fujita, T., Takeuchi, O., and Akira, S. (2005) Immunity 23 19-28 [DOI] [PubMed] [Google Scholar]
- 19.Akira, S., and Takeda, K. (2004) Nat. Rev. Immunol. 4 499-511 [DOI] [PubMed] [Google Scholar]
- 20.Honda, K., Yanai, H., Negishi, H., Asagiri, M., Sato, M., Mizutani, T., Shimada, N., Ohba, Y., Takaoka, A., Yoshida, N., and Taniguchi, T. (2005) Nature 434 772-777 [DOI] [PubMed] [Google Scholar]
- 21.Osterlund, P. I., Pietila, T. E., Veckman, V., Kotenko, S. V., and Julkunen, I. (2007) J. Immunol. 179 3434-3442 [DOI] [PubMed] [Google Scholar]
- 22.Roux, P. P., and Blenis, J. (2004) Microbiol. Mol. Biol. Rev. 68 320-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueda, T., Watanabe-Fukunaga, R., Fukuyama, H., Nagata, S., and Fukunaga, R. (2004) Mol. Cell Biol. 24 6539-6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotlyarov, A., Neininger, A., Schubert, C., Eckert, R., Birchmeier, C., Volk, H. D., and Gaestel, M. (1999) Nat. Cell Biol. 1 94-97 [DOI] [PubMed] [Google Scholar]
- 25.Gaestel, M. (2006) Nat. Rev. Mol. Cell Biol. 7 120-130 [DOI] [PubMed] [Google Scholar]
- 26.Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005) Cell 122 669-682 [DOI] [PubMed] [Google Scholar]
- 27.Kawai, T., Takahashi, K., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981-988 [DOI] [PubMed] [Google Scholar]
- 28.Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Bartenschlager, R., and Tschopp, J. (2005) Nature 437 1167-1172 [DOI] [PubMed] [Google Scholar]
- 29.Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z., and Shu, H. B. (2005) Mol. Cell 19 727-740 [DOI] [PubMed] [Google Scholar]
- 30.Oganesyan, G., Saha, S. K., Guo, B., He, J. Q., Shahangian, A., Zarnegar, B., Perry, A., and Cheng, G. (2005) Nature 439 211. [DOI] [PubMed] [Google Scholar]
- 31.Saha, S. K., Pietras, E. M., He, J. Q., Kang, J. R., Liu, S. Y., Oganesyan, G., Shahangian, A., Zarnegar, B., Shiba, T. L., Wang, Y., and Cheng, G. (2006) EMBO J. 25 3257-3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michallet, M. C., Meylan, E., Ermolaeva, M. A., Vazquez, J., Rebsamen, M., Curran, J., Poeck, H., Bscheider, M., Hartmann, G., Konig, M., Kalinke, U., Pasparakis, M., and Tschopp, J. (2008) Immunity 28 651-661 [DOI] [PubMed] [Google Scholar]
- 33.Balachandran, S., Thomas, E., and Barber, G. N. (2004) Nature 432 401-405 [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, K., Kawai, T., Kumar, H., Sato, S., Yonehara, S., and Akira, S. (2006) J. Immunol. 176 4520-4524 [DOI] [PubMed] [Google Scholar]
- 35.Yeh, W.-C., Shahinian, A., Speiser, D., Kraunus, J., Billia, F., Wakeham, A., de la Pompa, J. L., Ferrick, D., Hum, B., Iscove, N., Ohashi, P., Rothe, M., Goeddel, D. V., and Mak, T. W. (1997) Immunity 7 715-725 [DOI] [PubMed] [Google Scholar]
- 36.Shim, J. H., Xiao, C., Paschal, A. E., Bailey, S. T., Rao, P., Hayden, M. S., Lee, K. Y., Bussey, C., Steckel, M., Tanaka, N., Yamada, G., Akira, S., Matsumoto, K., and Ghosh, S. (2005) Genes Dev. 19 2668-2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen, M. S., Toldbod, H. E., Holtz, S., Hokland, M., Bolund, L., and Agger, R. (2000) Scand. J. Immunol. 51 586-594 [DOI] [PubMed] [Google Scholar]
- 38.Pirhonen, J., Sareneva, T., Kurimoto, M., Julkunen, I., and Matikainen, S. (1999) J. Immunol. 162 7322-7329 [PubMed] [Google Scholar]
- 39.Osterlund, P., Veckman, V., Siren, J., Klucher, K. M., Hiscott, J., Matikainen, S., and Julkunen, I. (2005) J. Virol. 79 9608-9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagawa, Y., Iijima, N., Iwabuchi, K., and Onoe, K. (2002) J. Leukocyte Biol. 71 125-132 [PubMed] [Google Scholar]
- 41.Ohmori, Y., and Hamilton, T. A. (1995) J. Immunol. 154 5235-5244 [PubMed] [Google Scholar]
- 42.Sato, S., Sanjo, H., Takeda, K., Ninomiya-Tsuji, J., Yamamoto, M., Kawai, T., Matsumoto, K., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 2668-2681 [DOI] [PubMed] [Google Scholar]
- 43.Ananieva, O., Darragh, J., Johansen, C., Carr, J. M., McIlrath, J., Park, J. M., Wingate, A., Monk, C. E., Toth, R., Santos, S. G., Iversen, L., and Arthur, J. S. (2008) Nat. Immunol. 9 1028-1036 [DOI] [PubMed] [Google Scholar]
- 44.Ge, B., Gram, H., Di Padova, F., Huang, B., New, L., Ulevitch, R. J., Luo, Y., and Han, J. (2002) Science 295 1291-1294 [DOI] [PubMed] [Google Scholar]
- 45.Kim, L., Del Rio, L., Butcher, B. A., Mogensen, T. H., Paludan, S. R., Flavell, R. A., and Denkers, E. Y. (2005) J. Immunol. 174 4178-4184 [DOI] [PubMed] [Google Scholar]
- 46.Guo, B., and Cheng, G. (2007) J. Biol. Chem. 282 11817-11826 [DOI] [PubMed] [Google Scholar]
- 47.Sasai, M., Shingai, M., Funami, K., Yoneyama, M., Fujita, T., Matsumoto, M., and Seya, T. (2006) J. Immunol. 177 8676-8683 [DOI] [PubMed] [Google Scholar]
- 48.Zhao, T., Yang, L., Sun, Q., Arguello, M., Ballard, D. W., Hiscott, J., and Lin, R. (2007) Nat. Immunol. 8 592-600 [DOI] [PubMed] [Google Scholar]
- 49.Yoshida, R., Takaesu, G., Yoshida, H., Okamoto, F., Yoshioka, T., Choi, Y., Akira, S., Kawai, T., Yoshimura, A., and Kobayashi, T. (2008) J. Biol. Chem. 283 36211-36220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashwell, J. D. (2006) Nat. Rev. Immunol. 6 532-540 [DOI] [PubMed] [Google Scholar]
- 51.Su, B., and Karin, M. (1996) Curr. Opin. Immunol. 8 402-411 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.