Abstract
The secreted glycoprotein sclerostin has recently emerged as a key negative regulator of Wnt signaling in bone and has stimulated considerable interest as a potential target for therapeutics designed to treat conditions associated with low bone mass, such as osteoporosis. We have determined the structure of sclerostin, which resulted in the identification of a previously unknown binding site for heparin, suggestive of a functional role in localizing sclerostin to the surface of target cells. We have also mapped the interaction site for an antibody that blocks the inhibition of Wnt signaling by sclerostin. This shows minimal overlap with the heparin binding site and highlights a key role for this region of sclerostin in protein interactions associated with the inhibition of Wnt signaling. The conserved N- and C-terminal arms of sclerostin were found to be unstructured, highly flexible, and unaffected by heparin binding, which suggests a role in stabilizing interactions with target proteins.
Sclerostin is a 190-residue secreted glycoprotein which is predicted to contain a cystine-knot motif and is a member of the DAN/Cerberus protein family. Recently, patients suffering from a rare inherited bone disorder (sclerosteosis) characterized by exceptionally high bone density (1) were found to be homozygous for a defective sclerostin gene (SOST) (2, 3). A similar high bone mass phenotype has been reported for sclerostin knock-out mice (4), and together, the human and mouse data clearly highlight the importance of sclerostin in the regulation of bone formation. Sclerostin is secreted by bone dwelling osteocytes and has been shown to down-regulate the synthesis of several markers of bone formation by osteogenic cells (5-7). A number of potential protein binding partners have been reported for sclerostin, including BMPs-2, -4, -5, -6, -7 and the BMP antagonist noggin as well as LRP5 and -6, which are co-receptors for Wnt proteins (6, 8-11). Evidence is accumulating that one of the important mechanisms of bone regulation by sclerostin is the modulation of Wnt/β-catenin signaling (12). This signaling pathway is now known to play a central role in the regulation of bone growth and remodeling. Activation of the pathway results in increased proliferation and differentiation of osteoblast precursor cells as well as reduced apoptosis of mature osteoblasts, which favors the deposition of new bone and increased bone density (for recent reviews, see Refs. 13-15). The role of sclerostin in regulating Wnt activity is supported by the recent discovery of a number of mutations in LRP5, which lead to a high bone mass phenotype with similarities to sclerosteosis and result in reduced binding of sclerostin (16-18).
Sclerosteosis is not associated with ectopic bone formation and the bone produced is of high quality (1). This has stimulated considerable interest in sclerostin as a therapeutic target for the treatment of a number of low bone mass disorders such as osteoporosis. Animal studies have shown that administration of sclerostin-specific antibodies results in significant increases in bone formation, bone density, and bone strength (19, 20). In addition, a recent study in human volunteers reported a dose-dependent increase in markers of bone formation after administration of a neutralizing antibody to sclerostin (21).
Structural studies of a number of members of the cystineknot family (transforming growth factor-β2, chorionic gonadotropin, IL17F, BMP-7, etc.) have revealed that the proteins adopt fairly similar backbone topologies, with two pairs of twisted antiparallel β-strands forming an extended finger like structure with the cystine-knot at the base. Within the cystine-knot motif four highly conserved cysteine residues form two intra-chain disulfide bonds to produce a ring structure, which typically contains 8-14 residues. The final pair of conserved cysteines form a third disulfide bond that threads back through the ring to produce an exceptionally stable structural motif (22, 23). The majority of cystine-knot proteins form functional homo- or heterodimers, but the complexes show no conservation in the orientation of the subunits nor in the surface regions involved in dimer interfaces. This variation in dimer structure almost certainly reflects the diverse functional roles of members of the cystine-knot family, which includes growth factors, hormones, cytokines, and also inhibitors of these classes of proteins (24).
In this communication we report the solution structure of sclerostin, which reveals a number of interesting features, such as the presence of a potential heparan sulfate binding site and highly flexible N- and C-terminal regions. The ability of sclerostin to specifically bind heparin was confirmed by NMR titration experiments. Similarly, mapping of the binding site for an antibody that antagonizes the effects of sclerostin on Wnt signaling in vitro and stimulates bone formation in vivo has identified a functionally important region of sclerostin, which is clearly involved in the modulation of Wnt signaling.
EXPERIMENTAL PROCEDURES
Expression and Purification of Sclerostin—The mature human sclerostin protein (residues Gln-1—Tyr-190) was expressed as a maltose-binding protein fusion (MBP-Scl) in Escherichia coli OrigamiB (DE3) pLysS using a modified pMal C2X vector (New England Biolabs). A tobacco etch virus protease site was inserted between the maltose-binding protein and sclerostin, resulting in a single amino acid (Gly) being added to the N terminus of sclerostin after cleavage from the fusion protein. Uniformly 15N- and 13C,15N-labeled sclerostin was produced from cells grown in minimal medium containing [15N]ammonium sulfate and, if appropriate, d-[13C]glucose as the sole nitrogen and carbon sources. To improve the quality of the 13C,1H HSQC3-NOESY spectra, 13C,15N-labeled samples of sclerostin were prepared that contained unlabeled aromatic residues (Phe, Tyr, Trp, and His). Additional details are given in the supplemental material. Mammalian cell derived recombinant human sclerostin was produced using the HEK293 cell line.
Mass Spectrometry—Purified sclerostin was digested with 1:50 w/w trypsin overnight at 37 °C and pH 7.5. Peptides produced were separated on C4 column using an Agilent 1100 HPLC system. 10 μl of digest was loaded onto the column equilibrated with water containing 0.1% trifluoroacetic acid (buffer A). Peptides were eluted with a linear gradient from 100% buffer A to 50% buffer B (acetonitrile containing 0.1% trifluoroacetic acid) over 23 min and then from 50 to 95% B over 10 min. Eluted peaks were monitored at 208 nm, collected manually, and analyzed by MALDI and nano-LCMS. Samples of purified sclerostin, its digests, or collected HPLC fractions were analyzed using an ABI Voyager DE-STR instrument in positive-ion reflectron mode. Fractions containing tryptic peptides eluted from the C4 column were also analyzed in positive-ion mode on a Waters Qtof mass spectrometer equipped with a z-spray source. Spectra were analyzed using Waters Masslynx and deconvoluted using the MaxEnt software.
T-cell Factor-luciferase Reporter Cell Line Development and Luciferase Assay—The mouse osteoblast cell line MC3T3-E1 was transfected with a Super-TOPFlash reporter construct (25), and stable cell lines were selected and evaluated (26). In these stably transfected cells luciferase expression was induced by adding either conditioned media from Wnt3a-transfected cells (CRL-2647, ATCC) or purified recombinant mouse Wnt3a (R & D Systems). Cells were incubated at 37 °C for 24 h either in the presence or absence of sclerostin, and luciferase was assayed using Luclite reagent according to manufacturer's recommendations (PerkinElmer Life Sciences).
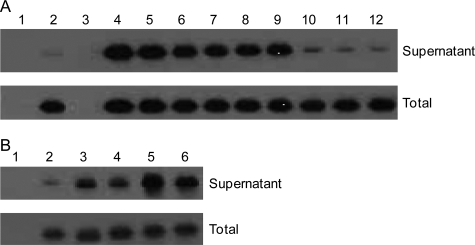
Sclerostin Binding to MC3T3-E1 Cells—The MC3T3-E1 reporter cell line described above was transfected with 0.1 μg of vector DNA per well (in a 96-well plate) encoding human sclerostin (or derivatives) using Lipofectamine (Invitrogen) according to the manufacturer's recommendations. After transfection the cells were incubated at 37 °C for 24 h, and 10 μl of supernatant was removed and pooled for Western blot analysis. One-fifth volume of lysis buffer (225 mm Tris-HCl, 50% glycerol, 5% SDS, 250 mm dithiothreitol, pH 6.8) was then added to each well (containing both supernatant and cells) to liberate cell-associated material and allow assessment of the total amount of sclerostin being produced. The plate was then frozen at -20 °C overnight before pooling of the lysate, and Western blot analysis was performed with an antibody to sclerostin. In some experiments heparin from porcine intestinal mucosa (Sigma) was added (at the concentrations indicated in the legend to Fig. 6) for the last 3 h of the incubation to displace any sclerostin bound to cell surface glycosaminoglycans before Western blot analysis of the supernatant. Western blotting was performed with a mouse monoclonal antibody to sclerostin and was revealed using a goat anti-mouse peroxide conjugate (Jackson ImmunoResearch Laboratories) and ECL (Amersham Biosciences). In each case the Western plot was performed on material pooled from eight separate transfections. Site-directed mutagenesis using the QuikChange II mutagenesis kit (Stratagene) and oligonucleotides from Sigma Genesys was carried out according to the manufacturer's recommendations to generate genes encoding forms of sclerostin mutated in the heparin binding site. The sequence of all sclerostin mutants was confirmed by DNA sequencing.
FIGURE 6.
Localization of sclerostin to the surface of cells. The top section of panel A shows a Western blot (with an antibody to sclerostin) of 24-h supernatants obtained from MC3T3-E1 cells transfected with wild type human sclerostin. The transfections and treatments were as follows: 1, empty vector; 2, sclerostin; 3, empty vector with heparin added to a final concentration of 500 μg/ml; 4-12, sclerostin with heparin added to final concentrations of 500, 250, 100, 50, 25, 12.5, 1, 0.5, and 0.25 μg/ml, respectively. The bottom section of panel A shows the Western blot of samples generated from corresponding wells incubated with a lysis buffer to determine the total amount of sclerostin being produced. The top section of panel B shows the Western blot (with an antibody to sclerostin) obtained for samples of 24-h supernatants from MC3T3-E1 cells transfected with vectors encoding wild type or mutant sclerostin. The transfections were as follows: 1, empty vector; 2, wild type sclerostin; 3, sclerostin R114A, R116A, and R119A; 4, sclerostin K134A and R136A; 5, sclerostin R97A, K99A, and R102A; 6, sclerostin K142A, K144A, and R145A. The bottom section of panel B shows the Western blot obtained for equivalent samples of total cell lysates.
NMR Spectroscopy—NMR spectra were acquired from 0.35-ml samples of sclerostin (200-310 μm) in a 25 mm sodium phosphate, 100 mm sodium chloride, and 10 μm EDTA buffer at pH 5.5. All the NMR experiments were collected at 35 °C on either 600 or 800 MHz Bruker Avance spectrometers equipped with a triple resonance (13C, 1H,15N) cryoprobe. A series of double- and triple-resonance spectra was recorded to determine essentially complete sequence-specific resonance assignments for sclerostin, as described previously (27-29). 1H-1H distance constraints required to calculate the structure of sclerostin were derived from NOEs identified in NOESY, 1H,15N NOESY-HSQC, and 1H,13C HSQC-NOESY spectra, which were acquired with an NOE mixing time of 100 ms. The specific binding of either heparin or an inhibitory Fab fragment to sclerostin was monitored by changes induced in the positions of signals from backbone amide groups of 15N-labeled sclerostin in 1H,15N HSQC spectra. Residues involved in stable backbone hydrogen bonds were identified by monitoring the rate of backbone amide exchange in two-dimensional 1H,15N HSQC spectra of sclerostin dissolved in D2O.
Protein Structural Calculations—The family of converged sclerostin structures was calculated in a two-stage process using the program Cyana 2.1 (30). Initially, the combined automated NOE assignment and structure determination protocol (Candid) was used to automatically assign the NOE cross-peaks identified in two-dimensional NOESY and three-dimensional 15N- and 13C-edited NOESY spectra and to produce preliminary structures. Subsequently, several cycles of simulated annealing combined with redundant dihedral angle constraints (Redac) were used to produce the final converged sclerostin structures (31). Analysis of the family of structures obtained was carried out using the programs Cyana, Molmol, and PyMOL (DeLano Scientific LLC) (30, 32).
Docking-based Prediction of Heparin Binding Sites on Sclerostin—The potential of sclerostin to bind heparin was assessed using a docking protocol previously shown to identify heparin binding sites on proteins with high confidence (34). The program Autodock Version 2.4.7 was used for the docking calculations; this keeps the protein rigid but allows for a degree of flexibility around exocyclic bonds in the heparin ligand. To provide some indication of the effect of protein flexibility on potential heparin binding, the 10 lowest energy NMR structures of sclerostin were docked with each of the three heparin oligosaccharides assessed. The flexible N- and C-terminal tails of sclerostin were not used in the docking calculations, which included the structured Phe-46—Arg-148 region only. Heparin oligosaccharides were derived from the published NMR structure (PDB code 1HPN), and separate docking calculations were carried out with an endecasaccharide and two pentasaccharides, one with internal 2-O-sulfated iduronate residues in the 1C4 chair conformation and one in the 2S0 skew-boat conformation (35). All the oligosaccharide ligand models had N-sulfated, 6-O-sulfated glucosamine as both reducing and non-reducing terminal residues.
Preparation of Heparin—Heparin fragments of defined length were prepared from partially heparinase-depolymerized porcine mucosal heparin as previously described (36).
Preparation of Inhibitory Antibodies to Sclerostin—A range of anti-sclerostin monoclonal antibodies was initially raised in rabbits using a modification of the selected lymphocyte antibody method approach (37) and assessed for biological activity. Chimeric antibody and Fab (rabbit V regions, mouse γ1 constant regions) expression vectors were then constructed for a number of parent antibodies with the ability to inhibit sclerostin and transiently expressed in Chinese hamster ovary cells. The expressed antibodies and Fabs were purified by protein G affinity chromatography using standard methods.
Effects of a Sclerostin-specific Antibody on Bone Formation and Bone Quality—Groups of 10 male Balb/c mice (∼22 g) were dosed with a 200-μl subcutaneous injection every 2 weeks. Group 1 received a phosphate-buffered saline (PBS) control, group 2 Scl-AbI at 25 mg/kg on day 0 and 42 and PBS at all other times, and group 3 received Scl-AbI at 25 mg/kg at all times. The bone mineral density of all animals was measured with a PIXIMus DEXA scan at time 0 and 2, 6, 8, and 10 weeks. Mice were maintained and studied according to UKHome Office regulations and housed in proximity to sentinels which were used for regular pathogen screening.
Analysis of NMR Binding Data—The minimal shift approach (38-40) was used to assess the changes in the positions of sclerostin backbone amide signals resulting from the binding of either heparin or the Fab fragment. A detailed description of the exact procedure used has been published recently (41, 42). To facilitate the identification of ligand binding sites on the surface of sclerostin, histograms of minimal combined shift versus the protein sequence were used to identify regions of the protein containing a number of significantly perturbed backbone amide signals. The affected residues within these regions were then assessed as possible interaction sites in the ligand binding site by examination of the solution structure determined for sclerostin. In this analysis only clusters of residues located on the surface of the protein were considered to be available for ligand binding.
Calculation of the Structure of the Sclerostin-Heparin Complex—The docking software package Haddock (43) was used to calculate the structure of the sclerostin-heparin complex using experimental NMR data. In this well validated approach the algorithm guiding the docking of the protein and ligand is driven by ambiguous distance constraints derived from combined minimal shift data. Precise details of the protocol used have been published recently (41). The complete family of converged solution structures obtained for sclerostin together with a dodecameric heparin molecule parameterized with the Xplo2D and Prodrg software (44, 45) were used as the starting point for the docking calculations. Ambiguous distance constraints were defined between all heparin atoms, and the residues on sclerostin were identified to be involved in the heparin binding, as described previously (41).
RESULTS AND DISCUSSION
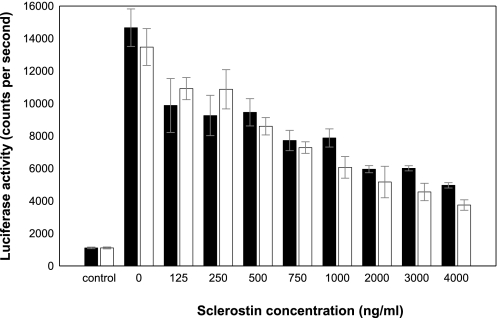
Sclerostin Characterization—Analysis of purified recombinant sclerostin using MALDI mass spectrometry identified a single major species, with an intact mass corresponding to that expected for the protein (observed mass of 21,579 ± 22 Da versus an expected value of 21,573 Da). The protein expressed in E. coli showed equivalent activity in a cell-based Wnt signaling assay to sclerostin produced in mammalian cells (Fig. 1) and was also recognized by a panel of antibodies with distinct epitopes on sclerostin (data not shown).
FIGURE 1.
Inhibition of Wnt3a-dependent signaling by recombinant sclerostin produced in mammalian cells and E. coli. MC3T3-E1 cells stably transfected with a Super-TOPFlash reporter construct were treated with Wnt3a (50 ng/ml) and purified human recombinant sclerostin expressed either in mammalian cells or E. coli. The data shown illustrate the comparable inhibition of Wnt-dependent signaling obtained with either mammalian cell-derived sclerostin (black bars) or E. coli-derived 15N-labeled sclerostin (white bars).
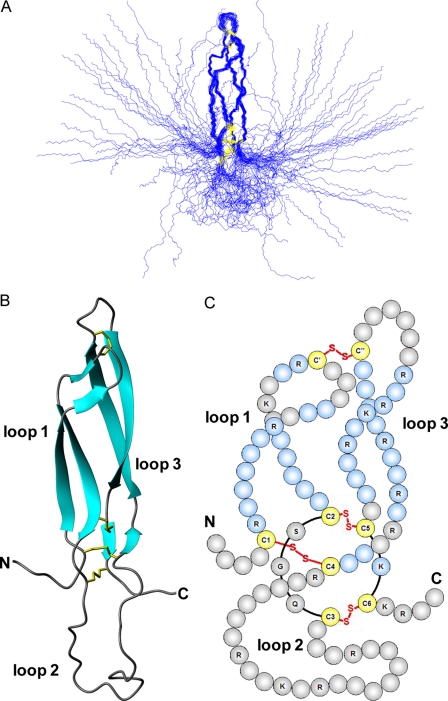
The pattern of the disulfide bonds in E. coli-expressed sclerostin was mapped by trypsin digestion of the protein followed by identification of the peptides produced using mass spectrometry (supplemental Tables 1 and 2). The mass spectrometry data are consistent with the following pairs of cysteines forming disulfide bonds: Cys-56—Cys-110 (C1-C4), Cys-81—Cys-141 (C2-C5), Cys-85—Cys-143 (C3-C6), and Cys-70—Cys-124 (C′-C″), which conforms to the pattern expected for a cystine-knot. The pairs C2-C5 and C3-C6 form part of an eight-member cystine ring, with the C1-C4 disulfide threading through the ring to form the cystine-knot. The final pair of cysteines (C′-C″) link loops 1 and 3, as shown schematically in Fig. 2C.
FIGURE 2.
Solution structure of sclerostin. Panel A shows a best-fit superposition of the protein backbone for the family of 36 converged structures obtained for sclerostin, whereas panel B contains a ribbon representation of the backbone topology of the structured core of the protein (residues 52 to 147) in the same orientation. Panel C shows a schematic representation of the protein, highlighting the positions of disulfide bonds, the resulting 3 loop structure, and residues involved in regular β-sheets (blue).
Solution Structure of Sclerostin—Comprehensive sequence-specific backbone and side-chain resonance assignments were obtained for sclerostin using a combination of triple-resonance experiments, as described in previously published work (27-29). The backbone amide signals (15N and 1H) were assigned for all residues except His-44, Phe-108, Arg-109, Arg-145, Asn-151, and Asn-187, whereas assignments obtained for non-exchangeable side-chain signals were more than 92% complete.
The completeness of the 15N, 13C, and 1H resonance assignments obtained for sclerostin allowed automated assignment of the NOEs identified in three-dimensional 1H,15N NOESY-HSQC and 1H,13C HMQC-NOESY and in the aromatic to aliphatic region of two-dimensional NOESY spectra using the Candid procedure implemented in Cyana (30). This approach proved very successful and yielded unique assignments for 93.3% (2974/3187) of the NOE peaks observed, which provided 1828 non-redundant 1H-1H distance constraints. In the final round of structural calculations 36 satisfactorily converged sclerostin structures were obtained from 100 random starting conformations using a total of 1974 NMR-derived structural constraints (an average of 22.4 constraints per residue for the well defined region corresponding to residues 56-144), including 1828 NOE-based upper distance limits (514 intra residue, 683 sequential (i, i +1), 111 medium range (i, i ≤ 4), and 520 long range (i, i ≥ 5)), 110 backbone torsion angle constraints (55ϕ and 55 ϕ), 12 hydrogen bond constraints (6 backbone hydrogen bonds), and 24 disulfide bond constraints (4 disulfide bonds).
The converged sclerostin structures contain no distance or van der Waals violations greater than 0.5 Å and no dihedral angle violations greater than 5°, with an average value for the Cyana target function of 0.97 ± 0.20 Å2 (30). The sums of the violations for upper distance limits, van der Waals contacts, and torsion angle constraints were 2.4 ± 0.6 Å, 3.4 ± 0.3 Å, and 8.4 ± 2.8°, respectively. Similarly, maximum violations for the converged structures were 0.32 ± 0.09 Å, 0.25 ± 0.03 Å, and 2.75 ± 1.05°, respectively. Analysis of the backbone dihedral angles for the family of converged structures revealed that nearly 64% of the residues adopt backbone conformations within the most favored regions of a Ramachandran plot, >33% lie within the additional allowed regions, and about 3% lie within the generously allowed regions, with no residues consistently found in disallowed regions. The family of sclerostin structures together with the NMR constraints have been deposited in the Protein Data Bank under accession number 2K8P.
The solution structure of sclerostin has been determined to relatively high precision, which is clearly evident from the overlay of the protein backbone shown for the family of converged structures in Fig. 2A. This is also reflected in reasonably low root mean squared deviation values to the mean structure for both the backbone and all heavy atoms (0.89 ± 0.21 and 1.50 ± 0.21 Å, respectively) of residues forming the well defined core region (residues 56-85 and 110-144).
The NMR data clearly indicate that the fairly long N- and C-terminal regions (residues 1-55 and 145-189) of sclerostin are highly flexible and completely disordered (Fig. 2A). The remaining residues form three loop-like structures (loop 1, Arg-57—Val-80; loop 2. Gly-86—Arg-109; loop 3. Ile-111—Ser-140), which are cross-linked by a network of disulfide bonds (C1-C4, Cys-56—Cys-110; C2-C5, Cys-81—Cys-141; C3-C6, Cys-85—Cys-143; C′-C″, Cys-70—Cys-124) as illustrated schematically in Fig. 2C. As expected, three of the disulfides form a cystine-knot motif. The residues in loops 1 and 3 form two pairs of twisted anti-parallel β-strands, which lie alongside each other and are joined at their tips by a single disulfide bond (C′-C″). This region of the structure resembles a pair of long, moderately curved fingers. In common with the N- and C-terminal regions, loop 2 appears to be relatively flexible and adopts a diverse range of conformations within the family of converged structures. The significant mobility in loop 2 and in the N- and C-terminal regions is clearly seen in 15N{1H} heteronuclear NOE measurements obtained for sclerostin (data not shown).
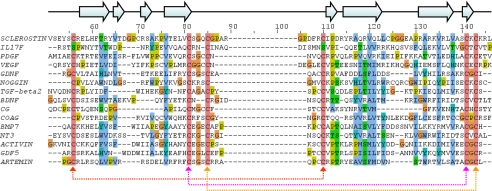
Sclerostin Is an Atypical Member of the Cystine-knot Family—Comparison of the backbone topology of sclerostin with all protein structures deposited in the Protein Data Bank (24) using Dali (46) identified 14 close structural homologues (backbone root mean squared deviation of 3.0-5.1 Å over 58 to 77 matching residues), which are members of the cystine-knot family. This includes a number of growth factors (platelet-derived growth factor, vascular endothelial growth factor, glial cell line-derived neurotrophic factor, transforming growth factor-β2, brain-derived neurotrophic factor, BMP-7, NT3, activin, GDF-5 and Artemin), the cytokine IL17F, the inhibitory protein Noggin, the glycoprotein hormone chorionic gonadotropin and a coagulation factor) (47-61). With the exception of the coagulation factor, all of these cystine-knot proteins are known to form functional homo- or heterodimers; however, there is no evidence to suggest that sclerostin dimerizes. Additionally, none of the identified structural homologues contain long flexible N and C termini.
A structure-based sequence alignment of sclerostin and its close structural homologues is shown in Fig. 3 and highlights the highly conserved cysteines that form the cystine-knot motif as well as a number of conserved hydrophobic residues. Analysis of the positions of these hydrophobic residues in sclerostin revealed that their side chains are mostly buried within the core of the protein and do not form a conserved hydrophobic patch on the surface of the protein. This clearly indicates that these residues are required to stabilize the shared protein structure rather than reflecting a common functional site. The relatively low degree of sequence conservation between sclerostin and its structural homologues (<22%) appears to be typical for members of the cystine-knot family (22).
FIGURE 3.
Sequence conservation between structural homologues in the cystine-knot family. The figure shows an optimized structure-based sequence alignment for the structured region of sclerostin against structurally related members of cystine-knot family. The highly conserved cysteines involved in formation of the cystine-knot motif are shaded in pink, and conserved or semi-conserved hydrophobic residues are highlighted in blue. The residue numbers and positions of regular secondary structure elements shown above the sequence alignment refer to sclerostin. The multiple alignment was produced with JalView (33) using an optimized structure based alignment determined by DALI (46). PDGF, platelet-derived growth factor; VEGF, vascular endothelial growth factor; TGF, transforming growth factor; GDNF, glial cell line-derived neurotrophic factor; COAG, coagulation factror; BDNF, brain-derived neurotrophic factor; CG, chorionic gonadotropin.
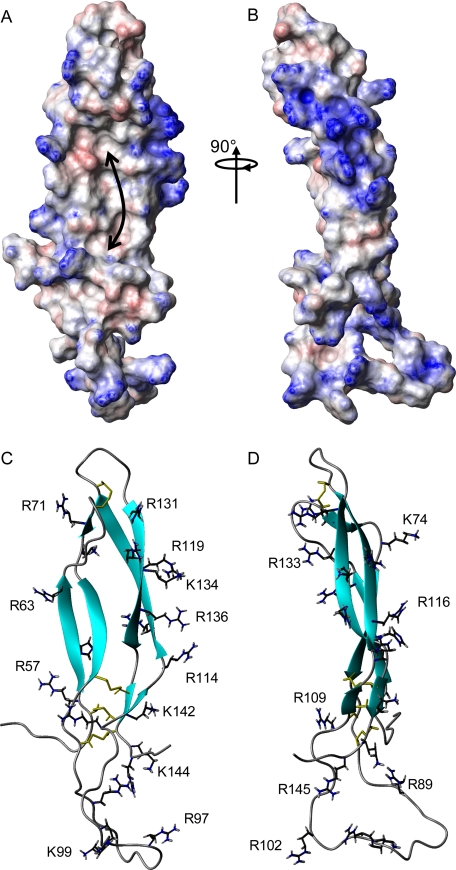
Potential Functional Sites on Sclerostin—The most striking feature on the surface of the high resolution structure obtained for sclerostin is an essentially linear, positively charged patch, which covers one entire side of the protein and is formed from the side chains of residues in loops 2 and 3 (Lys-99, Arg-102, Arg-114, Arg-116, Arg-119, Arg-131, Arg-133, Lys-134, Arg-136, Lys-142, Lys-144, and Arg-145), as illustrated in Fig. 4. This type of extended basic surface is very reminiscent of known heparin binding sites on a number of proteins and may point to a functional association between sclerostin and heparin. Analysis of the surface of sclerostin also revealed a significant hydrophobic patch (∼1771 Å2) on the concave face of the two fingers formed by loops 1 and 3, which primarily involves the side chains of Leu-59, Phe-61, Val-65, Thr-66, Val-76, Val-80, Tyr-115, Val-135, Leu-137, and Val-138 (Fig. 4A) and may represent a potential protein binding site.
FIGURE 4.
Surface features of sclerostin. Panels A and B show contact surface views of sclerostin, which are colored according to electrostatic potential, with areas of significant positive charge shown in blue, significant negative charge in red, and neutral in white. The orientation of the protein in panel A is equivalent to the ribbon representation in panel C. The location of the hydrophobic patch on the concave surface of the extended finger-like structures is indicated by the arrow. Panels C and D show a ribbon representation of sclerostin, with the positions of the basic side chains from arginine and lysine residues highlighted.
The potential of sclerostin to interact with heparin was assessed by a proven procedure that employs a rigid body docking protocol to identify heparin binding sites on proteins (62). This analysis strongly suggested the presence of an extensive heparin binding site. The predicted structures for heparin endecasaccharide-sclerostin complexes show a highly consistent orientation of the heparin with respect to the protein, in which the oligosaccharide interacts with a linear surface of basic residues running parallel to the long axis of the protein. The putative heparin binding site on sclerostin extends over a surface of 52 ± 14 Å in length, with the variability arising from the flexibility in loop 2. This corresponds quite closely to the length expected for a dodecameric (12-mer) heparin molecule (≈48 Å) and formed the basis for the use of heparin 12-mer in NMR binding experiments.
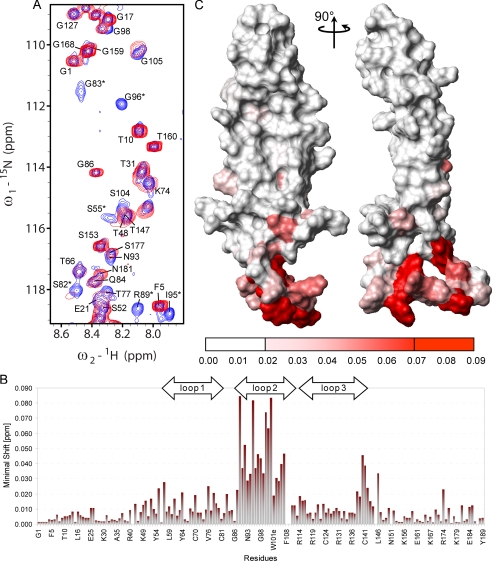
Mapping of the Heparin Binding Site on Sclerostin and Calculation of the Structure of the Sclerostin-Heparin Complex—The predicted binding of heparin to sclerostin was confirmed by detecting changes in the positions of backbone amide NMR signals (15N and 1H) induced by binding of a dodecameric heparin fragment (Fig. 5A). The binding of the heparin fragment clearly results in very significant shifts in the positions of backbone amide signals from residues located mainly in loops 2 and 3, in particular, Glu-53, Ser-55, Arg-57, His-60, Ser-82, Gly-83, Leu-91, Trp-100, Trp-101, Arg-102, Ser-104, Asp-113, Arg-114, Tyr-115, Arg-116, Ala-117, Arg-119, Val-120, Gln-121, Leu-122, Leu-123, Glu-128, Ala-129, Arg-131, Ala-132, Val-135, Leu-137, Val-138, Cys-141, Lys-144, and Leu-146 (Fig. 5B). The residues affected by heparin binding form a distinct patch on the surface of sclerostin (Fig. 5C), which closely matches the positively charged region previously identified as the potential binding site and confirms the presence of a specific heparin interaction site on sclerostin.
FIGURE 5.
Characterization of the interaction of heparin with sclerostin. Panel A shows an overlay of selected regions from 1H,15N HSQC spectra of uniformly 15N-labeled sclerostin (200 μm) acquired in the absence (blue) and presence (red) of an equimolar concentration of heparin 12-mer. Changes in the positions of individual backbone amide peaks illustrate the shifts induced by heparin binding, which are summarized by the histogram of backbone amide chemical shift change versus sequence shown in panel B. Panel C shows a surface view of sclerostin, in which residues are colored according to the perturbation of their backbone amide signals induced by heparin binding, as indicated by the scale below. The protein is shown in a similar orientation to the ribbon representation contained in Fig. 2. Panel D shows a representative view of heparin docked onto its binding site on the surface of sclerostin using ambiguous interaction restraints derived from the NMR data.
The significant shifts in specific backbone NH signals of sclerostin induced by heparin binding were used to generate structures of the complex consistent with the NMR data using the ambiguous interaction restraint-based docking protocol implemented in Haddock (41, 43). Visual inspection of the solution structure of sclerostin revealed an extensive patch of residues with substantially solvent exposed side chains and significantly perturbed NMR signals (Glu-53, Ser-55, Arg-57, His-60, Trp-101, Arg-102, Arg-114, Tyr-115, Arg-116, Ala-117, Arg-119, Gln-121, Leu-123, Ala-129, Arg-131, Ala-132, Lys-144, and Leu-146), which were included as active residues in the docking procedure. In addition, surface residues located adjacent to this group and with the potential to contribute to heparin binding (Tyr-54, Glu-58, Leu-79, Arg-89, Leu-91, Lys-99, Gln-118, Pro-130, Arg-133, Lys-134, Arg-136, Ala-139, and Lys-142) were defined as passive residues in the docking protocol.
The docking calculations produced a fairly closely related family of structures for the sclerostin-heparin complex (cluster of 14 structures with backbone root mean squared deviation of 2.6 ± 0.8 Å), which are represented by the structure shown in Fig. 5D. The entire length of the heparin 12-mer makes favorable contacts with the positively charged face on the protein and results in only minor changes in the conformation of the oligosaccharide. The conformation of the protein is similarly unaffected by heparin binding. As expected, detailed analysis of the structure of the complex reveals that specific binding is predominantly stabilized by favorable electrostatic interactions between negatively charged sulfate groups on heparin and positively charged arginine and lysine side on sclerostin, which are also supported by a network of hydrogen bonds. Critical electrostatic interactions are made by the side chains of Arg-102, Arg-114, Arg-116, Arg-119, Arg-131, Arg-133, Lys-134, Arg-136, Lys-142, Lys-144, Arg-145, and Arg-148, which form the extended basic ridge illustrated in Fig. 4. In addition, the binding is enhanced by several hydrogen bonds between polar groups on heparin and backbone amide groups of sclerostin (Tyr-115, Ala-117, Ala-132, and Lys-134). The average buried surface area for the sclerostin-heparin interface is about 1450 Å2.
Heparin Mediates the Localization of Sclerostin at the Cell Surface—The mouse pre-osteoblast reporter cell line MC3T3-E1 was transiently transfected with an expression vector encoding human wild type sclerostin. Additions of heparin to the cell culture for 3 h before the harvest of the supernatant resulted in a striking dose-dependent increase in the amount of sclerostin detected in the supernatant without affecting the total amount of sclerostin produced by the cells, as shown in Fig. 6A. Similar transfection experiments were also carried out to determine the localization of several mutant forms of sclerostin in which groups of positively charged residues involved in heparin binding had been changed to alanine residues (group A: Arg-114, Arg-116, and Arg-119; group B: Lys-134 and Arg-136; group C: Arg-97, Lys-99, and Arg-102; group D: Arg-142, Lys-144, and Arg-145). The mutant forms of sclerostin were found at much higher levels in the supernatant of transfected cells compared with wild type sclerostin, as illustrated in Fig. 6B, although the total amount of sclerostin produced by the cells remained unchanged. Co-transfection experiments with vectors encoding wild type or mutant forms of sclerostin and a vector encoding Wnt3a showed that impaired heparin binding had only minor effects on the ability of sclerostin to inhibit Wnt signaling (data not shown).
Heparan sulfates have been shown to play key roles in the regulation of several important signaling pathways, including some mediated by Wnt proteins. This regulation appears to be mediated via a number of mechanisms, including localization of proteins to specific cell surfaces and by direct involvement in ligand-receptor interaction. Interestingly, both the osteoblasts and osteoclasts have been shown to secrete heparan sulfates which localize to the cell surfaces and surrounding bone matrix (63-65). The experimental data reported here clearly indicates that the interaction between heparan sulfate and sclerostin will result in a significantly higher concentration of the protein at the surface of responsive cells, which may facilitate its regulation of the Wnt/β-catenin signaling pathway in a more spatially controlled and temporally dependent manner. In addition, heparan sulfate binding may ensure that this important regulator of Wnt signaling in bone is retained in osseous tissue and cannot down-regulate Wnt signaling systemically.
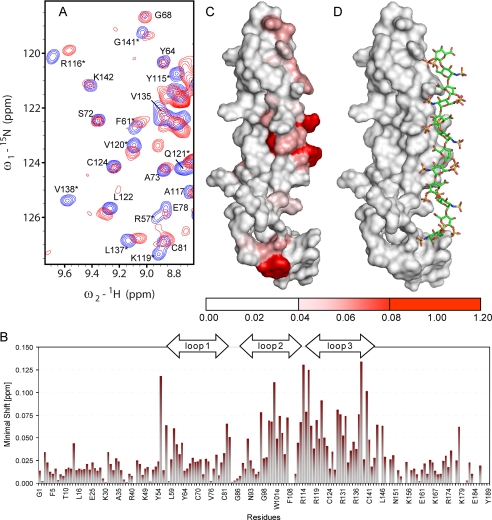
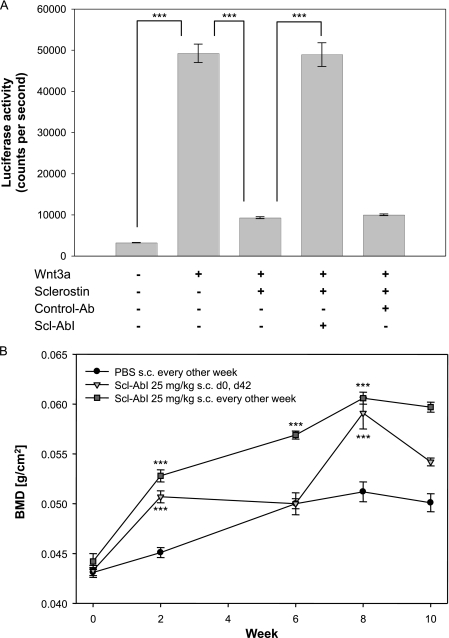
Identification of the Binding Site of an Antibody That Antagonizes the Inhibition of Wnt Signaling by Sclerostin—One of a series of antibodies (Scl-AbI) raised against sclerostin was found to block the protein inhibition of the Wnt/β-catenin signaling pathway in cell culture, as illustrated in Fig. 7A. Mice treated with this antibody showed a significant increase in bone mineral density (Fig. 7B), highlighting the role that sclerostin plays in regulating bone formation. As described for heparin, the location of the inhibitory antibody binding site on sclerostin was mapped by following the changes in the positions of backbone amide NMR signals (15N and 1H) induced by the binding of the Fab fragment of the inhibitory antibody. Examples of significantly affected amide signals of sclerostin are highlighted in the selected regions of 1H,15N HSQC spectra shown in Fig. 8A. The combined minimal shift values for the backbone amide signals versus protein sequence are shown in the histogram in Fig. 8B, which indicates that the majority of the perturbed residues are located within the flexible and solvent exposed loop 2 region of sclerostin (Fig. 8C). This clearly highlights an essential role for the loop 2 region of sclerostin in perturbing Wnt signaling. Comparison of the regions of sclerostin affected by heparin and inhibitory Fab binding reveal little, if any, overlap in the interaction sites, which supports the suggestion of heparin-mediated localization to cell surfaces facilitating the inhibition of Wnt signaling by sclerostin.
FIGURE 7.
Blocking of the inhibition of the Wnt/β-catenin signaling pathway by sclerostin. Panel A shows the luciferase activity of MC3T3-E1 cells stably transfected with a Super-TOPFlash reporter construct, which were treated with Wnt3a conditioned medium and 15N-labeled human recombinant sclerostin (2 μg/ml) in the presence or absence of Scl-AbI or an isotype-matched control antibody (Ab, both 100 μg/ml). Data are presented as the mean ± S.E. (n = 8). Statistical analysis was carried out by one way analysis of variance with a Bonferroni post hoc test (***, p < 0.001). Panel B shows the effects of Scl-AbI on total area bone mineral density (BMD) of mice as measured by DEXA scans. The antibody was administered (25 mg/kg subcutaneously (s.c.)) at either days 0 or 42 (triangles) or once every 2 weeks throughout the experiment (squares). Results obtained from animals treated with a phosphate-buffered saline (PBS) control alone are shown for comparison (circles). Data are presented as the mean ± S.E. (n = 8-10). Statistical analysis was carried out by one way analysis of variance with a Bonferroni post hoc test (***, p < 0.001). d, day.
FIGURE 8.
Identification of the binding site of an antibody that blocks the sclerostin-mediated inhibition of Wnt signaling. Panel A shows selected regions from overlaid 1H,15N HSQC spectra of uniformly 15N labeled sclerostin (200 μm) acquired in the absence (blue) and presence (red) of an equimolar amount of an antibody (Fab fragment of Scl-AbI) known to block the inhibition of Wnt signaling by sclerostin. The minimal backbone amide chemical shift changes observed for sclerostin on Fab binding are summarized in the histogram shown in panel B. Panel C contains a surface view of sclerostin in which residues are colored according to the perturbation of their backbone amide signals induced by Fab binding, as indicated by the scale below. The orientation of the protein in panel C is equivalent to the ribbon representation in Fig. 2.
Conclusions—Sclerostin displays an interesting range of structural features and properties, including highly flexible N- and C-terminal arms, a well defined structured core involved in heparin binding, and a semi-flexible loop involved in the inhibition of Wnt signaling. The N- and C-terminal arms of the protein show a similar level of sequence conservation to that seen for the functional and structural regions determined by this study, which strongly suggests one or more essential roles, perhaps in stabilizing interactions with functional partners. There is also a significant hydrophobic patch on the concave face of the extended fingers formed by loops 1 and 3, which may form a potential protein interaction site.
The identification of the loop 2 region of sclerostin as the binding site for an antibody that modulates sclerostin function clearly highlights its importance. This conclusion is further supported by the identification of this region as the target site for several other antibodies that block the inhibition of Wnt signaling by sclerostin.4 The loop 2 region is fairly flexible in solution, and the NMR data are consistent with significant conformational variability; however, loop 2 region is likely to adopt a well defined structure upon binding to functional partners. Modulation of the essential biological functions of sclerostin in bone may have therapeutic utility, and in this regard the structural work described here provides a firm foundation for knowledge-based drug development.
Supplementary Material
The atomic coordinates and structure factors (code 2K8P) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by UCB-Celltech Ltd and the Wellcome Trust.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
Footnotes
The abbreviations used are: HSQC, heteronuclear single quantum correlation; HPLC, high performance liquid chromatography; MALDI, matrix-assisted laser desorption ionization; NOESY, two-dimensional nuclear Overhauser effect (NOE) spectroscopy.
V. Veverka, A. J. Henry, P. M. Slocombe, A. Ventom, B. Mulloy, F. W. Muskett, M. Muzylak, K. Greenslade, A. Moore, L. Zhang, J. Gong, X. Qian, C. Paszty, R. J. Taylor, M. K. Robinson, and M. D. Carr, unpublished data.
References
- 1.Hamersma, H., Gardner, J., and Beighton, P. (2003) Clin. Genet. 63 192-197 [DOI] [PubMed] [Google Scholar]
- 2.Brunkow, M. E., Gardner, J. C., Van Ness, J., Paeper, B. W., Kovacevich, B. R., Proll, S., Skonier, J. E., Zhao, L., Sabo, P. J., Fu, Y. H., Alisch, R. S., Gillett, L., Colbert, T., Tacconi, P., Galas, D., Hamersma, H., Beighton, P., and Mulligan, J. T. (2001) Am. J. Hum. Genet. 68 577-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balemans, W., Ebeling, M., Patel, N., Van Hul, E., Olson, P., Dioszegi, M., Lacza, C., Wuyts, W., Van den Ende, J., Willems, P., Paes-Alves, A. F., Hill, S., Bueno, M., Ramos, F. J., Tacconi, P., Dikkers, F. G., Stratakis, C., Lindpaintner, K., Vickery, B., Foernzler, D., and Van Hul, W. (2001) Hum. Mol. Genet. 10 537-543 [DOI] [PubMed] [Google Scholar]
- 4.Li, X., Ominsky, M. S., Niu, Q. T., Sun, N., Daugherty, B., D'Agostin, D., Kurahara, C., Gao, Y., Cao, J., Gong, J., Asuncion, F., Barrero, M., Warmington, K., Dwyer, D., Stolina, M., Morony, S., Sarosi, I., Kostenuik, P. J., Lacey, D. L., Simonet, W. S., Ke, H. Z., and Paszty, C. (2008) J. Bone Miner. Res. 23 860-869 [DOI] [PubMed] [Google Scholar]
- 5.Poole, K. E. S., van Bezooijen, R. L., Loveridge, N., Hamersma, H., Papapoulos, S. E., Lowik, C. W., and Reeve, J. (2005) FASEB J. 19 1842-1844 [DOI] [PubMed] [Google Scholar]
- 6.Winkler, D. G., Sutherland, M. K., Geoghegan, J. C., Yu, C. P., Hayes, T., Skonier, J. E., Shpektor, D., Jonas, M., Kovacevich, B. R., Staehling-Hampton, K., Appleby, M., Brunkow, M. E., and Latham, J. A. (2003) EMBO J. 22 6267-6276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Bezooijen, R. L., Roelen, B. A. J., Visser, A., van der Wee-Pals, L., de Wilt, E., Karperien, M., Hamersma, H., Papapoulos, S. E., ten Dijke, P., and Lowik, C. W. G. M. (2004) J. Exp. Med. 199 805-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler, D. G., Yu, C. P., Geoghegan, J. C., Ojala, E. W., Skonier, J. E., Shpektor, D., Sutherland, M. K., and Latham, J. A. (2004) J. Biol. Chem. 279 36293-36298 [DOI] [PubMed] [Google Scholar]
- 9.Kusu, N., Laurikkala, J., Imanishi, M., Usui, H., Konishi, M., Miyake, A., Thesleff, I., and Itoh, N. (2003) J. Biol. Chem. 278 24113-24117 [DOI] [PubMed] [Google Scholar]
- 10.Li, X. F., Zhang, Y. Z., Kang, H. S., Liu, W. Z., Liu, P., Zhang, J. G., Harris, S. E., and Wu, D. Q. (2005) J. Biol. Chem. 280 19883-19887 [DOI] [PubMed] [Google Scholar]
- 11.Semenov, M., Tamai, K., and Xi, H. (2005) J. Biol. Chem. 280 26770-26775 [DOI] [PubMed] [Google Scholar]
- 12.van Bezooijen, R. L., Svensson, J. P., Eefting, D., Visser, A., van der Horst, G., Karperien, M., Quax, P. H., Vrieling, H., Papapoulos, S. E., ten Dijke, P., and Lowik, C. W. (2007) J. Bone Miner. Res. 22 19-28 [DOI] [PubMed] [Google Scholar]
- 13.Yavropoulou, M. P., and Yovos, J. G. (2007) Hormones (Athens) 6 279-294 [DOI] [PubMed] [Google Scholar]
- 14.Clevers, H. (2006) Cell 127 469-480 [DOI] [PubMed] [Google Scholar]
- 15.Johnson, M. L., and Kamel, M. A. (2007) Curr. Opin Rheumatol. 19 376-382 [DOI] [PubMed] [Google Scholar]
- 16.Van Wesenbeeck, L., Cleiren, E., Gram, J., Beals, R. K., Benichou, O., Scopelliti, D., Key, L., Renton, T., Bartels, C., Gong, Y. Q., Warman, M. L., de Vernejoul, M. C., Bollerslev, J., and Van Hul, W. (2003) Am. J. Hum. Genet. 72 763-771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semenov, M. V., and He, X. (2006) J. Biol. Chem. 281 38276-38284 [DOI] [PubMed] [Google Scholar]
- 18.Ellies, D. L., Viviano, B., McCarthy, J., Rey, J. P., Itasaki, N., Saunders, S., and Krumlauf, R. (2006) J. Bone Miner. Res. 21 1738-1749 [DOI] [PubMed] [Google Scholar]
- 19.Ominsky, M., Stouch, B., Doellgast, G., Gong, L., Cao, J., Gao, Y., Tipton, B., Haldankar, R., Winters, A., Chen, Q., Graham, K., Zhou, L., Hale, M., Henry, A., Lightwood, D., Moore, A., Popplewell, A., Robinson, M., Vlasseros, F., Jolette, J., Smith, S. Y., Kostenuik, P. J., Simonet, W. S., Lacey, D. L., and Paszty, C. (2006) J. Bone Miner. Res. 21 Suppl. 1, S44 (Abstr. 1162) [Google Scholar]
- 20.Ominsky, M. S., Warmington, K. S., Asuncion, F. J., Tan, H. L., Grisanti, M. S., Geng, Z., Stephens, T., Henry, A., Lawson, A., Lightwood, D., Perkins, V., Kirby, H., Moore, A., Robinson, M., Li, X., Kostenuik, P. J., Simonet, Y. S., Lacey, D. L., and Paszty, C. (2006) J. Bone Miner. Res. 21 Suppl. 1, S44 [Google Scholar]
- 21.Padhi, D., Stouch, B., Jang, G., Fang, L., Darling, M., Glise, H., Robinson, M., Harris, S., and Posvar, E. (2007) J. Bone Miner. Res. 21 Suppl. 1, S37 (Abstr. 1161) [Google Scholar]
- 22.McDonald, N. Q., and Hendrickson, W. A. (1993) Cell 73 421-424 [DOI] [PubMed] [Google Scholar]
- 23.Avsian-Kretchmer, O., and Hsueh, A. J. W. (2004) Mol. Endocrinol 18 1-12 [DOI] [PubMed] [Google Scholar]
- 24.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E. (2000) Nucleic Acids Res. 28 235-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veeman, M. T., Slusarski, D. C., Kaykas, A., Louie, S. H., and Moon, R. T. (2003) Curr. Biol. 13 680-685 [DOI] [PubMed] [Google Scholar]
- 26.Zhang, L., You, A., Gong, H., Manoukian, R., Elliott, G., Stevens, J., Damore, M., Paszty, C., and Qian, X. (2006) J. Bone Miner. Res. 21 Suppl. 1, S259 (Abstr. 1129) [Google Scholar]
- 27.Veverka, V., Lennie, G., Crabbe, T., Bird, I., Taylor, R. J., and Carr, M. D. (2006) J. Biomol. NMR 36 3-3 [DOI] [PubMed] [Google Scholar]
- 28.Waters, L. C., Bohm, M., Veverka, V., Muskett, F. W., Frenkiel, T. A., Kelly, G. P., Prescott, A., Dosanjh, N. S., Klempnauer, K. H., and Carr, M. D. (2006) J. Biomol. NMR 36 18. [DOI] [PubMed] [Google Scholar]
- 29.Renshaw, P. S., Veverka, V., Kelly, G., Frenkiel, T. A., Williamson, R. A., Gordon, S. V., Hewinson, R. G., and Carr, M. D. (2004) J. Biomol. NMR 30 225-226 [DOI] [PubMed] [Google Scholar]
- 30.Herrmann, T., Guntert, P., and Wuthrich, K. (2002) J. Mol. Biol. 319 209-227 [DOI] [PubMed] [Google Scholar]
- 31.Guntert, P., and Wuthrich, K. (1991) J. Biomol. NMR 1 447-456 [DOI] [PubMed] [Google Scholar]
- 32.Koradi, R., Billeter, M., and Wuthrich, K. (1996) J. Mol. Graph. 14 51-55 [DOI] [PubMed] [Google Scholar]
- 33.Clamp, M., Cuff, J., Searle, S. M., and Barton, G. J. (2004) Bioinformatics 20 426-427 [DOI] [PubMed] [Google Scholar]
- 34.Forster, M., and Mulloy, B. (2006) Biochem. Soc. Trans. 34 431-434 [DOI] [PubMed] [Google Scholar]
- 35.Mulloy, B., Forster, M. J., Jones, C., and Davies, D. B. (1993) Biochem. J. 293 849-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulloy, B., Gee, C., Wheeler, S. F., Wait, R., Gray, E., and Barrowcliffe, T. W. (1997) Thromb. Haemostasis 77 668-674 [PubMed] [Google Scholar]
- 37.Babcook, J. S., Leslie, K. B., Olsen, O. A., Salmon, R. A., and Schrader, J. W. (1996) Proc. Natl. Acad Sci. U. S. A. 93 7843-7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farmer, B. T., Constantine, K. L., Goldfarb, V., Friedrichs, M. S., Wittekind, M., Yanchunas, J., Robertson, J. G., and Mueller, L. (1996) Nat. Struct. Mol. Biol. 3 995-997 [DOI] [PubMed] [Google Scholar]
- 39.Muskett, F. W., Frenkiel, T. A., Feeney, J., Freedman, R. B., Carr, M. D., and Williamson, R. A. (1998) J. Biol. Chem. 273 21736-21743 [DOI] [PubMed] [Google Scholar]
- 40.Williamson, R. A., Carr, M. D., Frenkiel, T. A., Feeney, J., and Freedman, R. B. (1997) Biochemistry 36 13882-13889 [DOI] [PubMed] [Google Scholar]
- 41.Veverka, V., Crabbe, T., Bird, I., Lennie, G., Muskett, F. W., Taylor, R. J., and Carr, M. D. (2008) Oncogene 27 585-595 [DOI] [PubMed] [Google Scholar]
- 42.Waters, L. C., Veverka, V., Bohm, M., Schmedt, T., Choong, P. T., Muskett, F. W., Klempnauer, K. H., and Carr, M. D. (2007) Oncogene 26 4941-4950 [DOI] [PubMed] [Google Scholar]
- 43.Dominguez, C., Boelens, R., and Bonvin, A. M. (2003) J. Am. Chem. Soc. 125 1731-1737 [DOI] [PubMed] [Google Scholar]
- 44.Kleywegt, G. J., and Jones, T. A. (1997) Method Enzymol. 277 208-230 [DOI] [PubMed] [Google Scholar]
- 45.vanAalten, D. M. F., Bywater, R., Findlay, J. B. C., Hendlich, M., Hooft, R. W. W., and Vriend, G. (1996) J. Comput. Aided Mol. Des. 10 255-262 [DOI] [PubMed] [Google Scholar]
- 46.Holm, L., and Sander, C. (1993) J. Mol. Biol. 233 123-138 [DOI] [PubMed] [Google Scholar]
- 47.Eigenbrot, C., and Gerber, N. (1997) Nat. Struct. Biol. 4 435-438 [DOI] [PubMed] [Google Scholar]
- 48.Oefner, C., Darcy, A., Winkler, F. K., Eggimann, B., and Hosang, M. (1992) EMBO J. 11 3921-3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller, Y. A., Li, B., Christinger, H. W., Wells, J. A., Cunningham, B. C., and DeVos, A. M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7192-7197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlunegger, M. P., and Grutter, M. G. (1992) Nature 358 430-434 [DOI] [PubMed] [Google Scholar]
- 51.Robinson, R. C., Radziejewski, C., Stuart, D. I., and Jones, E. Y. (1995) Biochemistry 34 4139-4146 [DOI] [PubMed] [Google Scholar]
- 52.Groppe, J., Greenwald, J., Wiater, E., Rodriguez-Leon, J., Economides, A. N., Kwiatkowski, W., Affolter, M., Vale, W. W., Belmonte, J. C. I., and Choe, S. (2002) Nature 420 636-642 [DOI] [PubMed] [Google Scholar]
- 53.Butte, M. J., Hwang, P. K., Mobley, W. C., and Fletterick, R. J. (1998) Biochemistry 37 16846-16852 [DOI] [PubMed] [Google Scholar]
- 54.Harrington, A. E., Morris-Triggs, S. A., Ruotolo, B. T., Robinson, C. V., Ohnuma, S., and Hyvonen, M. (2006) EMBO J. 25 1035-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schreuder, H., Liesum, A., Pohl, J., Kruse, B., and Koyama, M. (2005) Biochem. Biophys. Res. Commun. 329 1076-1086 [DOI] [PubMed] [Google Scholar]
- 56.Wang, X. Q., Baloh, R. H., Milbrandt, J., and Garcia, K. C. (2006) Structure 14 1083-1092 [DOI] [PubMed] [Google Scholar]
- 57.Hymowitz, S. G., Filvaroff, E. H., Yin, J. P., Lee, J., Cai, L. P., Risser, P., Maruoka, M., Mao, W. G., Foster, J., Kelley, R. F., Pan, G. H., Gurney, A. L., de Vos, A. M., and Starovasnik, M. A. (2001) EMBO J. 20 5332-5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, H., Lustbader, J. W., Liu, Y., Canfield, R. E., and Hendrickson, W. A. (1994) Structure 2 545-558 [DOI] [PubMed] [Google Scholar]
- 59.Fan, Q. R., and Hendrickson, W. A. (2005) Nature 433 269-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bergner, A., Oganessyan, V., Muta, T., Iwanaga, S., Typke, D., Huber, R., and Bode, W. (1996) EMBO J. 15 6789-6797 [PMC free article] [PubMed] [Google Scholar]
- 61.Greenwald, J., Groppe, J., Gray, P., Wiater, E., Kwiatkowski, W., Vale, W., and Choe, S. (2003) Mol. Stimulat. 11 605-617 [DOI] [PubMed] [Google Scholar]
- 62.Mulloy, B., and Forster, M. J. (2008) Mol. Simulations 34 481-489 [Google Scholar]
- 63.Jackson, R. A., Nurcombe, V., and Cool, S. M. (2006) Gene (Amst.) 379 79-91 [DOI] [PubMed] [Google Scholar]
- 64.Lin, X. (2004) Development 131 6009-6021 [DOI] [PubMed] [Google Scholar]
- 65.Cattaruzza, S., and Perris, R. (2006) Macromol. Biosci. 6 667-680 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.