Abstract
Huntingtin proteolysis is implicated in Huntington disease pathogenesis, yet, the nature of huntingtin toxic fragments remains unclear. Huntingtin undergoes proteolysis by calpains and caspases within an N-terminal region between amino acids 460 and 600. We have focused on proteolytic steps producing shorter N-terminal fragments, which we term cp-1 and cp-2 (distinct from previously described cp-A/cp-B). We used HEK293 cells to express the first 511 residues of huntingtin and further define the cp-1 and cp-2 cleavage sites. Based on epitope mapping with huntingtin-specific antibodies, we found that cp-1 cleavage occurs between residues 81 and 129 of huntingtin. Affinity and size exclusion chromatography were used to further purify huntingtin cleavage products and enrich for the cp-1/cp-2 fragments. Using mass spectrometry, we found that the cp-2 fragment is generated by cleavage of huntingtin at position Arg167. This site was confirmed by deletion analysis and specific detection with a custom-generated cp-2 site neo-epitope antibody. Furthermore, alterations of this cleavage site resulted in a decrease in toxicity and an increase in aggregation of huntingtin in neuronal cells. These data suggest that cleavage of huntingtin at residue Arg167 may mediate mutant huntingtin toxicity in Huntington disease.
Huntington disease (HD)3 is a progressive neurodegenerative disorder caused by a polyglutamine (poly(Q)) expansion within the coding region of the HD gene product, huntingtin (Htt) (1, 2). The mechanisms of Htt induced toxicity are still largely unknown; however, there is mounting evidence that toxic poly(Q)-expanded Htt fragments are formed from full-length Htt via proteolysis (3-18). Thus, the Htt cleavage pathway and the molecules involved in it may represent potential therapeutic targets for intervention in HD.
There are at least two domains susceptible to proteolysis in the 345-kDa Htt protein (Fig. 1). Htt can be cleaved by caspases and calpains at multiple sites within the region between residues 460 and 600 (14, 19-29). Cleavage of Htt at position 586 by caspase 6 is of particular importance for HD pathogenesis, as alterations of this site in a YAC128 HD mouse model strikingly ameliorate the phenotype (25). Another cleavage-prone region lies near the N terminus of Htt. Lunkes and co-workers (5) described two short N-terminal Htt cleavage products, cp-A and cp-B, and mapped the cp-A cleavage site to residues 105-114. We have recently characterized two short N-terminal fragments of similar size (cp-1 and cp-2) generated in a stable inducible PC12 cell model engineered to express full-length normal and expanded Htt (30). We found that deletion of amino acids 105-114 failed to prevent the formation of either fragment, suggesting that cp-1 is distinct from previously described cp-A. Furthermore, in previous reports cp-A/B fragments were observed only in the presence of proteasome inhibitors, whereas cp-1/2 fragments are easily detectable without proteasome inhibition in transient and stably transfected cells in our systems, suggesting that these fragments may be distinct from cp-A/B. Using the PC12 cell model we found that cp-1 and cp-2 fragments accumulate within nuclear and cytoplasmic inclusions, and can be generated via a caspase independent pathway (30).
FIGURE 1.
The schematics of Htt proteolysis. HEAT repeats (blue boxes) are indicated within the Htt sequence, N-terminal and caspase/calpain putative proteolytic domains are shown (yellow boxes). Putative cp-1 and confirmed cp-2 sites are indicated by arrows. Epitopes recognized by Htt antibodies are marked with red lines.
Although Htt cleavage may play an important role in HD, it is possible that not all Htt proteolytic fragments contribute to toxicity. In fact, there is growing evidence for a role of fragments of specific size in the pathogenic process. For example, YAC128 mouse HD models indicate that caspase 2- and caspase 3-generated expanded Htt fragments (552 and 513 amino acids in length, respectively) are not involved in pathogenesis, whereas the caspase 6-generated 586-amino acid fragment is pathogenic (25). Furthermore, endogenous caspase-generated Htt fragments have different cellular distributions: caspase 2/3-generated fragments localize to the perinuclear region, whereas caspase 6-generated fragments are enriched in the nucleus (31). Other mouse models of HD demonstrate that the exon 1 Htt fragment (R6/2 mice) (15) and the N-terminal 171-amino acid Htt fragment (N171-82Q mice) (16) are toxic, whereas the N-terminal 117-amino acid fragment (N117) does not convey the HD phenotype in the “shortstop” mice (32). Thus, it is of particular interest to map the precise cleavage sites in expanded Htt and to evaluate toxic properties of specific Htt fragments formed by proteolysis.
Here we employed a combination of mass spectrometry and site-directed mutagenesis to define the cleavage events producing short N-terminal fragments of Htt. We found a potential new site of cleavage, generating the cp-2 fragment, at position Arg167. To determine a role for cp-2 in aggregation and toxicity, we generated mutations within the potential cp-2 cleavage site. Based on these studies, the presence of the cp-2 fragment is involved in Htt-induced toxicity in neuronal HT22 cells. In contrast, the N117 (shortstop) fragment is less toxic. These results support the idea that cleavage of Htt at specific sites may mediate mutant Htt toxicity in HD.
EXPERIMENTAL PROCEDURES
Plasmids and Mutagenesis—All Htt constructs used represent N-terminal fragments and are referred to as N followed by a number of amino acids present (e.g. N511). Truncated Htt expression constructs N511-8Q/82Q were generated from previously described (11) full-length Htt constructs (HD-FL-23/82Q) by an introduction of a stop codon after amino acid 511 of Htt. Htt-N511-32Q/52Q constructs were produced from N511-82Q by random contractions of the poly(Q) repeat in bacterial cells. Truncated Htt expression constructs N90-8Q/82Q, N117-8Q/82Q, N167-8Q/82Q, and N171-8Q/82Q were generated from Htt-N511-8Q/82Q by an introduction of a stop codon after the indicated amino acid. The stop codons and all deletions used in this study were introduced by site-directed mutagenesis using the QuikChange II XL kit (Stratagene) according to the manufacturer's protocol. Htt-N586-82Q plasmid was a gift from David Borchelt (University of Florida, Gainesville, FL) and Htt-N586-17Q/128Q plasmids were previously described (25). The Htt-N586-52Q construct was produced from N511-82Q by random contractions of poly(Q) repeat in bacterial cells.
Cell Culture and Transfection—Human embryonic kidney (HEK) 293FT cells (Invitrogen) were grown in Dulbecco's modified Eagle's medium (with 4.5 g/liter d-glucose, Invitrogen) supplemented with 10% fetal bovine serum, 100 μg/ml Geneticin, 100 units/ml penicillin, and 100 units/ml streptomycin. Mouse hippocampal HT22 cells were maintained in Dulbecco's modified Eagle's medium (with 1 g/liter d-glucose, Invitrogen) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 units/ml streptomycin. Mouse neuroblastoma 2a (N2a) cells (ATCC, CCL-131) were maintained in 50% Opti-MEM (Invitrogen), 50% Dulbecco's modified Eagle's medium (with 4.5 g/liter d-glucose), supplemented with 5% fetal bovine serum, 100 units/ml penicillin, and 100 units/ml streptomycin. All cells were transfected with Htt constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Purification of Htt Fragments for Mass Spectrometry—HEK293 cells were transfected with either Htt-N511-52Q or Htt-N511-8Q constructs, and lysed in M-PER buffer (Pierce) with protease inhibitors (Protease Inhibitor Mixture III, Calbiochem). The lysates were diluted 1:1 with phosphate-buffered saline and NaCl was added to a final concentration of 150 mm. FLAG-Htt fusion proteins were immunoprecipitated from a large amount of material (15-20 mg of total protein per immunoprecipitation) using anti-FLAG M2 affinity gel (Sigma) according to the manufacturer's protocol, followed by elution with 100 μg/ml of FLAG peptide. Eluted proteins were further purified by size-exclusion chromatography on a Superose-12 column (Amersham Biosciences) under denaturing conditions (phosphate-buffered saline, 2 m guanidine hydrochloride, 5 mm dithiothreitol) to disrupt Htt protein complexes. Collected fractions were concentrated and purified using the PAGEprep Advance kit (Pierce), and aliquots were analyzed by Western blotting with an antibody to FLAG. Fractions containing the cp-1/cp-2 fragments, or those containing only Htt-N511 were loaded on the NuPAGE 4-12% BisTris polyacrylamide gel, and proteins were visualized with silver stain (SilverQuest kit, Invitrogen) for subsequent mass spectrometric analysis.
In-gel Digestion of Htt Proteins—The bands, containing Htt proteins were manually cut out for in-gel digestion and further mass spectrometric analysis. The gel pieces were destained with a 1:1 mixture of 30 mm potassium ferricyanide and 100 mm sodium thiosulfate, rinsed 3 times with water, and incubated in 20 mm ammonium bicarbonate for 10 min, followed by dehydration with acetonitrile. The whole cycle was repeated two more times, and the gel pieces were then dried in a SpeedVac. For in-gel digestion, gel pieces were incubated overnight at 37 °C with 10 ng/μl chymotrypsin (Roche), in 20 mm bicarbonate. The peptides were extracted with 50% acetonitrile and 2% formic acid (2 times). The extracts were pooled and evaporated to dryness.
Mass Spectrometry of Htt Proteins: LC-MS/MS Analysis—The peptides were analyzed using QSTAR Pulsar (Applied Biosystems-MDS Sciex) interfaced with an Eksigent nano-LC system. Peptides were dissolved in 10 μl of 0.2% formic acid and 5 μl of the solution was loaded on a peptide trap and then separated on a 360 × 75-μm reverse-phase column packed with 10 cm of C18 beads (5 μm, 120 Å, YMC ODS-AQ, Waters) and a 10-μm emitter tip (New Objective). The high pressure liquid chromatography gradient was 5-40% B for 25 min (A, 0.1% formic acid; B, 90% acetonitrile in 0.1% formic acid) and the flow rate was 300 nl/min. Survey scans were acquired from m/z 350-1200 with up to three precursors selected for MS/MS using a dynamic exclusion of 30 s. Rolling collision energy was used to promote fragmentation. The synthetic peptide MDSNLPR was analyzed in nanospray mode using Protana nanospray tips. The electrospray voltage was 900 V and MS/MS spectra of ion 416.7 m/z were acquired for 3 min.
The MS/MS spectra were searched against NCBInr data base for all species, with no enzyme and 2 missed cleavages using the in-house Mascot server and Mascot Daemon as an interface. Proteins with two peptides having scores higher than probability threshold (usually 50) were considered good hits.
Western Blot Analysis, Immunoprecipitation, and Antibodies—Polyclonal antibodies to Htt, htt-(55-65) and htt-(81-90), were described previously (7). Goat polyclonal antibody, prepared against the N-terminal Htt exon-1 fragment was described previously (33). Htt-(115-129) antibody (against residues 115-129 of Htt) and Htt-(1-82) antibody (against residues 1-82 of Htt) were from Chemicon International. The antibodies to the neo-epitope Arg167 of Htt were produced in rabbit and affinity purified against two synthetic peptides: Ac-CMDSNLPR-OH (NE167) and Ac-CGGGGNLPR-OH (Covance). For Western blotting analysis, HEK293 cells, expressing indicated constructs, were lysed (48 h after transfection) in M-PER buffer (Pierce) with protease inhibitors (Protease Inhibitor Mixture III, Calbiochem), and protein concentrations were estimated using the BCA method (Bio-Rad). Lysates were fractionated on NuPAGE 4-12% BisTris polyacrylamide gels (Invitrogen), transferred to nitrocellulose membranes, and probed with antibodies to Htt, FLAG (M2, Sigma), or actin (Sigma) for loading control. For immunoprecipitation, HT22 cells (1 × 150 mm dish), transfected with the indicated constructs, were lysed in M-PER buffer (Pierce) with protease inhibitors (Protease Inhibitor Mixture III). The lysates were diluted 1:1 with phosphate-buffered saline, and NaCl was added to a final concentration of 150 mm. Htt-FLAG fusion proteins were immunoprecipitated using anti-FLAG M2 affinity gel (Sigma) according to the manufacturer's protocol, followed by fractionation on NuPAGE 4-12% BisTris polyacrylamide gels (Invitrogen), and detection with an antibody to exon 1 of Htt.
For cell fractionation in HEK293 cells, M-PER lysates were centrifuged at 15,000 × g. Supernatants (soluble cytoplasmic fractions) were fractionated on NuPAGE 4-12% BisTris polyacrylamide gels (Invitrogen) or on native PAGE 4-16% BisTris gels (Invitrogen) according to the manufacturer's protocol. The pellets were resuspended in 2% SDS with Benzonase nuclease (Sigma) and sonicated. The resulting suspensions were either fractionated on SDS-PAGE (NuPAGE 4-12%), or further centrifuged at 15,000 × g to obtain the pellet of SDS-insoluble aggregates, which was than treated with formic acid at 37 °C for 30 min, as previously described (30). Formic acid-soluble aggregates were analyzed on SDS-PAGE as above. Immunoblots were developed with peroxidase-conjugated secondary antibodies (Amersham Biosciences), and enhanced chemiluminescence (ECL-Plus detection reagent, Amersham Biosciences).
Immunofluorescence—HEK293 or HT22 cells, expressing the indicated constructs, were fixed (48 h after transfection) with 4% paraformaldehyde for 15 min, permeabilized with 0.5% Triton X-100 (Sigma) for 10 min, blocked in 10% normal goat serum (Sigma) for 30 min, and incubated with monoclonal antibody to Htt epitope 115-129, and polyclonal antibody to the neo-epitope Arg167 of Htt, NE167 (1 h at room temperature), followed by goat anti-mouse IgG FITC-conjugated (Vector), and goat anti-rabbit Cy3-conjugated (Vector) secondary antibodies (1 h at room temperature). Nuclei were stained with 4′,6-diamidino-2-phenylindole (Vector). Microscopy was performed using a Zeiss Axiovert 200M microscope equipped with a ×40 objective.
Htt Aggregation and Cell Viability Measurements—For evaluation of Htt aggregation, N2a or HT22 cells, expressing the indicated Htt constructs, were stained with an antibody to FLAG or Htt-(1-82) antibody, followed by anti-mouse IgG FITC-conjugated antibody, and the aggregates were counted in 24-well plates in duplicate wells. 200-400 cells were counted for each construct. For cell viability assay, HT22 cells, grown in 24-well plates, were co-transfected with the indicated Htt constructs and luciferase reporter vector (pGL3-Control, Promega) in a ratio 20:1. 48 h after transfection cells from triplicate wells were lysed in 200 μl of M-PER buffer (Pierce), and luciferase activity was measured in 1/5 of the lysate using the Luciferase Assay System (Promega) according to the manufacturer's protocol. For caspase 3 activation cytotoxicity assays, HT22 cells were transiently transected with the indicated Htt constructs. 24 h post-transfection, cells were cultured in the absence of serum for an additional 8 h. Immunofluoresence of Htt fragments and active caspase 3 was performed as described above. Htt expression was detected using an antibody to residues 1-82 of Htt (Millipore), and active caspase 3 was detected using anti-active caspase 3 antibody (Promega). Microscopy was performed using a Zeiss Axiovert 200M microscope equipped with a ×40 objective. In total, more than 100 transfected cells were counted per condition and the number of cells expressing both htt and active caspase 3 were presented as a percentage of the total number of htt positive cells.
RESULTS
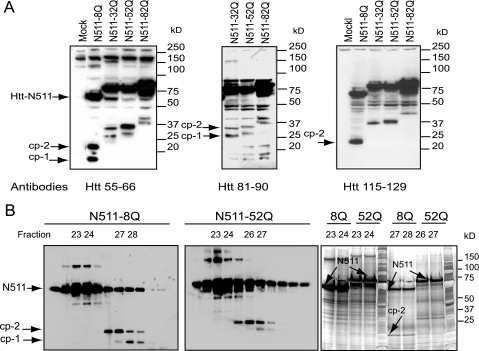
HEK293 Cells, Ectopically Expressing Truncated Htt Produce the cp-1 and cp-2 Fragments—To produce sufficient quantities of the cp-1 and cp-2 fragments for mass spectrometry, portions of normal and expanded Htt (N511-8Q and N-511-52Q) were expressed in HEK293 cells. Because Htt-N511 does not contain caspase cleavage sites, it may be cleaved into short N-terminal fragments via a caspase-independent pathway (30). We found that cp-1/cp-2-like N-terminal fragments are produced from various Htt-N511 constructs and migrate on a gel in a poly(Q)-dependent manner (Fig. 2A). Both fragments were detected with an antibody recognizing an N-terminal FLAG tag (data not shown), and with two Htt peptide antibodies generated against residues 55-66 and 81-90. However, only cp-2 was detected with an antibody recognizing the epitope between residues 115 and 129. Based on epitope mapping, the cp-1 cleavage site is located between residues 81 and 129 of Htt. Thus, cp-1 and cp-2 fragments, generated from Htt-N511 cleavage in HEK293 cells appear comparable with those resulting from Htt-N1212 and full-length Htt cleavage in HEK293 and stable PC12 cells (30).
FIGURE 2.
Expression and purification of Htt fragments in HEK293 cells. A, epitope mapping of the cp-1/cp-2 fragments. Western blot of total cell extracts (30 μg/lane) from HEK293 cells transfected with either N511-Htt constructs with different poly(Q) lengths, or cells transfected with empty vector (Mock). Htt-N511 and cp-1/cp-2 fragments (arrows) were detected with an antibody to residues 55-66 of Htt (left panel), with antibody to residues 81-90 of Htt (middle panel), or with antibody to residues 115-129 of Htt (right panel). HEK293 cells transfected with expanded Htt constructs produce fragments similar to the cp-1/cp-2 fragments observed previously in PC12 cells (30). B, purification of the cp-1/cp-2 fragments for mass spectrometry. Western blotting analysis of 1/10 aliquots of fractions from size chromatography of FLAG immunoprecipitates of either Htt-N511-8Q (left panel), or Htt-N511-52Q (middle panel) are expressed in HEK293 cells. Htt proteins, marked by arrows, were detected with antibody to FLAG. Indicated fractions containing either the cp-1/cp-2 fragments or in Htt-N511, were separated on SDS-PAGE, and stained with silver protein stain (right panel). Htt bands (indicated with arrows) corresponding to cp-1/cp-2 fragments, and bands containing only Htt-N511 were excised from the gel for mass spectrometry.
Purification of the cp-1 and cp-2 fragments for mass spectrometry was carried out in two steps. In the first step, FLAG-tagged Htt proteins were isolated by immunoprecipitation from HEK293 cell lysates using FLAG-agarose and a large quantity of starting material (15-20 mg of total protein per immunoprecipitation). Following elution with a FLAG peptide, the proteins were further purified using size exclusion chromatography. This step was performed under denaturing conditions to disrupt Htt protein complexes and enrich for the cp-1/cp-2 fragments (see “Experimental Procedures”). Analysis of sizing column fractions by Western blotting with an antibody to the FLAG demonstrated that the cp-1/cp-2 fragments were enriched in fractions 26 through 28, whereas Htt-N511 was predominantly found in fractions 23 and 24 (Fig. 2B, left panels). These fractions were then separated on the gel and proteins were visualized with silver stain (Fig. 2B, right panel). Htt bands corresponding to the cp-1 and cp-2 fragments, as well as bands containing only Htt-N511, were processed for mass spectrometry.
Detection of the Cp-2 Cleavage Site by Mass Spectrometry—Following excision from the gel (Fig. 2B, right panel), Htt-8Q and Htt-52Q protein bands, described above, were subjected to in-gel digestion with chymotrypsin. The peptides were eluted from the gel and processed by LC-MS/MS (see “Experimental Procedures”) for peptide identification. Chymotrypsin cleaves proteins after the following residues: phenylalanine, tyrosine, tryptophan, methionine, leucine, and histidine. As result of our experiments, we identified a number of Htt peptides produced by chymotrypsin cleavage of the N511 band, and 2 peptides resulting from digestion of the cp-2 band (Fig. 3A, data shown for Htt-8Q). One of these peptides, MDSNLPR, ending at arginine (Arg167 of Htt), cannot be generated by chymotrypsin and may represent the C terminus of the cp-2 fragment generated by an endogenous cleavage event. Furthermore, this peptide was only detected in the digest of the cp-2 band (Fig. 3B). We failed to detect MDSNLPR peptide in digests of the N511 band from fractions 26 to 28, or in fractions 23-24 lacking any cp-1/cp-2 fragments, even though the N511 band contains ∼10 times more Htt protein than the band corresponding to the cp-2 fragment. We found, however, four longer peptides produced by chymotrypsin digest of the N511 band within the same region of Htt (Table 1). These data strongly suggest that the cp-2 cleavage site is located at position Arg167 of Htt.
FIGURE 3.
Identification of the cp-2 cleavage site by mass spectrometry. A, peptide coverage obtained from chymotrypsin digest of huntingtin bands (N511 and cp-2). B, extracted ion chromatogram of MDSNPLR ion (m/z 416.7) and full spectrum at 17.7 min for fragment cp-2 (showing the presence of the peptide). C, confirmation of sequence identification of peptide MDSNLPR (comparison of fragmentation spectra obtained from the cp-2 fragment, produced from Htt-N511-8Q (top), and from synthesized peptide MDSNLPR (bottom)). The tables below show the matched fragments in bold. The two fragmentation spectra are very similar, which confirms that the peptide detected in the cp-2 fragment digest is indeed MDSNLPR.
TABLE 1.
Huntingtin peptides identified around sequence MDSNLPR
| N511 band | Peptide score | Cp-2 fragment band | Peptide score |
|---|---|---|---|
| MDSNLPRL | 26 | MDSNLPR | 41 |
| MDSNLPRLQ | 59 | ||
| MDSNLPRLQL | 37 | ||
| MDSNLPRLQLELY | 30 |
Because the Mascot score (41) for the MDSNLPR peptide was below the 95% confidence threshold, we obtained additional confirmation of the peptide sequence: the MDSNLPR peptide fragmentation spectrum from the digest of cp-2 was compared with that of a synthetic peptide of the same sequence (Fig. 3C). The two fragmentation spectra patterns and masses matched closely, confirming that the peptide detected in the cp-2 fragment digest is indeed MDSNLPR. The data shown were generated for the cp-2 fragment derived from Htt-N511-8Q. A peptide of the same mass and with the same retention time was also detected in the digest of cp-2 derived from Htt-511-52Q (data not shown). However, we failed to detect any Htt peptides indicative of potential cp-1 cleavage from trypsin or chymotrypsin digests of either Htt-8Q or Htt-52Q bands (data not shown).
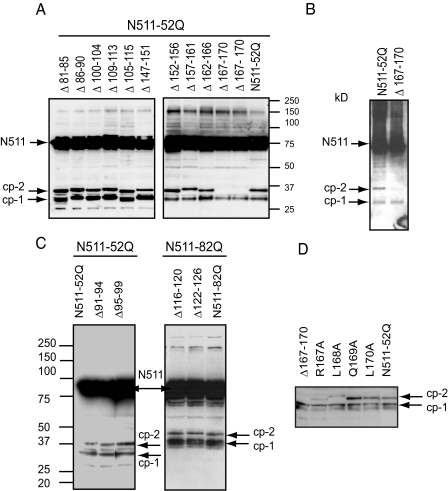
Mapping of the Cp-2 Cleavage Site by Deletion Analysis—A complementary approach was used to confirm the position of the cp-2 cleavage, detected by mass spectrometry, and to further define the cp-1 site. Based on epitope mapping with Htt-specific antibodies, the region for the potential cp-1 cleavage site should be located between residues 81 and 129 of Htt (Figs. 1 and 2A), whereas the cp-2 cleavage site was mapped by mass spectrometry to position 167 of Htt. Therefore, the entire region of Htt from residue 81 to 170 was subjected to systematic deletion analysis, with deletions of 4 to 5 amino acids per deletion mutant. Htt-N511-52Q deletion constructs were expressed in HEK293 cells and analyzed by Western blotting with an antibody to the FLAG epitope (Fig. 4, A and C, and data not shown). Deletion of residues 167-170 (including Arg167) prevented the generation of cp-2 from Htt-N511-52Q expressed in HEK293 (Fig. 4A) or mouse hippocampal HT22 (Fig. 4B) cells, confirming the mass spectrometry results. (It should be noted, however, that in some experiments with the Δ167-170 construct we observed very low levels of an alternative fragment migrating more slowly than cp-2 (Fig. 5A).)
FIGURE 4.
Mapping of the cp-2 cleavage site by deletion analysis. A, C, D, Htt constructs with indicated deletions (A and C) or point mutations (D), or unaltered Htt with expanded poly(Q) (N511-52Q or N511-82Q) were expressed in HEK293 cells, and Htt-FLAG fusion proteins (indicated by arrows) were detected by Western blotting with an antibody to FLAG. B, Htt constructs with either Δ167-170 deletion or unaltered Htt with expanded poly(Q) (N511-52Q) were expressed in neuronal HT22 cells, FLAG-Htt fusion proteins (indicated by arrows) were immunoprecipitated with FLAG-agarose (∼2 mg of protein per immunoprecipitation), and detected with an antibody to exon 1 of Htt. Deletion of residues 167-170 greatly reduces generation of the cp-2 fragment from Htt-N511-52Q construct expressed in HEK293 or HT22 cells.
FIGURE 5.
Detection of the cp-2 fragment by a specific antibody to the neo-epitope Arg167 of Htt in HEK293 cells. A, Western blot of total cell extracts (30 μg/lane) from HEK293 cells transfected with the indicated Htt constructs, or an empty vector (Mock). Truncated FLAG-Htt fusion proteins and cp-1/cp-2 fragments (marked by arrows) were detected with antibodies to FLAG (left panel). The Htt-N167-82Q fragment migrates on a gel more slowly than the cp-2 fragment derived from Htt-N511-52Q, consistent with the length of poly(Q). Antibody to Arg167 neo-epitope detects the Htt-N167 fragment and the cp-2 fragment, has low cross-reactivity with the Htt-N171 fragment, and fails to detect Htt-N511 fragment or any fragments of Htt-Δ167-170 (right panel, data shown for NE167 antibody). B, Western blot of total cell extracts from HEK293 cells transfected with the indicated Htt constructs or an empty vector (Mock). Htt proteins (indicated by arrows) were detected with an antibody to exon 1 of Htt (left panel, 20 μg per lane). The cp-2 fragments produced from Htt-N511 or Htt-N586 were specifically detected by an antibody to the Arg167 neo-epitope (right panel, 40 μg/lane). Blots, shown in A and B, were re-probed with an antibody to actin for loading control. C, Western blot of formic acid-soluble aggregate fractions from HEK293 cells transfected with indicated constructs or an empty vector (Mock). Htt fragments were detected with antibodies to exon 1 (left panel) or to the Arg167 neo-epitope (right panel) of Htt.
The decreased levels of the cp-2 found in SDS soluble fractions (Fig. 4A) were not due to a decrease in solubility of cp-2 formed from the Δ167-170 mutant (see below). The analysis of the SDS-insoluble material, treated with formic acid known to disrupt the aggregates, showed the lack of immunoreactivity for the Δ167-170 construct (Fig. 8B). This supports the conclusion that reduced levels of cp-2 shown on Fig. 4A are likely to reflect reduced cleavage of Htt.
FIGURE 8.
Alteration of cp-2 cleavage site changes biochemical properties of Htt protein. A, the procedure used to fractionate and dissociate Htt proteins (see “Experimental Procedures”). B, Western blots of subcellular fractions from HEK293 cells transfected with the indicated constructs: 1, native PAGE of soluble cytoplasmic fractions; 2, SDS-PAGE of soluble cytoplasmic fractions; 3, SDS-PAGE of the pellet, with SDS-insoluble material detected on top of the gel; 4, SDS-PAGE of formic acid-soluble aggregate fractions. Htt fragments were detected with antibodies to exon 1. *, minimal immunoreactivity is observed for Htt-N511-52Q-Δ167-170 following formic acid treatment. **, less SDS-insoluble material is detected for the Δ167-170 mutant than for unaltered N511-52Q. ***, new high molecular weight soluble complexes are detected for Htt-N511-52Q-Δ167-170 in native conditions.
In an attempt to confirm Arg167 as the cp-2 cleavage site, we have also introduced point mutations of the four residues within the 167-170 region (Fig. 4D). None of the single mutations on its own completely eliminated the production of the cp-2. However, Arg167-Ala substitution caused a substantial decrease in the levels of cp-2. Leu168-Ala substitution also decreased cleavage and led to generation of an alternative fragment (migrating slightly slower than the cp-2). This is consistent with the observation that many proteases recognize in their substrates a three-dimensional epitope composed of several residues, and the deletion of the whole cleavage region may be required to completely block the cleavage at certain sites. Surprisingly, none of the deletions across the region seem to significantly affect formation of the cp-1 fragment (Fig. 4, A and C, and data not shown).
The Cp-2 Fragment Generated in HEK293 and Neuronal HT22 Cells Is Detected by a Specific Antibody to the Neo-epitope Arg167 of Htt—To further verify the identity of the cp-2 fragment produced in our cell models, we generated two antibodies aimed for a specific recognition of the neo-epitope generated following Htt cleavage at position Arg167 (see “Experimental Procedures”). Both antibodies were highly reactive to the N167 fragment of Htt, had low cross-reactivity with the N171 fragment, and failed to detect the N511 fragment (Fig. 5A, right panel, data shown for NE167 antibody). In addition, both antibodies specifically recognized the cp-2 fragment derived from Htt-N511-52 in HEK293 cells, and, as expected, did not detect any fragments of Htt Δ167-170 mutant.
Next we tested whether constructs expressing a physiologically relevant caspase 6 fragment of Htt can also produce cp-1 and cp-2 (Fig. 5B). We found that expression of Htt-N586 and Htt-N511 with different poly(Q) lengths (normal and expanded) resulted in formation of cp-1 and cp-2 fragments. These fragments were present in the soluble cytoplasmic fraction, as shown by detection with an antibody to exon 1 of Htt (Fig. 5B, left panel). The cp-2 fragments were also specifically detected with an antibody to the Arg167 epitope (Fig. 5B, right panel). The levels of N586/N511 and cp-1/cp-2 htt fragments found in the SDS-soluble fraction decreased with an increase in poly(Q) tracts, probably due to an increase in aggregation of htt proteins with the longer poly(Q) expansions. To test this hypothesis we transfected HEK293 cells with normal and expanded N586 constructs and isolated insoluble material 48 h after transfection. The insoluble fractions were then treated with formic acid to disrupt SDS-insoluble Htt aggregates. Analysis of this material by Western blotting with an antibody to Htt exon 1 demonstrated that cp-1/cp-2-like fragments, derived from expanded poly(Q) Htt, were enriched in the aggregate fraction (Fig. 5C, left panel). Furthermore, an increase in accumulation of N586 and cp-1/cp-2 fragments was observed with longer poly(Q) repeats. However, analysis of this material with an Arg167 neo-epitope antibody showed decreased levels of cp-2 with an increased poly(Q) length. These data suggest that the cp-2 fragment ending at Arg167 may represent one component of Htt-586-derived aggregates, with other expanded Htt fragments of similar length making up for the observed increase. Another possibility is that incorporation of cp-2 into SDS-insoluble aggregates masks the Arg167 epitope, resulting in poor detection by the neo-epitope antibody.
Next, subcellular localization of the cp-2 (N167) fragments was investigated by immunofluorescent microscopy (Fig. 6). HEK293 cells transfected with normal and expanded Htt-N511 and Htt-N568 constructs were co-stained with Htt peptide antibody 115-129 (green) and the antibody to Arg167 neo-epitope (NE167, red). Diffuse cytoplasmic staining representing soluble Htt was observed with both antibodies in cells expressing normal poly(Q) length Htt (Fig. 6A, two top panels). In contrast, cp-2 generated in HEK293 cells expressing expanded Htt formed mostly cytoplasmic inclusions. The presence of cp-2 in these cells is highlighted by the yellow staining in the merged images (Fig. 6A, right panels).
FIGURE 6.
Immunofluorescent detection of the cp-2 fragment by a specific antibody to the neo-epitope Arg167 of Htt. HEK293 (A) or HT22 (B) cells were transfected with indicated Htt constructs. Epitope between residues 115-129 of Htt is shown in green (FITC); Arg167 neo-epitope is shown in red (Cy 3); the nuclear staining (4′,6-diamidino-2-phenylindole) is shown in blue. Yellow staining in merged images (marked by arrows) demonstrates the presence of cp-2.
Detection of cp-1 and cp-2 fragments in neuronal HT22 cells was difficult due to a low transfection efficiency (∼10% compared with ∼90% in HEK293 cells), and possibly a lower extent of Htt cleavage (Fig. 4B). We, therefore, failed to detect cp-2 in HT22 cell lysate by Western blotting with Htt neoepitope antibody NE167. However, the same antibody used in immunofluorescence recognized cp-2 in the form of cytoplasmic aggregates in a small percentage of transfected HT22 cells expressing expanded Htt (Fig. 6B, red and yellow staining). Cp-2 fragment derived from normal length poly(Q) Htt was not detectable in HT22 cells by the same method. As predicted, cp-2 was also not detected in cells transfected with a Htt internal deletion construct lacking amino acids 167-170 (Htt-Δ167-170, data not shown).
Htt N-terminal Fragments of Specific Size Have Different Aggregation and Toxic Properties—To investigate a possible effect of the cp-2 fragment on aggregation and toxicity we used two mouse neuronal cell lines: N2a neuroblastoma and hippocampal HT22 cells. The aggregate formation was evaluated in both cell lines, expressing Htt-N511-8Q, Htt-N511-52Q, or Htt-Δ167-170 with altered cp-2 sites. Transfected cells were analyzed 48 h following transfection by immunofluorescent microscopy with an antibody to the FLAG or to the 1-82 epitope of Htt. Aggregates of Htt-N511-52Q were observed in ∼5% of transfected N2a cells, and in about 30% of transfected HT22 cells. Interestingly, deletion of the cp-2 cleavage site (Htt-N511-52Q-Δ167-170) led to a robust increase in aggregation in both cell lines (Fig. 7B). However, the aggregates formed from the Δ167-170 construct appeared morphologically different from N511-52Q aggregates (Fig. 7A, data shown for HT22 cells).
FIGURE 7.
Alteration of cp-2 cleavage site results in an increase in aggregation of Htt in neuronal cells. A, aggregate formation in HT22 cells expressing either Htt-N511-52Q or Htt-N511-52Q-Δ167-170. Cells were labeled with an antibody to FLAG, followed by anti-mouse FITC-conjugated antibody. B, aggregate formation in N2a and HT22 cells expressing Htt-N511-8Q, Htt-N511-52Q, or Htt-N511-52Q-Δ167-170. Cells were labeled with Htt-(1-82) antibody, followed by anti-mouse FITC-conjugated antibody and the aggregates were counted in 24-well plates in ∼200-400 cells for each construct (n = 2, *, p = 0.02 N511-52Q versus N511-52Q-Δ167-170 for N2a cells; **, n = 3, p = 0.07 N511-52Q versus N511-52Q-Δ167-170 for HT22 cells). One representative experiment of two is shown.
To assess biochemical properties of Htt proteins formed from different constructs, the lysates of transfected HEK293 cells were fractionated as shown on Fig. 8A (see “Experimental Procedures”), and the fractions were analyzed by Western blotting with an antibody to exon 1 of Htt (Fig. 8B). Similar to N586 constructs (Fig. 5C), cp-1/cp-2-like fragments, derived from the expanded poly(Q) Htt-N511, were enriched in the aggregate fraction solubilized with formic acid, with an increase in accumulated fragments with longer poly(Q) repeat lengths (Fig. 8B, panel 4). However, as described above, the lack of immunoreactivity was observed for the N511-52Q-Δ167-170 construct following formic acid treatment. This construct also produced less SDS-insoluble material detected on the top of the gel, when M-PER-insoluble pellet fractions were analyzed on SDS-PAGE (Fig. 8B, panel 3). This indicates that the aggregates formed in cells expressing poly(Q)-expanded Htt-Δ167-170 are more soluble in SDS, than those formed from non-altered Htt. In addition, new high molecular weight soluble complexes of Htt-N511-52Q-Δ167-170 were detected on native PAGE (Fig. 8B, panel 1). These data suggest that the expanded Htt with the deleted cp-2 site may acquire altered conformation with different biochemical properties, and may form unusual protein interactions.
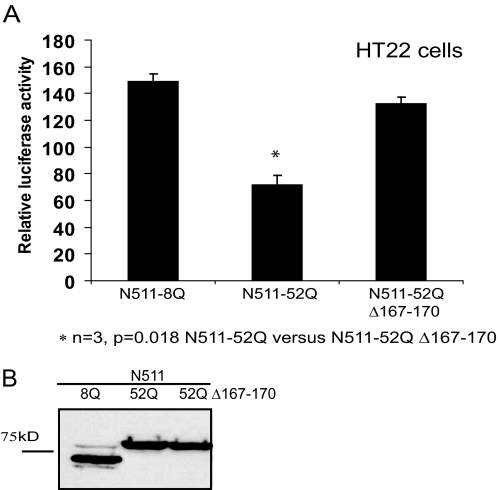
Next, we examined how alteration of the cp-2 site affects HT22 cell viability (Fig. 9). Because of the low efficiency of transfection of HT22 cells, we used the viability assay based on a co-transfection of luciferase and Htt constructs. This method has been previously established and is widely used for evaluation of cell death (34-37). Because luciferase is only expressed in transfected cells and because it is rapidly degraded following cell death, its levels can be used as a measure of transfected cell viability. As expected, cells expressing expanded Htt-N511-52Q were less viable than cells transfected with normal Htt-N511-8Q. Deletion of the cp-2 site almost completely ameliorated expanded Htt-N511-induced cell toxicity. These data indicate that the cp-2 fragment derived from expanded Htt-N511 is toxic to cultured HT22 cells.
FIGURE 9.
Alteration of cp-2 cleavage site results in a decrease in toxicity of Htt protein in neuronal cells. A, cell viability (measured by luciferase assay, see “Experimental Procedures”) of HT22 cells, expressing the indicated Htt constructs co-transfected with the luciferase construct (n = 3, **, p = 0.018 N511-52Q versus N511-52Q-Δ167-170). One representative experiment of three for each set of constructs is shown. B, the expression levels of the indicated constructs were verified by Western blotting with an antibody to FLAG.
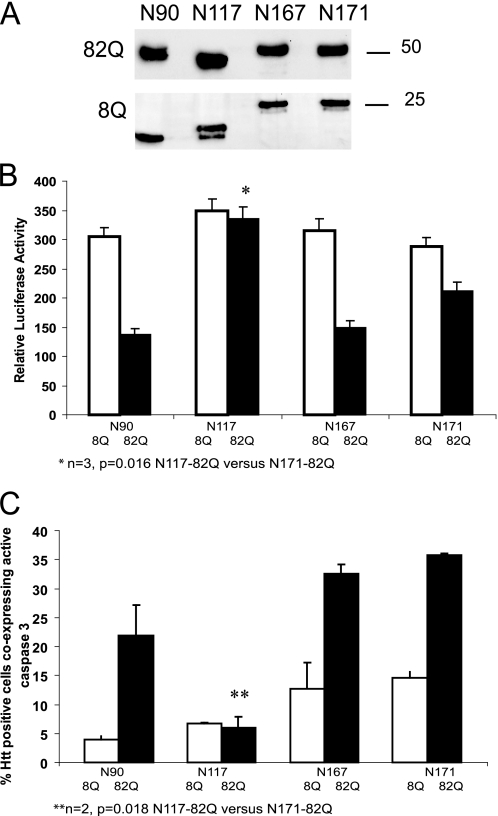
To extend these findings, we have compared toxic and aggregation properties of individually expressed Htt fragments of different lengths including N167 (Fig. 10). As shown by the luciferase-based viability assay, an expanded Htt construct analogous to cp-2 (N167-82Q) was nearly as toxic as a shorter Htt-N90-82Q Htt fragment (corresponding to Htt exon 1), when expressed in HT22 cells (Fig. 10B). In contrast, the N117-82Q-Htt fragment (also known as shortstop), previously observed to be not pathogenic in a transgenic mouse model (32), had a only mild effect on HT22 cell viability (Fig. 10B). Similar results were obtained using caspase 3 activation as a measure of cytotoxicity: HT22 cells expressing Htt fragments were co-stained with an antibody to the active caspase 3 and an antibody to Htt, and caspase 3 positive cells expressing Htt were counted (Fig. 10C).
FIGURE 10.
Htt N-terminal fragments of specific size have different toxic properties. A, Western blot of total cell extracts from HT22 cells transfected with the indicated Htt constructs. Htt proteins were detected with an antibody to exon 1 of Htt. B, cell viability, measured by luciferase assay (see “Experimental Procedures”) of HT22 cells, expressing the indicated Htt construct co-transfected with the luciferase construct (n = 3, *, p = 0.016 N117-82Q versus N171-82Q, p < 0.05 for all normal repeat versus expanded repeat constructs). One representative experiment of three for each set of constructs is shown. C, cytotoxicity, measured by caspase 3 activation (see “Experimental Procedures”) of HT22 cells, expressing the indicated Htt construct. Results are presented as a percentage of Htt positive cells co-expressing active caspase 3 (n = 2, over 100 cells were counted for each condition, **, p < 0.05 N117-82Q versus N171-82Q). One representative experiment of two is shown.
In relation to aggregation, we found that HT22 cells expressing N117-82Q formed ∼30-40% more aggregates than cells expressing either N167-82Q or N171-82Q (data not shown). Thus, our data support the idea that Htt fragments of specific size may contribute differently to Htt-induced toxicity.
DISCUSSION
The nature of the toxic Htt fragments relevant to HD pathogenesis remains unknown. Multiple reports support the role of Htt proteolysis in the pathogenic cascade (3-18); however, it is not clear where expanded Htt is cleaved within its N terminus (N-terminal to the caspase 6 site) to contribute to its toxicity. We applied multiple approaches to map the exact sites of proteolysis producing short N-terminal fragments of Htt. Based on mass spectrometry of Htt proteins and deletion analysis of Htt constructs, we found the cp-2 site at position Arg167. Although we failed to determine the exact position of the cp-1 site, epitope mapping with Htt-specific antibodies allowed us to locate it to the region between residues 81 and 129 of Htt. In HEK293 and neuronal HT22 cells, the cp-2 fragment can be detected by a specific antibody to the Arg167 neo-epitope. Alteration of this site leads to a reduction in toxicity and an increase in aggregation in neuronal cells. Collectively, these results demonstrate that Htt can be endogenously cleaved at position Arg167, and that this proteolytic step may be relevant to Htt-induced pathogenesis.
The analysis using mass spectrometry identified a fragment corresponding to the cp-2 site with a C terminus at residue Arg167 of Htt. The peptide MDSNLPR was detected following chymotrypsin digest of the cp-2 fragment band but not in the band corresponding to N511, despite the fact that the N511 band contains 10-fold more protein (Table 1). The identity of this peptide was further confirmed by comparison of its fragmentation spectrum to that of a synthetic peptide of the same sequence (Fig. 3C). This evidence strongly suggests that Htt undergoes endoproteolysis at position Arg167 in HEK293 cells.
However, the mass spectrometry approach employed here has some limitations in detecting potential cleavage sites in Htt. For example, this approach has not yet been useful for detection of the cp-1 site. The probability of detecting specific peptides is largely affected by peptide size and biochemical characteristics, both of which can influence ionization efficiency and subsequent detection in a mass spectrometer. In fact, we have achieved limited coverage of the Htt sequence within the region of potential N-terminal cleavage sites (Fig. 3A). The possible peptides resulting from trypsin or chymotrypsin cleavage between residues 81 and 105 of Htt are too short to be identified by mass spectrometry. This fact may explain our inability to map the precise cp-1 site using this method. Future studies aimed at providing a more complete coverage of the Htt sequence in the region of interest will involve using different enzymes for in-gel digestion of Htt fragments, such as Asp-N endopeptidase.
Mutagenesis has been widely used for mapping of cleavage sites. However, this method did not allow us to locate the cp-1 site. One possible explanation is that many proteases (e.g. calpains) recognize in their substrates a three-dimensional epitope composed of several amino acids that may be positioned apart from each other (38, 39). Thus deletions of short linear stretches of 4-5 residues may not be sufficient to prevent proteolysis by certain proteases. Another consideration is that mutations and deletions of the Htt sequence may facilitate cleavage at alternative sites that are normally not favorable within the context of wild type Htt. This also seems to be the case with the internal deletion of the cp-2 site (Δ167-170): although this alteration prevents production of the cp-2 fragment, a slower migrating alternative fragment not recognized by the cp-2 neo-epitope antibody was detected in some experiments. It is also possible that endoproteolytic cleavage occurs at other amino acids near Arg167, and that the apparent cp-2 band contains several fragments differing by just a few amino acids.
The relevance of caspase 6-mediated cleavage of Htt (at position 586) to Htt pathology has been demonstrated in vivo (25). We find that the cp-1 and cp-2 fragments may be generated from either N511 or N586 fragments of the expanded Htt (Fig. 5B). Further in vivo experiments will be designed to establish a possible relationship between caspase 6-mediated cleavage of Htt, and its N-terminal proteolysis that may produce potentially toxic cp-1 and cp-2 fragments.
Transgenic mouse models of HD generally indicate that shorter expanded Htt fragments cause more severe phenotypes (15-18). However, a short N-terminal fragment of Htt, N117, expressed by shortstop mice, did not produce an HD phenotype (32). This suggests that the specific size of the fragments, rather than overall length, may determine their toxic potential in HD. We used a cell viability assay to directly compare the toxicity of expanded Htt fragments corresponding in size to those previously used in truncated Htt mouse models. Our results are consistent with mouse phenotypes, and demonstrate that the exon 1 Htt (R6/2 mice) (15) and N171 fragments (N171-82Q mice) (16) are toxic when expressed in neuronal HT22 cells. In contrast, the expanded shortstop N117 fragment was nontoxic to HT22 cells. The cp-2 (N167-82Q) fragment of Htt appears to be comparable in toxicity to exon 1 and N171 Htt fragments (Fig. 10, B and C). Furthermore, deletion of the cp-2 site from Htt-N511-52Q ameliorates the toxicity of this construct (Fig. 9A). Thus, exact mapping of the cp-2 site allowed us to evaluate the effect of alteration of this site on Htt-induced toxicity, suggesting that this cleavage event could in part mediate toxicity in neuronal cells.
Although deletion of the cp-2 cleavage site from Htt-N511 prevented generation of the cp-2 fragment, it also resulted in an increase in Htt-N511 aggregation (Fig. 7, A and B). One possible explanation for these findings is that internal deletion of residues 167-170 alters the conformation of the Htt-N511 protein, making other potential proteolytic sites more accessible. Htt fragments generated from such alternative cleavage events may have different biochemical properties (Fig. 8) resulting in an increase in large Htt aggregates observed by immunofluorescence (Fig. 7, A and B). An increase in the formation of large aggregates might be expected to decrease toxicity (40, 41), and could explain the increase in cell viability in cells expressing Δ167-170 (Fig. 9A). A moderate increase in aggregation was also observed in HT22 cells expressing a nontoxic shortstop N117 fragment (compared with cells expressing either N167 or N171 Htt). This is consistent with the large number of inclusions found in the brain of shortstop mice, and supports the idea that inclusions formed in HD neurons might be neuroprotective.
Short N-terminal fragments have been consistently observed in a variety of cell lines: NG108-15 (5), mouse striatal cells (42), PC12 (30), HEK293 and HT22 cells (this report), as well as mouse models and human HD postmortem material. These data are all consistent with a role for proteolytic cleavage in generation of toxic fragments of expanded Htt. In this study we have defined one such fragment, cp-2, produced by Htt cleavage at position Arg167. Analysis of cp-2 cellular distribution by immunofluorescence demonstrated that in both HEK293 and neuronal HT22 cells this particular fragment comprises only a portion of the aggregates (Fig. 6). Similar data were obtained using biochemical analysis: formic acid-soluble aggregate fractions from HEK293 cells showed the presence of multiple cp-1 and cp-2-like fragments of similar size, detected with an antibody to exon 1 of Htt. The N167 fragment appears to be one component of this fraction (Fig. 5C). Taken together, these observations indicate that multiple cleavage events may yield different fragments of expanded Htt, accumulating in nuclear and cytoplasmic inclusions. Another possibility is the loss of Arg167 epitope accessibility, and therefore poor detection of the expanded N167 fragment due to its increased aggregation.
Thus, Htt N-terminal proteolysis appears to be a heterogeneous process, generating fragments of several different lengths. Another example of heterogeneous cleavage in neurons is the processing of amyloid precursor protein by γ-secretase, which produces several forms of amyloid β peptide (including Aβ40 and Aβ42). The importance of these peptides, especially Aβ42, for Alzheimer disease pathogenesis has prompted therapeutic approaches for Alzheimer disease to focus on specific elimination of Aβ42 either by inhibiting its production, or inducing its clearance (43). Htt cleavage variability may result from specific cell types and environments and possibly is mediated by differential post-translation modifications of Htt. Although challenging, it is essential to understand if such variability occurs in vivo, and to characterize the underlying proteolytic pathways, which may produce specific therapeutic targets for HD.
Acknowledgments
We thank David Borchelt (University of Florida, Gainesville, FL) for providing expression constructs and valuable discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants 16375 from the NINDS and NS038144-08. This work was also supported by the Huntington Disease Society of America Coalition for the Cure, High Q Foundation, Canadian Institutes of Health Research, and the HSC (Huntington Society of Canada).
Footnotes
The abbreviations used are: HD, Huntington disease; Htt, huntingtin; poly(Q), polyglutamine; HEK, human embryonic kidney; N2a, Neuro-2a; NG108-15, neuroblastoma-glioma; PC12, pheochromocytoma; MS, mass spectrometry; LC, liquid chromatography; FITC, fluorescein isothiocyanate; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1.The Huntington's Disease Collaborative Research Group (1993) Cell 72 971-983 [DOI] [PubMed] [Google Scholar]
- 2.Ross, C. A., Margolis, R. L., Rosenblatt, A., Ranen, N. G., Becher, M. W., and Aylward, E. (1997) Medicine (Baltimore) 76 305-338 [DOI] [PubMed] [Google Scholar]
- 3.Becher, M. W., Kotzuk, J. A., Sharp, A. H., Davies, S. W., Bates, G. P., Price, D. L., and Ross, C. A. (1998) Neurobiol. Dis. 4 387-397 [DOI] [PubMed] [Google Scholar]
- 4.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P., and Aronin, N. (1997) Science 277 1990-1993 [DOI] [PubMed] [Google Scholar]
- 5.Lunkes, A., Lindenberg, K. S., Ben-Haiem, L., Weber, C., Devys, D., Land-wehrmeyer, G. B., Mandel, J. L., and Trottier, Y. (2002) Mol. Cell 10 259-269 [DOI] [PubMed] [Google Scholar]
- 6.Mende-Mueller, L. M., Toneff, T., Hwang, S. R., Chesselet, M. F., and Hook, V. Y. (2001) J. Neurosci. 21 1830-1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schilling, G., Klevytska, A., Tebbenkamp, A. T., Juenemann, K., Cooper, J., Gonzales, V., Slunt, H., Poirer, M., Ross, C. A., and Borchelt, D. R. (2007) J. Neuropathol. Exp. Neurol. 66 313-320 [DOI] [PubMed] [Google Scholar]
- 8.Sun, B., Fan, W., Balciunas, A., Cooper, J. K., Bitan, G., Steavenson, S., Denis, P. E., Young, Y., Adler, B., Daugherty, L., Manoukian, R., Elliott, G., Shen, W., Talvenheimo, J., Teplow, D. B., Haniu, M., Haldankar, R., Wypych, J., Ross, C. A., Citron, M., and Richards, W. G. (2002) Neurobiol. Dis. 11 111-122 [DOI] [PubMed] [Google Scholar]
- 9.Kim, M., Lee, H. S., LaForet, G., McIntyre, C., Martin, E. J., Chang, P., Kim, T. W., Williams, M., Reddy, P. H., Tagle, D., Boyce, F. M., Won, L., Heller, A., Aronin, N., and DiFiglia, M. (1999) J. Neurosci. 19 964-973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunkes, A., and Mandel, J. L. (1998) Hum. Mol. Genet. 7 1355-1361 [DOI] [PubMed] [Google Scholar]
- 11.Cooper, J. K., Schilling, G., Peters, M. F., Herring, W. J., Sharp, A. H., Kaminsky, Z., Masone, J., Khan, F. A., Delanoy, M., Borchelt, D. R., Dawson, V. L., Dawson, T. M., and Ross, C. A. (1998) Hum. Mol. Genet. 7 783-790 [DOI] [PubMed] [Google Scholar]
- 12.Igarashi, S., Morita, H., Bennett, K. M., Tanaka, Y., Engelender, S., Peters, M. F., Cooper, J. K., Wood, J. D., Sawa, A., and Ross, C. A. (2003) Neuroreport 14 565-568 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, H., Cao, F., Wang, Z., Yu, Z. X., Nguyen, H. P., Evans, J., Li, S. H., and Li, X. J. (2003) J. Cell Biol. 163 109-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gafni, J., and Ellerby, L. M. (2002) J. Neurosci. 22 4842-4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangiarini, L., Sathasivam, K., Seller, M., Cozens, B., Harper, A., Hetherington, C., Lawton, M., Trottier, Y., Lehrach, H., Davies, S. W., and Bates, G. P. (1996) Cell 87 493-506 [DOI] [PubMed] [Google Scholar]
- 16.Schilling, G., Becher, M. W., Sharp, A. H., Jinnah, H. A., Duan, K., Kotzuk, J. A., Slunt, H. H., Ratovitski, T., Cooper, J. K., Jenkins, N. A., Copeland, N. G., Price, D. L., Ross, C. A., and Borchelt, D. R. (1999) Hum. Mol. Genet. 8 397-407 [DOI] [PubMed] [Google Scholar]
- 17.Rubinsztein, D. C. (2002) Trends Genet. 18 202-209 [DOI] [PubMed] [Google Scholar]
- 18.Tanaka, Y., Igarashi, S., Nakamura, M., Gafni, J., Torcassi, C., Schilling, G., Crippen, D., Wood, J. D., Sawa, A., Jenkins, N. A., Copeland, N. G., Borchelt, D. R., Ross, C. A., and Ellerby, L. M. (2006) Neurobiol. Dis. 21 381-391 [DOI] [PubMed] [Google Scholar]
- 19.Wellington, C. L., Ellerby, L. M., Hackam, A. S., Margolis, R. L., Trifiro, M. A., Singaraja, R., McCutcheon, K., Salvesen, G. S., Propp, S. S., Bromm, M., Rowland, K. J., Zhang, T., Rasper, D., Roy, S., Thornberry, N., Pinsky, L., Kakizuka, A., Ross, C. A., Nicholson, D. W., Bredesen, D. E., and Hayden, M. R. (1998) J. Biol. Chem. 273 9158-9167 [DOI] [PubMed] [Google Scholar]
- 20.Wellington, C. L., Singaraja, R., Ellerby, L., Savill, J., Roy, S., Leavitt, B., Cattaneo, E., Hackam, A., Sharp, A., Thornberry, N., Nicholson, D. W., Bredesen, D. E., and Hayden, M. R. (2000) J. Biol. Chem. 275 19831-19838 [DOI] [PubMed] [Google Scholar]
- 21.Kim, Y. J., Yi, Y., Sapp, E., Wang, Y., Cuiffo, B., Kegel, K. B., Qin, Z. H., Aronin, N., and DiFiglia, M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12784-12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wellington, C. L., Ellerby, L. M., Gutekunst, C. A., Rogers, D., Warby, S., Graham, R. K., Loubser, O., van Raamsdonk, J., Singaraja, R., Yang, Y. Z., Gafni, J., Bredesen, D., Hersch, S. M., Leavitt, B. R., Roy, S., Nicholson, D. W., and Hayden, M. R. (2002) J. Neurosci. 22 7862-7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermel, E., Gafni, J., Propp, S. S., Leavitt, B. R., Wellington, C. L., Young, J. E., Hackam, A. S., Logvinova, A. V., Peel, A. L., Chen, S. F., Hook, V., Singaraja, R., Krajewski, S., Goldsmith, P. C., Ellerby, H. M., Hayden, M. R., Bredesen, D. E., and Ellerby, L. M. (2004) Cell Death Differ. 11 424-438 [DOI] [PubMed] [Google Scholar]
- 24.Sawa, A., Nagata, E., Sutcliffe, S., Dulloor, P., Cascio, M. B., Ozeki, Y., Roy, S., Ross, C. A., and Snyder, S. H. (2005) Neurobiol. Dis. 20 267-274 [DOI] [PubMed] [Google Scholar]
- 25.Graham, R. K., Deng, Y., Slow, E. J., Haigh, B., Bissada, N., Lu, G., Pearson, J., Shehadeh, J., Bertram, L., Murphy, Z., Warby, S. C., Doty, C. N., Roy, S., Wellington, C. L., Leavitt, B. R., Raymond, L. A., Nicholson, D. W., and Hayden, M. R. (2006) Cell 125 1179-1191 [DOI] [PubMed] [Google Scholar]
- 26.Gafni, J., Hermel, E., Young, J. E., Wellington, C. L., Hayden, M. R., and Ellerby, L. M. (2004) J. Biol. Chem. 279 20211-20220 [DOI] [PubMed] [Google Scholar]
- 27.Kim, M., Roh, J. K., Yoon, B. W., Kang, L., Kim, Y. J., Aronin, N., and DiFiglia, M. (2003) Exp. Neurol. 183 109-115 [DOI] [PubMed] [Google Scholar]
- 28.Bizat, N., Hermel, J. M., Boyer, F., Jacquard, C., Creminon, C., Ouary, S., Escartin, C., Hantraye, P., Kajewski, S., and Brouillet, E. (2003) J. Neurosci. 23 5020-5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goffredo, D., Rigamonti, D., Tartari, M., De Micheli, A., Verderio, C., Matteoli, M., Zuccato, C., and Cattaneo, E. (2002) J. Biol. Chem. 277 39594-39598 [DOI] [PubMed] [Google Scholar]
- 30.Ratovitski, T., Nakamura, M., D'Ambola, J., Chighladze, E., Liang, Y., Wang, W., Graham, R., Hayden, M. R., Borchelt, D. R., Hirschhorn, R. R., and Ross, C. A. (2007) Cell Cycle 6 2970-2981 [DOI] [PubMed] [Google Scholar]
- 31.Warby, S. C., Doty, C. N., Graham, R. K., Carroll, J. B., Yang, Y. Z., Singaraja, R. R., Overall, C. M., and Hayden, M. R. (2008) Hum. Mol. Genet. 17 2390-2404 [DOI] [PubMed] [Google Scholar]
- 32.Slow, E. J., Graham, R. K., Osmand, A. P., Devon, R. S., Lu, G., Deng, Y., Pearson, J., Vaid, K., Bissada, N., Wetzel, R., Leavitt, B. R., and Hayden, M. R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 11402-11407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters, M. F., and Ross, C. A. (2001) J. Biol. Chem. 276 3188-3194 [DOI] [PubMed] [Google Scholar]
- 34.Coombe, D. R., Nakhoul, A. M., Stevenson, S. M., Peroni, S. E., and Sanderson, C. J. (1998) J. Immunol. Methods 215 145-150 [DOI] [PubMed] [Google Scholar]
- 35.Zha, J., Harada, H., Osipov, K., Jockel, J., Waksman, G., and Korsmeyer, S. J. (1997) J. Biol. Chem. 272 24101-24104 [DOI] [PubMed] [Google Scholar]
- 36.Jiang, H., Nucifora, F. C., Jr., Ross, C. A., and DeFranco, D. B. (2003) Hum. Mol. Genet. 12 1-12 [DOI] [PubMed] [Google Scholar]
- 37.Wen, Y., Hu, M. C., Makino, K., Spohn, B., Bartholomeusz, G., Yan, D. H., and Hung, M. C. (2000) Cancer Res. 60 6841-6845 [PubMed] [Google Scholar]
- 38.Cuerrier, D., Moldoveanu, T., and Davies, P. L. (2005) J. Biol. Chem. 280 40632-40641 [DOI] [PubMed] [Google Scholar]
- 39.Stabach, P. R., Cianci, C. D., Glantz, S. B., Zhang, Z., and Morrow, J. S. (1997) Biochemistry 36 57-65 [DOI] [PubMed] [Google Scholar]
- 40.Muchowski, P. J., Ning, K., D'Souza-Schorey, C., and Fields, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 727-732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross, C. A., and Poirier, M. A. (2005) Nat. Rev. Mol. Cell Biol. 6 891-898 [DOI] [PubMed] [Google Scholar]
- 42.Kim, Y. J., Sapp, E., Cuiffo, B. G., Sobin, L., Yoder, J., Kegel, K. B., Qin, Z. H., Detloff, P., Aronin, N., and DiFiglia, M. (2006) Neurobiol. Dis. 22 346-356 [DOI] [PubMed] [Google Scholar]
- 43.Findeis, M. A. (2007) Pharmacol. Ther. 116 266-286 [DOI] [PubMed] [Google Scholar]