Abstract
Growth of Saccharomyces cerevisiae following glucose depletion (the diauxic shift) depends on a profound metabolic adaptation accompanied by a global reprogramming of gene expression. In this study, we provide evidence for a heretofore unsuspected role for Isc1p in mediating this reprogramming. Initial studies revealed that yeast cells deleted in ISC1, the gene encoding inositol sphingolipid phospholipase C, which resides in mitochondria in the post-diauxic phase, showed defective aerobic respiration in the post-diauxic phase but retained normal intrinsic mitochondrial functions, including intact mitochondrial DNA, normal oxygen consumption, and normal mitochondrial polarization. Microarray analysis revealed that the Δisc1 strain failed to up-regulate genes required for nonfermentable carbon source metabolism during the diauxic shift, thus suggesting a mechanism for the defective supply of respiratory substrates into mitochondria in the post-diauxic phase. This defect in regulating nuclear gene induction in response to a defect in a mitochondrial enzyme raised the possibility that mitochondria may initiate diauxic shift-associated regulation of nucleus-encoded genes. This was established by demonstrating that in respiratory-deficient petite cells these genes failed to be up-regulated across the diauxic shift in a manner similar to the Δisc1 strain. Isc1p- and mitochondrial function-dependent genes significantly overlapped with Adr1p-, Snf1p-, and Cat8p-dependent genes, suggesting some functional link among these factors. However, the retrograde response was not activated in Δisc1, suggesting that the response of Δisc1 cannot be simply attributed to mitochondrial dysfunction. These results suggest a novel role for Isc1p in allowing the reprogramming of gene expression during the transition from anaerobic to aerobic metabolism.
Mitochondria are essential organelles whose primary function is the synthesis of ATP by oxidative phosphorylation. Mitochondria are also the site of metabolic and biosynthetic reactions, such as the tricarboxylic acid cycle (1), lipids (2, 3), amino acids (4), and iron metabolism (5). Mitochondrial proteins consist of those encoded by both the nuclear and mitochondrial genomes. The mitochondrial genome encodes 12 proteins, most of which are components of the oxidative phosphorylation apparatus (6). However, most of the mitochondrial proteins are encoded by nuclear genes (7). Moreover, alterations in mitochondrial function and mitochondrial damage have been linked to a variety of cellular and organismal responses, including apoptosis, neuromuscular disease, tumor pathogenesis, and aging (8-11).
Isc1p, the homolog of mammalian neutral sphingomyelinases, was recently shown to localize to mitochondria in the post-diauxic phase of yeast growth (3, 12), although its function there remains undetermined. Isc1p produces ceramide from the complex sphingolipids inositol phosphorylceramide, mannosylinositol phosphorylceramide, and mannosyldiinositol phosphorylceramide (13-16), modulates iron levels and apoptosis (17), and confers resistance to the antifungal effects of macrophages in Cryptococcus neoformans (18). Although a Δisc1 strain grew very slowly in the post-diauxic phase or when grown in media containing nonfermentable carbon sources (12, 13), the cause of this growth defect had not been elucidated; however, because utilization of nonfermentable carbon sources requires intact mitochondrial function, the findings suggested a role for Isc1p in mitochondrial function.
In this study, we initially sought to elucidate the cause of the growth defect of Δisc1 in the post-diauxic phase and on nonfermentable carbon sources. During this pursuit, it was found that the mitochondrial DNA, respiratory chain, and reactive oxygen species from Δisc1 were not significantly compromised; however, the results revealed a defect in induction of those genes involved in nonfermentable carbon source metabolism, including glucose-repressed genes, probably resulting in defective supply of respiratory substrates into mitochondria. In turn, these results led to the identification of a novel and unexpected role for mitochondria in regulating expression of nuclear genes during the diauxic shift. This role was established using petite cells, which have lost mitochondrial DNA and integrity. These cells, as compared with grande cells, failed to execute the regulation of a large spectrum of genes, including glucose-repressed genes required for executing the diauxic shift. These results indicate that Isc1p provides an indispensable role in the global metabolic adaptation in this phase of yeast growth.
EXPERIMENTAL PROCEDURES
Yeast Strains, Media, and Culture Conditions—JK9-3dα WT2 and JK9-3dα Δisc1 (MATα leu2-3, 112 ura3-52 rme1 trp1 his4 Δisc1::kanMX) (16) were used in this study. Standard yeast culture medium was prepared using Difco YPD broth. Plates were prepared using Difco YPD agar and stored at 4 °C. YPethanol contained 1% yeast extract, 2% bactopeptone, and 2% ethanol. YPGE plate contained 1% yeast extract, 2% bactopeptone, 2% ethanol, 2% glycerol, and 2% agar. Yeast cells were cultured with shaking at 30 °C otherwise indicated. Exponentially growing cells were inoculated to fresh media, incubated for 4 or 24 h with shaking, and used as pre-diauxic and post-diauxic phase cells.
RNA Preparation—Yeast cells were cultured in medium as described above, centrifuged at 3,000 × g at 4 °C, and snapfrozen with liquid nitrogen, and RNA was isolated from cell pellets using the RNeasy RNA Isolation Kit from Qiagen following the enzymatic lysis protocol.
Microarray Procedures—Sample preparation for microarray hybridization was carried out essentially as described in the Affymetrix Expression Analysis Technical Manual and previous reports (19, 20). In brief, isolated total RNA was subjected to reverse transcription using the Superscript Choice System (Invitrogen), and the derived double-stranded cDNA was purified using phase-lock gel columns (Eppendorf) followed by ethanol precipitation or the cDNA clean up columns provided by Affymetrix. The purified cDNA was then subjected to in vitro transcription using biotinylated ribonucleotides in the Bio-Array™ High Yield™ RNA transcript labeling kit (Enzo Diagnostics). The biotinylated transcript was purified using the RNeasy Mini Kit (Qiagen) and then fragmented using 0.2 m Tris acetate, pH 8.1, containing 0.5 m potassium acetate and 0.15 m magnesium acetate. Fragmentation was confirmed by agarose gel electrophoresis using ethidium bromide for visualization. Samples were then hybridized to the YG-S98 yeast genomic chip (Affymetrix), stained with streptavidin-phycoerythrin, and washed in the Affymetrix Fluidics Station exactly as indicated by the Affymetrix Expression Analysis Technical Manual.
Data Analysis—Gene chips were scanned using the Affymetrix scanner. Fluorescence values were statistically analyzed to yield gene expression signal values using Microarray Suite 5.0. Expression integrity was confirmed by verification of expected values for internal control probe sets, including sets for “spikes” (BioA, BioB, and BioC), ratios of signals between probe sets designed with 3′ or 5′ bias for ACT1 (β-actin), and absence of internal negative controls, including murine genes. Absolute analysis was conducted for each analysis with scaling to a target value of 2500 to facilitate comparison between experiments. Microarray data were derived from two independent experiments for WT and Δisc1 grande and from one experiment for petite. In Fig. 3 and Fig. 6A, genes whose expression at 24 h in WT was greater than 300 were selected. Among the selected genes, the genes whose ratio of WT-24 h/WT-4 h above 2.0 were further selected, and the gene expression ratio of WT-24 h/WT-4 h and mutant-24 h/mutant-4 h was plotted along the log axis. Statistical analyses, including regression and slope analysis of these genes, were performed with a statistical software KaleidaGraph 4 (Hulinks). In Fig. 5A and Fig. 6B, the ratio of mutant-24 h/mutant-4 h/WT-24 h/WT-4 h of the carbohydrate metabolism pathway genes was calculated, and the ratio was expressed by color and thickness of the boxes (red; 24 h/4 h ratio is bigger in mutant than WT; blue, 24 h/4 h ratio is smaller in mutant than WT). The whole set of microarray data are available at Medical University of South Carolina web site.
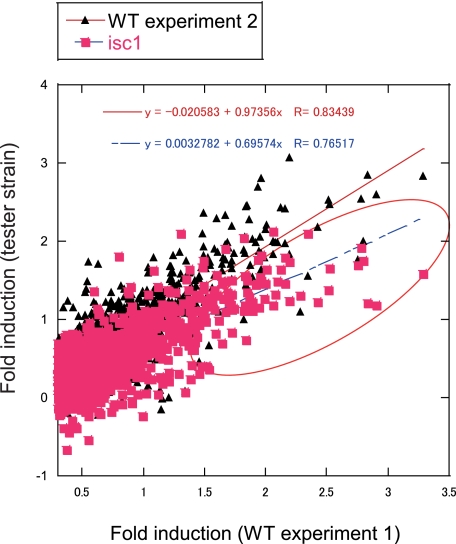
FIGURE 3.
Diauxic shift of gene expression is suppressed in Δisc1. Comparison of gene expression change across diauxic shift in WT and Δisc1 is shown. The ratio of gene expression was calculated from the data obtained from microarray result. The genes that are up-regulated more than 2-fold in WT grande experiment 1 at 24 h relative to 4 h were selected. The log10 value of the induction ratio of gene expression at 24 h to that at 4 h in WT grande experiment 2 and Δisc1 grande were plotted against that in WT grande experiment 1.
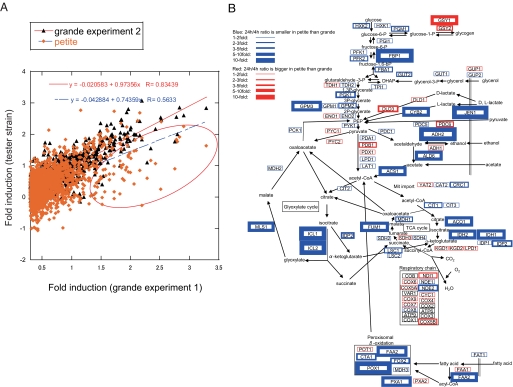
FIGURE 6.
Defective up-regulation of nonfermentable carbon source metabolism genes in petite. Comparison of gene expression change across the diauxic shift in WT and petite. The ratio of gene expression was calculated from the data obtained from microarray result. The genes that are up-regulated more than 2-fold in grande experiment 1 at 24 h relative to 4 h were selected. A, log10 value of the induction ratio of gene expression at 24 h to that at 4 h in grande experiment 2 and petite were plotted against that in grande experiment 1. B, induction ratio of carbon source metabolism pathway genes across the diauxic shift in WT and petite. Red boxes indicate the genes more induced across the diauxic shift in the petite relative to grande according to the ratio of grande (24 h)/grande (4 h)/petite (24 h)/petite (4 h). Blue boxes indicate the genes less induced across the diauxic shift in petite relative to grande. Thickness of the boxes indicates the fold change as shown in the figure.
FIGURE 5.
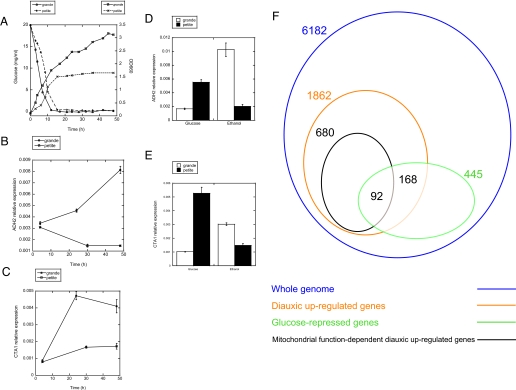
Defective respiration of Δisc1 in the post-diauxic phase. A, induction ratio of carbon source metabolism pathway genes across diauxic shift in WT and Δisc1. Carbon source metabolism pathway was modified from a review by Schuller (45). Red boxes indicate the genes more induced across the diauxic shift in Δisc1 relative to WT according to the ratio of WT (24 h)/WT (4 h)/Δisc1 (24 h)/Δisc1 (4 h). Blue boxes indicate the genes less induced across the diauxic shift in Δisc1 relative to WT. Thickness of the boxes indicates the fold change as shown in the figure. The results are representatives of at least two independent experiments. B, Venn diagram of the number of whole genome (blue), diauxic up-regulated (orange), glucose-repressed (green), and Isc1p-dependent diauxic up-regulated (purple) genes. A cohort of glucose-repressed genes was extracted from glucose pulse experiment (33). C, growth of WT and Δisc1 in ethanol-containing media. WT and Δisc1 cells in the post-diauxic phase were inoculated to fresh YPethanol and incubated at 30 °C for the indicated times. D, oxygen consumption rate of whole cells (WT and Δisc1) in the post-diauxic phase was measured during incubation with 2% ethanol. The results are represented as mean ± S.E. of at least two independent experiments.
Real Time RT-PCR—Total RNA was harvested using RNeasy Mini Kit (Qiagen). First strand cDNA was produced using 1 μg of total RNA and Superscript II reverse transcription kit (Invitrogen). Real time analysis was performed using iCycler IQ Real Time PCR System (Bio-Rad) and SYBR Green Supermix protocol (Bio-Rad). In brief, primers specific for a 75-150-bp region within the target gene were added to a final core of 12.5 mm to a master mix of sterile water and SYBR Green Supermix. The cDNA template was then diluted 1:50 and added to the mix. The final mixture was pipetted in triplicate into a 96-well plate (Bio-Rad), sealed with iCycler tape (Bio-Rad), and real time PCR performed. Primers used in the real time PCR are shown in supplemental Table SI. The expression of the target genes was normalized to the expression of RDN1.
Isolation of Mitochondria—A 6-liter flask containing 1.25 liters of YPD was inoculated with the exponentially growing culture and incubated at 30 °C for 24 h with vigorous aeration. Yeast cells were centrifuged at 2,000 × g for 5 min; washed with water; resuspended in 12.5 ml of 100 mm Tris-SO4, pH 9.4, 10 mm dithiothreitol (2 ml/g wet cell); incubated at 30 °C for 15 min; centrifuged at 2,000 × g for 5 min; washed with buffer SP (1.2 m sorbitol, 20 mm potassium/phosphate buffer, pH 7.4); dissolved in buffer SP containing Zymolyase 100T (Seikagaku Corp., 7 ml/g cells, 1.5 mg/g cells); incubated at 30 °C for 60 min; centrifuged at 2,500 × g for 5 min; washed twice with buffer SP (7 ml/g wet cell); resuspended in homogenization buffer (0.6 m sorbitol, 10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride) (6.5 ml/g cells); homogenized in a 40-ml Dounce homogenizer using 15 strokes; diluted 2-fold with homogenizing buffer; centrifuged for 5 min at 1,500 × g to remove cell nuclei and debris; and centrifuged again 2,500 × g for 5 min. The supernatant was centrifuged at 12,000 × g for 15 min. The pellet was resuspended in 30 ml of SM buffer (250 mm sucrose, 1 mm EDTA, 10 mm MOPS/KOH, pH 7.2) and centrifuged at 2,500 × g for 5 min. The supernatant was then centrifuged at 12,000 × g for 15 min, and the pellet was resuspended in 10 ml of SM buffer as crude mitochondria.
Oxygen Consumption Rate—Exponentially growing yeast cells were inoculated at A600 of 0.1 and incubated 24 h in YPD medium, and 4.4 × 107 cells were washed and resuspended in a total 20 μl of solution A (10 mm HEPES, 25 mm K2HPO4, pH 7.0). Five microliters of the cell suspension was added to 0.6 ml of solution A in the chamber followed by 10 μl of ethanol. Oxygen consumption rate was measured using Dual Oxygen Electrode Amplifier model 203 (Instech). Alternatively, 10 mg of isolated mitochondria were washed and resuspended in a total 200 μl of 0.6 m sorbitol, 10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride. After adding a final 10 μm NADH, 5 μl of the mitochondrial suspension was added to 0.6 ml of 0.6 m sorbitol, 10 mm HEPES, pH 7.0, 25 mm K2HPO4, 2 mm MgCl2, 1 mm EDTA in the chamber, followed by adding a final 2 μm ADP. Oxygen consumption rate was measured using the same machine described above. The oxygen consumption rate after addition of mitochondria was calculated as state 4 respiration rate, that after addition of ADP as state 3 respiration rate, and that after ADP exhaustion as state 4 (2) respiration rate.
Measurement of Reactive Oxygen Species—Yeast cells grown to 3 × 106 cells/ml were washed, dissolved in distilled water (1 × 106 cells in 1 ml), and incubated in final 40 μm 2′,7′-dichlorofluorescein diacetate (Sigma) for 1 h at room temperature in a dark room. Cells were washed with distilled water two times, and the fluorescence was measured using FACSCalibur (BD Biosciences) with 488-nm argon laser and 530/30BP filter. As a positive control, WT cells treated with α-factor (final 11.9 μm) for 1.5 h were applied to the same analysis.
Petite Induction—Petite induction was performed by the ethidium bromide method described by Schneider-Berlin et al. (21). Briefly, logarithmically grown yeast cells were diluted to 2 × 106 cells/ml in 1% yeast extract, 2% peptone, 2% dextrose, 50 mm potassium phosphate, pH 6.24. Ethidium bromide was added to a final concentration of 50 μm, and the cultures were incubated at 30 °C in the dark for 8 h with shaking and then plated onto YPD or YPGE medium to confirm inability to grow on nonfermentable medium.
Glucose Concentration—The concentration of glucose was measured with Glucose CII-test Wako (Wako Chemicals) according to the manufacturer's instructions. Briefly, 1.0 ml of the reaction mixture was mixed with 6.7 μl of the sample and incubated at 37 °C for 5 min, and absorbance at 530 nm was measured. The absorbance of the blank sample and that without the reaction mixture was subtracted from the absorbance of the sample, and concentration of glucose was calculated from the dilution ratio and the calibration curve.
RESULTS AND DISCUSSION
Δisc1 Displays Defective Growth on Nonfermentable Carbon Sources but No Defects in Mitochondrial DNA—The Δisc1 strain showed hardly any defect of growth in the pre-diauxic phase in glucose-containing medium and a modest defect in the post-diauxic phase (Fig. 1A), consistent with previous results (12, 13). On the other hand, Δisc1 showed strikingly defective growth in medium containing nonfermentable carbon sources such as glycerol (Fig. 1B), thus demonstrating a clear-cut preference for fermentable carbon sources. Because Isc1p associated with mitochondria in the post-diauxic growth phase (3, 12), it seemed plausible that these phenotypes may arise from a mitochondrial defect in Δisc1.
FIGURE 1.
Δisc1 exhibits a defective phenotype in the post-diauxic phase. A and B, WT and Δisc1 cells were grown in YPD medium and inoculated to fresh YPD (A) or YPglycerol medium (B), and the absorbance was measured at different time points. A, glucose concentration was also measured at each time point.
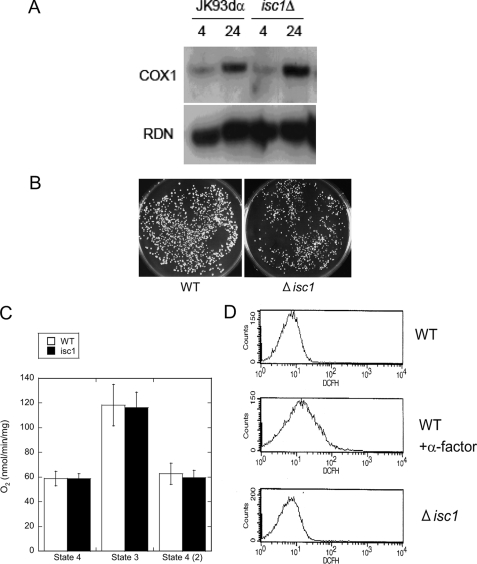
Defects in mitochondria such as petite formation often indicate defects in mitochondrial DNA. Thus, yeast strains require WT mitochondrial DNA (rho+) to respire and grow on nonfermentable media (22), whereas cells in which part or all mitochondrial DNA is lost (rho- or rho0, respectively) lose functional mitochondrial DNA-encoded subunits of both the electron transport chain and the ATP synthase, fail to grow on nonfermentable media, and form “petite” colonies on fermentable media. Moreover, mutants that have lost mitochondrial integrity often show decreased content of their mitochondrial DNA (23). Therefore, the mitochondrial DNA content in the Δisc1 strain was determined by Southern blotting using a probe directed to the COX1 gene, which is encoded in the mitochondrial genome. The results showed that Δisc1 did not contain less mitochondrial DNA relative to WT, and if any, there was an increase in total mitochondrial DNA (Fig. 2A), suggesting that the growth defect of Δisc1 is not due to loss of mitochondrial DNA.
FIGURE 2.
Lack of intrinsic mitochondrial defects in Δisc1. A, mitochondrial DNA content of WT and Δisc1. The genomic DNA of WT and Δisc1 was obtained from 4 and 24 h after inoculation and analyzed by Southern blotting with the mitochondrial COX1 and nuclear RDN1 probe. B, WT and Δisc1 were incubated in medium containing ethidium bromide for 8 h and plated onto YPD plates. The plates were incubated at 30 °C for 2 days. Plating onto YPGE plates did not produce any colonies (data not shown). C, oxygen consumption rate of the isolated mitochondria measured as described under “Experimental Procedures.” The results are represented as the mean ± S.E. of at least two independent experiments. D, reactive oxygen species of WT cells with or without treatment with α-factor (final 11.9 μm at 30 °C for 1.5 h) and Δisc1 cells. The cells were mixed with 40 μm 2′,7′-dichlorofluorescein diacetate and applied to FACS analysis.
To further confirm this hypothesis, the petite-forming ability of Δisc1 was analyzed. Saccharomyces cerevisiae can usually grow without mitochondrial DNA in fermentable carbon medium (but not in nonfermentable carbon medium). However, mutants that cannot grow without mitochondrial DNA even in fermentable carbon medium are called “petite-negative” mutants and are characterized by the inability to form colonies when incubated in fermentable carbon medium containing ethidium bromide (24). Δisc1 cells were incubated with 50 μm ethidium bromide in the dark at 30 °C for 8 h to generate petites, and then colony formation on YPD was evaluated. As shown in Fig. 2B, colonies were distinctly formed in Δisc1 as well as in WT (Fig. 2B), indicating that the Δisc1 mutant is not petite-negative, as described previously (25). These results argue against a major defect in mitochondrial DNA underlying the growth defect of the Δisc1 strain.
Isolated Mitochondria from Δisc1 Display an Intact Respiration Rate—Defective mitochondrial phenotypes and the very slow growth of the Δisc1 in nonfermentable carbon source medium suggested the possibility that the respiration rate of mitochondria from the Δisc1 may be lower than that of the WT. WT and Δisc1 cells were incubated in YPD medium and grown to the post-diauxic phase, and mitochondria were isolated as described under “Experimental Procedures.” Oxygen consumption rate of the isolated mitochondria was measured by incubating 500 μg of the mitochondria with 10 μm NADH and 2 μm ADP and measuring the decrease in the oxygen concentration in the chamber. Unexpectedly, the oxygen consumption rate of isolated mitochondria from Δisc1 in the post-diauxic phase was similar to that of WT (Fig. 2C), suggesting that a critical defect does not exist in the respiratory chain of this mutant.
Next, the levels of reactive oxygen species were evaluated because defects in mitochondria often result in an increase in the concentration of the reactive oxygen species of the cells (26). For these experiments, logarithmically growing WT and Δisc1 cells were suspended in 40 μm 2′,7′-dichlorofluorescein diacetate and incubated for 1 h at room temperature in a dark room, and the fluorescence was measured by FACS equipped with a 488-nm argon laser and a 530/30BP filter as an indication of the amount of reactive oxygen species in the cells. The strength of the fluorescence of Δisc1 was not significantly different from that of WT, indicating that the amount of reactive oxygen species in the Δisc1 did not differ significantly from that in the WT (Fig. 2D). The sensitivity of this assay was verified by measuring the fluorescence of WT cells treated with α-factor (Fig. 2D), which are known to generate increased amounts of reactive oxygen species (27). Together, these results strongly support that mitochondrial DNA, respiratory chain, and reactive oxygen species are not significantly compromised; and thus it seems unlikely that an intrinsic functional defect of respiration of mitochondria could be responsible for the very slow growth observed for the Δisc1 strain in nonfermentable medium.
Defects in Transcriptional Adaptation of Metabolic Enzymes in Δisc1—Because the above studies unexpectedly suggested normal mitochondrial DNA, respiratory chain, and reactive oxygen species, it became important to determine the key defects that prevent the Δisc1 strain from normal growth in the post-diauxic phase. Therefore, microarrays were used to determine gene expression levels in the Δisc1 and its parental strain during log phase growth as well as in the post-diauxic phase. WT and Δisc1 cells were grown in YPD medium at 30 °C; RNA was extracted, and gene expression was analyzed. Analysis of the microarray profiles of the WT revealed major changes in gene induction or repression, consistent with previous results (28) such that more than 1850 genes showed greater than 2-fold induction, and more than 987 genes showed more than 50% repression. Importantly, examination of gene induction in the Δisc1 strain revealed major aberrancies in gene regulation in the post-diauxic phase. For initial assessment of the global changes, we plotted the ratio of induction of up-regulated genes in Δisc1 versus the ratio of gene expression in WT across the diauxic shift. In this representation, genes that show failure of induction in the strain appear below the red line of identity to the right of the X = 0.3 line (red circle in Fig. 3). When the induction of all microarray genes of WT experiment 1 were plotted against the induction of WT experiment 2 (black triangles in Fig. 3), there was little scatter beyond the red line of identity (slope = 0.97), confirming the reproducibility of the microarray experiments. However, when gene induction of Δisc1 was plotted against gene induction in WT (pink squares in Fig. 3), the slope was 0.70 (blue line in Fig. 3), indicating that enough of the genome was aberrantly regulated to cause an overall shift in the slope and demonstrating faulty regulation of the diauxic shift on a global scale in the Δisc1 strain.
To determine the relationship between the aberrant gene regulation observed in the Δisc1 strain with its apparent growth defect during the diauxic shift, the set of genes that largely failed to be up-regulated in the Δisc1 was grouped by functional categories. This analysis revealed that more than 488 genes failed to be induced (less than 50% of WT) in the Δisc1 strain. The aberrantly regulated genes are predominantly involved in the utilization of a nonfermentable carbon source, especially those involved in glyoxylate cycle, peroxisomal β-oxidation, tricarboxylic acid cycle, gluconeogenesis, ethanol utilization, and mitochondrial import genes (supplemental Table SII). Notably, this cohort included a significant portion of the genes involved in gluconeogenesis (MLS1, CAT2, IDP2, SFC1, JEN1, and FBP1), suggesting a defect of generating glucose of Δisc1 in the post-diauxic phase. Interestingly, there were no significant changes in the nucleotide/nucleoside processing and protein catabolism genes in the Δisc1 strain compared with WT (supplemental Table SIII), indicating some specificity of the failed gene regulation in Δisc1 for metabolic pathways required for adaptation to growth on ethanol. On the other hand, the genes that failed to be down-regulated in Δisc1 (113 genes) were largely involved in the uptake of nutrients and amino acid metabolisms (supplemental Table SIV), again supporting a specific role for Isc1p in the metabolic adaptation of yeast to the post-diauxic phase. Thus taken together, the results from the microarray studies disclosed important defects in gene regulation in the Δisc1 in the post-diauxic phase that center on genes involved in metabolism of nonfermentable carbon source.
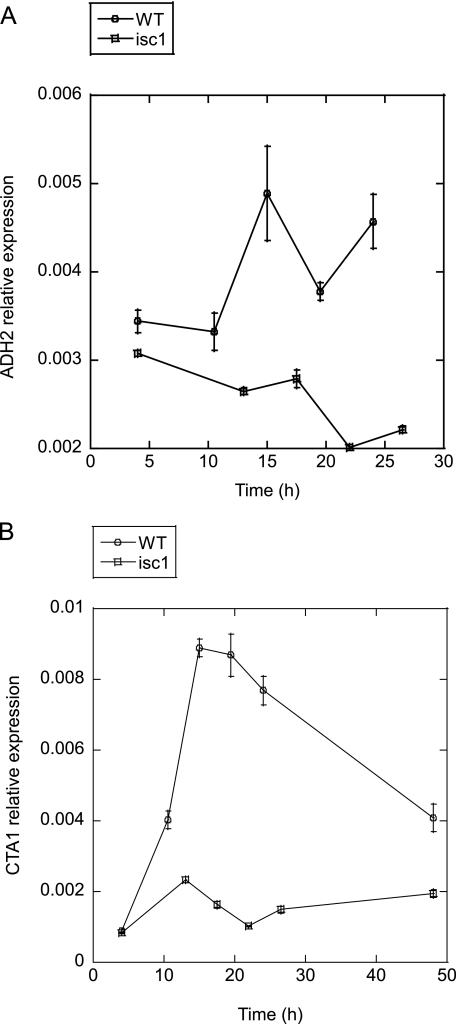
Gene expression during the diauxic shift has been reported to change drastically according to the growth phases (28). To rule out the possibility that the difference in the gene expression profiles between WT and Δisc1 was caused by differences in growth, we quantitated gene expression changes of ADH2 and CTA1 as representative genes involved in nonfermentable carbon source metabolism that have been reported to change significantly during the diauxic shift. WT and Δisc1 cells were cultured in YPD medium over a time course that included pre- and post-diauxic growth as monitored by changes in glucose concentration in the medium (Fig. 1A), and the expression of these two genes was investigated by real time RT-PCR. The results shown in Fig. 4, A and B, show that both ADH2 and CTA1 were induced significantly in the WT cells during the post-diauxic phase. Importantly, however, there was near total loss of their induction in the Δisc1. These gene expression profiles are consistent with the gene expression profile obtained by microarray analysis, confirming the conclusions drawn from the microarray analysis and indicating that the failed gene regulation was not merely a by-product of delayed growth phase in the Δisc1 strain.
FIGURE 4.
Up-regulation of genes involved in nonfermentable carbon source metabolism is suppressed in Δisc1. Logarithmically growing cells were inoculated to YPD medium and incubated for the indicated times. A and B, expression of ADH2 (A) and CTA1 (B) quantitated by real time RT-PCR in WT and Δisc1. The results are means of relative expression of at least three experiments with error bars showing S.E.
Next, to gain functional insight into the metabolic consequences of the dysregulation of the induction of genes involved in carbon source metabolism, the genes of the carbon source metabolism pathway were superimposed on a metabolic pathway map according to the fold change of Δisc1/WT across the diauxic shift (Fig. 5A). This revealed that the genes involved in oxidative growth, such as ethanol, lactate, acetate, glycerol, and pyruvate metabolism genes, tricarboxylic acid cycle genes, glyoxylate cycle genes, mitochondrial import genes, gluconeogenesis genes, and peroxisomal genes, failed to be up-regulated, whereas genes involved in fermentative growth such as glycolysis genes were not altered in Δisc1 (Fig. 5A). These results suggested that proliferation and development of organelles, which enable cells to grow in nonfermentable carbon source such as mitochondria and peroxisome, the supply of substrates into mitochondria and peroxisomes required for respiration and utilization of nonfermentable carbon sources, and the gluconeogenesis required for generation of sugars in the absence of glucose in the medium were not functional enough to support oxidative growth in the post-diauxic phase, and thus the transition from fermentative to oxidative growth upon glucose exhaustion is defective in Δisc1.
The diauxic shift has been described to be closely related to glucose derepression (28-33). To enlighten how glucose-derepressed genes are regulated in Δisc1 across diauxic shift, we first investigated the expression of glucose-derepressed genes in the post-diauxic phase. A cohort of glucose-repressed genes was extracted from a published gene expression profile of cells shifted from galactose chemostat to 2 g/liter glucose for 30 min, which was deposited in the GEO database (33). Indeed, 58% of total glucose-repressed genes were contained in diauxic-up-regulated genes in our experiment (Fig. 5B, 260 genes out of 445 genes). A significant portion of the aberrantly regulated genes in Δisc1 was composed of glucose-repressible genes (Fig. 5B, 91 genes, 19% of total Isc1p-dependent diauxic up-regulated genes), indicating that Isc1p significantly regulates glucose-repressible genes across the diauxic shift, although its influence is not restricted to glucose repression because the majority of Isc1p-dependent genes are not glucose-derepressed genes.
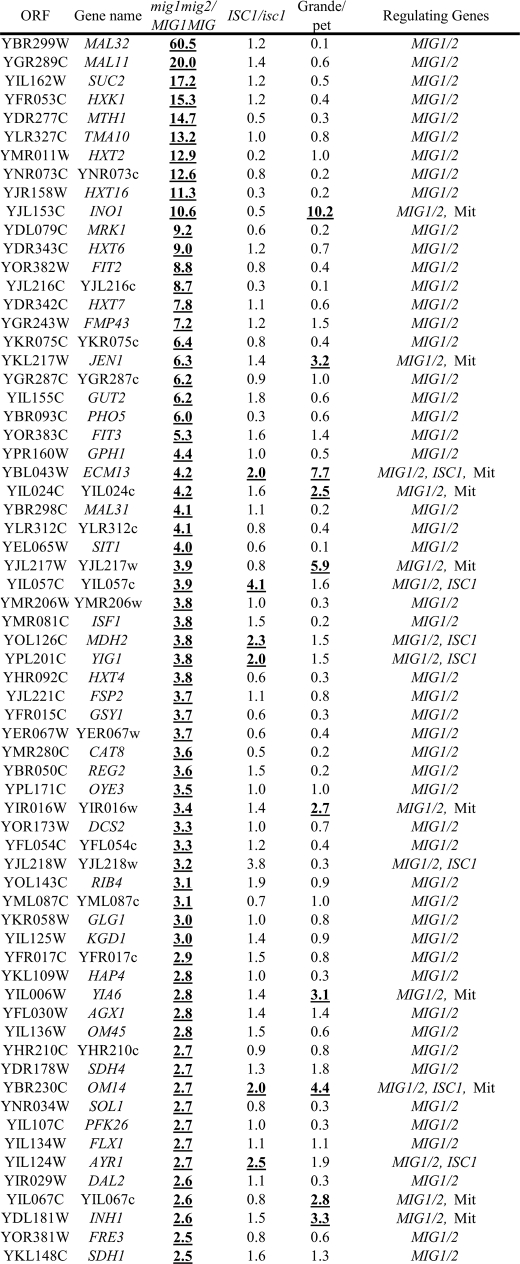
Because the above results indicated that Isc1p regulates a significant portion of glucose-repressible genes, we next compared the subgroup of Isc1p-dependent genes with those of Adr1p-, Snf1p-, and Cat8p-regulated genes (Table 1). Adr1p, Snf1p, and Cat8p have been reported to play critical roles in glucose repression (29). The result showed that Isc1p regulates a significant portion of most highly glucose-repressed genes (16 genes out of the 40 most highly glucose-derepressed genes). Although this number was smaller than Snf1p-regulated genes (27 genes out of the 40 most highly glucose-derepressed genes) and Adr1p (26 genes), it was similar to Cat8p (16 genes), further enforcing the hypothesis that Isc1p is a new player in the regulation of glucose repression. Isc1p-regulated genes significantly overlapped with those regulated by Adr1p (13 genes out of 16 Isc1p-regulated genes), but slightly less than those regulated by Snf1p (10 genes out of 16 Isc1p-regulated genes) or Cat8p (7 genes out of 16 Isc1p-regulated genes), suggesting a functional link between Isc1p and these pathways. Together, these analyses reveal that Isc1p regulates the diauxic shift, including glucose repression pathway with possibly some functional interaction with Snf1p, Adr1p, and Cat8p.
TABLE 1.
Regulation of the 40 most highly glucose repressed genes by Isc1p, mitochondrial function, Adr1p, Snf1p, and Cat8p
The genes whose expression ratio(s) in the parental strain relative to the mutant strain are more than 2-fold were described as genes that are regulated by the factor deficient in the mutant strain and expressed as boldface and underlined letters.
| Open reading frame | Gene name | I/R | ISC1/isc1 | Grande/petite | I/R | ADR1/adr1 | SNF1/snf1 | CAT8/cat8 | Regulating genes |
|---|---|---|---|---|---|---|---|---|---|
| YLR377c | FBP1 | 98.7 | 1.1 | 13.9 | 130.0 | 0.8 | 36.0 | 7.4 | Mit, SNF1, CAT8 |
| YKL217w | JEN1 | 1921.4 | 1.4 | 3.2 | 98.0 | 3.3 | 28.0 | 2.1 | Mit, ADR1, SNF1, CAT8 |
| YGR236c | SPG1 | 144.6 | 1.6 | 0.4 | 92.0 | 15.0 | 5.0 | 0.5 | ADR1, SNF1 |
| YKR097w | PCK1 | 67.3 | 0.9 | 0.5 | 92.0 | 1.4 | 180.0 | 3.6 | SNF1, CAT8 |
| YJR095w | SFC1 | 102.0 | 1.2 | 18.1 | 78.0 | 1.1 | 170.0 | 6.2 | Mit, SNF1, CAT8 |
| YIL057c | YIL057c | 2.4 | 0.6 | 0.4 | 77.0 | 16.0 | 8.9 | 0.8 | ADR1, SNF1 |
| YMR107w | SPG4 | 85.5 | 1.2 | 0.7 | 75.0 | 2.3 | 18.0 | 0.5 | ADR1, SNF1 |
| YCR010c | ADY2 | 144.6 | 2.9 | 26.0 | 72.0 | 20.0 | 32.0 | 9.3 | ISC1, Mit, ADR1, SNF1, CAT8 |
| YDR384c | ATO3 | 2.3 | 5.9 | 62.0 | 55.0 | 6.4 | 2.0 | 4.6 | ISC1, Mit, ADR1, CAT8 |
| YPL276w | YPL276w | 685.5 | 59.7 | 8.3 | 50.0 | 64.0 | 5.1 | 1.8 | ISC1, Mit, ADR1, SNF1 |
| YPR001w | CIT3 | 88.9 | 3.3 | 0.5 | 50.0 | 10.0 | 1.4 | 0.7 | ISC1, ADR1 |
| YGL205w | POX1 | 566.1 | 2.4 | 10.4 | 44.0 | 55.0 | 0.8 | 2.3 | ISC1, Mit, ADR1, CAT8 |
| YPR002w | PDH1 | 98.1 | 3.6 | 1.5 | 44.0 | 2.1 | 1.1 | 1.0 | ISC1, ADR1 |
| YAL054c | ACS1 | 87.0 | 2.8 | 6.1 | 43.0 | 9.0 | 30.0 | 3.5 | ISC1, Mit, ADR1, SNF1, CAT8 |
| YOR388c | FDH1 | 797.6 | 39.0 | 5.5 | 35.0 | 52.0 | 4.6 | 1.8 | ISC1, Mit, ADR1, SNF1 |
| YKL187c | YKL187c | 23.5 | 3.0 | 0.5 | 34.0 | 2.7 | 6.2 | 3.2 | ISC1, ADR1, SNF1, CAT8 |
| YDR256c | CTA1 | 114.8 | 3.6 | 7.0 | 34.0 | 16.0 | 7.6 | 3.3 | ISC1, Mit, ADR1, SNF1, CAT8 |
| YKR009c | FOX2 | 24.6 | 2.1 | 2.8 | 33.0 | 16.0 | 2.4 | 1.5 | ISC1, Mit, ADR1, SNF1 |
| YGR067c | YGR067c | 82.1 | 1.5 | 3.2 | 33.0 | 0.9 | 39.0 | 7.1 | Mit, SNF1, CAT8 |
| YNL195c | YNL195c | 22.2 | 0.6 | 0.3 | 29.0 | 3.0 | 7.7 | 0.8 | ADR1, SNF1 |
| YER065c | ICL1 | 43.5 | 1.3 | 18.8 | 28.0 | 0.4 | 77.0 | 20.0 | Mit, SNF1, CAT8 |
| YER024w | YAT2 | 44.5 | 1.4 | 0.8 | 26.0 | 0.6 | 5.6 | 2.0 | SNF1 |
| YGR243w | FMP43 | 23.8 | 1.2 | 1.5 | 26.0 | 2.8 | 6.1 | 0.6 | ADR1, SNF1 |
| YAR035w | YAT1 | 90.3 | 2.2 | 1.6 | 25.0 | 0.9 | 7.4 | 1.5 | ISC1, SNF1 |
| YMR206w | YMR206w | 69.4 | 1.0 | 0.3 | 25.0 | 1.2 | 5.9 | 1.0 | SNF1 |
| YIL160c | POT1 | 46.4 | 1.8 | 1.5 | 22.0 | 14.0 | 1.5 | 0.4 | ADR1 |
| YPR150w | YPR150w | 5.4 | 0.8 | 0.5 | 22.0 | 4.3 | 2.0 | 1.3 | ADR1, SNF1 |
| YLR174w | IDP2 | 92.0 | 1.4 | 2.2 | 21.0 | 0.9 | 16.0 | 2.5 | Mit, SNF1, CAT8 |
| YLR126c | YLR126c | 0.8 | 0.6 | 2.1 | 21.0 | 7.7 | 1.5 | 1.0 | Mit, ADR1 |
| YBR050c | REG2 | 22.2 | 1.5 | 0.2 | 21.0 | 1.5 | 42.0 | 5.0 | SNF1, CAT8 |
| YEL008w | YEL008w | 8.9 | 10.0 | 4.1 | 21.0 | 1.1 | 7.5 | 1.5 | ISC1, Mit, SNF1 |
| YPR006c | ICL2 | 24.9 | 4.2 | 13.4 | 21.0 | 7.0 | 4.0 | 2.1 | ISC1, Mit, ADR1, SNF1, CAT8 |
| YHL032c | GUT1 | 13.1 | 2.0 | 1.4 | 20.0 | 2.4 | 6.8 | 2.3 | ADR1, SNF1, CAT8 |
| YHR139c | SPS100 | 6.3 | 0.2 | 0.3 | 19.0 | 0.5 | 1.0 | 0.7 | |
| YPR151c | SUE1 | 2.3 | 0.6 | 0.9 | 18.0 | 3.8 | 1.9 | 1.2 | ADR1 |
| YLR267w | BOP2 | 17.3 | 0.6 | 0.2 | 17.0 | 0.4 | 0.9 | 0.1 | |
| YNR002c | ATO2 | 78.4 | 2.2 | 1.4 | 16.0 | 1.0 | 1.3 | 0.6 | ISC1 |
| YNL009w | IDP3 | 10.5 | 1.5 | 1.4 | 15.0 | 5.0 | 2.0 | 1.6 | ADR1 |
| YNL013c | YNL013c | 2.4 | 0.6 | 0.4 | 15.0 | 3.0 | 1.6 | 0.1 | ADR1 |
| YER179w | DMC1 | 7.8 | 2.3 | 3.9 | 14.0 | 7.0 | 1.8 | 0.9 | ISC1, Mit, ADR1 |
Glucose repression is also negatively regulated by Mig1p and Mig2p (34). Therefore, to investigate the relationship between Isc1p, Mig1p, and Mig2p, the genes regulated by Mig1p and Mig2p (34) were compared with those regulated by Isc1p. The analysis showed that only 9 genes out of 102 Mig1p/Mig2p-regulated genes are regulated by Isc1p, and thus the association of Isc1p with Mig1p and Mig2p in glucose signaling pathway remains unclear.
Defect in Ethanol Metabolism in the Δisc1—The above results suggested that a major mechanism for failure of the metabolic switch from glucose to ethanol in the Δisc1 strain is a result of failure of up-regulation of key enzymes in ethanol metabolism and a subsequent defective supply of substrates into mitochondria, rather than an intrinsic mitochondrial defect. Therefore, to investigate whether these large scale defects in gene regulation would culminate in an inability to metabolize ethanol, the utilization of ethanol by Δisc1 was determined by measuring growth in ethanol-containing medium. WT and Δisc1 cells incubated in YPD medium to the post-diauxic phase were inoculated to ethanol-containing medium and incubated aerobically with shaking, and growth was monitored at different time points. The results showed that Δisc1 shows very slow growth relative to WT in ethanol-containing medium (Fig. 5C), indicating that Δisc1 harbors severe intrinsic defects in ethanol-metabolizing ability in their post-diauxic phase. To further confirm the defect of Δisc1 in utilizing ethanol, WT and Δisc1 cells were incubated in YPD medium to the post-diauxic phase, and O2 consumption rate of the cells in the presence of ethanol was measured. Indeed, the Δisc1 strain demonstrated a significantly lower respiration rate (63% of WT) (Fig. 5D). Together, these data demonstrate failure of the Δisc1 to properly utilize ethanol as a carbon source.
Requirement for Intact Mitochondria in Induction of Nuclear Genes during the Diauxic Shift—The Δisc1 strain, although not showing any significantly defective mitochondrial DNA, defective function of the respiratory chain, or altered amount of reactive oxygen species, failed to utilize ethanol as a carbon source because of severely aberrant gene expression during the diauxic shift. These observations, coupled with the previous finding that Isc1p has a mitochondrial localization, raised the intriguing possibility that functional mitochondria may be required for proper execution of the reprogramming of nuclear gene expression during the diauxic shift. If this hypothesis is true, then gene expression associated with the diauxic shift should be compromised in respiratory-deficient petite cells.
To test this hypothesis, we sought to determine whether intact mitochondria are required for the global gene induction observed in the post-diauxic phase, which is required for optimal metabolic function. Therefore, microarray studies were performed at 4 and 24 h in petite cells and analyzed for genome-wide gene expression changes with respect to grande cells. First, the induction ratios of gene expression of up-regulated genes at 24 to 4 h of grande experiment 2 (black triangles) and petite (orange diamonds) were plotted against that of grande experiment 1 (Fig. 6A). In this representation, genes that show failure of induction in the petite strain appear below the red line of identity (Fig. 6A). Contrary to grande cells, the points of petite cells were scattered downward from the trend line of the grande cells (red circle, Fig. 6A). This was quantitated by the decreased correlation coefficient from 0.83 (grande, black triangles in Fig. 6A) to 0.56 (petite, orange diamonds in Fig. 6A) and the decreased slope of the dots from 0.97 (grande, red line in Fig. 6A) to 0.74 (petite, blue line in Fig. 6A), clearly indicating a global defect in gene regulation across the diauxic shift in petite cells. Next, and to gain functional insight into the metabolic consequences of the dysregulation of induction of genes involved in carbon source metabolism, the genes of the carbon source metabolism pathway were superimposed on a metabolic pathway map according to the fold change of petite/grande across the diauxic shift (Fig. 6B). As in Δisc1, the genes involved in utilization of nonfermentable carbon source, such as glyoxylate cycle, peroxisomal β-oxidation, tricarboxylic acid cycle, gluconeogenesis, ethanol utilization, and mitochondrial import genes, failed to be up-regulated, whereas those involved in fermentative growth such as glycolysis genes remained similar to grande cells, as shown by the color and the thickness of the boxes. Notably, similarly to Δisc1, the genes that failed to be up-regulated in the post-diauxic phase of petite cells included a significant portion of the genes involved in gluconeogenesis such as MLS1, IDP2, SFC1, JEN1, and FBP1, again suggesting a defect of generating glucose of petite cells in the post-diauxic phase. This result also shows that petite cells are defective in up-regulation of genes involved in utilization of nonfermentable carbon source.
Also, to determine whether the defects that occurred in Δisc1 and petite cells are similar, we investigated how those genes that failed to be up-regulated in Δisc1 behave in petite cells. As shown in supplemental Table SII, most of the genes that failed to be up-regulated in Δisc1 also failed to be up-regulated in petite cells, indicating a high degree of similarity in the aberrant gene regulation between Δisc1 and petite cells.
Because, as discussed above, the gene expression profile has been reported to drastically change according to the growth phases, the expression of ADH2 and CTA1 was measured across the diauxic shift in respiratory-competent cells and in petite cells by real time RT-PCR. The growth of the cells was monitored by A660 and glucose concentration (Fig. 7A). The results showed that these two genes failed to be fully induced across the diauxic shift in the petite cells up to 48 h (Fig. 7, B and C), which is consistent with the conclusion obtained from microarray analysis. This result also rules out the possibility that the difference in gene expression profile between grande and petite cells obtained by microarray analysis is merely due to difference in growth phase, and further supports the hypothesis that intact mitochondria are required for proper execution of the genetic reprogramming of the diauxic shift.
FIGURE 7.
Suppression of induction of ADH2 and CTA1 in respiratory-deficient petite cells during the diauxic shift and upon transfer to nonfermentable medium. A, growth curve of grande and petite cells in YPD medium as monitored by glucose concentration and A660. The expression of ADH2 (B) and CTA1 (C) was quantitated by real time RT-PCR at the indicated times. D and E, logarithmically growing cells were transferred from YPD to YPethanol and incubated for 2 h, and the expression of ADH2 (D) and CTA1 (E) was quantitated by real time RT-PCR. The results are represented as means ± S.E. of relative expression of at least three experiments. F, Venn diagram of the number of whole genome (blue), diauxic up-regulated (orange), glucose-repressed (green), and mitochondrial function-dependent diauxic up-regulated (brown) genes. A cohort of glucose-repressed genes was extracted from glucose pulse experiment (33).
Because these results suggested a role for mitochondria in the induction of these genes during the diauxic shift, we next employed a direct and acute shift of growth from glucose to ethanol, a model that recapitulates this metabolic component of the diauxic shift. Grande and petite cells were grown to exponential phase in YPD and then incubated in YPethanol for 2 h. RNA was extracted, and the gene expression of ADH2 and CTA1 was analyzed by real time RT-PCR. The results showed that these genes also failed to be up-regulated when transferred from YPD to YPethanol during logarithmic growth (Fig. 7, D and E). These results demonstrate that the gene expression essential for executing the acute shift of growth from glucose to ethanol requires normal mitochondrial function.
To investigate how glucose-repressible genes behave in petite cells across the diauxic shift, induction of glucose-repressible genes during the diauxic shift was analyzed by comparing with a previous study (33). First, analysis of the mitochondrial function-dependent genes revealed that 680 genes failed to become up-regulated more than 2-fold in the petite strain across diauxic shift relative to grande cells. A significant portion of the aberrantly regulated genes in petite were glucose-repressible genes (Fig. 7F, 92 genes, 14% of total mitochondrial function-dependent diauxic up-regulated genes), indicating that mitochondrial function significantly regulates glucose repression, although its influence is not restricted to glucose repression.
Because the above analysis indicated the involvement of mitochondrial function in glucose repression, the mitochondrial function-regulated genes were compared with Adr1p-, Snf1p-, and Cat8p-regulated genes (29), as was performed above with Δisc1 (Table 1). The results were similar to those obtained for Δisc1, and they showed that mitochondrial function actually regulates a significant cohort of genes that are up-regulated upon glucose derepression (18 genes out of the 40 most highly glucose-derepressed genes), further confirming the role of mitochondrial function in glucose repression. Mitochondrial function-regulated genes significantly overlapped with those regulated by Snf1p (14 genes out of 18 mitochondrial function-regulated genes), but overlapped less with Cat8p-regulated genes (12 genes out of 18) or Adr1p (12 genes out of 18), suggesting that some functional link exists between respiratory function and Snf1p, and some unknown glucose-signaling pathway is regulated by respiratory function. To the contrary, only 19 genes out of 102 Mig1p- and Mig2p-regulated genes overlapped with mitochondrial function-regulated genes (Table 2), and thus the association of mitochondrial function and Mig1p/Mig2p-regulated glucose signaling pathway remains unclear. Together, these results indicate that respiratory competence is required for global gene regulation of nuclear genes associated with the diauxic shift, including glucose repression through some functional association with Snf1p, Cat8p and Adr1p, which has, to our knowledge, not been reported previously. These two characteristics, defective mitochondrial function and failure of up-regulation of genes across the diauxic shift shared between Δisc1 and petite cells, link the mitochondrial state, specifically respiratory competence of mitochondria, to the nuclear gene expression required to mediate the transition from anaerobic to aerobic metabolism, which is a novel observation.
TABLE 2.
Comparison of Isc1p- and mitochondrial function-regulated genes with Mig1p- and Mig2p-regulated genes
ORF means open reading frame; pet means petite. The genes whose expression ratio(s) in Δmig1 Δmig2 relative to the parental strain are more than 2-fold were extracted as genes that are regulated by Mig1P, Mig2P (34), Isc1p, or mitochondrial function (this study) and expressed as boldface and underlined letters.
Relationship of the Mitochondrial Function in the Post-diauxic Induction of Nuclear Genes to the Retrograde Response—It should be noted that several recent reports have provided evidence that compromising mitochondria, such as by treatment with antimycin A or generation of respiratory-deficient petite cells, leads to a compensatory genome response that has been termed the “retrograde” response (35-44). However, the genome-wide response in petite and Δisc1 elucidated in this study appears distinct from the retrograde response in several aspects. First, the retrograde response functions in the logarithmic phase; in contrast, the response elucidated in this study functions during the diauxic shift and the shift from glucose to ethanol. Indeed, examination of results shown in Fig. 7, D and E, confirms induction of ADH2 and CTA1 in the pre-diauxic phase in petite cells that is characteristic of the retrograde response. It is the failure of post-diauxic induction of genes that differentiates the response elucidated in this study from the retrograde response. Second, whereas the retrograde response up-regulates expression of genes of the peroxisomal and metabolite restoration (anapleurotic) pathway, the response elucidated in this study suppresses up-regulation of genes of nonfermentable carbon source metabolism during the diauxic shift and upon transfer from the exponentially grown state in YPD to YPethanol. Third, the overall function of the retrograde response appears to mitigate the loss of a competent tricarboxylic acid cycle, whereas the function of the response elucidated in this study is suggested to prevent entrance into the post-diauxic (aerobic) phase if mitochondria are not respiratory-competent. Finally, clear-cut differentiation came from studies utilizing the Δisc1 strain.
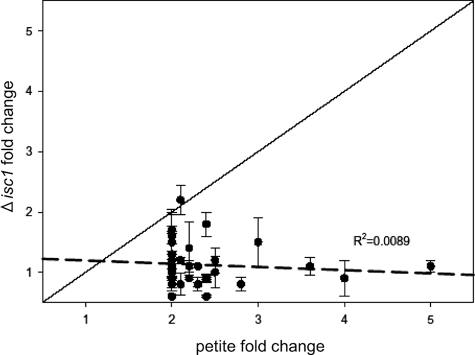
Because Δisc1 displayed a similar defective response as respiratory-deficient petite cells while apparently maintaining normal “intrinsic” function of the respiratory chain (Fig. 2C), we hypothesized that this deletion mutant could provide direct evidence to distinguish the retrograde response from the response elucidated in this study. Therefore, we examined how genes known to be regulated by the retrograde response are regulated in WT and Δisc1 because induction of the retrograde-response genes in Δisc1 would indicate activation of this program. Thus, the fold change in expression of these retrograde-specific genes in the Δisc1 strain versus the parental strain (incubated in YPD) was determined and graphed versus induction in petite cells, which exhibit the retrograde response (Fig. 8 and supplemental Table SV) (44). The analysis indicated no induction of these genes in Δisc1 (less than 2-fold changes), and there was no correlation between retrograde gene induction and gene induction in the Δisc1 strain (R2 = 0.0089). These findings indicate that the retrograde response is not activated in the Δisc1 strain, demonstrating that deletion of ISC1 does not generate a signal for tricarboxylic acid cycle deficiency required for the retrograde response. Thus, the suppression of expression of genes essential for diauxic shift is distinct from activation of the retrograde response. The results show that the Δisc1 does not activate the retrograde response, which has been proposed as a sine que non of mitochondrial dysfunction. Together with the results that the mitochondria of the Δisc1 strain do not show intrinsic functional defects (DNA levels, oxygen consumption, and reactive oxygen species), these results raise the intriguing possibility that Isc1p may function more in the “signaling” of mitochondrial health rather than play a direct role in mitochondrial integrity/functions, thus enabling the supply of substrates of respiration into mitochondria. This hypothesis is worthy of further investigation. In conclusion, the results from this study disclose a novel role for Isc1p in regulating the post-diauxic induction of genes involved in aerobic carbon metabolism. The results also demonstrate a new link between mitochondrial state and gene induction during the diauxic shift.
FIGURE 8.
Retrograde response is not apparently up-regulated in pre-diauxic phase of Δisc1. The plot of fold change in expression of the genes in the Δisc1 strain versus the parental strain when grown in YPD was determined and graphed versus induction in petite cells (which exhibit the retrograde response) (44). Regression analysis was used to establish a trend line (dashed line), which indicated little to no correlation between retrograde gene induction and gene induction in the Δisc1 strain (R2 = 0.0089). Fold change is shown as the mean of two independent experiments ± S.E.
Supplementary Material
Acknowledgments
We thank Dr. Hiroko Hama, Dr. Kazuyuki Kitatani, Dr. Chiara Luberto, and Dr. Fernando Alvarez-Vasquez for helpful advice. The microarray studies were performed at the Microarray and Bioinformatics Core Facility at the Medical University of South Carolina, which is supported by National Institutes of Health Grants P01CA095841 and P20RR016434.
This work was supported, in whole or in part, by National Institutes of Health Grant GM43825 (to Y. A. H.) and Grant R01 AG016583 (to L. M. O.). This work was also supported by a Veterans Affairs Merit Award (to L. M. O.), a Merit Review Entry Program Award (to L. A. C.) from the Department of Veterans Affairs, Office of Research and Development, and in part by an NISR research grant from the Noda Institute for Scientific Research (to H. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables SI-SV.
Footnotes
The abbreviations used are: WT, wild type; FACS, fluorescence-activated cell sorter; RT, reverse transcription; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Regev-Rudzki, N., and Pines, O. (2007) BioEssays 29 772-782 [DOI] [PubMed] [Google Scholar]
- 2.Schlame, M., and Greenberg, M. L. (1997) Biochim. Biophys. Acta 1348 201-206 [DOI] [PubMed] [Google Scholar]
- 3.Kitagaki, H., Cowart, L. A., Matmati, N., Vaena de Avalos, S., Novgorodov, S. A., Zeidan, Y. H., Bielawski, J., Obeid, L. M., and Hannun, Y. A. (2007) Biochim. Biophys. Acta 1768 2849-2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trotter, P. J., Adamson, A. L., Ghrist, A. C., Rowe, L., Scott, L. R., Sherman, M. P., Stites, N. C., Sun, Y., Tawiah-Boateng, M. A., Tibbetts, A. S., Wadington, M. C., and West, A. C. (2005) Arch. Biochem. Biophys. 442 21-32 [DOI] [PubMed] [Google Scholar]
- 5.Lill, R., Dutkiewicz, R., Elsässer, H. P., Hausmann, A., Netz, D. J., Pierik, A. J., Stehling, O., Urzica, E., and Mühlenhoff, U. (2006) Biochim. Biophys. Acta 1763 652-667 [DOI] [PubMed] [Google Scholar]
- 6.Foury, F., Roganti, T., Lecrenier, N., and Purnelle, B. (1998) FEBS Lett. 440 325-331 [DOI] [PubMed] [Google Scholar]
- 7.Sickmann, A., Reinders, J., Wagner, Y., Joppich, C., Zahedi, R., Meyer, H. E., Schonfisch, B., Perschil, I., Chacinska, A., Guiard, B., Rehling, P., Pfanner, N., and Meisinger, C. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13207-13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, D. R., and Reed, J. C. (1998) Science 281 1309-1312 [DOI] [PubMed] [Google Scholar]
- 9.Schapira, A. H. (2006) Lancet 368 70-82 [DOI] [PubMed] [Google Scholar]
- 10.Singh, K. K. (2006) Ann. N. Y. Acad. Sci. 1067 182-190 [DOI] [PubMed] [Google Scholar]
- 11.Trifunovic, A., Wredenberg, A., Falkenberg, M., Spelbrink, J. N., Rovio, A. T., Bruder, C. E., Bohlooly-Y, M., Gidlof, S., Oldfors, A., Wibom, R., Tornell, J., Jacobs, H. T., and Larsson, N. G. (2004) Nature 429 417-423 [DOI] [PubMed] [Google Scholar]
- 12.Vaena de Avalos, S., Okamoto, Y., and Hannun, Y. A. (2004) J. Biol. Chem. 279 11537-11545 [DOI] [PubMed] [Google Scholar]
- 13.Vaena de Avalos, S., Su, X., Zhang, M., Okamoto, Y., Dowhan, W., and Hannun, Y. A. (2005) J. Biol. Chem. 280 7170-7177 [DOI] [PubMed] [Google Scholar]
- 14.Okamoto, Y., Vaena De Avalos, S., and Hannun, Y. A. (2002) J. Biol. Chem. 277 46470-46477 [DOI] [PubMed] [Google Scholar]
- 15.Okamoto, Y., Vaena de Avalos, S., and Hannun, Y. A. (2003) Biochemistry 42 7855-7862 [DOI] [PubMed] [Google Scholar]
- 16.Sawai, H., Okamoto, Y., Luberto, C., Mao, C., Bielawska, A., Domae, N., and Hannun, Y. A. (2000) J. Biol. Chem. 275 39793-39798 [DOI] [PubMed] [Google Scholar]
- 17.Almeida, T., Marques, M., Mojzita, D., Amorim, M. A., Silva, R. D., Almeida, B., Rodrigues, P., Ludovico, P., Hohmann, S., Moradas-Ferreira, P., Côrte-Real, M., and Costa, V. (2008) Mol. Biol. Cell 19 865-876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shea, J. M., Kechichian, T. B., Luberto, C., and Del Poeta, M. (2006) Infect. Immun. 74 5977-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowart, L. A., Okamoto, Y., Pinto, F. R., Gandy, J. L., Almeida, J. S., and Hannun, Y. A. (2003) J. Biol. Chem. 278 30328-30338 [DOI] [PubMed] [Google Scholar]
- 20.Cowart, L. A., Okamoto, Y., Lu, X., and Hannun, Y. A. (2006) Biochem. J. 393 733-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider-Berlin, K. R., Bonilla, T. D., and Rowe, T. C. (2005) Mutat. Res. 572 84-97 [DOI] [PubMed] [Google Scholar]
- 22.Tzagoloff, A., and Dieckmann, C. L. (1990) Microbiol. Rev. 54 211-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foury, F., and Cazzalini, O. (1997) FEBS Lett. 411 373-377 [DOI] [PubMed] [Google Scholar]
- 24.Giraud, M. F., and Velours, J. (1997) Eur. J. Biochem. 245 813-818 [DOI] [PubMed] [Google Scholar]
- 25.Dunn, C. D., Lee, M. S., Spencer, F. A., and Jensen, R. E. (2005) Mol. Biol. Cell 17 213-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machida, K., Tanaka, T., Fujita, K., and Taniguchi, M. (1998) J. Bacteriol. 180 4460-4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pozniakovsky, A. I., Knorre, D. A., Markova, O. V., Hyman, A. A., Skulachev, V. P., and Severin, F. F. (2005) J. Cell Biol. 168 257-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeRisi, J. L., Iyer, V. R., and Brown, P. O. (1997) Science 278 680-686 [DOI] [PubMed] [Google Scholar]
- 29.Young, E. T., Dombek, K. M., Tachibana, C., and Ideker, T. (2003) J. Biol. Chem. 278 26146-26158 [DOI] [PubMed] [Google Scholar]
- 30.Nishizawa, M., Katou, Y., Shirahige, K., and Toh-e, A. (2004) Yeast 21 903-918 [DOI] [PubMed] [Google Scholar]
- 31.Haurie, V., Boucherie, H., and Sagliocco, F. (2003) J. Biol. Chem. 278 45391-45396 [DOI] [PubMed] [Google Scholar]
- 32.Haurie, V., Perrot, M., Mini, T., Jenö, P., Sagliocco, F., and Boucherie, H. (2001) J. Biol. Chem. 276 76-85 [DOI] [PubMed] [Google Scholar]
- 33.Ronen, M., and Botstein, D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 389-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westergaard, S. L., Oliveira, A. P., Bro, C., Olsson, L., and Nielsen, J. (2007) Biotechnol. Bioeng. 96 134-145 [DOI] [PubMed] [Google Scholar]
- 35.Epstein, C. B., Waddle, J. A., Hale, W., IV, Dave, V., Thornton, J., Macatee, T. L., Garner, H. R., and Butow, R. A. (2001) Mol. Biol. Cell 12 297-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butow, R. A. (2002) Cell Death Differ. 9 1043-1045 [DOI] [PubMed] [Google Scholar]
- 37.Butow, R. A., and Avadhani, N. G. (2004) Mol. Cell 14 1-15 [DOI] [PubMed] [Google Scholar]
- 38.Liao, X., and Butow, R. A. (1993) Cell 72 61-71 [DOI] [PubMed] [Google Scholar]
- 39.Sekito, T., Thornton, J., and Butow, R. A. (2000) Mol. Biol. Cell 11 2103-2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parikh, V. S., Morgan, M. M., Scott, R., Clements, L. S., and Butow, R. A. (1987) Science 235 576-580 [DOI] [PubMed] [Google Scholar]
- 41.Borghouts, C., Benguria, A., Wawryn, J., and Jazwinski, S. M. (2004) Genetics 166 765-777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jazwinski, S. M. (2005) Gene (Amst.) 354 22-27 [DOI] [PubMed] [Google Scholar]
- 43.Jazwinski, S. M. (2005) FEMS Yeast Res. 5 1253-1259 [DOI] [PubMed] [Google Scholar]
- 44.Traven, A., Wong, J. M., Xu, D., Sopta, M., and Ingles, C. J. (2001) J. Biol. Chem. 276 4020-4027 [DOI] [PubMed] [Google Scholar]
- 45.Schuller, H. J. (2003) Curr. Genet. 43 139-160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.